The Role of Runx2 in Microtubule Acetylation in Bone Metastatic Breast Cancer Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Immunofluorescence
2.3. Western Blotting
2.4. Co-Immunoprecipitation
2.5. Microtubule Polymer Mass Isolation
2.6. Viral Transduction
2.7. qPCR Array Analysis
2.8. Statistical Analysis
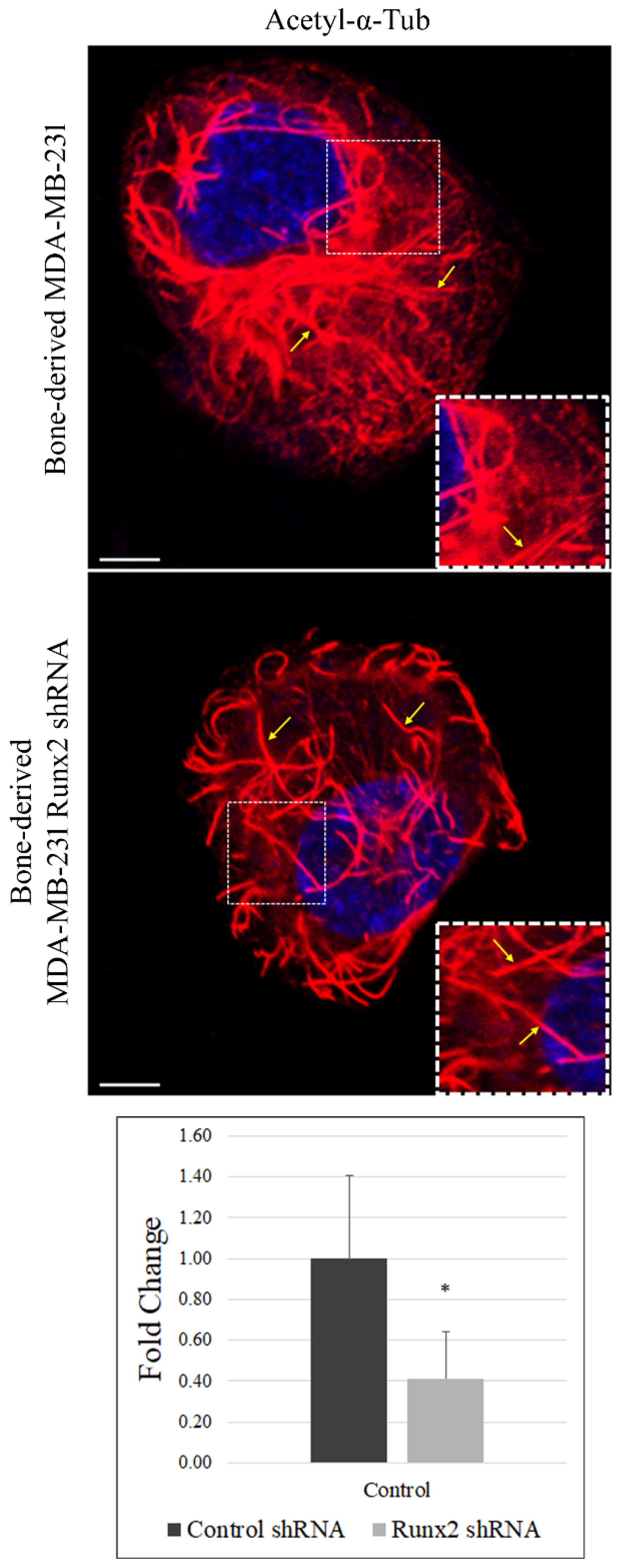
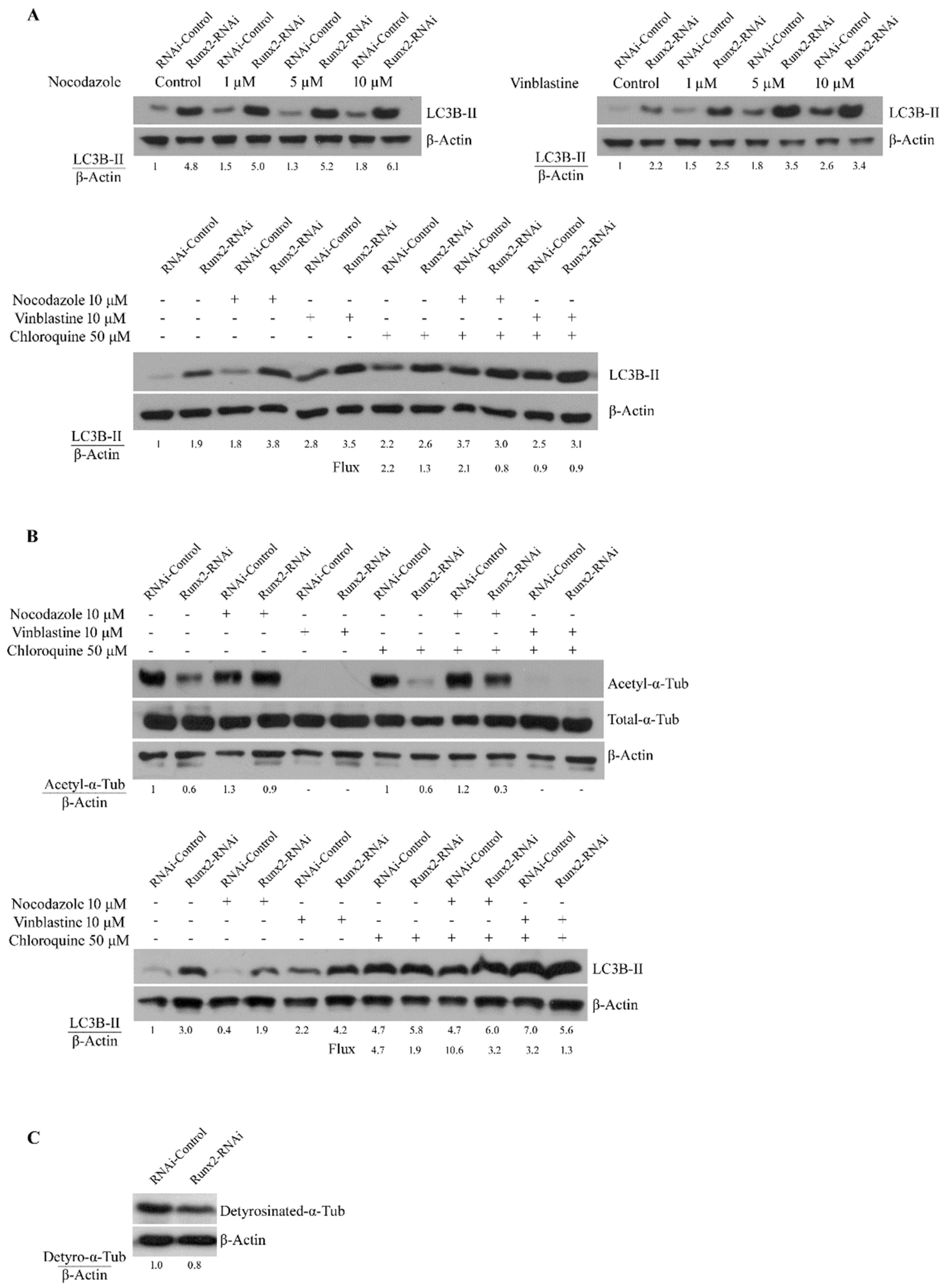
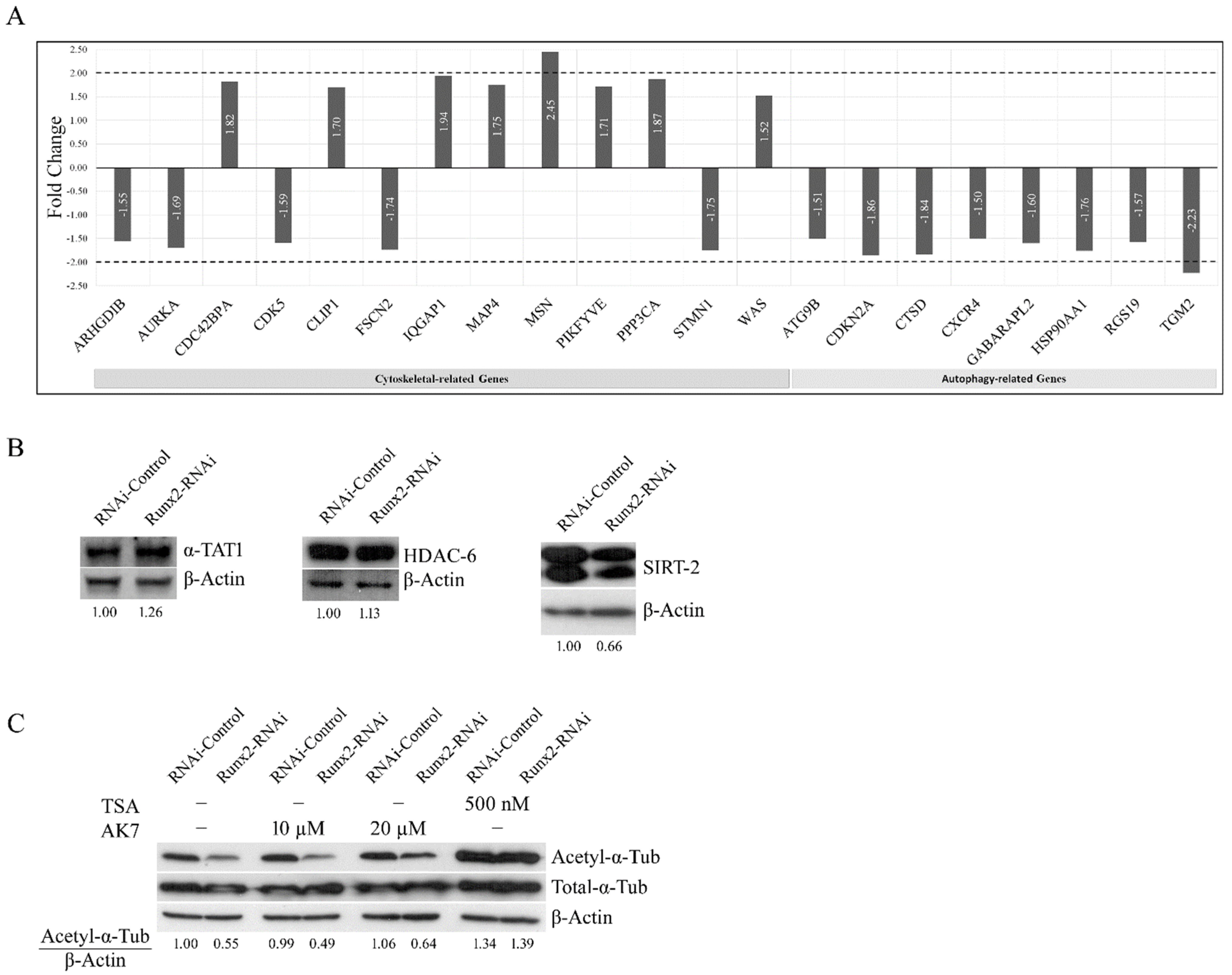
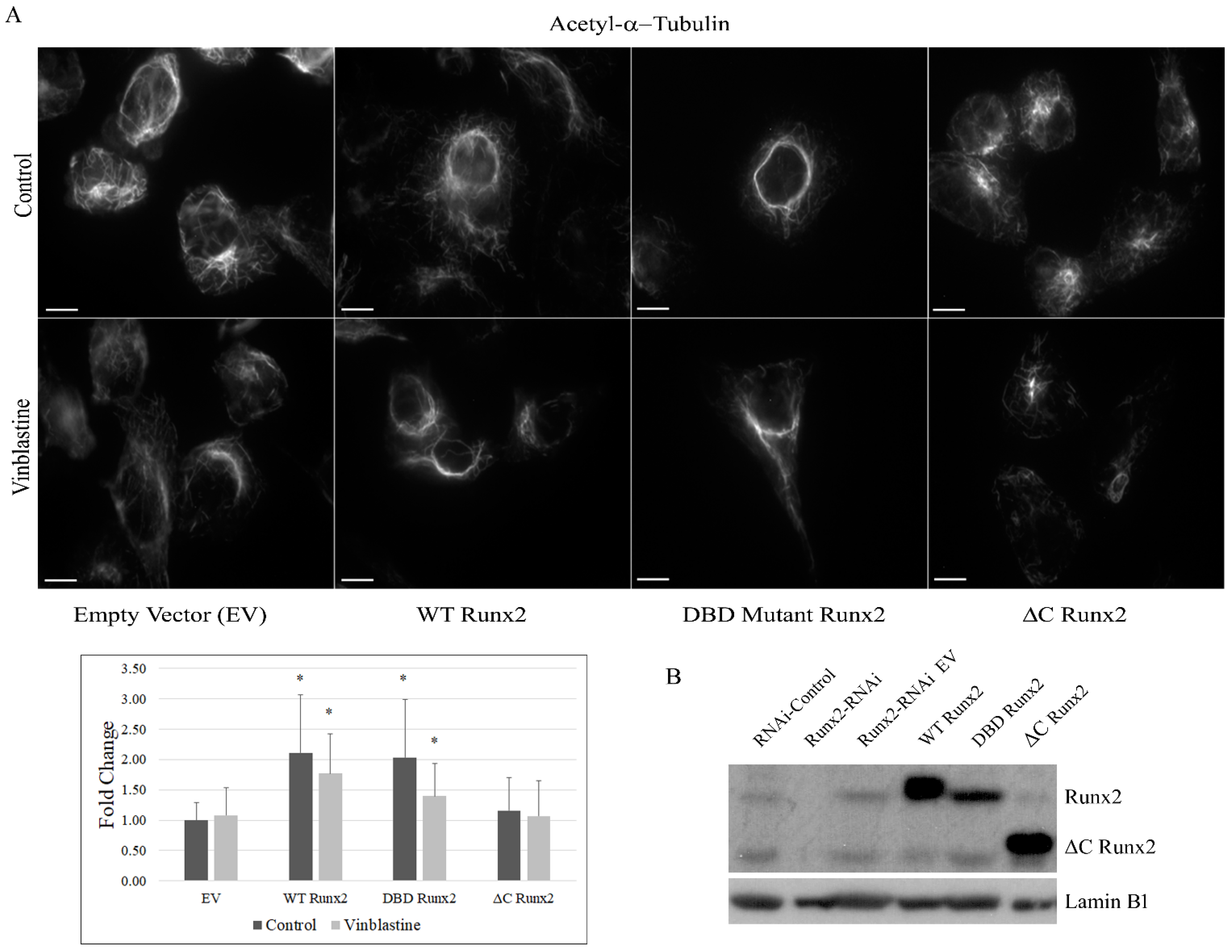
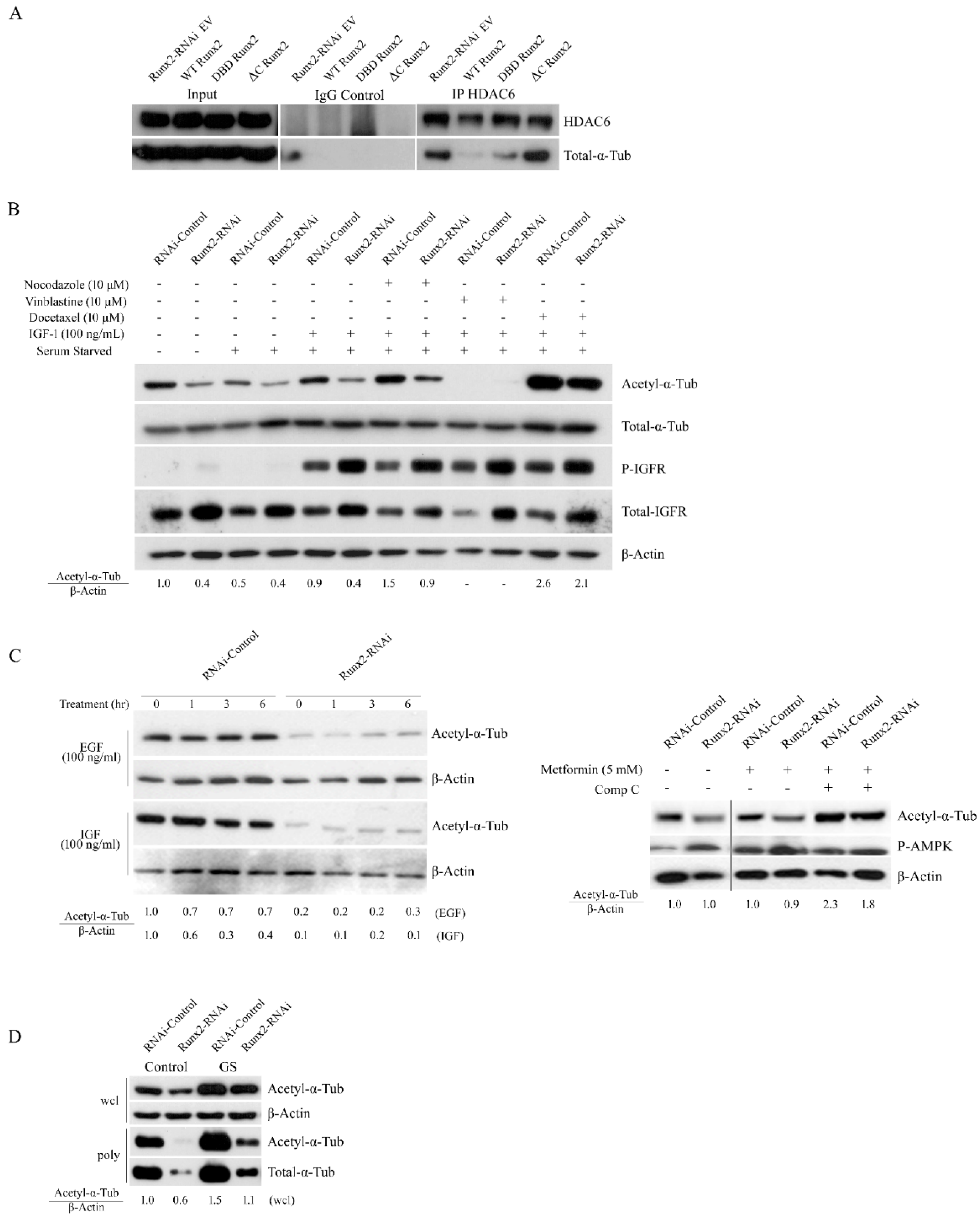
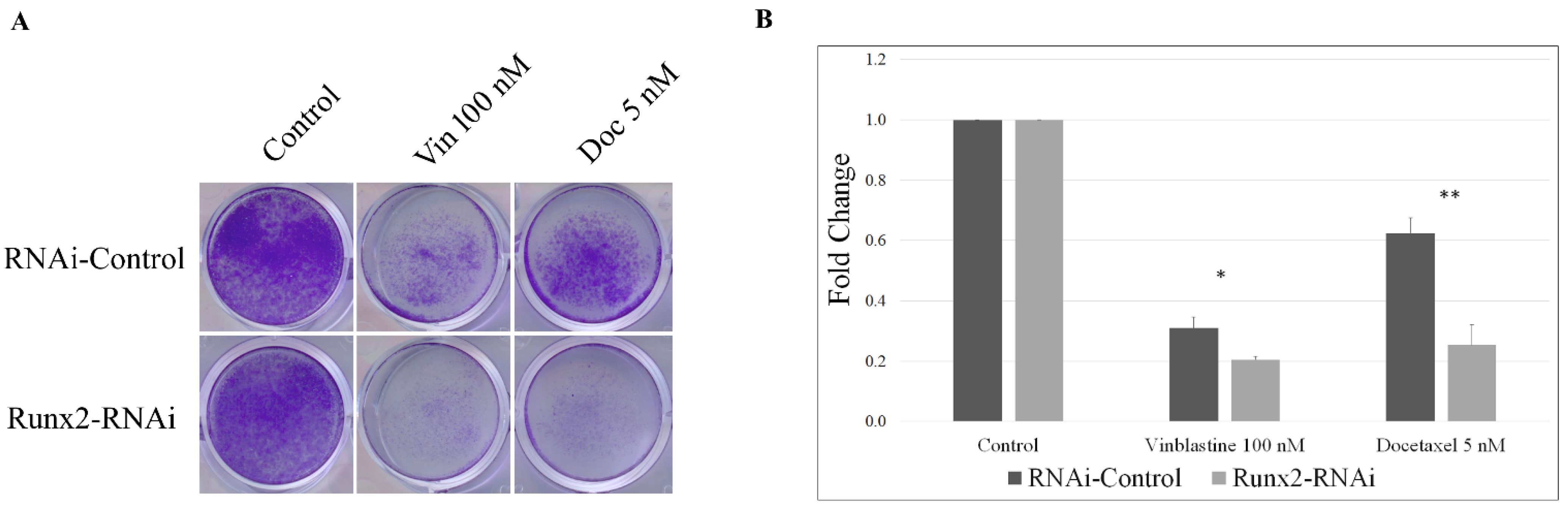
3. Results
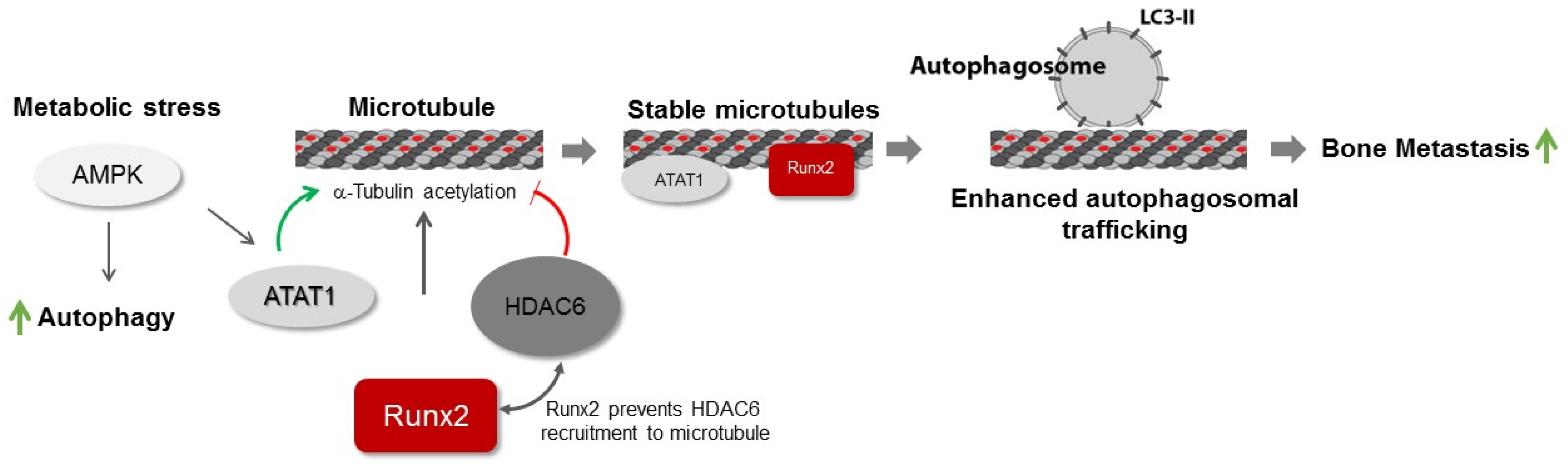
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA A Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Tandon, M.; Chen, Z.; Othman, A.H.; Pratap, J. Role of Runx2 in IGF-1Rβ/Akt-and AMPK/Erk-dependent growth, survival and sensitivity towards metformin in breast cancer bone metastasis. Oncogene 2016, 35, 4730–4740. [Google Scholar] [CrossRef] [PubMed]
- Tandon, M.; Othman, A.H.; Ashok, V.; Stein, G.S.; Pratap, J. The role of Runx2 in facilitating autophagy in metastatic breast cancer cells. J. Cell. Physiol. 2018, 233, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Roodman, G.D. Biology of osteoclast activation in cancer. J. Clin. Oncol. 2001, 19, 3562–3571. [Google Scholar] [CrossRef]
