A Novel ALDH1A1 Inhibitor Blocks Platinum-Induced Senescence and Stemness in Ovarian Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemical Reagents
2.2. Protein Purification and Enzymatic Assays
2.3. X-ray Crystallography
2.4. Cell Culture
2.5. Flow Cytometry
2.6. Quantitative PCR
2.7. Senescence Beta (β)-Gal Assay
2.8. RNA Sequencing and Bioinformatic Analysis
2.9. Mouse Xenograft Experiment
2.10. Colony Formation and Tumorsphere Assay
2.11. MTT Cell Proliferation Assay
2.12. Cell Transfection and Plasmids
2.13. Western Blotting
2.14. Statistical Analysis
2.15. Data Availability
3. Results
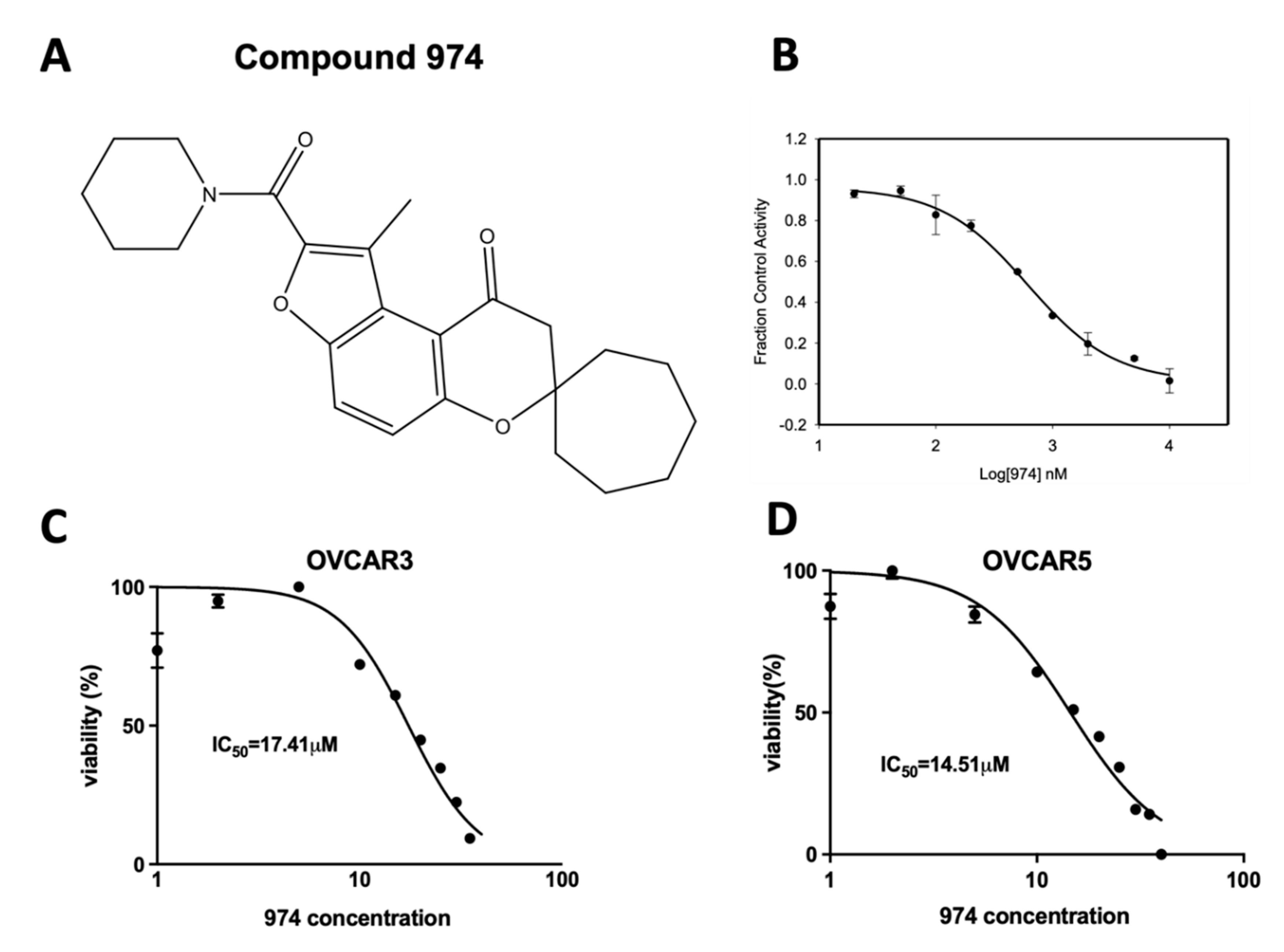
3.1. Discovery of 974, a Novel ALDH1A1-Specific Small-Molecule Inhibitor
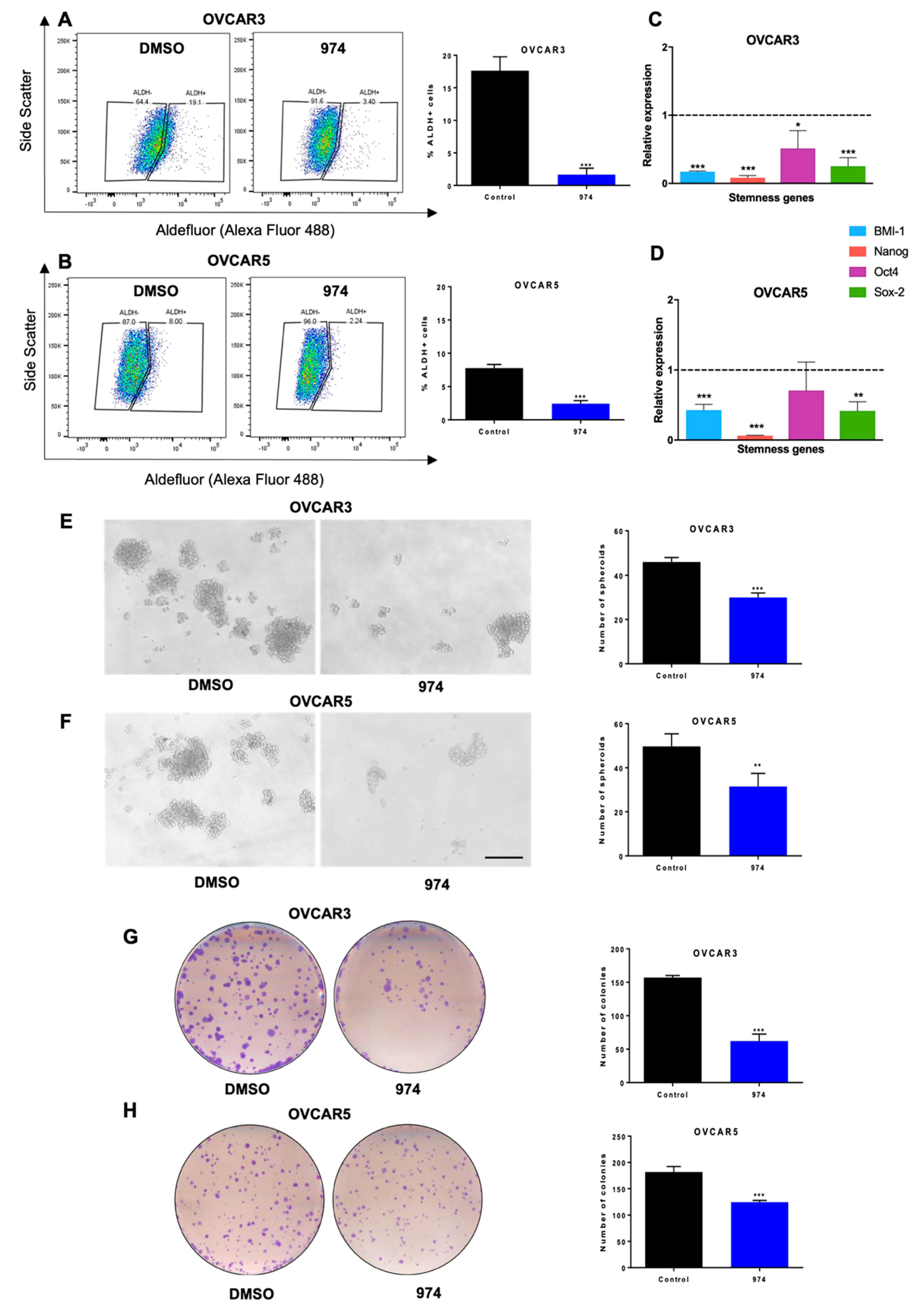
3.2. ALDH1A1 Inhibition Suppresses Stemness Phenotypes in Ovarian Cancer Cells
3.3. ALDH1A1 Inhibition Suppresses Ovarian Cancer Stemness In Vivo
3.4. ALDH1A1 Inhibition Downregulates Key Stemness and Chemoresistance Pathways
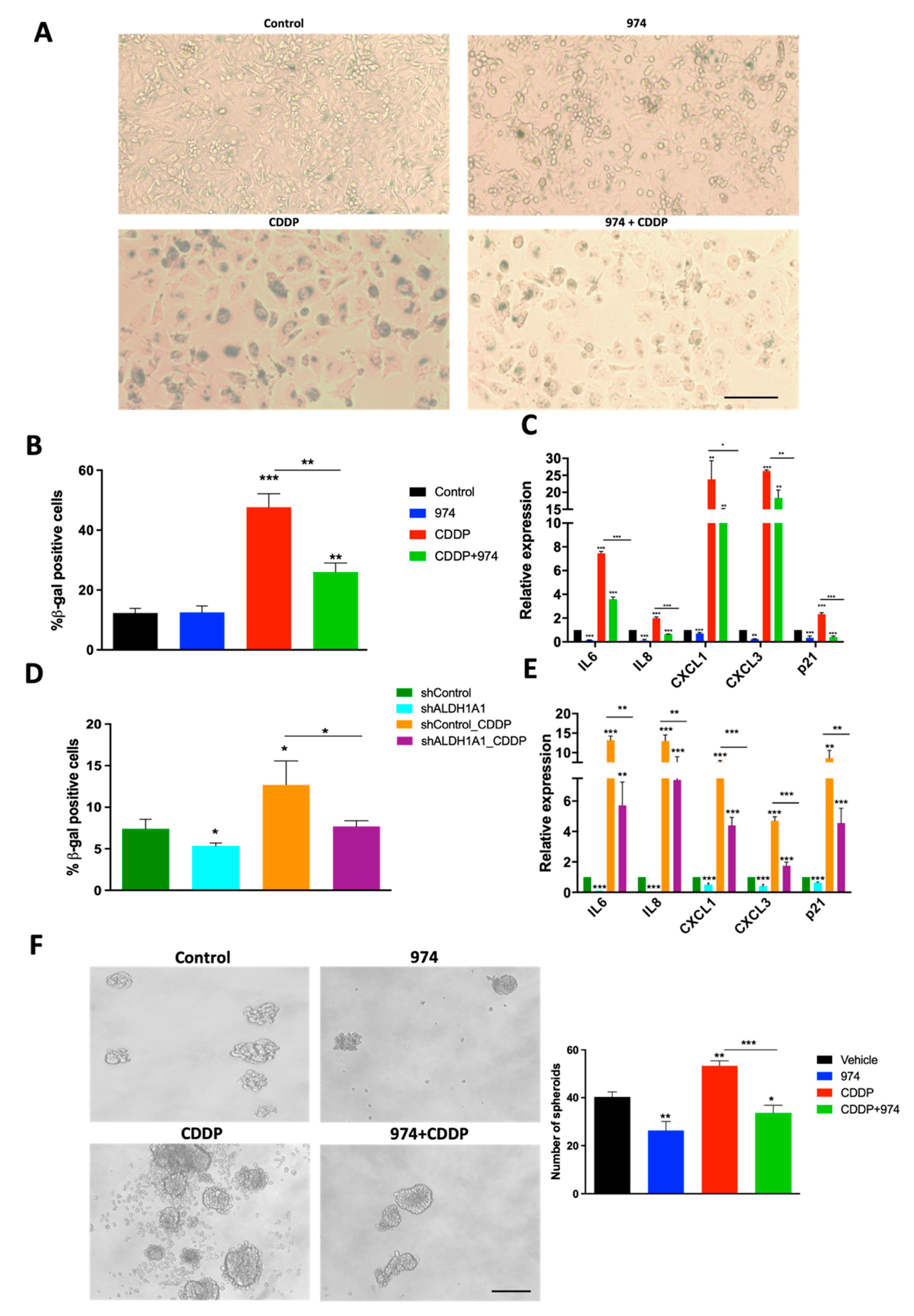
3.5. Inhibition of ALDH1A1 Suppresses Chemotherapy-Induced Senescence and Stemness
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Kurman, R.J.; Shih Ie, M. The Dualistic Model of Ovarian Carcinogenesis: Revisited, Revised, and Expanded. Am. J. Pathol. 2016, 186, 733–747. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.; Tinker, A.V.; Friedlander, M. “Platinum resistant” ovarian cancer: What is it, who to treat and how to measure benefit? Gynecol. Oncol. 2014, 133, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Steg, A.D.; Bevis, K.S.; Katre, A.A.; Ziebarth, A.; Dobbin, Z.C.; Alvarez, R.D.; Zhang, K.; Conner, M.; Landen, C.N. Stem cell pathways contribute to clinical chemoresistance in ovarian cancer. Clin. Cancer Res. 2012, 18, 869–881. [Google Scholar] [CrossRef]
- Al-Alem, L.F.; Pandya, U.M.; Baker, A.T.; Bellio, C.; Zarrella, B.D.; Clark, J.; DiGloria, C.M.; Rueda, B.R. Ovarian cancer stem cells: What progress have we made? Int. J. Biochem. Cell Biol. 2019, 107, 92–103. [Google Scholar] [CrossRef]
