Circulating Exosome Cargoes Contain Functionally Diverse Cancer Biomarkers: From Biogenesis and Function to Purification and Potential Translational Utility
Abstract
:Simple Summary
Abstract
1. Introduction
2. Extracellular Vesicles (EVs): A Journey from Discovery to Clinical Utility
3. Current Exosome Isolation Techniques—Advantages and Limitations
3.1. Ultracentrifugation-Based Isolation
3.2. Size-Based Separation and Isolation
3.3. Polyethylene Glycol (PEG) Precipitation-Based Isolation
3.4. Immunoaffinity-Based Isolation
3.5. Microfluidics-Based Isolation
3.6. Exosome Sorting by Fluorescence Activated Cell Sorting (FACS)
4. Biogenesis and Function of Exosomes in Normal and Pathological Processes
4.1. Biogenesis
4.1.1. The Formation of Early Endosomes
4.1.2. The Maturation of Late Endosomes and Formation of Exosomes
4.1.3. MVB Trafficking and Exosome Release
4.2. Biological Functions of Exosomes
4.3. Alteration of Exosome Biogenesis in Tumor Cells
5. Exosomes: A Source of Tumor Biomarkers
5.1. MicroRNA Biomarkers
5.1.1. Current Technologies for Quantification of Exosomal MicroRNAs
High-Throughput Expression Analyses
Quantitative PCR (qPCR) Analyses
Droplet Digital PCR (ddPCR) Analysis
5.1.2. Function of MicroRNA in Cancer Exosomes
Regulation of Tumor Growth
Evasion from Host Immune Responses
Tumor Microenvironment Remodeling and Metastasis
5.2. Protein Biomarkers
5.2.1. Protein Extraction from Exosomes and EVs Prior to Analysis
5.2.2. Exosome and EV Protein Processing Approaches
Traditional Methods
Filter-Aided Sample Preparation
5.2.3. Mass Spectrometric Analysis of EV and Exosomal Proteins
Top-Down vs. Bottom-Up Proteomics
5.2.4. Mass Spectrometric Analysis of EV and Exosomal Protein Modifications
5.3. Lipid Biomarkers
5.3.1. Methods for the Analysis of Lipids from Exosomes
Analysis of Intact Exosomes
Direct MS Analysis of Lipid Extracts
Indirect MS Analysis of Lipid Extracts
5.3.2. Lipidomic Profiling of Exosomes from Cancer Cells and Cancer Cell Lines
5.3.3. Lipidomic Profiling of Exosomes from Liquid Biopsies
5.3.4. Lipidomic-Specific Exosome Isolation for Use as Cancer Biomarkers
6. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Weiderpass, E.; Soerjomataram, I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer 2021, 127, 3029–3030. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Global Health Estimates 2020: Deaths by Cause, Age, Sex, by Country and by Region, 2000–2019. WHO. 2020. Available online: Who.int/data/gho/data/themes/mortality-and-global-health-estimates/ghe-leading-causes-of-death (accessed on 1 March 2022).
- The Centers for Disease Control and Prevention (CDC). Trends in Breast Cancer Incidence, by Race, Ethnicity, and aGE aMONG Women Aged >20 Years—United Steates, 1999–2018. Available online: https://www.cdc.gov/mmwr/volumes/71/wr/mm7102a2.htm (accessed on 10 May 2022).
- Wender, R.C.; Brawley, O.W.; Fedewa, S.A.; Gansler, T.; Smith, R.A. A blueprint for cancer screening and early detection: Advancing screening’s contribution to cancer control. CA Cancer J. Clin. 2019, 69, 50–79. [Google Scholar] [CrossRef] [PubMed]
- Weigelt, B.; Peterse, J.L.; van’t Veer, L.J. Breast cancer metastasis: Markers and models. Nat. Rev. Cancer 2005, 5, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Mehlen, P.; Puisieux, A. Metastasis: A question of life or death. Nat. Rev. Cancer 2006, 6, 449–458. [Google Scholar] [CrossRef]
- Dillekås, H.; Rogers, M.S.; Straume, O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019, 8, 5574–5576. [Google Scholar] [CrossRef] [Green Version]
- Thalanayar, P.M.; Altintas, N.; Weissfeld, J.L.; Fuhrman, C.R.; Wilson, D.O. Indolent, Potentially Inconsequential Lung Cancers in the Pittsburgh Lung Screening Study. Ann. Am. Thorac. Soc. 2015, 12, 1193–1196. [Google Scholar] [CrossRef] [Green Version]
- Zmrzljak, U.P.; Košir, R.; Krivokapić, Z.; Radojković, D.; Nikolić, A. Detection of Somatic Mutations with ddPCR from Liquid Biopsy of Colorectal Cancer Patients. Genes 2021, 12, 289. [Google Scholar] [CrossRef]
- Zou, W.; Yaung, S.J.; Fuhlbrück, F.; Ballinger, M.; Peters, E.; Palma, J.F.; Shames, D.S.; Gandara, D.; Jiang, Y.; Patil, N.S. ctDNA Predicts Overall Survival in Patients With NSCLC Treated with PD-L1 Blockade or with Chemotherapy. JCO Precis. Oncol. 2021, 5, 827–838. [Google Scholar] [CrossRef]
- Omri, N.; Kamil, M.; Alexander, K.; Alexander, K.; Edmond, S.; Ariel, Z.; David, K.; Gilad, A.E.; Azik, H. Association between PSA density and pathologically significant prostate cancer: The impact of prostate volume. Prostate 2020, 80, 1444–1449. [Google Scholar] [CrossRef]
- Wu, L.X.; Li, X.F.; Chen, H.F.; Zhu, Y.C.; Wang, W.X.; Xu, C.W.; Xie, D.F.; Wan, Y.; Du, K.Q. Combined detection of CEA and CA125 for the diagnosis for lung cancer: A meta-analysis. Cell Mol. Biol. 2018, 64, 67–70. [Google Scholar] [CrossRef]
- Charkhchi, P.; Cybulski, C.; Gronwald, J.; Wong, F.O.; Narod, S.A.; Akbari, M.R. CA125 and Ovarian Cancer: A Comprehensive Review. Cancers 2020, 12, 3730. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, X.; Tian, B.; Wang, Y.; Du, L.; Sun, T.; Shi, Y.; Zhao, X.; Jing, J. The diagnostic value of serum tumor markers CEA, CA19-9, CA125, CA15-3, and TPS in metastatic breast cancer. Clin. Chim. Acta 2017, 470, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Diamandis, E.P. Towards identification of true cancer biomarkers. BMC Med. 2014, 12, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maestroni, U.; Cavalieri, D.M.; Campobasso, D.; Guarino, G.; Ziglioli, F. PSA-IgM and iXip in the diagnosis and management of prostate cancer: Clinical relevance and future potential. A review. Acta Biomed. 2022, 92, e2021344. [Google Scholar] [CrossRef]
- Zhang, M.; Cheng, S.; Jin, Y.; Zhao, Y.; Wang, Y. Roles of CA125 in diagnosis, prediction, and oncogenesis of ovarian cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188503. [Google Scholar] [CrossRef]
- Zhang, B.; Sun, Z.; Song, M.; Ma, S.; Tian, Y.; Kong, Q. Ultrasound/CT combined with serum CEA/CA19.9 in the diagnosis and prognosis of rectal cancer. J. Buon. 2018, 23, 592–597. [Google Scholar]
- Taksler, G.B.; Keating, N.L.; Rothberg, M.B. Implications of false-positive results for future cancer screenings. Cancer 2018, 124, 2390–2398. [Google Scholar] [CrossRef] [Green Version]
- Palsdottir, T.; Nordstrom, T.; Karlsson, A.; Grönberg, H.; Clements, M.; Eklund, M. The impact of different prostate-specific antigen (PSA) testing intervals on Gleason score at diagnosis and the risk of experiencing false-positive biopsy recommendations: A population-based cohort study. BMJ Open 2019, 9, e027958. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Kumar-Sinha, C.; Chinnaiyan, A.M. Precision oncology in the age of integrative genomics. Nat. Biotechnol. 2018, 36, 46–60. [Google Scholar] [CrossRef] [PubMed]
- Krol, I.; Schwab, F.D.; Carbone, R.; Ritter, M.; Picocci, S.; De Marni, M.L.; Stepien, G.; Franchi, G.M.; Zanardi, A.; Rissoglio, M.D.; et al. Detection of clustered circulating tumour cells in early breast cancer. Br. J. Cancer 2021, 125, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Ried, K.; Tamanna, T.; Matthews, S.; Eng, P.; Sali, A. New Screening Test Improves Detection of Prostate Cancer Using Circulating Tumor Cells and Prostate-Specific Markers. Front. Oncol. 2020, 10, 582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, R.; Cai, Z.; Li, S.; Cheng, Y.; Gao, H.; Liu, F.; Wu, S.; Liu, S.; Dong, Y.; Zheng, L.; et al. Expression and clinical relevance of epithelial and mesenchymal markers in circulating tumor cells from colorectal cancer. Oncotarget 2017, 8, 9293–9302. [Google Scholar] [CrossRef] [Green Version]
- Ehlers, M.; Allelein, S.; Schwarz, F.; Hautzel, H.; Kuebart, A.; Schmidt, M.; Haase, M.; Dringenberg, T.; Schott, M. Increased Numbers of Circulating Tumor Cells in Thyroid Cancer Patients. Horm. Metab. Res. 2018, 50, 602–608. [Google Scholar] [CrossRef]
- Sequist, L.V.; Nagrath, S.; Toner, M.; Haber, D.A.; Lynch, T.J. The CTC-chip: An exciting new tool to detect circulating tumor cells in lung cancer patients. J. Thorac. Oncol. 2009, 4, 281–283. [Google Scholar] [CrossRef] [Green Version]
- Aaltonen, K.E.; Novosadová, V.; Bendahl, P.O.; Graffman, C.; Larsson, A.M.; Rydén, L. Molecular characterization of circulating tumor cells from patients with metastatic breast cancer reflects evolutionary changes in gene expression under the pressure of systemic therapy. Oncotarget 2017, 8, 45544–45565. [Google Scholar] [CrossRef]