- Weilbaecher, K.N.; Guise, T.A.; McCauley, L.K. Cancer to bone: A fatal attraction. Nat. Rev. Cancer 2011, 11, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Javed, A.; Barnes, G.L.; Pratap, J.; Antkowiak, T.; Gerstenfeld, L.C.; van Wijnen, A.J.; Stein, J.L.; Lian, J.B.; Stein, G.S. Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc. Natl. Acad. Sci. USA 2005, 102, 1454–1459. [Google Scholar] [CrossRef] [Green Version]
- Akech, J.; Wixted, J.J.; Bedard, K.; Van der Deen, M.; Hussain, S.; Guise, T.A.; Van Wijnen, A.J.; Stein, J.L.; Languino, L.R.; Altieri, D.C. Runx2 association with progression of prostate cancer in patients: Mechanisms mediating bone osteolysis and osteoblastic metastatic lesions. Oncogene 2010, 29, 811–821. [Google Scholar] [CrossRef] [Green Version]
- Barnes, G.L.; Hebert, K.E.; Kamal, M.; Javed, A.; Einhorn, T.A.; Lian, J.B.; Stein, G.S.; Gerstenfeld, L.C. Fidelity of Runx2 Activity in Breast Cancer Cells Is Required for the Generation of Metastases-Associated Osteolytic Disease. Cancer Res. 2004, 64, 4506–4513. [Google Scholar] [CrossRef] [Green Version]
- Westendorf, J.J.; Zaidi, S.K.; Cascino, J.E.; Kahler, R.; van Wijnen, A.J.; Lian, J.B.; Yoshida, M.; Stein, G.S.; Li, X. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol. Cell. Biol. 2002, 22, 7982–7992. [Google Scholar] [CrossRef] [Green Version]