- Dean, M.; Fojo, T.; Bates, S. Tumour stem cells and drug resistance. Nat. Rev. Cancer 2005, 5, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Convery, P.A.; Matsumura, N.; Whitaker, R.S.; Kondoh, E.; Perry, T.; Huang, Z.; Bentley, R.C.; Mori, S.; Fujii, S.; et al. Epigenetic regulation of CD133 and tumorigenicity of CD133+ ovarian cancer cells. Oncogene 2009, 28, 209–218. [Google Scholar] [CrossRef]
- Zhang, S.; Balch, C.; Chan, M.W.; Lai, H.C.; Matei, D.; Schilder, J.M.; Yan, P.S.; Huang, T.H.; Nephew, K.P. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008, 68, 4311–4320. [Google Scholar] [CrossRef]
- Yu, S.; Mulero, M.C.; Chen, W.; Shang, X.; Tian, S.; Watanabe, J.; Watanabe, A.; Vorberg, T.; Wong, C.; Gately, D.; et al. Therapeutic Targeting of Tumor Cells Rich in LGR Stem Cell Receptors. Bioconjug. Chem. 2021, 32, 376–384. [Google Scholar] [CrossRef]
- Landen, C.N., Jr.; Goodman, B.; Katre, A.A.; Steg, A.D.; Nick, A.M.; Stone, R.L.; Miller, L.D.; Mejia, P.V.; Jennings, N.B.; Gershenson, D.M.; et al. Targeting aldehyde dehydrogenase cancer stem cells in ovarian cancer. Mol. Cancer Ther. 2010, 9, 3186–3199. [Google Scholar] [CrossRef]
- Muralikrishnan, V.; Hurley, T.D.; Nephew, K.P. Targeting Aldehyde Dehydrogenases to Eliminate Cancer Stem Cells in Gynecologic Malignancies. Cancers 2020, 12, 961. [Google Scholar] [CrossRef] [PubMed]
- Nwani, N.G.; Condello, S.; Wang, Y.; Swetzig, W.M.; Barber, E.; Hurley, T.; Matei, D. A Novel ALDH1A1 Inhibitor Targets Cells with Stem Cell Characteristics in Ovarian Cancer. Cancers 2019, 11, 502. [Google Scholar] [CrossRef] [PubMed]
- Milanovic, M.; Yu, Y.; Schmitt, C.A. The Senescence-Stemness Alliance—A Cancer-Hijacked Regeneration Principle. Trends Cell Biol. 2018, 28, 1049–1061. [Google Scholar] [CrossRef]