- Qi, L.N.; Xiang, B.D.; Wu, F.X.; Ye, J.Z.; Zhong, J.H.; Wang, Y.Y.; Chen, Y.Y.; Chen, Z.S.; Ma, L.; Chen, J.; et al. Circulating Tumor Cells Undergoing EMT Provide a Metric for Diagnosis and Prognosis of Patients with Hepatocellular Carcinoma. Cancer Res. 2018, 78, 4731–4744. [Google Scholar] [CrossRef] [Green Version]
- Gorges, T.M.; Riethdorf, S.; von Ahsen, O.; Nastał, Y.P.; Röck, K.; Boede, M.; Peine, S.; Kuske, A.; Schmid, E.; Kneip, C.; et al. Heterogeneous PSMA expression on circulating tumor cells: A potential basis for stratification and monitoring of PSMA-directed therapies in prostate cancer. Oncotarget 2016, 7, 34930–34941. [Google Scholar] [CrossRef] [Green Version]
- Braig, D.; Becherer, C.; Bickert, C.; Braig, M.; Claus, R.; Eisenhardt, A.E.; Heinz, J.; Scholber, J.; Herget, G.W.; Bronsert, P.; et al. Genotyping of circulating cell-free DNA enables noninvasive tumor detection in myxoid liposarcomas. Int. J. Cancer 2019, 145, 1148–1161. [Google Scholar] [CrossRef]
- Lv, J.; Chen, Y.; Zhou, G.; Qi, Z.; Tan, K.R.L.; Wang, H.; Lin, L.; Chen, F.; Zhang, L.; Huang, X.; et al. Liquid biopsy tracking during sequential chemo-radiotherapy identifies distinct prognostic phenotypes in nasopharyngeal carcinoma. Nat. Commun. 2019, 10, 3941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrmann, S.; Zhan, T.; Betge, J.; Rauscher, B.; Belle, S.; Gutting, T.; Schulte, N.; Jesenofsky, R.; Härtel, N.; Gaiser, T.; et al. Detection of mutational patterns in cell-free DNA of colorectal cancer by custom amplicon sequencing. Mol. Oncol. 2019, 13, 1669–1683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, R.H.; Wei, W.; Krawczyk, M.; Wang, W.; Luo, H.; Flagg, K.; Yi, S.; Shi, W.; Quan, Q.; Li, K.; et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 2017, 16, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Onstenk, W.; Sieuwerts, A.M.; Weekhout, M.; Mostert, B.; Reijm, E.A.; van Deurzen, C.H.; Bolt-de Vries, J.B.; Peeters, D.J.; Hamberg, P.; Seynaeve, C.; et al. Gene expression profiles of circulating tumor cells versus primary tumors in metastatic breast cancer. Cancer Lett. 2015, 362, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lin, X.; Huang, Y.; Wang, M.; Cen, C.; Tang, S.; Dique, M.R.; Cai, L.; Luis, M.A.; Smollar, J.; et al. Detection Methods and Clinical Applications of Circulating Tumor Cells in Breast Cancer. Front. Oncol. 2021, 11, 652253. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Veiga, T.; Martínez-Fernández, M.; Abuin, C.; Piñeiro, R.; Cebey, V.; Cueva, J.; Palacios, P.; Blanco, C.; Muinelo-Romay, L.; Abalo, A.; et al. CTCs Expression Profiling for Advanced Breast Cancer Monitoring. Cancers 2019, 11, 1941. [Google Scholar] [CrossRef] [Green Version]
- Auer, M.; Heitzer, E.; Ulz, P.; Geigl, J.B.; Speicher, M.R. Single circulating tumor cell sequencing for monitoring. Oncotarget 2013, 4, 812–813. [Google Scholar] [CrossRef] [Green Version]
- Zinggeler, M.; Brandstetter, T.; Rühe, J. Biophysical Insights on the Enrichment of Cancer Cells from Whole Blood by (Affinity) Filtration. Sci. Rep. 2019, 9, 1246. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Wu, D.; Li, S.; Han, Y.; Xiang, N.; Wang, C.; Ni, Z. A polymer-film inertial microfluidic sorter fabricated by jigsaw puzzle method for precise size-based cell separation. Anal. Chim. Acta 2021, 1143, 306–314. [Google Scholar] [CrossRef]
- Chen, K.; Amontree, J.; Varillas, J.; Zhang, J.; George, T.J.; Fan, Z.H. Incorporation of lateral microfiltration with immunoaffinity for enhancing the capture efficiency of rare cells. Sci. Rep. 2020, 10, 14210. [Google Scholar] [CrossRef]
- Ates, H.C.; Ozgur, E.; Kulah, H. Comparative study on antibody immobilization strategies for efficient circulating tumor cell capture. Biointerphases 2018, 13, 021001. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, R.; Zhang, L.; Guo, S. Nanomaterial-Based Immunocapture Platforms for the Recognition, Isolation, and Detection of Circulating Tumor Cells. Front. Bioeng. Biotechnol. 2022, 10, 850241. [Google Scholar] [CrossRef]
- Mohme, M.; Riethdorf, S.; Pantel, K. Circulating and disseminated tumour cells—Mechanisms of immune surveillance and escape. Nat. Rev. Clin. Oncol. 2017, 14, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Riethdorf, S.; O’Flaherty, L.; Hille, C.; Pantel, K. Clinical applications of the CellSearch platform in cancer patients. Adv. Drug Deliv. Rev. 2018, 125, 102–121. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Liu, Y.; Jenkins, B.D.; Cheng, R.; Harris, B.N.; Zhang, W.; Xie, J.; Murrow, J.R.; Hodgson, J.; Egan, M.; et al. Tumor antigen-independent and cell size variation-inclusive enrichment of viable circulating tumor cells. Lab. Chip. 2019, 19, 1860–1876. [Google Scholar] [CrossRef]
- Gao, W.; Yuan, H.; Jing, F.; Wu, S.; Zhou, H.; Mao, H.; Jin, Q.; Zhao, J.; Cong, H.; Jia, C. Analysis of circulating tumor cells from lung cancer patients with multiple biomarkers using high-performance size-based microfluidic chip. Oncotarget 2017, 8, 12917–12928. [Google Scholar] [CrossRef] [Green Version]
- Ziegler, A.; Zangemeister-Wittke, U.; Stahel, R.A. Circulating DNA: A new diagnostic gold mine? Cancer Treat. Rev. 2002, 28, 255–271. [Google Scholar] [CrossRef]
- Yi, Z.; Ma, F.; Li, C.; Chen, R.; Yuan, L.; Sun, X.; Guan, X.; Li, L.; Liu, B.; Guan, Y.; et al. Landscape of somatic mutations in different subtypes of advanced breast cancer with circulating tumor DNA analysis. Sci. Rep. 2017, 7, 5995. [Google Scholar] [CrossRef] [Green Version]
- Chin, R.I.; Chen, K.; Usmani, A.; Chua, C.; Harris, P.K.; Binkley, M.S.; Azad, T.D.; Dudley, J.C.; Chaudhuri, A.A. Detection of Solid Tumor Molecular Residual Disease (MRD) Using Circulating Tumor DNA (ctDNA). Mol. Diagn. Ther. 2019, 23, 311–331. [Google Scholar] [CrossRef]
- Zhang, S.; Brazel, D.; Kumar, P.; Schafer, L.N.; Eidenschink, B.; Senthil, M.; Dayyani, F. Utility of tumor-informed circulating tumor DNA in the clinical management of gastrointestinal malignancies. J. Gastrointest. Oncol. 2021, 12, 2643–2652. [Google Scholar] [CrossRef]
- Zugazagoitia, J.; Ramos, I.; Trigo, J.M.; Palka, M.; Gómez-Rueda, A.; Jantus-Lewintre, E.; Camps, C.; Isla, D.; Iranzo, P.; Ponce-Aix, S.; et al. Clinical utility of plasma-based digital next-generation sequencing in patients with advance-stage lung adenocarcinomas with insufficient tumor samples for tissue genotyping. Ann. Oncol. 2019, 30, 290–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagahashi, M.; Shimada, Y.; Ichikawa, H.; Kameyama, H.; Takabe, K.; Okuda, S.; Wakai, T. Next generation sequencing-based gene panel tests for the management of solid tumors. Cancer Sci. 2019, 110, 6–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.T.; Abelson, S.; Zou, J.; Li, T.; Zhao, Z.; Dick, J.E.; Shlush, L.I.; Pugh, T.J.; Bratman, S.V. High efficiency error suppression for accurate detection of low-frequency variants. Nucleic Acids Res. 2019, 47, e87. [Google Scholar] [CrossRef] [PubMed]
- Ignatiadis, M.; Lee, M.; Jeffrey, S.S. Circulating Tumor Cells and Circulating Tumor DNA: Challenges and Opportunities on the Path to Clinical Utility. Clin. Cancer Res. 2015, 21, 4786–4800. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; He, Q.; Liang, H.; Cheng, B.; Li, J.; Xiong, S.; Zhao, Y.; Guo, M.; Liu, Z.; He, J.; et al. Diagnostic Accuracy of Droplet Digital PCR and Amplification Refractory Mutation System PCR for Detecting EGFR Mutation in Cell-Free DNA of Lung Cancer: A Meta-Analysis. Front. Oncol. 2020, 10, 290. [Google Scholar] [CrossRef] [Green Version]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Villaflor, V.; Won, B.; Nagy, R.; Banks, K.; Lanman, R.B.; Talasaz, A.; Salgia, R. Biopsy-free circulating tumor DNA assay identifies actionable mutations in lung cancer. Oncotarget 2016, 7, 66880–66891. [Google Scholar] [CrossRef] [Green Version]
- Mego, M.; De Giorgi, U.; Dawood, S.; Wang, X.; Valero, V.; Andreopoulou, E.; Handy, B.; Ueno, N.T.; Reuben, J.M.; Cristofanilli, M. Characterization of metastatic breast cancer patients with nondetectable circulating tumor cells. Int. J. Cancer 2011, 129, 417–423. [Google Scholar] [CrossRef]
- Keller, L.; Belloum, Y.; Wikman, H.; Pantel, K. Clinical relevance of blood-based ctDNA analysis: Mutation detection and beyond. Br. J. Cancer 2021, 124, 345–358. [Google Scholar] [CrossRef]
