Delta-Radiomics Based on Dynamic Contrast-Enhanced MRI Predicts Pathologic Complete Response in Breast Cancer Patients Treated with Neoadjuvant Chemotherapy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
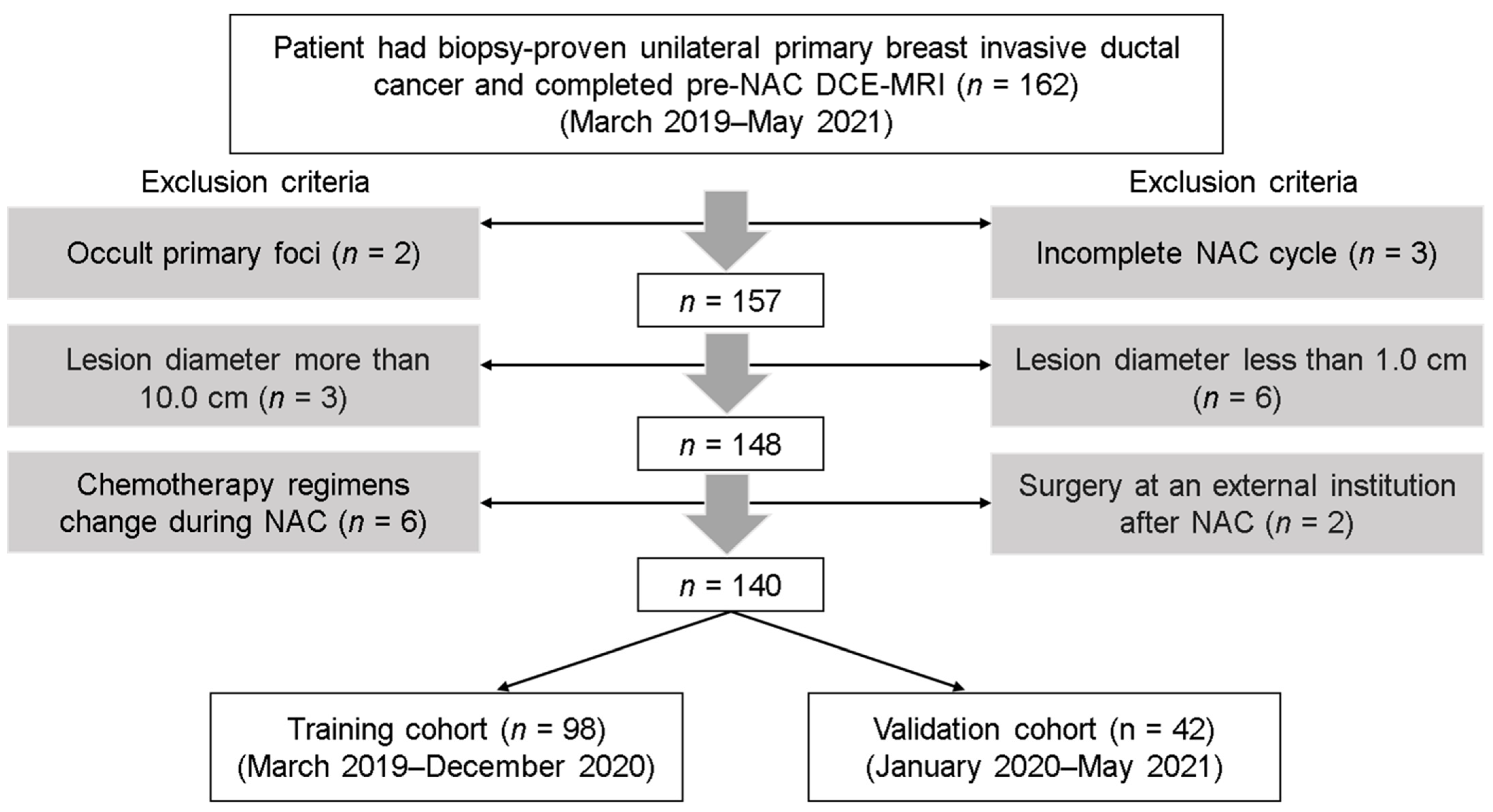
2.1. Patients
2.2. DCE-MRI Data Acquisition
2.3. Clinicopathologic Features
2.4. NAC Regimen and Criteria for pCR
2.5. Tumor Segmentation and Repeatability Analysis
2.6. Radiomic Features and Their Changes
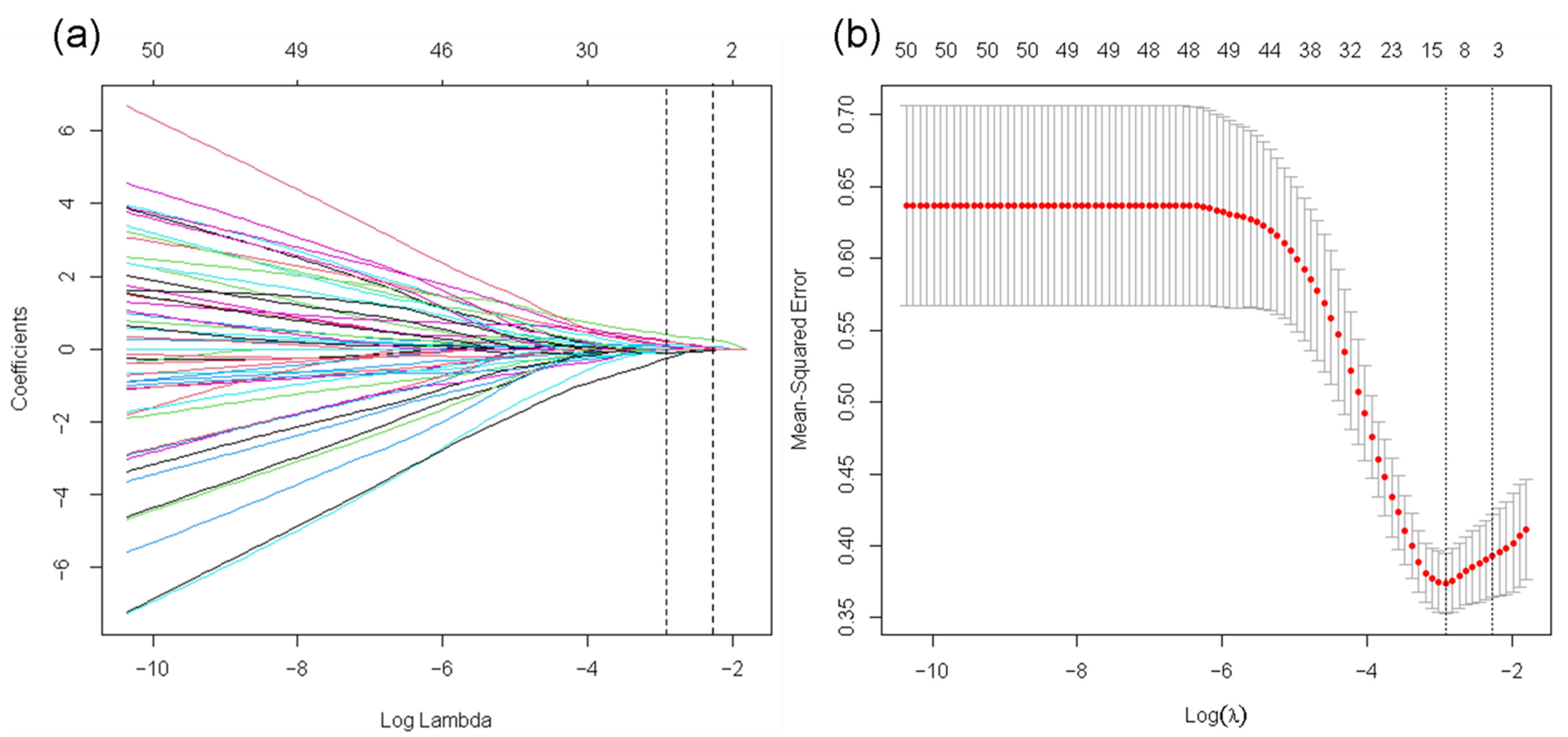
2.7. Feature Selection
2.8. Establishment and Performance of Models
2.8.1. Separate Radiomic Models
2.8.2. Longitudinal Fusion Radiomic Models
2.8.3. Clinical-Radiomics Models
2.9. Statistical Analysis
3. Results
3.1. Clinical Characteristics
3.2. Repeatability Analysis
3.3. Separate Radiomic Models
3.4. Determination of the Optimal Contrast Phase
3.5. Longitudinal Fusion Radiomic Model under the Optimal Contrast Phase
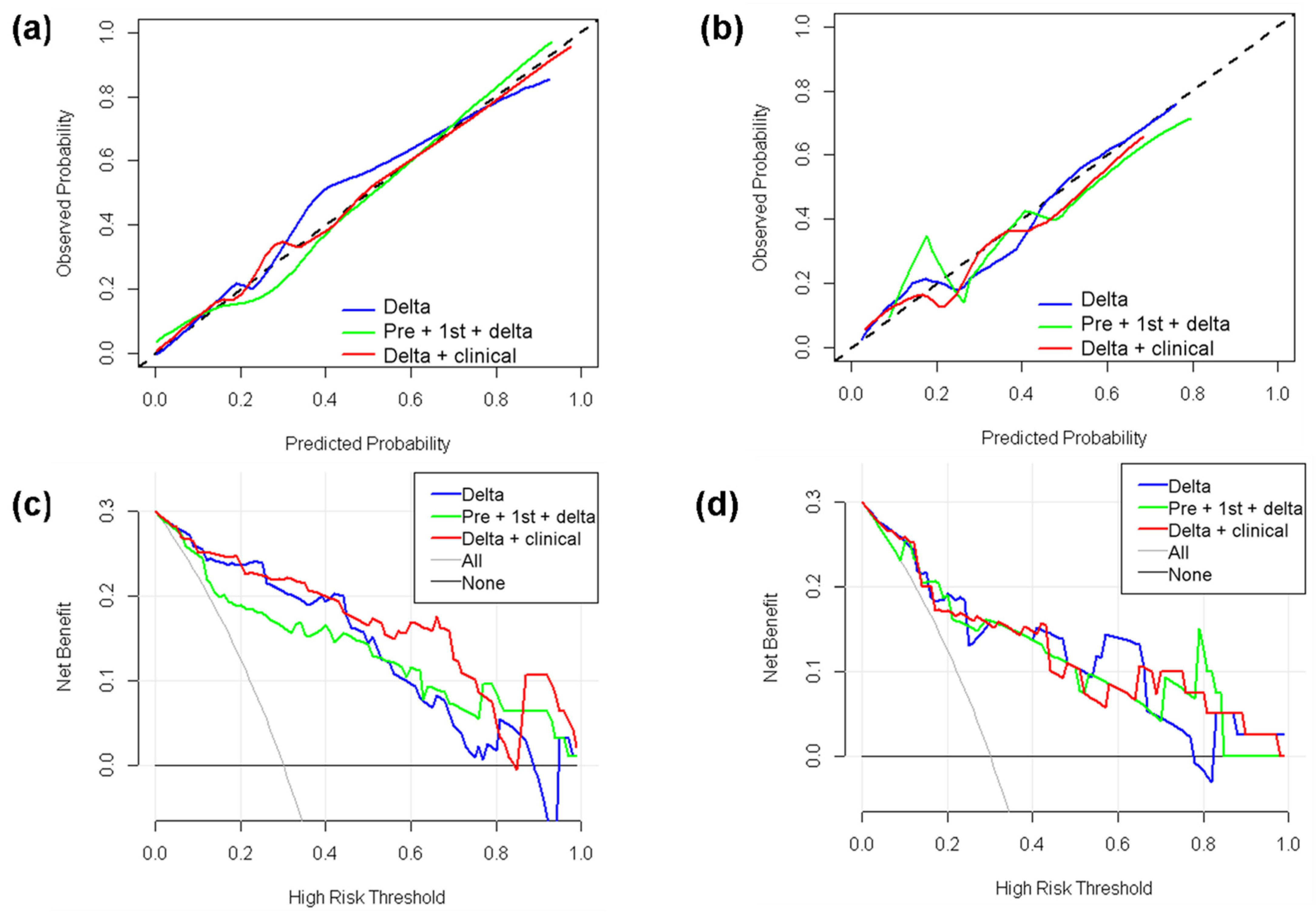
3.6. Clinical-Radiomics Models
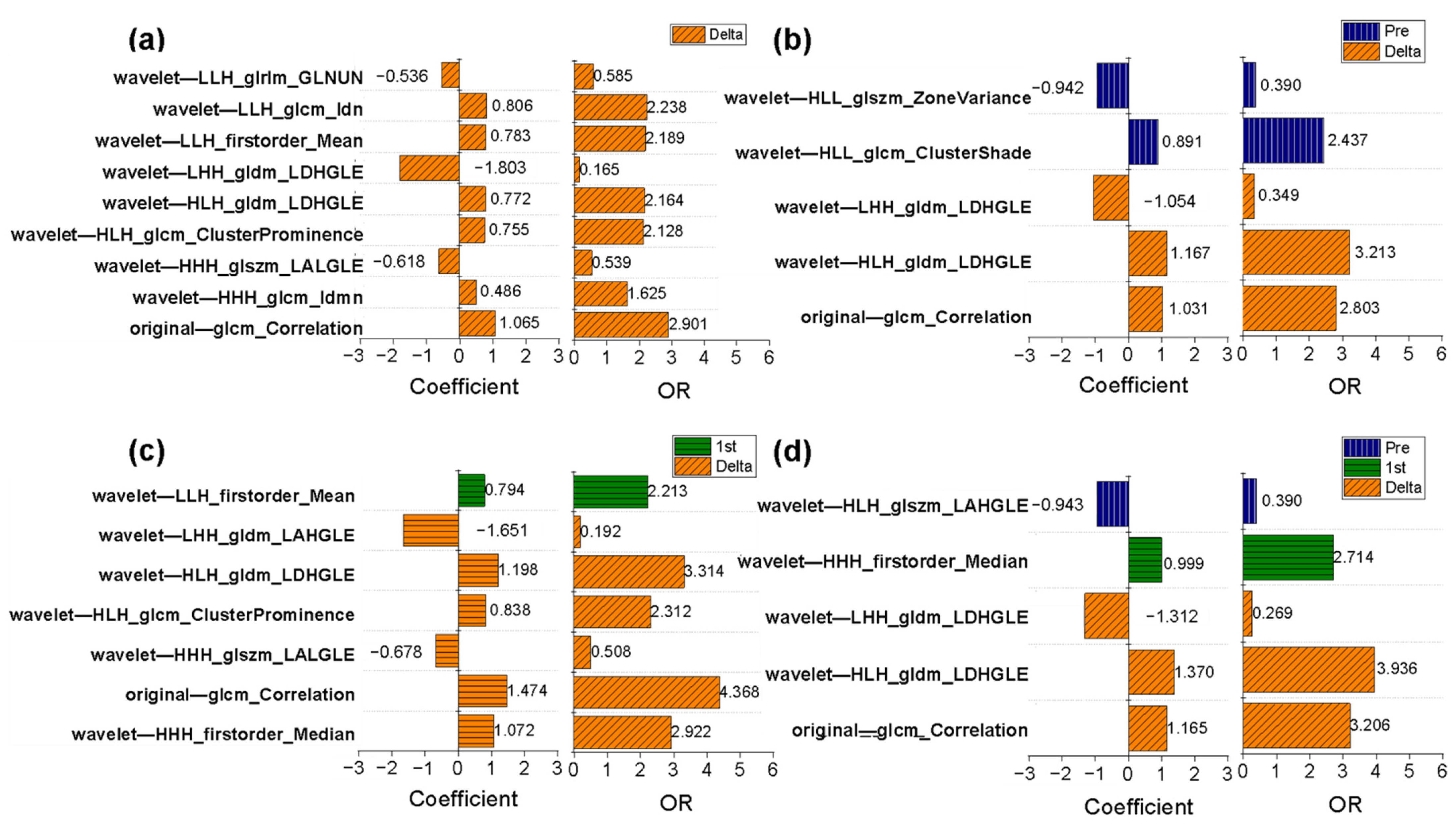
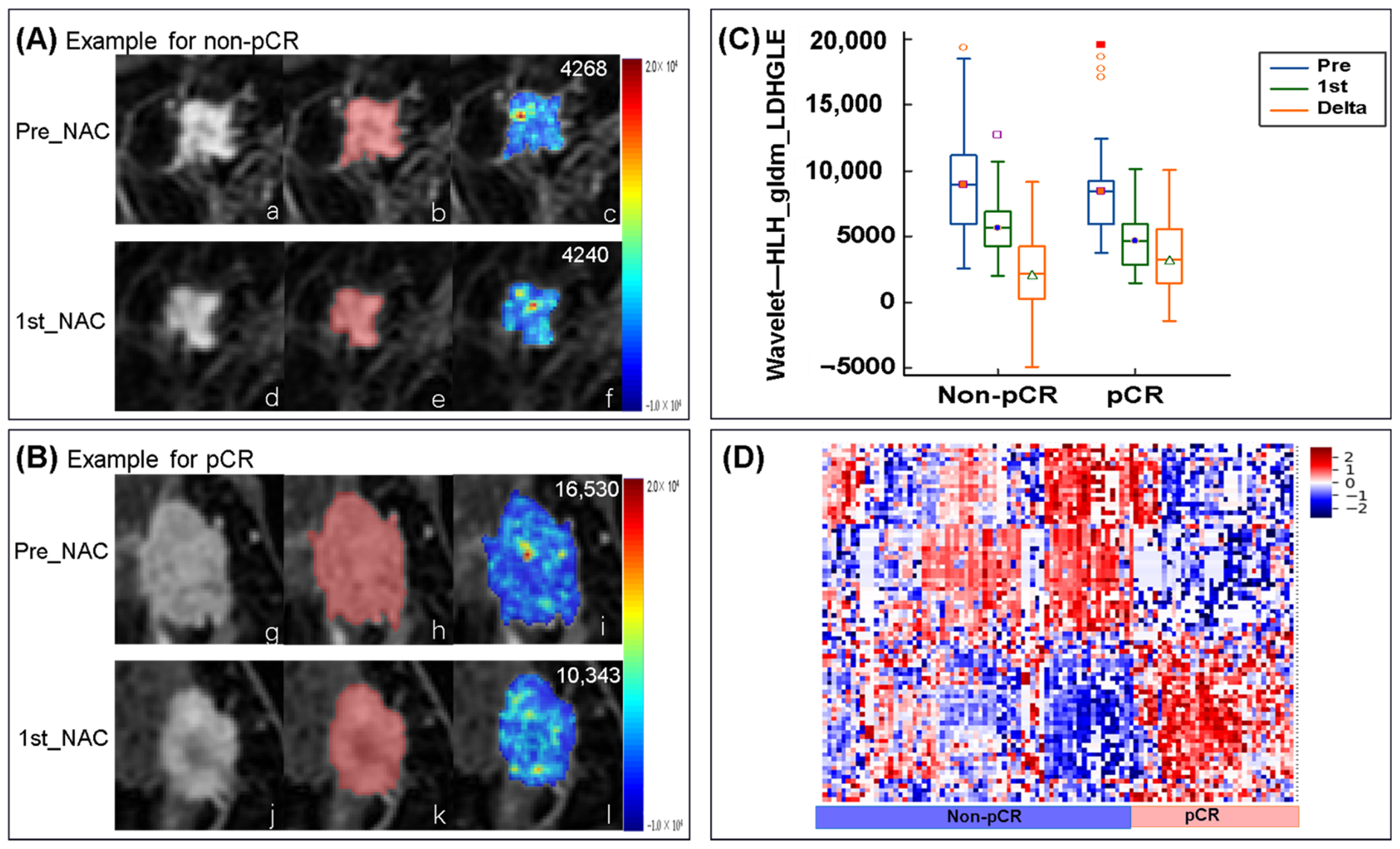
3.7. Changes in Radiomic Features in the Delta-Radiomics Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Derks, M.G.M.; van de Velde, C.J.H. Neoadjuvant chemotherapy in breast cancer: More than just downsizing. Lancet Oncol. 2017, 19, 2–3. [Google Scholar] [CrossRef] [Green Version]
- Spring, L.; Greenup, R.; Reynolds, K.; Smith, B.L.; Moy, B.; Bardia, A. Abstract 1439: Pathological complete response after neoadjuvant chemotherapy predicts improved survival in all major subtypes of breast cancer: Systematic review and me-ta-analyses of over 18,000 patients. Cancer Res. 2016, 76, 1439. [Google Scholar] [CrossRef]
- Haque, W.; Verma, V.; Hatch, S.; Klimberg, V.S.; Butler, E.B.; Teh, B.S. Response rates and pathologic complete response by breast cancer molecular subtype following neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2018, 170, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.G.; Kerin, E.; O’Flaherty, C.; Maher, E.; Richard, V.; McAnena, P.F.; McLaughlin, R.P.; Sweeney, K.J.; Barry, M.K.; Malone, C.M.; et al. Clinicopathological response to neoadjuvant therapies and pathological complete response as a biomarker of survival in human epidermal growth factor receptor-2 enriched breast cancer—A retrospective cohort study. Breast 2021, 59, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.M.; Kuhl, C.K.; Kinkel, K.; Boetes, C. Breast MRI: Guidelines from the European Society of Breast Imaging. Eur. Radiol. 2008, 18, 1307–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fowler, A.M.; Mankoff, D.A.; Joe, B.N. Imaging Neoadjuvant Therapy Response in Breast Cancer. Radiology 2017, 285, 358–375. [Google Scholar] [CrossRef] [Green Version]