- Herranz, N.; Gil, J. Mechanisms and functions of cellular senescence. J. Clin. Investig. 2018, 128, 1238–1246. [Google Scholar] [CrossRef]
- Milanovic, M.; Fan, D.N.Y.; Belenki, D.; Dabritz, J.H.M.; Zhao, Z.; Yu, Y.; Dorr, J.R.; Dimitrova, L.; Lenze, D.; Monteiro Barbosa, I.A.; et al. Senescence-associated reprogramming promotes cancer stemness. Nature 2018, 553, 96–100. [Google Scholar] [CrossRef]
- Wang, Y.; Cardenas, H.; Fang, F.; Condello, S.; Taverna, P.; Segar, M.; Liu, Y.; Nephew, K.P.; Matei, D. Epigenetic targeting of ovarian cancer stem cells. Cancer Res. 2014, 74, 4922–4936. [Google Scholar] [CrossRef]
- House, C.D.; Jordan, E.; Hernandez, L.; Ozaki, M.; James, J.M.; Kim, M.; Kruhlak, M.J.; Batchelor, E.; Elloumi, F.; Cam, M.C.; et al. NFκB Promotes Ovarian Tumorigenesis via Classical Pathways That Support Proliferative Cancer Cells and Alternative Pathways That Support ALDH(+) Cancer Stem-like Cells. Cancer Res. 2017, 77, 6927–6940. [Google Scholar] [CrossRef]
- Wang, Y.; Zong, X.; Mitra, S.; Mitra, A.K.; Matei, D.; Nephew, K.P. IL-6 mediates platinum-induced enrichment of ovarian cancer stem cells. JCI Insight 2018, 3, e122360. [Google Scholar] [CrossRef]
- Chambers, C.R.; Ritchie, S.; Pereira, B.A.; Timpson, P. Overcoming the senescence-associated secretory phenotype (SASP): A complex mechanism of resistance in the treatment of cancer. Mol. Oncol. 2021, 15, 3242–3255. [Google Scholar] [CrossRef]
- Nacarelli, T.; Fukumoto, T.; Zundell, J.A.; Fatkhutdinov, N.; Jean, S.; Cadungog, M.G.; Borowsky, M.E.; Zhang, R. NAMPT Inhibition Suppresses Cancer Stem-like Cells Associated with Therapy-Induced Senescence in Ovarian Cancer. Cancer Res. 2020, 80, 890–900. [Google Scholar] [CrossRef] [PubMed]
- Hammen, P.K.; Allali-Hassani, A.; Hallenga, K.; Hurley, T.D.; Weiner, H. Multiple conformations of NAD and NADH when bound to human cytosolic and mitochondrial aldehyde dehydrogenase. Biochemistry 2002, 41, 7156–7168. [Google Scholar] [CrossRef]