- Wang, Q.; Zhao, L.; Han, L.; Tuo, X.; Ma, S.; Wang, Y.; Feng, X.; Liang, D.; Sun, C.; Wang, Q.; et al. The Discordance of Gene Mutations between Circulating Tumor Cells and Primary/Metastatic Tumor. Mol. Ther. Oncolytics 2019, 15, 21–29. [Google Scholar] [CrossRef] [Green Version]
- Wong, C.H.; Chen, Y.C. Clinical significance of exosomes as potential biomarkers in cancer. World J. Clin. Cases 2019, 7, 171–190. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Achreja, A.; Iessi, E.; Logozzi, M.; Mizzoni, D.; Di Raimo, R.; Nagrath, D.; Fais, S. The key role of extracellular vesicles in the metastatic process. Biochim. Biophys. Acta Rev. Cancer 2018, 1869, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ji, X.; Liu, J.; Fan, D.; Zhou, Q.; Chen, C.; Wang, W.; Wang, G.; Wang, H.; Yuan, W.; et al. Effects of exosomes on pre-metastatic niche formation in tumors. Mol. Cancer 2019, 18, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, C.; Chen, M.; Jiang, R.; Guo, Y.; Wu, M.; Zhang, X. Exosome-related tumor microenvironment. J. Cancer 2018, 9, 3084–3092. [Google Scholar] [CrossRef] [Green Version]
- Maia, J.; Caja, S.; Strano Moraes, M.C.; Couto, N.; Costa-Silva, B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front. Cell Dev. Biol. 2018, 6, 18. [Google Scholar] [CrossRef]
- Konoshenko, M.; Sagaradze, G.; Orlova, E.; Shtam, T.; Proskura, K.; Kamyshinsky, R.; Yunusova, N.; Alexandrova, A.; Efimenko, A.; Tamkovich, S. Total Blood Exosomes in Breast Cancer: Potential Role in Crucial Steps of Tumorigenesis. Int. J. Mol. Sci. 2020, 21, 7341. [Google Scholar] [CrossRef] [PubMed]
- Willms, A.; Müller, C.; Julich, H.; Klein, N.; Schwab, R.; Güsgen, C.; Richardsen, I.; Schaaf, S.; Krawczyk, M.; Krawczyk, M.; et al. Tumour-associated circulating microparticles: A novel liquid biopsy tool for screening and therapy monitoring of colorectal carcinoma and other epithelial neoplasia. Oncotarget 2016, 7, 30867–30875. [Google Scholar] [CrossRef] [Green Version]
- Torrano, V.; Royo, F.; Peinado, H.; Loizaga-Iriarte, A.; Unda, M.; Falcón-Perez, J.M.; Carracedo, A. Vesicle-MaNiA: Extracellular vesicles in liquid biopsy and cancer. Curr. Opin. Pharmacol. 2016, 29, 47–53. [Google Scholar] [CrossRef] [Green Version]
- Ko, J.; Bhagwat, N.; Black, T.; Yee, S.S.; Na, Y.J.; Fisher, S.; Kim, J.; Carpenter, E.L.; Stanger, B.Z.; Issadore, D. miRNA Profiling of Magnetic Nanopore-Isolated Extracellular Vesicles for the Diagnosis of Pancreatic Cancer. Cancer Res. 2018, 78, 3688–3697. [Google Scholar] [CrossRef] [Green Version]
- Chargaff, E.; West, R. The biological significance of the thromboplastic protein of blood. J. Biol. Chem. 1946, 166, 189–197. [Google Scholar] [CrossRef]
- Hargett, L.A.; Bauer, N.N. On the origin of microparticles: From “platelet dust” to mediators of intercellular communication. Pulm. Circ. 2013, 3, 329–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chargaff, E. Cell structure and the problem of blood coagulation. J. Biol. Chem. 1945, 160, 351–359. [Google Scholar] [CrossRef]
- Harding, C.; Heuser, J.; Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983, 97, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Mathew, A.; Mason, A.B.; Teng, K. Exosome formation during maturation of mammalian and avian reticulocytes: Evidence that exosome release is a major route for externalization of obsolete membrane proteins. J. Cell Physiol. 1991, 147, 27–36. [Google Scholar] [CrossRef]
- Gawrisch, K.; Stibenz, D.; Möps, A.; Arnold, K.; Linss, W.; Halbhuber, K.J. The rate of lateral diffusion of phospholipids in erythrocyte microvesicles. Biochim. Biophys. Acta 1986, 856, 443–447. [Google Scholar] [CrossRef]
- Vidal, M.; Sainte-Marie, J.; Philippot, J.R.; Bienvenue, A. Asymmetric distribution of phospholipids in the membrane of vesicles released during in vitro maturation of guinea pig reticulocytes: Evidence precluding a role for “aminophospholipid translocase”. J. Cell Physiol. 1989, 140, 455–462. [Google Scholar] [CrossRef]
- Vidal, M.J.; Stahl, P.D. The small GTP-binding proteins Rab4 and ARF are associated with released exosomes during reticulocyte maturation. Eur. J. Cell Biol. 1993, 60, 261–267. [Google Scholar]
- Lee, Y.J.; Jy, W.; Horstman, L.L.; Janania, J.; Reyes, Y.; Kelley, R.E.; Ahn, Y.S. Elevated platelet microparticles in transient ischemic attacks, lacunar infarcts, and multiinfarct dementias. Thromb. Res. 1993, 72, 295–304. [Google Scholar] [CrossRef]
- Singh, N.; Gemmell, C.H.; Daly, P.A.; Yeo, E.L. Elevated platelet-derived microparticle levels during unstable angina. Can. J. Cardiol. 1995, 11, 1015–1021. [Google Scholar]
- Powell, J.J.; Harvey, R.S.; Thompson, R.P. Microparticles in Crohn’s disease—Has the dust settled? Gut 1996, 39, 340–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalra, H.; Adda, C.G.; Liem, M.; Ang, C.-S.; Mechler, A.; Simpson, R.J.; Hulett, M.D.; Mathivanan, S. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics 2013, 13, 3354–3364. [Google Scholar] [CrossRef] [PubMed]
- Wubbolts, R.; Leckie, R.S.; Veenhuizen, P.T.; Schwarzmann, G.; Möbius, W.; Hoernschemeyer, J.; Slot, J.W.; Geuze, H.J.; Stoorvogel, W. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem. 2003, 278, 10963–10972. [Google Scholar] [CrossRef] [Green Version]
- Bard, M.P.; Hegmans, J.P.; Hemmes, A.; Luider, T.M.; Willemsen, R.; Severijnen, L.A.; van Meerbeeck, J.P.; Burgers, S.A.; Hoogsteden, H.C.; Lambrecht, B.N. Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am. J. Respir. Cell Mol. Biol. 2004, 31, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Subra, C.; Laulagnier, K.; Perret, B.; Record, M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 2007, 89, 205–212. [Google Scholar] [CrossRef]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandão, B.B.; Kahn, C.R. Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21, Erratum in Gynecol. Oncol. 2010, 116, 153. [Google Scholar] [CrossRef]
- Rosell, R.; Wei, J.; Taron, M. Circulating MicroRNA Signatures of Tumor-Derived Exosomes for Early Diagnosis of Non-Small-Cell Lung Cancer. Clin. Lung Cancer 2009, 10, 8–9. [Google Scholar] [CrossRef]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Janowska-Wieczorek, A.; Wysoczynski, M.; Kijowski, J.; Marquez-Curtis, L.; Machalinski, B.; Ratajczak, J.; Ratajczak, M.Z. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int. J. Cancer 2005, 113, 752–760. [Google Scholar] [CrossRef]
- Hao, S.; Ye, Z.; Li, F.; Meng, Q.; Qureshi, M.; Yang, J.; Xiang, J. Epigenetic transfer of metastatic activity by uptake of highly metastatic B16 melanoma cell-released exosomes. Exp. Oncol. 2006, 28, 126–131. [Google Scholar] [PubMed]
- Kosaka, N.; Iguchi, H.; Ochiya, T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010, 101, 2087–2092. [Google Scholar] [CrossRef] [PubMed]
- Iguchi, H.; Kosaka, N.; Ochiya, T. Secretory microRNAs as a versatile communication tool. Commun. Integr. Biol. 2010, 3, 478–481. [Google Scholar] [CrossRef] [PubMed]
- Jung, T.; Castellana, D.; Klingbeil, P.; Cuesta Hernández, I.; Vitacolonna, M.; Orlicky, D.J.; Roffler, S.R.; Brodt, P.; Zöller, M. CD44v6 dependence of premetastatic niche preparation by exosomes. Neoplasia 2009, 11, 1093–1105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.E.; Tan, H.S.; Datta, A.; Lai, R.C.; Zhang, H.; Meng, W.; Lim, S.K.; Sze, S.K. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol. Cell Proteom. 2010, 9, 1085–1099. [Google Scholar] [CrossRef] [Green Version]
- Paget, S. The distribution of secondary growths in cancer of the breast. Lancet 1889, 133, 571–573. [Google Scholar] [CrossRef] [Green Version]
- Mathias, R.A.; Gopal, S.K.; Simpson, R.J. Contribution of cells undergoing epithelial-mesenchymal transition to the tumour microenvironment. J. Proteom. 2013, 78, 545–557. [Google Scholar] [CrossRef]
- Liu, C.M.; Hsieh, C.L.; Shen, C.N.; Lin, C.C.; Shigemura, K.; Sung, S.Y. Exosomes from the tumor microenvironment as reciprocal regulators that enhance prostate cancer progression. Int. J. Urol. 2016, 23, 734–744. [Google Scholar] [CrossRef]
- Halvaei, S.; Daryani, S.; Eslami, S.Z.; Samadi, T.; Jafarbeik-Iravani, N.; Bakhshayesh, T.O.; Majidzadeh, A.K.; Esmaeili, R. Exosomes in Cancer Liquid Biopsy: A Focus on Breast Cancer. Mol. Ther. Nucleic Acids 2018, 10, 131–141. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Xu, X.; Qian, Z.; Zhang, C.; Niu, Y.; Wang, Z.; Sun, J.; Zhang, X.; Yu, Y. The biological functions and clinical applications of exosomes in lung cancer. Cell Mol. Life Sci. 2019, 76, 4613–4633. [Google Scholar] [CrossRef]