- Pockwinse, S.M.; Rajgopal, A.; Young, D.W.; Mujeeb, K.A.; Nickerson, J.; Javed, A.; Redick, S.; Lian, J.B.; Van Wijnen, A.J.; Stein, J.L. Microtubule-dependent nuclear-cytoplasmic shuttling of Runx2. J. Cell. Physiol. 2006, 206, 354–362. [Google Scholar] [CrossRef]
- Webster, D.R.; Borisy, G.G. Microtubules are acetylated in domains that turn over slowly. J. Cell Sci. 1989, 92, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Eshun-Wilson, L.; Zhang, R.; Portran, D.; Nachury, M.V.; Toso, D.B.; Lohr, T.; Vendruscolo, M.; Bonomi, M.; Fraser, J.S.; Nogales, E. Effects of alpha-tubulin acetylation on microtubule structure and stability. Proc. Natl. Acad. Sci. USA 2019, 116, 10366–10371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Even, A.; Morelli, G.; Broix, L.; Scaramuzzino, C.; Turchetto, S.; Gladwyn-Ng, I.; Le Bail, R.; Shilian, M.; Freeman, S.; Magiera, M.M. ATAT1-enriched vesicles promote microtubule acetylation via axonal transport. Sci. Adv. 2019, 5, eaax2705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, R.; Nguyen, S.; McKeehan, W.L.; Liu, L. Acetylated microtubules are required for fusion of autophagosomes with lysosomes. BMC