- Padhani, A.R.; Hayes, C.; Assersohn, L.; Powles, T.; Makris, A.; Suckling, J.; Leach, M.; Husband, J.E. Prediction of Clinicopathologic Response of Breast Cancer to Primary Chemotherapy at Contrast-enhanced MR Imaging: Initial Clinical Results. Radiology 2006, 239, 361–374. [Google Scholar] [CrossRef]
- Minarikova, L.; Bogner, W.; Pinker, K.; Valkovič, L.; Zaric, O.; Bago-Horvath, Z.; Bartsch, R.; Helbich, T.H.; Trattnig, S.; Gruber, S. Investigating the prediction value of multiparametric magnetic resonance imaging at 3 T in response to neoadjuvant chemotherapy in breast cancer. Eur. Radiol. 2016, 27, 1901–1911. [Google Scholar] [CrossRef] [Green Version]
- Hylton, N.M.; Blume, J.D.; Bernreuter, W.K.; Pisano, E.D.; Rosen, M.A.; Morris, E.A.; Weatherall, P.T.; Lehman, C.D.; Newstead, G.M.; Polin, S.; et al. ACRIN 6657 Trial Team and I-SPY 1 TRIAL Investigators. Locally advanced breast cancer _MRI imaging for prediction of response to neoadjuvant chemotherapy-results from ACRIN 6657 or 1-SPY TRIAL. Radiology 2012, 263, 663–672. [Google Scholar] [CrossRef] [Green Version]
- Tudorica, A.; Oh, K.Y.; Chui, S.Y.-C.; Roy, N.; Troxell, M.L.; Naik, A.; Kemmer, K.A.; Chen, Y.; Holtorf, M.L.; Afzal, A.; et al. Early Prediction and Evaluation of Breast Cancer Response to Neoadjuvant Chemotherapy Using Quantitative DCE-MRI. Transl. Oncol. 2016, 9, 8–17. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Arlinghaus, L.R.; Ayers, G.D.; Chakravarthy, A.B.; Abramson, R.G.; Abramson, V.G.; Atuegwu, N.; Farley, J.; Mayer, I.A.; Kelley, M.C.; et al. DCE-MRI analysis methods for predicting the response of breast cancer to neoadjuvant chemotherapy: Pilot study findings. Magn. Reson. Med. 2013, 71, 1592–1602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dogan, B.E.; Yuan, Q.; Bassett, R.; Guvenc, I.; Jackson, E.F.; Cristofanilli, M.; Whitman, G.J. Comparing the Performances of Magnetic Resonance Imaging Size vs Pharmacokinetic Parameters to Predict Response to Neoadjuvant Chemotherapy and Survival in Patients With Breast Cancer. Curr. Probl. Diagn. Radiol. 2019, 48, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Pereira, N.P.; Curi, C.; Osório, C.A.B.T.; Marques, E.F.; Makdissi, F.B.; Pinker, K.; Bitencourt, A.G.V. Diffusion-Weighted Magnetic Resonance Imaging of Patients with Breast Cancer Following Neoadjuvant Chemotherapy Provides Early Prediction of Pathological Response—A Prospective Study. Sci. Rep. 2019, 9, 16372. [Google Scholar] [CrossRef]
- Eun, N.L.; Kang, D.; Son, E.J.; Park, J.S.; Youk, J.H.; Kim, J.-A.; Gweon, H.M. Texture Analysis with 3.0-T MRI for Association of Response to Neoadjuvant Chemotherapy in Breast Cancer. Radiology 2020, 294, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Henderson, S.; Purdie, C.; Michie, C.; Evans, A.; Lerski, R.; Johnston, M.; Vinnicombe, S.; Thompson, A.M. Interim heterogeneity changes measured using entropy texture features on T2-weighted MRI at 3.0 T are associated with pathological response to neoadjuvant chemotherapy in primary breast cancer. Eur. Radiol. 