- Jia, S.; Zhang, R.; Li, Z.; Li, J. Clinical and biological significance of circulating tumor cells, circulating tumor DNA, and exosomes as biomarkers in colorectal cancer. Oncotarget 2017, 8, 55632–55645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, B.; Li, Y.; Zhou, Y.; Ng, T.K.; Zhao, C.; Gan, Q.; Gu, X.; Xiang, J. Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J. Cell Physiol. 2019, 234, 1416–1425. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Risch, H.A. Exosomes: Potential for early detection in pancreatic cancer. Future Oncol. 2016, 12, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Wang, Q.; Kornmann, M.; Tian, X.; Yang, Y. The Role of Exosomes in Pancreatic Cancer from Bench to Clinical Application: An Updated Review. Front. Oncol. 2021, 11, 644358. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef]
- Zhang, P.; Zhou, X.; He, M.; Shang, Y.; Tetlow, A.L.; Godwin, A.K.; Zeng, Y. Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat. Biomed. Eng. 2019, 3, 438–451. [Google Scholar] [CrossRef]
- Ludwig, N.; Whiteside, T.L.; Reichert, T.E. Challenges in Exosome Isolation and Analysis in Health and Disease. Int. J. Mol. Sci. 2019, 20, 4684. [Google Scholar] [CrossRef] [Green Version]
- Lobb, R.J.; Becker, M.; Wen, S.W.; Wong, C.S.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef]
- Liangsupree, T.; Multia, E.; Riekkola, M.-L. Modern isolation and separation techniques for extracellular vesicles. J. Chromatogr. A 2020, 1636, 461773. [Google Scholar] [CrossRef]
- Li, W.J.; Chen, H.; Tong, M.L.; Niu, J.J.; Zhu, X.Z.; Lin, L.R. Comparison of the yield and purity of plasma exosomes extracted by ultracentrifugation, precipitation, and membrane-based approaches. Open Chem. 2022, 20, 182–191. [Google Scholar] [CrossRef]
- Livshits, M.A.; Khomyakova, E.; Evtushenko, E.G.; Lazarev, V.N.; Kulemin, N.A.; Semina, S.E.; Generozov, E.V.; Govorun, V.M. Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci. Rep. 2015, 5, 17319, Erratum in Sci. Rep. 2016, 6, 21447. Livshts, Mikhail A [corrected to Livshits, Mikhail A]. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, A. Exosomes from Cell Culture-Conditioned Medium: Isolation by Ultracentrifugation and Characterization. Methods Mol. Biol. 2019, 1952, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Kumeda, N.; Ogawa, Y.; Akimoto, Y.; Kawakami, H.; Tsujimoto, M.; Yanoshita, R. Characterization of Membrane Integrity and Morphological Stability of Human Salivary Exosomes. Biol. Pharm. Bull. 2017, 40, 1183–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Wang, C.; Li, T.; Liu, Z.; Li, L. Comparison of ultracentrifugation and density gradient separation methods for isolating Tca8113 human tongue cancer cell line-derived exosomes. Oncol. Lett. 2014, 8, 1701–1706. [Google Scholar] [CrossRef] [Green Version]
- Brakke, M.K. Density gradient centrifugation. A new centrifugation technique. J. Am. Chem. Soc. 1951, 73, 1847. [Google Scholar] [CrossRef]
- Dhondt, B.; Lumen, N.; De Wever, O.; Hendrix, A. Preparation of Multi-omics Grade Extracellular Vesicles by Density-Based Fractionation of Urine. STAR Protoc. 2020, 1, 100073. [Google Scholar] [CrossRef]
- Yang, D.; Zhang, W.; Zhang, H.; Zhang, F.; Chen, L.; Ma, L.; Larcher, L.M.; Chen, S.; Liu, N.; Zhao, Q.; et al. Progress, opportunity, and perspective on exosome isolation—Efforts for efficient exosome-based theranostics. Theranostics 2020, 10, 3684–3707. [Google Scholar] [CrossRef]
- Ayala-Mar, S.; Donoso-Quezada, J.; Gallo-Villanueva, R.C.; Perez-Gonzalez, V.H.; González-Valdez, J. Recent advances and challenges in the recovery and purification of cellular exosomes. Electrophoresis 2019, 40, 3036–3049. [Google Scholar] [CrossRef]
- Mol, E.A.; Goumans, M.J.; Doevendans, P.A.; Sluijter, J.P.G.; Vader, P. Higher functionality of extracellular vesicles isolated using size-exclusion chromatography compared to ultracentrifugation. Nanomedicine 2017, 13, 2061–2065. [Google Scholar] [CrossRef]
- Konoshenko, M.Y.; Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed. Res. Int. 2018, 2018, 8545347. [Google Scholar] [CrossRef]
- Haraszti, R.A.; Miller, R.; Stoppato, M.; Sere, Y.Y.; Coles, A.; Didiot, M.C.; Wollacott, R.; Sapp, E.; Dubuke, M.L.; Li, X.; et al. Exosomes Produced from 3D Cultures of MSCs by Tangential Flow Filtration Show Higher Yield and Improved Activity. Mol. Ther. 2018, 26, 2838–2847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stranska, R.; Gysbrechts, L.; Wouters, J.; Vermeersch, P.; Bloch, K.; Dierickx, D.; Andrei, G.; Snoeck, R. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J. Transl. Med. 2018, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lian, J.Q.; Wang, P.Z.; Pan, L.; Ji, X.Y.; Bai, X.F.; Jia, Z.S. Isolation of exosomes derived from dendritic cells by ultrafiltration centrifugalization and their morphologic characteristics. J. Cell. Mol. Immunol. 2007, 23, 1119–1121. (In Chinese) [Google Scholar]
- Krug, A.K.; Enderle, D.; Karlovich, C.; Priewasser, T.; Bentink, S.; Spiel, A.; Brinkmann, K.; Emenegger, J.; Grimm, D.G.; Castellanos-Rizaldos, E.; et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann. Oncol. 2018, 29, 700–706. [Google Scholar] [CrossRef] [Green Version]
- Heinemann, M.L.; Ilmer, M.; Silva, L.P.; Hawke, D.H.; Recio, A.; Vorontsova, M.A.; Alt, E.; Vykoukal, J. Benchtop isolation and characterization of functional exosomes by sequential filtration. J. Chromatogr. A 2014, 1371, 125–135. [Google Scholar] [CrossRef]
- van Reis, R.; Gadam, S.; Frautschy, L.N.; Orlando, S.; Goodrich, E.M.; Saksena, S.; Kuriyel, R.; Simpson, C.M.; Pearl, S.; Zydney, A.L. High performance tangential flow filtration. Biotechnol. Bioeng. 1997, 56, 71–82. [Google Scholar] [CrossRef]
- Xu, R.; Greening, D.W.; Zhu, H.J.; Takahashi, N.; Simpson, R.J. Extracellular vesicle isolation and characterization: Toward clinical application. J. Clin. Investig. 2016, 126, 1152–1162. [Google Scholar] [CrossRef] [Green Version]
- An, M.; Wu, J.; Zhu, J.; Lubman, D.M. Comparison of an Optimized Ultracentrifugation Method versus Size-Exclusion Chromatography for Isolation of Exosomes from Human Serum. J. Proteome Res. 2018, 17, 3599–3605. [Google Scholar] [CrossRef]
- Gaspar, L.S.; Santana, M.M.; Henriques, C.; Pinto, M.M.; Ribeiro-Rodrigues, T.M.; Girão, H.; Nobre, R.J.; Pereira de Almeida, L. Simple and Fast SEC-Based Protocol to Isolate Human Plasma-Derived Extracellular Vesicles for Transcriptional Research. Mol. Ther. Methods Clin. Dev. 2020, 18, 723–737. [Google Scholar] [CrossRef]
- Sidhom, K.; Obi, P.O.; Saleem, A. A Review of Exosomal Isolation Methods: Is Size Exclusion Chromatography the Best Option? Int. J. Mol. Sci. 2020, 21, 6466. [Google Scholar] [CrossRef]
- Wei, R.; Zhao, L.; Kong, G.; Liu, X.; Zhu, S.; Zhang, S.; Min, L. Combination of Size-Exclusion Chromatography and Ultracentrifugation Improves the Proteomic Profiling of Plasma-Derived Small Extracellular Vesicles. Biol. Proced. Online 2020, 22, 12. [Google Scholar] [CrossRef] [PubMed]
- Welton, J.L.; Loveless, S.; Stone, T.; von Ruhland, C.; Robertson, N.P.; Clayton, A. Cerebrospinal fluid extracellular vesicle enrichment for protein biomarker discovery in neurological disease; multiple sclerosis. J. Extracell. Vesicles 2017, 6, 1369805. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, K.; Mitchell, M.D.; Holland, O.J.; Qin Koh, Y.; Hill, R.J.; Harb, T.; Davies, P.S.W.; Peiris, H. A Method for the Isolation of Exosomes from Human and Bovine Milk. J. Nutr. Metab. 2019, 2019, 5764740. [Google Scholar] [CrossRef] [PubMed]
- Aqrawi, L.A.; Galtung, H.K.; Vestad, B.; Øvstebø, R.; Thiede, B.; Rusthen, S.; Young, A.; Guerreiro, E.M.; Utheim, T.P.; Chen, X.; et al. Identification of potential saliva and tear biomarkers in primary Sjögren’s syndrome, utilising the extraction of extracellular vesicles and proteomics analysis. Arthritis Res. Ther. 2017, 19, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oeyen, E.; Van Mol, K.; Baggerman, G.; Willems, H.; Boonen, K.; Rolfo, C.; Pauwels, P.; Jacobs, A.; Schildermans, K.; Cho, W.C.; et al. Ultrafiltration and size exclusion chromatography combined with asymmetrical-flow field-flow fractionation for the isolation and characterisation of extracellular vesicles from urine. J. Extracell. Vesicles 2018, 7, 1490143. [Google Scholar] [CrossRef]