Cell Biol. 2010, 11, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mackeh, R.; Lorin, S.; Ratier, A.; Mejdoubi-Charef, N.; Baillet, A.; Bruneel, A.; Hamaï, A.; Codogno, P.; Poüs, C.; Perdiz, D. Reactive oxygen species, AMP-activated protein kinase, and the transcription cofactor p300 regulate α-tubulin acetyltransferase-1 (αTAT-1/MEC-17)-dependent microtubule hyperacetylation during cell stress. J. Biol. Chem. 2014, 289, 11816–11828. [Google Scholar] [CrossRef] [Green Version]
- Boggs, A.E.; Vitolo, M.I.; Whipple, R.A.; Charpentier, M.S.; Goloubeva, O.G.; Ioffe, O.B.; Tuttle, K.C.; Slovic, J.; Lu, Y.; Mills, G.B.; et al. alpha-Tubulin acetylation elevated in metastatic and basal-like breast cancer cells promotes microtentacle formation, adhesion, and invasive migration. Cancer Res. 2015, 75, 203–215. [Google Scholar] [CrossRef] [Green Version]
- Debnath, J.; Muthuswamy, S.K.; Brugge, J.S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 2003, 30, 256–268. [Google Scholar] [CrossRef]
- Tandon, M.; Chen, Z.; Pratap, J. Runx2 activates PI3K/Akt signaling via mTORC2 regulation in invasive breast cancer cells. Breast Cancer Res. 2014, 16, R16. [Google Scholar] [CrossRef] [Green Version]