- Parajuli, B.; Kimble-Hill, A.C.; Khanna, M.; Ivanova, Y.; Meroueh, S.; Hurley, T.D. Discovery of novel regulators of aldehyde dehydrogenase isoenzymes. Chem. Biol. Interact. 2011, 191, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, B.; Georgiadis, T.M.; Fishel, M.L.; Hurley, T.D. Development of selective inhibitors for human aldehyde dehydrogenase 3A1 (ALDH3A1) for the enhancement of cyclophosphamide cytotoxicity. Chembiochem 2014, 15, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Buchman, C.D.; Hurley, T.D. Inhibition of the Aldehyde Dehydrogenase 1/2 Family by Psoralen and Coumarin Derivatives. J. Med. Chem. 2017, 60, 2439–2455. [Google Scholar] [CrossRef]
- Morgan, C.A.; Hurley, T.D. Development of a high-throughput in vitro assay to identify selective inhibitors for human ALDH1A1. Chem. Biol. Interact. 2015, 234, 29–37. [Google Scholar] [CrossRef]
- Tang, J.; Pulliam, N.; Ozes, A.; Buechlein, A.; Ding, N.; Keer, H.; Rusch, D.; O’Hagan, H.; Stack, M.S.; Nephew, K.P. Epigenetic Targeting of Adipocytes Inhibits High-Grade Serous Ovarian Cancer Cell Migration and Invasion. Mol. Cancer Res. 2018, 16, 1226–1240. [Google Scholar] [CrossRef]
- Fang, F.; Munck, J.; Tang, J.; Taverna, P.; Wang, Y.; Miller, D.F.; Pilrose, J.; Choy, G.; Azab, M.; Pawelczak, K.S.; et al. The novel, small-molecule DNA methylation inhibitor SGI-110 as an ovarian cancer chemosensitizer. Clin. Cancer Res. 2014, 20, 6504–6516. [Google Scholar] [CrossRef]
- Zong, X.; Wang, W.; Ozes, A.; Fang, F.; Sandusky, G.E.; Nephew, K.P. EZH2-Mediated Downregulation of the Tumor Suppressor DAB2IP Maintains Ovarian Cancer Stem Cells. Cancer Res. 2020, 80, 4371–4385. [Google Scholar] [CrossRef]
- Connor, E.V.; Saygin, C.; Braley, C.; Wiechert, A.C.; Karunanithi, S.; Crean-Tate, K.; Abdul-Karim, F.W.; Michener, C.M.; Rose, P.G.; Lathia, J.D.; et al. Thy-1 predicts poor prognosis and is associated with self-renewal in ovarian cancer. J. Ovarian Res. 2019, 12, 112. [Google Scholar] [CrossRef]