- Foers, A.D.; Chatfield, S.; Dagley, L.F.; Scicluna, B.J.; Webb, A.I.; Cheng, L.; Hill, A.F.; Wicks, I.P.; Pang, K.C. Enrichment of extracellular vesicles from human synovial fluid using size exclusion chromatography. J. Extracell. Vesicles 2018, 7, 1490145. [Google Scholar] [CrossRef]
- Bartel, S.; La Grutta, S.; Cilluffo, G.; Perconti, G.; Bongiovanni, A.; Giallongo, A.; Behrends, J.; Kruppa, J.; Hermann, S.; Chiang, D.; et al. Human airway epithelial extracellular vesicle miRNA signature is altered upon asthma development. Allergy 2020, 75, 346–356. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Caro, H.; Dragovic, R.; Shen, M.; Dombi, E.; Mounce, G.; Field, K.; Meadows, J.; Turner, K.; Lunn, D.; Child, T.; et al. In vitro decidualisation of human endometrial stromal cells is enhanced by seminal fluid extracellular vesicles. J. Extracell. Vesicles 2019, 8, 1565262. [Google Scholar] [CrossRef] [Green Version]
- Takov, K.; Yellon, D.M.; Davidson, S.M. Comparison of small extracellular vesicles isolated from plasma by ultracentrifugation or size-exclusion chromatography: Yield, purity and functional potential. J. Extracell. Vesicles 2018, 8, 1560809. [Google Scholar] [CrossRef]
- Buschmann, D.; Mussack, V.; Byrd, J.B. Separation, characterization, and standardization of extracellular vesicles for drug delivery applications. Adv. Drug Deliv. Rev. 2021, 174, 348–368. [Google Scholar] [CrossRef]
- Fang, X.; Duan, Y.; Adkins, G.B.; Pan, S.; Wang, H.; Liu, Y.; Zhong, W. Highly Efficient Exosome Isolation and Protein Analysis by an Integrated Nanomaterial-Based Platform. Anal. Chem. 2018, 90, 2787–2795. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.; Sui, Z.; Shan, Y.; Hu, Y.; Chen, Y.; Zhang, L.; Zhang, Y. Effective isolation of exosomes with polyethylene glycol from cell culture supernatant for in-depth proteome profiling. Analyst 2016, 141, 4640–4646. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.M.; Bothe, S.; Gutmann, T.; Buntkowsky, G. Unusual local molecular motions in the solid state detected by dynamic nuclear polarization enhanced NMR spectroscopy. J. Phys. Chem. C 2017, 121, 22948–22957. [Google Scholar] [CrossRef]
- Shu, S.; Yang, Y.; Allen, C.L.; Hurley, E.; Tung, K.H.; Minderman, H.; Wu, Y.; Ernstoff, M.S. Purity and yield of melanoma exosomes are dependent on isolation method. J. Extracell. Vesicles 2019, 9, 1692401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Li, P.; Zhang, T.; Xu, Z.; Huang, X.; Wang, R.; Du, L. Review on Strategies and Technologies for Exosome Isolation and Purification. Front. Bioeng. Biotechnol. 2022, 9, 811971. [Google Scholar] [CrossRef]
- Mitchell, M.I.; Ben-Dov, I.Z.; Liu, C.; Ye, K.; Chow, K.; Kramer, Y.; Gangadharan, A.; Park, S.; Fitzgerald, S.; Ramnauth, A.; et al. Extracellular Vesicle Capture by AnTibody of CHoice and Enzymatic Release (EV-CATCHER): A customizable purification assay designed for small-RNA biomarker identification and evaluation of circulating small-EVs. J. Extracell. Vesicles 2021, 10, e12110. [Google Scholar] [CrossRef]
- Wu, D.; Yan, J.; Shen, X.; Sun, Y.; Thulin, M.; Cai, Y.; Wik, L.; Shen, Q.; Oelrich, J.; Qian, X.; et al. Profiling surface proteins on individual exosomes using a proximity barcoding assay. Nat. Commun. 2019, 10, 3854. [Google Scholar] [CrossRef]
- Hu, Q.; Su, H.; Li, J.; Lyon, C.; Tang, W.; Wan, M.; Hu, T.Y. Clinical applications of exosome membrane proteins. Precis. Clin. Med. 2020, 3, 54–66. [Google Scholar] [CrossRef]
- Jankovičová, J.; Sečová, P.; Michalková, K.; Antalíková, J. Tetraspanins, More than Markers of Extracellular Vesicles in Reproduction. Int. J. Mol. Sci. 2020, 21, 7568. [Google Scholar] [CrossRef]
- Zhou, Y.G.; Mohamadi, R.M.; Poudineh, M.; Kermanshah, L.; Ahmed, S.; Safaei, T.S.; Stojcic, J.; Nam, R.K.; Sargent, E.H.; Kelley, S.O. Interrogating Circulating Microsomes and Exosomes Using Metal Nanoparticles. Small 2016, 12, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Davey, M.; Chute, I.C.; Griffiths, S.G.; Lewis, S.; Chacko, S.; Barnett, D.; Crapoulet, N.; Fournier, S.; Joy, A.; et al. Rapid isolation of extracellular vesicles from cell culture and biological fluids using a synthetic peptide with specific affinity for heat shock proteins. PLoS ONE 2014, 9, e110443. [Google Scholar] [CrossRef] [Green Version]
- Balaj, L.; Atai, N.A.; Chen, W.; Mu, D.; Tannous, B.A.; Breakefield, X.O.; Skog, J.; Maguire, C.A. Heparin affinity purification of extracellular vesicles. Sci. Rep. 2015, 5, 10266. [Google Scholar] [CrossRef] [Green Version]
- Koliha, N.; Wiencek, Y.; Heider, U.; Jüngst, C.; Kladt, N.; Krauthäuser, S.; Johnston, I.C.; Bosio, A.; Schauss, A.; Wild, S. A novel multiplex bead-based platform highlights the diversity of extracellular vesicles. J. Extracell. Vesicles 2016, 5, 29975. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.; Xie, X.; Zhang, D.; Zhou, Y.; Li, B.; Li, F.; Li, F.; Cheng, Y.; Mei, H.; Meng, H.; et al. Use of lung-specific exosomes for miRNA-126 delivery in non-small cell lung cancer. Nanoscale 2020, 12, 877–887. [Google Scholar] [CrossRef] [PubMed]
- Gotanda, K.; Hirota, T.; Saito, J.; Fukae, M.; Egashira, Y.; Izumi, N.; Deguchi, M.; Kimura, M.; Matsuki, S.; Irie, S.; et al. Circulating intestine-derived exosomal miR-328 in plasma, a possible biomarker for estimating BCRP function in the human intestines. Sci. Rep. 2016, 6, 32299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.; Meng, Q.; Shi, C.; Yang, H.; Li, X.; Wu, S.; Familiari, G.; Relucenti, M.; Aschner, M.; Wang, X.; et al. Hypoxia-Inducible Exosomes Facilitate Liver-Tropic Premetastatic Niche in Colorectal Cancer. Hepatology 2021, 74, 2633–2651. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.F.; Hannafon, B.N.; Zhao, Y.D.; Postier, R.G.; Ding, W.Q. Plasma exosome miR-196a and miR-1246 are potential indicators of localized pancreatic cancer. Oncotarget 2017, 8, 77028–77040. [Google Scholar] [CrossRef]
- Dilsiz, N. Role of exosomes and exosomal microRNAs in cancer. Future Sci. OA 2020, 6, FSO465. [Google Scholar] [CrossRef] [Green Version]
- Padda, J.; Khalid, K.; Khedr, A.; Patel, V.; Al-Ewaidat, O.A.; Tasnim, F.; Padda, S.; Cooper, A.C.; Jean-Charles, G. Exosome-Derived microRNA: Efficacy in Cancer. Cureus 2021, 13, e17441. [Google Scholar] [CrossRef]
- Lou, D.; Wang, Y.; Yang, Q.; Hu, L.; Zhu, Q. Ultrafiltration combing with phospholipid affinity-based isolation for metabolomic profiling of urinary extracellular vesicles. J. Chromatogr. A 2021, 1640, 461942. [Google Scholar] [CrossRef] [PubMed]
- Salafi, T.; Zeming, K.K.; Zhang, Y. Advancements in microfluidics for nanoparticle separation. Lab. Chip. 2016, 17, 11–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.; Ouyang, Y.; Wang, Z.; Zhang, R.; Huang, P.H.; Chen, C.; Li, H.; Li, P.; Quinn, D.; Dao, M.; et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. USA 2017, 114, 10584–10589, Erratum in Proc. Natl. Acad. Sci. USA 2020, 117, 28525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Vermesh, O.; Mani, V.; Ge, T.J.; Madsen, S.J.; Sabour, A.; Hsu, E.C.; Gowrishankar, G.; Kanada, M.; Jokerst, J.V.; et al. The Exosome Total Isolation Chip. ACS Nano 2017, 11, 10712–10723. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, S.S.; Dunlay, C.J.; Simeone, D.M.; Nagrath, S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab. Chip. 2014, 14, 1891–1900. [Google Scholar] [CrossRef]
- Lin, S.; Yu, Z.; Chen, D.; Wang, Z.; Miao, J.; Li, Q.; Zhang, D.; Song, J.; Cui, D. Progress in Microfluidics-Based Exosome Separation and Detection Technologies for Diagnostic Applications. Small 2020, 16, e1903916. [Google Scholar] [CrossRef]
- Gou, Y.; Jia, Y.; Wang, P.; Sun, C. Progress of Inertial Microfluidics in Principle and Application. Sensors 2018, 18, 1762. [Google Scholar] [CrossRef] [Green Version]
- Ding, L.; Yang, X.; Gao, Z.; Effah, C.Y.; Zhang, X.; Wu, Y.; Qu, L. A Holistic Review of the State-of-the-Art Microfluidics for Exosome Separation: An Overview of the Current Status, Existing Obstacles, and Future Outlook. Small 2021, 17, e2007174. [Google Scholar] [CrossRef]
- Yang, F.; Liao, X.; Tian, Y.; Li, G. Exosome separation using microfluidic systems: Size-based, immunoaffinity-based and dynamic methodologies. Biotechnol. J. 2017, 12, 1600699. [Google Scholar] [CrossRef]