- Pratap, J.; Imbalzano, K.M.; Underwood, J.M.; Cohet, N.; Gokul, K.; Akech, J.; Van Wijnen, A.J.; Stein, J.L.; Imbalzano, A.N.; Nickerson, J.A. Ectopic runx2 expression in mammary epithelial cells disrupts formation of normal acini structure: Implications for breast cancer progression. Cancer Res. 2009, 69, 6807–6814. [Google Scholar] [CrossRef] [Green Version]
- Tandon, M.; Chen, Z.; Pratap, J. Role of Runx2 in crosstalk between Mek/Erk and PI3K/Akt signaling in MCF-10A cells. J. Cell. Biochem. 2014, 115, 2208–2217. [Google Scholar] [CrossRef]
- Geiger, R.C.; Taylor, W.; Glucksberg, M.R.; Dean, D.A. Cyclic stretch-induced reorganization of the cytoskeleton and its role in enhanced gene transfer. Gene Ther. 2006, 13, 725–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaughan, E.E.; Geiger, R.C.; Miller, A.M.; Loh-Marley, P.L.; Suzuki, T.; Miyata, N.; Dean, D.A. Microtubule acetylation through HDAC6 inhibition results in increased transfection efficiency. Molecular Ther. 2008, 16, 1841–1847. [Google Scholar] [CrossRef] [PubMed]
- Young, D.W.; Hassan, M.Q.; Pratap, J.; Galindo, M.; Zaidi, S.K.; Lee, S.H.; Yang, X.; Xie, R.; Javed, A.; Underwood, J.M.; et al. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature 2007, 445, 442–446. [Google Scholar] [CrossRef]