2017, 27, 4602–4611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadrljanski, M.M.; Milosevic, Z.C. Tumor texture parameters of invasive ductal breast carcinoma in neoadjuvant chemotherapy: Early identification of non-responders on breast MRI. Clin. Imaging 2020, 65, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Granzier, R.; van Nijnatten, T.; Woodruff, H.; Smidt, M.; Lobbes, M. Exploring breast cancer response prediction to neoadjuvant systemic therapy using MRI-based radiomics: A systematic review. Eur. J. Radiol. 2019, 121, 108736. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Li, Z.; Qu, J.; Zhang, R.; Zhou, X.; Li, L.; Sun, K.; Tang, Z.; Jiang, H.; Li, H.; et al. Radiomics of Multiparametric MRI for Pretreatment Prediction of Pathologic Complete Response to Neoadjuvant Chemotherapy in Breast Cancer: A Multicenter Study. Clin. Cancer Res. 2019, 25, 3538–3547. [Google Scholar] [CrossRef] [Green Version]
- Braman, N.M.; Etesami, M.; Prasanna, P.; Dubchuk, C.; Gilmore, H.; Tiwari, P.; Plecha, D.; Madabhushi, A. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 2017, 19, 57. [Google Scholar] [CrossRef]
- Cain, E.H.; Saha, A.; Harowicz, M.R.; Marks, J.R.; Marcom, P.K.; Mazurowski, M.A. Multivariate machine learning models for prediction of pathologic response to neoadjuvant therapy in breast cancer using MRI features: A study using an independent validation set. Breast Cancer Res. Treat. 2018, 173, 455–463. [Google Scholar] [CrossRef]
- Shur, J.D.; Doran, S.J.; Kumar, S.; ap Dafydd, D.; Downey, K.; O’Connor, J.P.B.; Papanikolaou, N.; Messiou, C.; Koh, D.-M.; Orton, M.R. Radiomics in Oncology: A Practical Guide. RadioGraphics 2021, 41, 1717–1732. [Google Scholar] [CrossRef] [PubMed]
- Teruel, J.R.; Heldahl, M.G.; Goa, P.E.; Pickles, M.; Lundgren, S.; Bathen, T.F.; Gibbs, P. Dynamic contrast-enhanced MRI texture analysis for pretreatment prediction of clinical and pathological response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. NMR Biomed. 2014, 27, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Wu, G.; Cheng, H.; Zhang, J.; Shao, G.; Li, L. Radiomic analysis of DCE-MRI for prediction of response to neoadjuvant chemotherapy in breast cancer patients. Eur. J. Radiol. 2017, 94, 140–147. [Google Scholar] [CrossRef]
- Yu, Y.; He, Z.; Ouyang, J.; Tan, Y.; Chen, Y.; Gu, Y.; Mao, L.; Ren, W.; Wang, J.; Lin, L.; et al. Magnetic resonance imaging radiomics predicts preoperative axillary lymph node metastasis to support surgical decisions and is associated with tumor microenvironment in invasive breast cancer: A machine learning, multicenter study. eBioMedicine 2021, 69, 103460. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Chen, H.; You, C.; Liu, L.; Gu, Y.; Peng, W.; Gao, X.; Li, L. Radiomics of Tumor Heterogeneity in Longitudinal Dynamic Contrast-Enhanced Magnetic Resonance Imaging for Predicting Response to Neoadjuvant Chemotherapy in Breast Cancer. Front. Mol. Biosci. 2021, 8, 622219. [Google Scholar] [CrossRef]