- Lee, H.; Lee, J.; Lee, S.G.; Doyle, P.S. Hydrogel-Based Colorimetric Assay for Multiplexed MicroRNA Detection in a Microfluidic Device. Anal. Chem. 2020, 92, 5750–5755. [Google Scholar] [CrossRef] [Green Version]
- Liao, X.; Makris, M.; Luo, X.M. Fluorescence-activated Cell Sorting for Purification of Plasmacytoid Dendritic Cells from the Mouse Bone Marrow. J. Vis. Exp. 2016, 117, e54641. [Google Scholar] [CrossRef] [PubMed]
- Kindlund, B.; Sjöling, Å.; Yakkala, C.; Adamsson, J.; Janzon, A.; Hansson, L.E.; Hermansson, M.; Janson, P.; Winqvist, O.; Lundin, S.B. CD4+ regulatory T cells in gastric cancer mucosa are proliferating and express high levels of IL-10 but little TGF-β. Gastric Cancer 2017, 20, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Malmberg, E.B.; Ståhlman, S.; Rehammar, A.; Samuelsson, T.; Alm, S.J.; Kristiansson, E.; Abrahamsson, J.; Garelius, H.; Pettersson, L.; Ehinger, M.; et al. Patient-tailored analysis of minimal residual disease in acute myeloid leukemia using next-generation sequencing. Eur. J. Haematol. 2017, 98, 26–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, A.J.; Staats, J.; Enzor, J.; McKinnon, K.; Frelinger, J.; Denny, T.N.; Weinhold, K.J.; Chan, C. Setting objective thresholds for rare event detection in flow cytometry. J. Immunol. Methods 2014, 409, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Morales-Kastresana, A.; Musich, T.A.; Welsh, J.A.; Telford, W.; Demberg, T.; Wood, J.C.S.; Bigos, M.; Ross, C.D.; Kachynski, A.; Dean, A.; et al. High-fidelity detection and sorting of nanoscale vesicles in viral disease and cancer. J. Extracell. Vesicles 2019, 19, 1597603. [Google Scholar] [CrossRef] [Green Version]
- Cao, Z.; Li, C.; Higginbotham, J.N.; Franklin, J.L.; Tabb, D.L.; Graves-Deal, R.; Hill, S.; Cheek, K.; Jerome, W.G.; Lapierre, L.A.; et al. Use of fluorescence-activated vesicle sorting for isolation of Naked2-associated, basolaterally targeted exocytic vesicles for proteomics analysis. Mol. Cell Proteom. 2008, 7, 1651–1667. [Google Scholar] [CrossRef] [Green Version]
- Poncelet, P.; Robert, S.; Bouriche, T.; Bez, J.; Lacroix, R.; Dignat-George, F. Standardized counting of circulating platelet microparticles using currently available flow cytometers and scatter-based triggering: Forward or side scatter? Cytom. A 2016, 89, 148–158. [Google Scholar] [CrossRef]
- Chandler, W.L.; Yeung, W.; Tait, J.F. A new microparticle size calibration standard for use in measuring smaller microparticles using a new flow cytometer. J. Thromb. Haemost. 2011, 9, 1216–1224. [Google Scholar] [CrossRef]
- Groot Kormelink, T.; Arkesteijn, G.J.; Nauwelaers, F.A.; van den Engh, G.; Nolte-‘t Hoen, E.N.; Wauben, M.H. Prerequisites for the analysis and sorting of extracellular vesicle subpopulations by high-resolution flow cytometry. Cytom. A 2016, 89, 135–147. [Google Scholar] [CrossRef]
- Padda, R.S.; Deng, F.K.; Brett, S.I.; Biggs, C.N.; Durfee, P.N.; Brinker, C.J.; Williams, K.C.; Leong, H.S. Nanoscale flow cytometry to distinguish subpopulations of prostate extracellular vesicles in patient plasma. Prostate 2019, 79, 592–603. [Google Scholar] [CrossRef]
- . Higginbotham, J.N.; Zhang, Q.; Jeppesen, D.K.; Scott, A.M.; Manning, H.C.; Ochieng, J.; Franklin, J.L.; Coffey, R.J. Identification and characterization of EGF receptor in individual exosomes by fluorescence-activated vesicle sorting. J Extracell. Vesicles 2016, 5, 29254. [Google Scholar] [CrossRef]
- Mastoridis, S.; Bertolino, G.M.; Whitehouse, G.; Dazzi, F.; Sanchez-Fueyo, A.; Martinez-Llordella, M. Multiparametric Analysis of Circulating Exosomes and Other Small Extracellular Vesicles by Advanced Imaging Flow Cytometry. Front. Immunol. 2018, 9, 1583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Löf, L.; Arngården, L.; Ebai, T.; Landegren, U.; Söderberg, O.; Kamali-Moghaddam, M. Detection of Extracellular Vesicles Using Proximity Ligation Assay with Flow Cytometry Readout-ExoPLA. Curr. Protoc. Cytom. 2017, 81, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Larssen, P.; Wik, L.; Czarnewski, P.; Eldh, M.; Löf, L.; Ronquist, K.G.; Dubois, L.; Freyhult, E.; Gallant, C.J.; Oelrich, J.; et al. Tracing Cellular Origin of Human Exosomes Using Multiplex Proximity Extension Assays. Mol. Cell Proteom. 2017, 16, 1547, Erratum in Mol. Cell. Proteom. 2017, 16, 502–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Löf, L.; Ebai, T.; Dubois, L.; Wik, L.; Ronquist, K.G.; Nolander, O.; Lundin, E.; Söderberg, O.; Landegren, U.; Kamali-Moghaddam, M. Detecting individual extracellular vesicles using a multicolor in situ proximity ligation assay with flow cytometric readout. Sci. Rep. 2016, 6, 34358. [Google Scholar] [CrossRef]
- Morita, E.; Sandrin, V.; Chung, H.-Y.; Morham, S.G.; Gygi, S.P.; Rodesch, C.K.; Sundquist, W.I. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007, 26, 4215–4227. [Google Scholar] [CrossRef] [Green Version]
- Théry, C.; Duban, L.; Segura, E.; Véron, P.; Lantz, O.; Amigorena, S. Indirect activation of naïve CD4+ T cells by dendritic cell-derived exosomes. Nat. Immunol. 2002, 3, 1156–1162. [Google Scholar] [CrossRef]
- Buschow, S.I.; Nolte-‘t Hoen, E.N.; van Niel, G.; Pols, M.S.; Ten Broeke, T.; Lauwen, M.; Ossendorp, F.; Melief, C.J.; Raposo, G.; Wubbolts, R.; et al. MHC II in dendritic cells is targeted to lysosomes or T cell-induced exosomes via distinct multivesicular body pathways. Traffic 2009, 10, 1528–1542. [Google Scholar] [CrossRef]
- Buschow, S.I.; van Balkom, B.W.; Aalberts, M.; Heck, A.J.; Wauben, M.; Stoorvogel, W. MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. Immunol. Cell Biol. 2010, 88, 851–856. [Google Scholar] [CrossRef]
- Newman, L.A.; Useckaite, Z.; Johnson, J.; Sorich, M.J.; Hopkins, A.M.; Rowland, A. Selective Isolation of Liver-Derived Extracellular Vesicles Redefines Performance of miRNA Biomarkers for Non-Alcoholic Fatty Liver Disease. Biomedicines 2022, 10, 195. [Google Scholar] [CrossRef]
- Chiasserini, D.; van Weering, J.R.; Piersma, S.R.; Pham, T.V.; Malekzadeh, A.; Teunissen, C.E.; de Wit, H.; Jiménez, C.R. Proteomic analysis of cerebrospinal fluid extracellular vesicles: A comprehensive dataset. J. Proteom. 2014, 106, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Pillay, P.; Maharaj, N.; Moodley, J.; Mackraj, I. Placental exosomes and pre-eclampsia: Maternal circulating levels in normal pregnancies and, early and late onset pre-eclamptic pregnancies. Placenta 2016, 46, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Miranda, J.; Paules, C.; Nair, S.; Lai, A.; Palma, C.; Scholz-Romero, K.; Rice, G.E.; Gratacos, E.; Crispi, F.; Salomon, C. Placental exosomes profile in maternal and fetal circulation in intrauterine growth restriction—Liquid biopsies to monitoring fetal growth. Placenta 2018, 64, 34–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seibold, T.; Schönfelder, J.; Weeber, F.; Lechel, A.; Armacki, M.; Waldenmaier, M.; Wille, C.; Palmer, A.; Halbgebauer, R.; Karasu, E.; et al. Small Extracellular Vesicles Propagate the Inflammatory Response after Trauma. Adv. Sci. 2021, 8, e2102381. [Google Scholar] [CrossRef]
- Valadi, H.; Eksröm, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Bissig, C.; Gruenberg, J. ALIX and the multivesicular endosome: ALIX in wonderland. Trends Cell Biol. 2014, 24, 19–25. [Google Scholar] [CrossRef]
- Alenquer, M.; Amorim, M.J. Exosome biogenesis, regulation, and function in viral infection. Viruses 2015, 7, 5066–5083. [Google Scholar] [CrossRef] [Green Version]
- Hemler, M.E. Targeting of tetraspanin proteins—Potential benefits and strategies. Nat. Rev. Drug Discov. 2008, 7, 747–758. [Google Scholar] [CrossRef] [Green Version]
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-Mediated Metastasis: Communication from a Distance. Dev. Cell 2019, 49, 347–360. [Google Scholar] [CrossRef]
- Helenius, A.; Mellman, I.; Wall, D.; Hubbard, A. Endosomes. Trends Biochem. Sci. 1983, 8, 245–250. [Google Scholar] [CrossRef]
- Elkin, S.R.; Lakoduk, A.M.; Schmid, S.L. Endocytic pathways and endosomal trafficking: A primer. Wien. Med. Wochenschr. 2016, 166, 196–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alberts, B.; Johnson, A.; Lewis, J. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002; Transport into the Cell from the Plasma Membrane: Endocytosis. Available online: https://www.ncbi.nlm.nih.gov/books/NBK26870/ (accessed on 26 May 2022).