- Caminear, M.W.; Harrington, B.S.; Kamdar, R.D.; Kruhlak, M.J.; Annunziata, C.M. Disulfiram Transcends ALDH Inhibitory Activity When Targeting Ovarian Cancer Tumor-Initiating Cells. Front. Oncol. 2022, 12, 762820. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, G.; Condello, S.; Huang, H.; Cardenas, H.; Tanner, E.J.; Wei, J.; Ji, Y.; Li, J.; Tan, Y.; et al. Frizzled-7 Identifies Platinum-Tolerant Ovarian Cancer Cells Susceptible to Ferroptosis. Cancer Res. 2021, 81, 384–399. [Google Scholar] [CrossRef]
- Ozes, A.R.; Miller, D.F.; Ozes, O.N.; Fang, F.; Liu, Y.; Matei, D.; Huang, T.; Nephew, K.P. NF-κB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene 2016, 35, 5350–5361. [Google Scholar] [CrossRef] [PubMed]
- Raspaglio, G.; Petrillo, M.; Martinelli, E.; Li Puma, D.D.; Mariani, M.; De Donato, M.; Filippetti, F.; Mozzetti, S.; Prislei, S.; Zannoni, G.F.; et al. Sox9 and Hif-2alpha regulate TUBB3 gene expression and affect ovarian cancer aggressiveness. Gene 2014, 542, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Condello, S.; Thomes-Pepin, J.; Ma, X.; Xia, Y.; Hurley, T.D.; Matei, D.; Cheng, J.X. Lipid Desaturation Is a Metabolic Marker and Therapeutic Target of Ovarian Cancer Stem Cells. Cell Stem Cell 2017, 20, 303–314.e305. [Google Scholar] [CrossRef]
- Hirschmann-Jax, C.; Foster, A.E.; Wulf, G.G.; Nuchtern, J.G.; Jax, T.W.; Gobel, U.; Goodell, M.A.; Brenner, M.K. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc. Natl. Acad. Sci. USA 2004, 101, 14228–14233. [Google Scholar] [CrossRef] [PubMed]
- Campisi, J.; d’Adda di Fagagna, F. Cellular senescence: When bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007, 8, 729–740. [Google Scholar] [CrossRef]
- Tomita, H.; Tanaka, K.; Tanaka, T.; Hara, A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget 2016, 7, 11018–11032. [Google Scholar] [CrossRef]