- Morrison, C.K.; Bancroft, L.H.; DeMartini, W.B.; Holmes, J.H.; Wang, K.; Bosca, R.J.; Korosec, F.R.; Strigel, R.M. Novel High Spatiotemporal Resolution Versus Standard-of-Care Dynamic Contrast-Enhanced Breast MRI. Investig. Radiol. 2017, 52, 198–205. [Google Scholar] [CrossRef] [Green Version]
- Hammond, M.E.H.; Hayes, D.F.; Dowsett, M.; Allred, D.C.; Hagerty, K.L.; Badve, S.; Fitzgibbons, P.L.; Francis, G.; Goldstein, N.S.; Hayes, M.; et al. American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for Immunohistochemical Testing of Estrogen and Progesterone Receptors in Breast Cancer. J. Clin. Oncol. 2010, 28, 2784–2795. [Google Scholar] [CrossRef] [Green Version]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.S.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018, 36, 2105–2122. [Google Scholar] [CrossRef] [Green Version]
- Bustreo, S.; Osella-Abate, S.; Cassoni, P.; Donadio, M.; Airoldi, M.; Pedani, F.; Papotti, M.; Sapino, A.; Castellano, I. Optimal Ki67 cut-off for luminal breast cancer prognostic evaluation: A large case series study with a long-term follow-up. Breast Cancer Res. Treat. 2016, 157, 363–371. [Google Scholar] [CrossRef] [Green Version]
- Curigliano, G.; Burstein, H.J.; Winer, E.P.; Gnant, M.; Dubsky, P.; Loibl, S.; Colleoni, M.; Regan, M.M.; Piccart-Gebhart, M.; Senn, H.-J.; et al. De-escalating and escalating treatments for early-stage breast cancer: The St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2017, 28, 1700–1712. [Google Scholar] [CrossRef]
- Gradishar, W.J.; Anderson, B.O.; Balassanian, R.; Blair, S.L.; Burstein, H.J.; Cyr, A.; Elias, A.D.; Farrar, W.B.; Forero, A.; Giordano, S.H.; et al. Breast Cancer, Version 4.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2018, 16, 310–320. [Google Scholar] [CrossRef]
- Zhou, J.; Lu, J.; Gao, C.; Zeng, J.; Zhou, C.; Lai, X.; Cai, W.; Xu, M. Predicting the response to neoadjuvant chemotherapy for breast cancer: Wavelet transforming radiomics in MRI. BMC Cancer 2020, 20, 100. [Google Scholar] [CrossRef]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. NeuroImage 2006, 31, 1116–1128. [Google Scholar] [CrossRef] [Green Version]
- Waugh, S.A.; Lerski, R.A.; Bidaut, L.; Thompson, A.M. The influence of field strength and different clinical breast MRI protocols on the outcome of texture analysis using foam phantoms. Med. Phys. 2011, 38, 5058–5066. [Google Scholar] [CrossRef] [Green Version]
- Tsarouchi, M.I.; Vlachopoulos, G.F.; Karahaliou, A.N.; Vassiou, K.G.; Costaridou, L.I. Multi-parametric MRI lesion heterogeneity biomarkers for breast cancer diagnosis. Phys. Med. 2020, 80, 101–110. [Google Scholar] [CrossRef]