- Kaur, G.; Lakkaraju, A. Early Endosome Morphology in Health and Disease. Adv. Exp. Med. Biol. 2018, 1074, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Hotaru, J.; Helenius, A. Endosome maturation. EMBO J. 2011, 30, 3481–3500. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.R. Multiple routes of protein transport from endosomes to the trans Golgi network. FEBS Lett. 2009, 583, 3811–3816. [Google Scholar] [CrossRef] [Green Version]
- Piper, R.C.; Katzmann, D.J. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 2007, 23, 519–547. [Google Scholar] [CrossRef] [Green Version]
- Lu, A.; Tebar, F.; Alvarez-Moya, B.; López-Alcalá, C.; Calvo, M.; Enrich, C.; Agell, N.; Nakamura, T.; Matsuda, M.; Bachs, O. A clathrin-dependent pathway leads to KRas signaling on late endosomes en route to lysosomes. J. Cell Biol. 2009, 184, 863–879. [Google Scholar] [CrossRef] [Green Version]
- Frankel, E.B.; Audhya, A. ESCRT-dependent cargo sorting at multivesicular endosomes. Semin. Cell Dev. Biol. 2018, 74, 4–10. [Google Scholar] [CrossRef]
- Katzmann, D.J.; Babst, M.; Emr, S.D. Ubiquitin-Dependent Sorting into the Multivesicular Body Pathway Requires the Function of a Conserved Endosomal Protein Sorting Complex, ESCRT-I. Cell 2001, 106, 145–155. [Google Scholar] [CrossRef] [Green Version]
- Piper, R.C.; Luzio, J.P. Ubiquitin-dependent sorting of integral membrane proteins for degradation in lysosomes. Curr. Opin. Cell Biol. 2008, 19, 459–465. [Google Scholar] [CrossRef] [Green Version]
- Peng, X.; Yang, L.; Ma, Y.; Li, Y.; Li, H. Focus on the morphogenesis, fate and the role in tumor progression of multivesicular bodies. Cell Commun. Signal. 2020, 18, 122. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Schapira, A.H.; Gardiner, C.; Sargent, I.L.; Wood, M.J.; Cooper, J.M. Lysosomal dysfunction increases exosome-mediated alpha-synuclein release and transmission. Neurobiol. Dis. 2011, 42, 360–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, Y.; Li, G.; Zhang, X.; Xu, H.; Abraham, S.N. A TRP Channel Senses Lysosome Neutralization by Pathogens to Trigger Their Expulsion. Cell 2015, 161, 1306–1319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, S.; Kovalenko, A.; Bogdanov, K.; Wallach, D. MLKL, the Protein that Mediates Necroptosis, Also Regulates Endosomal Trafficking and Extracellular Vesicle Generation. Immunity 2017, 47, 51–65.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olenick, M.A.; Holzbaur, E.L.F. Dynein activators and adaptors at a glance. J. Cell Sci. 2019, 132, jcs227132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutagalung, A.H.; Novick, P.J. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 2011, 91, 119–149. [Google Scholar] [CrossRef] [Green Version]
- Koike, S.; Jahn, R. SNARE proteins: Zip codes in vesicle targeting? Biochem. J. 2022, 479, 273–288. [Google Scholar] [CrossRef]
- Lorentz, A.; Baumann, A.; Vitte, J.; Blank, U. The SNARE Machinery in Mast Cell Secretion. Front. Immunol. 2012, 3, 143. [Google Scholar] [CrossRef] [Green Version]
- Homma, Y.; Hiragi, S.; Fukuda, M. Rab family of small GTPases: An updated view on their regulation and functions. FEBS J. 2021, 288, 36–55. [Google Scholar] [CrossRef]
- Yu, X.; Prekeris, R.; Gould, G.W. Role of endosomal Rab GTPases in cytokinesis. Eur. J. Cell Biol. 2007, 86, 25–35. [Google Scholar] [CrossRef]
- Ramel, D.; Wang, X.; Laflamme, C.; Montell, D.J.; Emery, G. Rab11 regulates cell-cell communication during collective cell movements. Nat. Cell Biol. 2013, 15, 317–324. [Google Scholar] [CrossRef] [Green Version]
- Heo, P.; Coleman, J.; Fleury, J.B.; Rothman, J.E.; Pincet, F. Nascent fusion pore opening monitored at single-SNAREpin resolution. Proc. Natl. Acad. Sci. USA 2021, 118, e2024922118. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Névo, N.; Jouve, M.; Valenzuela, J.I.; Maurin, M.; Verweij, F.J.; Palmulli, R.; Lankar, D.; Dingli, F.; Loew, D.; et al. Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nat. Commun. 2021, 12, 4389. [Google Scholar] [CrossRef] [PubMed]
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and function of extracellular vesicles in cancer. Pharmacol. Ther. 2018, 188, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, P.; Schuller, J.M.; Bonneau, F.; Basquin, J.; Reichelt, P.; Falk, S.; Conti, E. Distinct and evolutionary conserved structural features of the human nuclear exosome complex. Elife 2018, 7, e38686. [Google Scholar] [CrossRef]
- Bahrami, A.; Moradi Binabaj, M.; AFerns, G. Exosomes: Emerging modulators of signal transduction in colorectal cancer from molecular understanding to clinical application. Biomed. Pharmacother. 2021, 141, 111882. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Gahan, P.B. MicroRNA Shuttle from Cell-To-Cell by Exosomes and Its Impact in Cancer. Noncoding RNA 2019, 5, 28. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Chen, W.; Zhi, X.; Chen, E.J.; Wei, T.; Zhang, J.; Shen, J.; Hu, L.Q.; Zhao, B.; Feng, X.H.; et al. Tumor-derived exosomes promote tumor self-seeding in hepatocellular carcinoma by transferring miRNA-25-5p to enhance cell motility. Oncogene 2018, 37, 4964–4978. [Google Scholar] [CrossRef]
- Iguchi, Y.; Eid, L.; Parent, M.; Soucy, G.; Bareil, C.; Riku, Y.; Kawai, K.; Takagi, S.; Yoshida, M.; Katsuno, M.; et al. Exosome secretion is a key pathway for clearance of pathological TDP-43. Brain 2016, 139 Pt 12, 3187–3201. [Google Scholar] [CrossRef] [Green Version]
- Vulpis, E.; Soriani, A.; Cerboni, C.; Santoni, A.; Zingoni, A. Cancer Exosomes as Conveyors of Stress-Induced Molecules: New Players in the Modulation of NK Cell Response. Int. J. Mol. Sci. 2019, 20, 611. [Google Scholar] [CrossRef] [Green Version]
- Troyer, Z.; Tilton, J.C. Extracellular vesicles as carriers of viruses. ExRNA 2021, 3, 13. [Google Scholar] [CrossRef]
- Göran Ronquist, K. Extracellular vesicles and energy metabolism. Clin. Chim. Acta 2019, 488, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.; Silva, J.; Herrera, A.; Herrera, M.; Peña, C.; Martín, P.; Gil-Calderón, B.; Larriba, M.J.; Coronado, M.J.; Soldevilla, B.; et al. Exosomes enriched in stemness/metastatic-related mRNAS promote oncogenic potential in breast cancer. Oncotarget 2015, 6, 40575–40587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borges, F.T.; Melo, S.A.; Özdemir, B.C.; Kato, N.; Revuelta, I.; Miller, C.A.; Gattone, V.H., 2nd; LeBleu, V.S.; Kalluri, R. TGF-β1-containing exosomes from injured epithelial cells activate fibroblasts to initiate tissue regenerative responses and fibrosis. J. Am. Soc. Nephrol. 2013, 24, 385–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, Y.; Miyaki, S.; Ishitobi, H.; Matsuyama, S.; Nakasa, T.; Kamei, N.; Akimoto, T.; Higashi, Y.; Ochi, M. Mesenchymal-stem-cell-derived exosomes accelerate skeletal muscle regeneration. FEBS Lett. 2015, 589, 1257–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gartz, M.; Strande, J.L. Examining the Paracrine Effects of Exosomes in Cardiovascular Disease and Repair. J. Am. Heart Assoc. 2018, 7, e007954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pegtel, D.M.; Peferoen, L.; Amor, S. Extracellular vesicles as modulators of cell-to-cell communication in the healthy and diseased brain. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bang, C.; Thum, T. Exosomes: New players in cell-cell communication. Int. J. Biochem. Cell Biol. 2012, 44, 2060–2064. [Google Scholar] [CrossRef]