- Janke, C. The tubulin code: Molecular components, readout mechanisms, and functions. J. Cell Biol. 2014, 206, 461–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Yang, X.-J. Tubulin acetylation: Responsible enzymes, biological functions and human diseases. Cell. Mol. Life Sci. 2015, 72, 4237–4255. [Google Scholar] [CrossRef]
- Shah, N.; Kumar, S.; Zaman, N.; Pan, C.C.; Bloodworth, J.C.; Lei, W.; Streicher, J.M.; Hempel, N.; Mythreye, K.; Lee, N.Y. TAK1 activation of alpha-TAT1 and microtubule hyperacetylation control AKT signaling and cell growth. Nat. Commun. 2018, 9, 1696. [Google Scholar] [CrossRef] [Green Version]
- Williams, K.A.; Zhang, M.; Xiang, S.; Hu, C.; Wu, J.-Y.; Zhang, S.; Ryan, M.; Cox, A.D.; Der, C.J.; Fang, B. Extracellular signal-regulated kinase (ERK) phosphorylates histone deacetylase 6 (HDAC6) at serine 1035 to stimulate cell migration. J. Biol. Chem. 2013, 288, 33156–33170. [Google Scholar] [CrossRef] [Green Version]
- Mercado-Matos, J.; Clark, J.L.; Piper, A.J.; Janusis, J.; Shaw, L.M. Differential involvement of the microtubule cytoskeleton in insulin receptor substrate 1 (IRS-1) and IRS-2 signaling to AKT determines the response to microtubule disruption in breast carcinoma cells. J. Biol. Chem. 2017, 292, 7806–7816. [Google Scholar] [CrossRef] [Green Version]