- Chinese Expert Group on Neoadjuvant Therapy for Breast Cancer. Expert consensus on neoadjuvant therapy for breast cancer in china (2019 Edition). Chin. J. Cancer 2019, 29, 390–400. [Google Scholar] [CrossRef]
- Pesapane, F.; Rotili, A.; Botta, F.; Raimondi, S.; Bianchini, L.; Corso, F.; Ferrari, F.; Penco, S.; Nicosia, L.; Bozzini, A.; et al. Radiomics of MRI for the Prediction of the Pathological Response to Neoadjuvant Chemotherapy in Breast Cancer Patients: A Single Referral Centre Analysis. Cancers 2021, 13, 4271. [Google Scholar] [CrossRef]
- Massafra, R.; Comes, M.C.; Bove, S.; Didonna, V.; Gatta, G.; Giotta, F.; Fanizzi, A.; La Forgia, D.; Latorre, A.; Pastena, M.I.; et al. Robustness Evaluation of a Deep Learning Model on Sagittal and Axial Breast DCE-MRIs to Predict Pathological Complete Response to Neoadjuvant Chemotherapy. J. Pers. Med. 2022, 12, 953. [Google Scholar] [CrossRef]
- Comes, M.C.; Fanizzi, A.; Bove, S.; Didonna, V.; Diotaiuti, S.; La Forgia, D.; Latorre, A.; Martinelli, E.; Mencattini, A.; Nardone, A.; et al. Early prediction of neoadjuvant chemotherapy response by exploiting a transfer learning approach on breast DCE-MRIs. Sci. Rep. 2021, 11, 14123. [Google Scholar] [CrossRef]
- La Forgia, D.; Vestito, A.; Lasciarrea, M.; Comes, M.; Diotaiuti, S.; Giotta, F.; Latorre, A.; Lorusso, V.; Massafra, R.; Palmiotti, G.; et al. Response Predictivity to Neoadjuvant Therapies in Breast Cancer: A Qualitative Analysis of Background Parenchymal Enhancement in DCE-MRI. J. Pers. Med. 2021, 11, 256. [Google Scholar] [CrossRef]
- Qi, T.H.; Hian, O.H.; Kumaran, A.M.; Tan, T.J.; Cong, T.R.Y.; Su-Xin, G.L.; Lim, E.H.; Ng, R.; Yeo, M.C.R.; Tching, F.L.L.W.; et al. Multi-center evaluation of artificial intelligent imaging and clinical models for predicting neoadjuvant chemotherapy response in breast cancer. Breast Cancer Res. Treat. 2022, 193, 121–138. [Google Scholar] [CrossRef]
- Saranathan, M.; Rettmann, D.W.; Hargreaves, B.A.; Lipson, J.A.; Daniel, B.L. Variable spatiotemporal resolution three-dimensional dixon sequence for rapid dynamic contrast-enhanced breast MRI. J. Magn. Reson. Imaging 2013, 40, 1392–1399. [Google Scholar] [CrossRef] [Green Version]
- Milon, A.; Perre, S.V.; Poujol, J.; Trop, I.; Kermarrec, E.; Bekhouche, A.; Thomassin-Naggara, I. Abbreviated breast MRI combining FAST protocol and high temporal resolution (HTR) dynamic contrast enhanced (DCE) sequence. Eur. J. Radiol. 2019, 117, 199–208. [Google Scholar] [CrossRef]
- Perre, S.V.; Duron, L.; Milon, A.; Bekhouche, A.; Balvay, D.; Cornelis, F.H.; Fournier, L.; Thomassin-Naggara, I. Radiomic analysis of HTR-DCE MR sequences improves diagnostic performance compared to BI-RADS analysis of breast MR lesions. Eur. Radiol. 2021, 31, 4848–4859. [Google Scholar] [CrossRef]