- Meng, E.; Mitra, A.; Tripathi, K.; Finan, M.A.; Scalici, J.; McClellan, S.; Madeira da Silva, L.; Reed, E.; Shevde, L.A.; Palle, K.; et al. ALDH1A1 maintains ovarian cancer stem cell-like properties by altered regulation of cell cycle checkpoint and DNA repair network signaling. PLoS ONE 2014, 9, e107142. [Google Scholar] [CrossRef]
- Kaipio, K.; Chen, P.; Roering, P.; Huhtinen, K.; Mikkonen, P.; Ostling, P.; Lehtinen, L.; Mansuri, N.; Korpela, T.; Potdar, S.; et al. ALDH1A1-related stemness in high-grade serous ovarian cancer is a negative prognostic indicator but potentially targetable by EGFR/mTOR-PI3K/aurora kinase inhibitors. J. Pathol. 2020, 250, 159–169. [Google Scholar] [CrossRef]
- Liu, L.; Cai, S.; Han, C.; Banerjee, A.; Wu, D.; Cui, T.; Xie, G.; Zhang, J.; Zhang, X.; McLaughlin, E.; et al. ALDH1A1 Contributes to PARP Inhibitor Resistance via Enhancing DNA Repair in BRCA2(−/−) Ovarian Cancer Cells. Mol. Cancer Ther. 2020, 19, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Levi, B.P.; Yilmaz, O.H.; Duester, G.; Morrison, S.J. Aldehyde dehydrogenase 1a1 is dispensable for stem cell function in the mouse hematopoietic and nervous systems. Blood 2009, 113, 1670–1680. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Cong, Y.; Qin, H.; Benveniste, E.N.; Elson, C.O. Generation of mucosal dendritic cells from bone marrow reveals a critical role of retinoic acid. J. Immunol. 2010, 185, 5915–5925. [Google Scholar] [CrossRef] [PubMed]
- Saw, Y.T.; Yang, J.; Ng, S.K.; Liu, S.; Singh, S.; Singh, M.; Welch, W.R.; Tsuda, H.; Fong, W.P.; Thompson, D.; et al. Characterization of aldehyde dehydrogenase isozymes in ovarian cancer tissues and sphere cultures. BMC Cancer 2012, 12, 329. [Google Scholar] [CrossRef] [PubMed]
- Huddle, B.C.; Grimley, E.; Buchman, C.D.; Chtcherbinine, M.; Debnath, B.; Mehta, P.; Yang, K.; Morgan, C.A.; Li, S.; Felton, J.; et al. Structure-Based Optimization of a Novel Class of Aldehyde Dehydrogenase 1A (ALDH1A) Subfamily-Selective Inhibitors as Potential Adjuncts to Ovarian Cancer Chemotherapy. J. Med. Chem. 2018, 61, 8754–8773. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.; Srivastava, A.K.; Han, C.; Wu, D.; Wani, N.; Liu, L.; Gao, Z.; Qu, M.; Zou, N.; Zhang, X.; et al. DDB2 represses ovarian cancer cell dedifferentiation by suppressing ALDH1A1. Cell Death Dis. 2018, 9, 561. [Google Scholar] [CrossRef]
- Uddin, M.H.; Kim, B.; Cho, U.; Azmi, A.S.; Song, Y.S. Association of ALDH1A1-NEK-2 axis in cisplatin resistance in ovarian cancer cells. Heliyon 2020, 6, e05442. [Google Scholar] [CrossRef]
- Yokoyama, Y.; Zhu, H.; Lee, J.H.; Kossenkov, A.V.; Wu, S.Y.; Wickramasinghe, J.M.; Yin, X.; Palozola, K.C.; Gardini, A.; Showe, L.C.; et al. BET Inhibitors Suppress ALDH Activity by Targeting ALDH1A1 Super-Enhancer in Ovarian Cancer. Cancer Res. 2016, 76, 6320–6330. [Google Scholar] [CrossRef]
- Ozes, A.R.; Wang, Y.; Zong, X.; Fang, F.; Pilrose, J.; Nephew, K.P. Therapeutic targeting using tumor specific peptides inhibits long non-coding RNA HOTAIR activity in ovarian and breast cancer. Sci. Rep. 2017, 7, 894. [Google Scholar] [CrossRef]
- Dey, D.K.; Kang, S.C. CopA3 peptide induces permanent cell-cycle arrest in colorectal cancer cells. Mech. Ageing Dev. 2021, 196, 111497. [Google Scholar] [CrossRef]
- Jordan, K.R.; Sikora, M.J.; Slansky, J.E.; Minic, A.; Richer, J.K.; Moroney, M.R.; Hu, J.; Wolsky, R.J.; Watson, Z.L.; Yamamoto, T.M.; et al. The Capacity of the Ovarian Cancer Tumor Microenvironment to Integrate Inflammation Signaling Conveys a Shorter Disease-free Interval. Clin. Cancer Res. 2020, 26, 6362–6373. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Torres, C.; Gaytan-Cervantes, J.; Vazquez-Santillan, K.; Mandujano-Tinoco, E.A.; Ceballos-Cancino, G.; Garcia-Venzor, A.; Zampedri, C.; Sanchez-Maldonado, P.; Mojica-Espinosa, R.; Jimenez-Hernandez, L.E.; et al. NF-κB Participates in the Stem Cell Phenotype of Ovarian Cancer Cells. Arch. Med. Res. 2017, 48, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Uruski, P.; Sepetowska, A.; Konieczna, C.; Pakula, M.; Wyrwa, M.; Tussupkaliyev, A.; Tykarski, A.; Mikula-Pietrasik, J.; Ksiazek, K. Primary high-grade serous ovarian cancer cells are sensitive to senescence induced by carboplatin and paclitaxel in vitro. Cell Mol. Biol. Lett. 2021, 26, 44. [Google Scholar] [CrossRef] [PubMed]
- Saleh, T.; Bloukh, S.; Carpenter, V.J.; Alwohoush, E.; Bakeer, J.; Darwish, S.; Azab, B.; Gewirtz, D.A. Therapy-Induced Senescence: An “Old” Friend Becomes the Enemy. Cancers 2020, 12, 822. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaissonet, M.; Gingeras, T. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muralikrishnan, V.; Fang, F.; Given, T.C.; Podicheti, R.; Chtcherbinine, M.; Metcalfe, T.X.; Sriramkumar, S.; O’Hagan, H.M.; Hurley, T.D.; Nephew, K.P. A Novel ALDH1A1 Inhibitor Blocks Platinum-Induced Senescence and Stemness in Ovarian Cancer. Cancers 2022, 14, 3437. https://doi.org/10.3390/cancers14143437
Muralikrishnan V, Fang F, Given TC, Podicheti R, Chtcherbinine M, Metcalfe TX, Sriramkumar S, O’Hagan HM, Hurley TD, Nephew KP. A Novel ALDH1A1 Inhibitor Blocks Platinum-Induced Senescence and Stemness in Ovarian Cancer. Cancers. 2022; 14(14):3437. https://doi.org/10.3390/cancers14143437
Chicago/Turabian StyleMuralikrishnan, Vaishnavi, Fang Fang, Tyler C. Given, Ram Podicheti, Mikhail Chtcherbinine, Tara X. Metcalfe, Shruthi Sriramkumar, Heather M. O’Hagan, Thomas D. Hurley, and Kenneth P. Nephew. 2022. "A Novel ALDH1A1 Inhibitor Blocks Platinum-Induced Senescence and Stemness in Ovarian Cancer" Cancers 14, no. 14: 3437. https://doi.org/10.3390/cancers14143437
APA StyleMuralikrishnan, V., Fang, F., Given, T. C., Podicheti, R., Chtcherbinine, M., Metcalfe, T. X., Sriramkumar, S., O’Hagan, H. M., Hurley, T. D., & Nephew, K. P. (2022). A Novel ALDH1A1 Inhibitor Blocks Platinum-Induced Senescence and Stemness in Ovarian Cancer. Cancers, 14(14), 3437. https://doi.org/10.3390/cancers14143437