- Onishi, K.; Higuchi, M.; Asakura, T.; Masuyama, N.; Gotoh, Y. The PI3K-Akt pathway promotes microtubule stabilization in migrating fibroblasts. Genes Cells 2007, 12, 535–546. [Google Scholar] [CrossRef]
- Chimge, N.-O.; Baniwal, S.K.; Little, G.H.; Chen, Y.-b.; Kahn, M.; Tripathy, D.; Borok, Z.; Frenkel, B. Regulation of breast cancer metastasis by Runx2 and estrogen signaling: The role of SNAI2. Breast Cancer Res. 2011, 13, R127. [Google Scholar] [CrossRef] [Green Version]
- Sierra, J.; Villagra, A.; Paredes, R.; Cruzat, F.; Gutierrez, S.; Javed, A.; Arriagada, G.; Olate, J.; Imschenetzky, M.; Van Wijnen, A.J. Regulation of the bone-specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyltransferase activity. Mol. Cell. Biol. 2003, 23, 3339–3351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lock, R.; Kenific, C.M.; Leidal, A.M.; Salas, E.; Debnath, J. Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov. 2014, 4, 466–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lock, R.; Roy, S.; Kenific, C.M.; Su, J.S.; Salas, E.; Ronen, S.M.; Debnath, J. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol. Biol. Cell 2011, 22, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Mahboubi, H.; Koromilas, A.E.; Stochaj, U. AMP Kinase Activation Alters Oxidant-Induced Stress Granule Assembly by Modulating Cell Signaling and Microtubule Organization. Mol. Pharmacol. 2016, 90, 460–468. [Google Scholar] [CrossRef] [Green Version]