- Romeo, V.; Cavaliere, C.; Imbriaco, M.; Verde, F.; Petretta, M.; Franzese, M.; Stanzione, A.; Cuocolo, R.; Aiello, M.; Basso, L.; et al. Tumor segmentation analysis at different post-contrast time points: A possible source of variability of quantitative DCE-MRI parameters in locally advanced breast cancer. Eur. J. Radiol. 2020, 126, 108907. [Google Scholar] [CrossRef]
- Mann, R.M.; Cho, N.; Moy, L. Breast MRI: State of the Art. Radiology 2019, 292, 520–536. [Google Scholar] [CrossRef]
- Macura, K.J.; Ouwerkerk, R.; Jacobs, M.A.; Bluemke, D. Patterns of Enhancement on Breast MR Images: Interpretation and Imaging Pitfalls. RadioGraphics 2006, 26, 1719–1734. [Google Scholar] [CrossRef] [Green Version]
- Jahani, N.; Cohen, E.; Hsieh, M.-K.; Weinstein, S.P.; Pantalone, L.; Hylton, N.; Newitt, D.; Davatzikos, C.; Kontos, D. Prediction of Treatment Response to Neoadjuvant Chemotherapy for Breast Cancer via Early Changes in Tumor Heterogeneity Captured by DCE-MRI Registration. Sci. Rep. 2019, 9, 12114. [Google Scholar] [CrossRef]


| Characteristics | Training Cohort (n = 98) | p | Validation Cohort (n = 42) | |||
|---|---|---|---|---|---|---|
| pCR (n = 28) | Non-pCR (n = 70) | pCR (n = 12) | Non-pCR (n = 30) | p | ||
| Age (years) | 51.0 ± 9.1 | 51.7 ± 10.5 | 0.601 | 50.7 ± 7.2 | 47.3 ± 10.7 | 0.242 |
| Tumor size (cm) | 4.2 (3.1, 5.6) | 4.8 (3.4, 6.1) | 0.276 | 3.4 (2.5, 5.2) | 5.0 (3.1, 5.7) | 0.177 |
| Enhancement type (%) | 0.442 | 0.657 | ||||
| Mass-like | 15 (53.6) | 42 (60) | 7 (58.3) | 14 (46.7) | ||
| Non-mass-like | 6 (21.4) | 8 (11.4) | 2 (16.7) | 4 (13.3) | ||
| Mass + non-mass | 7 (25.0) | 20 (28.6) | 3 (25.0) | 12 (40.0) | ||
| Type (%) | 0.236 | 0.469 | ||||
| Multi-lesion | 8 (28.6) | 29 (41.4) | 3 (25.0) | 11 (36.7) | ||
| Single-lesion | 20 (71.4) | 41 (58.6) | 9 (75.0) | 19 (63.3) | ||
| TNM (%) | 0.276 | 0.368 | ||||
| Ⅱ A | 7 (25.0) | 9 (12.9) | 3 (25.0) | 7 (23.3) | ||
| Ⅱ B | 5 (17.9) | 18 (25.7) | 5 (41.7) | 5 (16.7) | ||
| Ⅲ A | 2 (7.1) | 11 (15.7) | 2 (16.7) | 5 (16.7) | ||
| Ⅲ B | 9 (32.1) | 26 (37.1) | 2 (16.7) | 10 (33.3) | ||
| Ⅲ C | 5 (17.9) | 6 (8.6) | 0 (0.0) | 3 (10.0) | ||
| Grades (%) | 0.104 | 0.205 | ||||
| 2 | 13 (46.4) | 45 (64.3) | 10 (83.3) | 19 (63.3) | ||
| 3 | 15 (53.6) | 25 (35.7) | 2 (16.7) | 11 (36.7) | ||
| ER status (%) | 0.001 * | 0.002 * | ||||
| Positive | 5 (17.9) | 46 (65.7) | 2 (16.7) | 21 (70.0) | ||
| Negative | 23 (82.1) | 24 (34.3) | 10 (83.3) | 9 (30.0) | ||
| PR status (%) | 0.001 * | 0.554 | ||||
| Positive | 7 (25.0) | 49 (70.0) | 7 (58.3) | 17 (56.7) | ||
| Negative | 21 (75.0) | 21 (30.0) | 5 (41.7) | 13 (43.3) | ||
| HER2 status (%) | 0.041 * | 0.008 * | ||||
| Positive | 15 (53.6) | 22 (31.4) | 8 (66.7) | 7 (23.3) | ||
| Negative | 13 (46.4) | 48 (68.6) | 4 (33.3) | 23 (76.7) | ||
| Ki-67 status (%) | 0.092 | 0.263 | ||||
| ≤20% | 2 (7.1) | 15 (21.4) | 1 (8.3) | 23 (76.7) | ||
| >20% | 26 (92.9) | 55 (78.6) | 11 (91.7) | 7 (23.3) | ||
| Molecular subtypes (%) | <0.001 * | 0.046 * | ||||
| Luminal A | 0 (0.0) | 5 (7.1) | 0 (0.0) | 2 (6.7) | ||
| Luminal B | 7 (25.0) | 48 (68.6) | 5 (41.7) | 19 (63.3) | ||
| HER2 enriched | 11 (39.3) | 5 (7.1) | 5 (41.7) | 2 (6.7) | ||
| TN | 10 (35.7) | 12 (17.2) | 2 (16.7) | 7 (23.3) | ||
| CEe | CEp | CEd | |
|---|---|---|---|
| Pre-radiomics model | |||
| No. of selected features | |||
| LASSO_ CV | 9 | 9 | 2 |
| Logistic_ CV | 3 | 5 | 1 |
| AUC (training/validation) | 0.759/0.617 | 0.827/0.694 | 0.649/0.539 |
| 95% CI of AUC | 0.647, 0.871/0.403, 0.830 | 0.723, 0.922/0.533, 0.856 | 0.527, 0.770/0.319, 0.759 |
| Sensitivity (training/validation) | 0.643/0.667 | 0.679/0.917 | 0.857/0.250 |
| Specificity (training/validation) | 0.800/0.633 | 0.857/0.500 | 0.386/0.967 |
| Accuracy (training/validation) | 0.755/0.643 | 0.806/0.619 | 0.520/0.762 |
| 1st-radiomics model | |||
| No. of selected features | |||
| LASSO_ CV | 10 | 9 | 13 |
| Logistic_ CV | 5 | 4 | 4 |
| AUC (training/validation) | 0.803/0.775 | 0.816/0.650 | 0.826/0.703 |
| 95% CI of AUC | 0.694, 0.913/0.627, 0.923 | 0.717, 0.915/0.432, 0.868 | 0.738, 0.914/0.514, 0.892 |
| Sensitivity (training/validation) | 0.756/0.667 | 0.786/0.667 | 0.821/0.417 |
| Specificity(training/validation) | 0.771/0.800 | 0.771/0.800 | 0.700/0.967 |
| Accuracy (training/validation) | 0.776/0.762 | 0.776/0.667 | 0.735/0.810 |
| Delta-radiomics model | |||
| No. of selected features | |||
| LASSO_ CV | 13 | 11 | 3 |
| Logistic_ CV | 9 | 7 | 1 |
| AUC (training/validation) | 0.917/0.842 | 0.803/0.764 | 0.708/0.697 |
| 95% CI of AUC | 0.861, 0.974/0.709, 0.974 | 0.64, 0.913/0.592, 0.936 | 0.594, 0.821/0.512, 0.883 |
| Sensitivity (training/validation) | 0.929/0.667 | 0.786/0.917 | 0.750/0.833 |
| Specificity (training/validation) | 0.829/0.900 | 0.771/0.667 | 0.629/0.700 |
| Accuracy (training/validation) | 0.857/0.833 | 0.776/0.738 | 0.663/0.738 |
| AUC | Sensitivity | Specificity | Accuracy | |
|---|---|---|---|---|
| Longitudinal fusion radiomic-models (training/validation) | ||||
| Pre + delta | 0.866/0.750 | 0.929/0.833 | 0.623/0.700 | 0.723/0.738 |
| 1st + delta | 0.903/0.839 | 0.821/0.667 | 0.871/0.933 | 0.857/0.857 |
| Pre + 1st + delta | 0.871/0.869 | 0.750/0.667 | 0.871/0.900 | 0.837/0.833 |
| Clinical-radiomic model (training/validation) | ||||
| Pre + clinical | 0.762/0.569 | 0.679/0.333 | 0.757/0.900 | 0.735/0.738 |
| 1st + clinical | 0.801/0.767 | 0.786/0.833 | 0.771/0.633 | 0.776/0.691 |
| Delta + clinical | 0.934/0.864 | 0.927/0.750 | 0.829/0.867 | 0.857/0.833 |
| Pre + delta + clinical | 0.870/0.736 | 0.821/0.667 | 0.757/0.800 | 0.776/0.762 |
| 1st + delta + clinical | 0.908/0.828 | 0.821/0.667 | 0.900/0.967 | 0.878/0.881 |
| Pre + 1st + delta + clinical | 0.866/0.864 | 0.714/0.667 | 0.886/0.900 | 0.837/0.833 |