- Harrison, R.E.; Turley, E.A. Active erk regulates microtubule stability in H-ras-transformed cells. Neoplasia 2001, 3, 385–394. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Othman, A.; Winogradzki, M.; Patel, S.; Holmes, W.; Blank, A.; Pratap, J. The Role of Runx2 in Microtubule Acetylation in Bone Metastatic Breast Cancer Cells. Cancers 2022, 14, 3436. https://doi.org/10.3390/cancers14143436
Othman A, Winogradzki M, Patel S, Holmes W, Blank A, Pratap J. The Role of Runx2 in Microtubule Acetylation in Bone Metastatic Breast Cancer Cells. Cancers. 2022; 14(14):3436. https://doi.org/10.3390/cancers14143436
Chicago/Turabian StyleOthman, Ahmad, Marcus Winogradzki, Shreya Patel, Waddell Holmes, Alan Blank, and Jitesh Pratap. 2022. "The Role of Runx2 in Microtubule Acetylation in Bone Metastatic Breast Cancer Cells" Cancers 14, no. 14: 3436. https://doi.org/10.3390/cancers14143436
APA StyleOthman, A., Winogradzki, M., Patel, S., Holmes, W., Blank, A., & Pratap, J. (2022). The Role of Runx2 in Microtubule Acetylation in Bone Metastatic Breast Cancer Cells. Cancers, 14(14), 3436. https://doi.org/10.3390/cancers14143436