| Median (IQR) | pCR | Non-pCR | Trend | ||||
|---|---|---|---|---|---|---|---|
| Pre | 1st | Delta | Pre | 1st | Delta | ||
| First order-related features | |||||||
| LLH_Mean (×101) | −7.2 (−9.5, 4.5) | −7.2 (−14.8, −5.2) | 2.6 (−0.5, 5.0) | −6.7 (−9.0, −5.0) | −7.7 (−10.6, 5.5) | 0.8 (−1.0, 2.6) | Down |
| GLCM-related features | |||||||
| Correlation (×10−1) | 7.2 (6.2, 8.0) | 6.2 (5.2, 7.5) | 0.9 (0.03, 1.7) | 7.4 (6.5, 8.0) | 7.0 (6.0, 7.8) | 0.3 (−0.3, 0.8) | Down |
| HHH_Idmn (×10−1) | 9.9 (9.9, 10.0) | 9.8 (9.8, 9.9) | 0.1 (0.03, 0.2) | 9.9 (9.9, 10.0) | 9.9 (9.8, 10.0) | 0.06 (−0.09, 0.08) | Down |
| HLH_ Cluster Prominence (×104) | 3.0 (0.9, 6.2) | 1.73 (0.4, 3.0) | 1.1 (0.1, 3.4) | 2.3 (1.1, 4.2) | 2.0 (0.8, 4.2) | 0.2 (−1.2, 1.4) | Down |
| LLH _ Idn (×10−1) | 9.2 (9.1, 9.3) | 9.1 (8.9, 9.2) | 0.1 (0.04, 0.2) | 9.2 (9.1, 9.3) | 9.2 (9.1, 9.3) | 0.05 (−0.02, 0.13) | Down |
| GLDM-related features | |||||||
| HLH_ LDHGLE (×103) | 8.5 (6.0, 12) | 4.7 (2.8, 7.6) | 3.5 (0.9, 6.0) | 9.5 (5.9, 16.0) | 6.6 (4.2, 11.2) | 2.4 (−0.6, 4.9) | Down |
| LHH_ LDHGLE (×104) | 1.0 (0.6, 1.6) | 0.6 (0.3, 1.0) | 0.3 (0.003, −1.1) | 1.2 (0.7, 1.8) | 0.7 (0.6, 2.7) | 0.3 (0.03, −0.7) | Down |
| GLSZM-related features | |||||||
| HHH_ LALGLE (×103) | 0.9 (0.2, 6.3) | 0.4 (0.05, 6.1) | 0.2 (0.04, 1.1) | 1.6 (0.4, 5.9) | 0.9 (0.2, 5.0) | 0.1 (−0.9, 1.3) | Down |
| GLRLM-related features | |||||||
| LLH_ GLNUN (×10−2) | 0.9 (0.7, 1.2) | 1.0 (0.8, 1.3) | −0.1 (−0.3, 0.02) | 0.9 (0.7, 1.1) | 0.9 (−0.8, 1.2) | −0.03 (−0.2, 0.1) | Up |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, L.; Du, S.; Gao, S.; Zhao, R.; Huang, G.; Jin, F.; Teng, Y.; Zhang, L. Delta-Radiomics Based on Dynamic Contrast-Enhanced MRI Predicts Pathologic Complete Response in Breast Cancer Patients Treated with Neoadjuvant Chemotherapy. Cancers 2022, 14, 3515. https://doi.org/10.3390/cancers14143515
Guo L, Du S, Gao S, Zhao R, Huang G, Jin F, Teng Y, Zhang L. Delta-Radiomics Based on Dynamic Contrast-Enhanced MRI Predicts Pathologic Complete Response in Breast Cancer Patients Treated with Neoadjuvant Chemotherapy. Cancers. 2022; 14(14):3515. https://doi.org/10.3390/cancers14143515
Chicago/Turabian StyleGuo, Liangcun, Siyao Du, Si Gao, Ruimeng Zhao, Guoliang Huang, Feng Jin, Yuee Teng, and Lina Zhang. 2022. "Delta-Radiomics Based on Dynamic Contrast-Enhanced MRI Predicts Pathologic Complete Response in Breast Cancer Patients Treated with Neoadjuvant Chemotherapy" Cancers 14, no. 14: 3515. https://doi.org/10.3390/cancers14143515
APA StyleGuo, L., Du, S., Gao, S., Zhao, R., Huang, G., Jin, F., Teng, Y., & Zhang, L. (2022). Delta-Radiomics Based on Dynamic Contrast-Enhanced MRI Predicts Pathologic Complete Response in Breast Cancer Patients Treated with Neoadjuvant Chemotherapy. Cancers, 14(14), 3515. https://doi.org/10.3390/cancers14143515