- Samanta, S.; Rajasingh, S.; Drosos, N.; Zhou, Z.; Dawn, B.; Rajasingh, J. Exosomes: New molecular targets of diseases. Acta Pharmacol. Sin. 2018, 39, 501–513. [Google Scholar] [CrossRef]
- Emmanouilidou, E.; Melachroinou, K.; Roumeliotis, T.; Garbis, S.D.; Ntzouni, M.; Margaritis, L.H.; Stefanis, L.; Vekrellis, K. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J. Neurosci. 2010, 30, 6838–6851. [Google Scholar] [CrossRef] [Green Version]
- Rajendran, L.; Honsho, M.; Zahn, T.R.; Keller, P.; Geiger, K.D.; Verkade, P.; Simons, K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11172–11177. [Google Scholar] [CrossRef] [Green Version]
- Saá, P.; Yakovleva, O.; de Castro, J.; Vasilyeva, I.; De Paoli, S.H.; Simak, J.; Cervenakova, L. First demonstration of transmissible spongiform encephalopathy-associated prion protein (PrPTSE) in extracellular vesicles from plasma of mice infected with mouse-adapted variant Creutzfeldt-Jakob disease by in vitro amplification. J. Biol. Chem. 2014, 289, 29247–29260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basso, M.; Pozzi, S.; Tortarolo, M.; Fiordaliso, F.; Bisighini, C.; Pasetto, L.; Spaltro, G.; Lidonnici, D.; Gensano, F.; Battaglia, E.; et al. Mutant copper-zinc superoxide dismutase (SOD1) induces protein secretion pathway alterations and exosome release in astrocytes: Implications for disease spreading and motor neuron pathology in amyotrophic lateral sclerosis. J. Biol. Chem. 2013, 288, 15699–15711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; Lucci, A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef] [Green Version]
- Psaila, B.; Kaplan, R.N.; Port, E.R.; Lyden, D. Priming the ‘soil’ for breast cancer metastasis: The pre-metastatic niche. Breast Dis. 2007, 26, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, J.; Garcia, V.; Rodriguez, M.; Compte, M.; Cisneros, E.; Veguillas, P.; Garcia, J.M.; Dominguez, G.; Campos-Martin, Y.; Cuevas, J.; et al. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer 2012, 51, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624, Erratum in Nat. Cell Biol. 2008, 10, 752. [Google Scholar] [CrossRef]
- Choi, D.; Montermini, L.; Kim, D.K.; Meehan, B.; Roth, F.P.; Rak, J. The Impact of Oncogenic EGFRvIII on the Proteome of Extracellular Vesicles Released from Glioblastoma Cells. Mol. Cell Proteom. 2018, 17, 1948–1964. [Google Scholar] [CrossRef] [Green Version]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126 Pt 24, 5553–5565. [Google Scholar] [CrossRef] [Green Version]
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [Green Version]
- Tamai, K.; Tanaka, N.; Nakano, T.; Kakazu, E.; Kondo, Y.; Inoue, J.; Shiina, M.; Fukushima, K.; Hoshino, T.; Sano, K.; et al. Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem. Biophys. Res. Commun. 2010, 399, 384–390. [Google Scholar] [CrossRef]
- Gross, J.C.; Chaudhary, V.; Bartscherer, K.; Boutros, M. Active Wnt proteins are secreted on exosomes. Nat. Cell Biol. 2012, 14, 1036–1045. [Google Scholar] [CrossRef]
- Valcz, G.; Galamb, O.; Krenács, T.; Spisák, S.; Kalmár, A.; Patai, Á.V.; Wichmann, B.; Dede, K.; Tulassay, Z.; Molnár, B. Exosomes in colorectal carcinoma formation: ALIX under the magnifying glass. Mod. Pathol. 2016, 29, 928–938. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, Y.; Gao, X.; Yuan, Y.; Zhao, J.; Zhou, S.; Wang, H.; Wang, L.; Xu, G.; Li, X.; et al. Plasma-Derived Exosomal ALIX as a Novel Biomarker for Diagnosis and Classification of Pancreatic Cancer. Front. Oncol. 2021, 11, 628346. [Google Scholar] [CrossRef]
- Larios, J.; Mercier, V.; Roux, A.; Gruenberg, J. ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J. Cell Biol. 2020, 219, e201904113. [Google Scholar] [CrossRef] [Green Version]
- Roucourt, B.; Meeussen, S.; Bao, J.; Zimmermann, P.; David, G. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 2015, 25, 412–428. [Google Scholar] [CrossRef] [Green Version]
- Monypenny, J.; Milewicz, H.; Flores-Borja, F.; Weitsman, G.; Cheung, A.; Chowdhury, R.; Burgoyne, T.; Arulappu, A.; Lawler, K.; Barber, P.R.; et al. ALIX Regulates Tumor-Mediated Immunosuppression by Controlling EGFR Activity and PD-L1 Presentation. Cell Rep. 2018, 24, 630–641. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Hong, Q.; Shi, P.; Liu, Z.; Luo, J.; Shao, Z. Elevated expression of syntenin in breast cancer is correlated with lymph node metastasis and poor patient survival. Breast Cancer Res. 2013, 15, R50. [Google Scholar] [CrossRef] [Green Version]
- Das, S.K.; Guo, C.; Pradhan, A.K.; Bhoopathi, P.; Talukdar, S.; Shen, X.N.; Emdad, L.; Subler, M.A.; Windle, J.J.; Sarkar, D.; et al. Knockout of MDA-9/Syntenin (SDCBP) expression in the microenvironment dampens tumor-supporting inflammation and inhibits melanoma metastasis. Oncotarget 2016, 7, 46848–46861. [Google Scholar] [CrossRef] [Green Version]
- Kim, O.; Hwangbo, C.; Tran, P.T.; Lee, J.H. Syntenin-1-mediated small extracellular vesicles promotes cell growth, migration, and angiogenesis by increasing onco-miRNAs secretion in lung cancer cells. Cell Death Dis. 2022, 13, 122. [Google Scholar] [CrossRef]
- Kugeratski, F.G.; Hodge, K.; Lilla, S.; McAndrews, K.M.; Zhou, X.; Hwang, R.F.; Zanivan, S.; Kalluri, R. Quantitative proteomics identifies the core proteome of exosomes with syntenin-1 as the highest abundant protein and a putative universal biomarker. Nat. Cell Biol. 2021, 23, 631–641. [Google Scholar] [CrossRef]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef]
- Tkachenko, E.; Rhodes, J.M.; Simons, M. Syndecans: New kids on the signaling block. Circ. Res. 2005, 96, 488–500. [Google Scholar] [CrossRef] [Green Version]
- David, G.; Zimmermann, P. Heparanase tailors syndecan for exosome production. Mol. Cell Oncol. 2015, 3, e1047556. [Google Scholar] [CrossRef] [Green Version]
- Gallagher, J. Fell-Muir Lecture: Heparan sulphate and the art of cell regulation: A polymer chain conducts the protein orchestra. Int. J. Exp. Pathol. 2015, 96, 203–231. [Google Scholar] [CrossRef] [Green Version]
- Hammond, E.; Khurana, A.; Shridhar, V.; Dredge, K. The Role of Heparanase and Sulfatases in the Modification of Heparan Sulfate Proteoglycans within the Tumor Microenvironment and Opportunities for Novel Cancer Therapeutics. Front. Oncol. 2014, 4, 195. [Google Scholar] [CrossRef]
- Ilan, N.; Elkin, M.; Vlodavsky, I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int. J. Biochem. Cell Biol. 2006, 38, 2018–2039. [Google Scholar] [CrossRef]
- Mytilinaiou, M.; Nikitovic, D.; Berdiaki, A.; Kostouras, A.; Papoutsidakis, A.; Tsatsakis, A.M.; Tzanakakis, G.N. Emerging roles of syndecan 2 in epithelial and mesenchymal cancer progression. IUBMB Life 2017, 69, 824–833. [Google Scholar] [CrossRef] [Green Version]
- Thompson, C.A.; Purushothaman, A.; Ramani, V.C.; Vlodavsky, I.; Sanderson, R.D. Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes. J. Biol. Chem. 2013, 288, 10093–10099. [Google Scholar] [CrossRef] [Green Version]
- Fares, J.; Kashyap, R.; Zimmermann, P. Syntenin: Key player in cancer exosome biogenesis and uptake? Cell Adh. Migr. 2017, 11, 124–126. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Batagov, A.O.; Kurochkin, I.V. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3’-untranslated regions. Biol. Direct. 2013, 8, 12. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Feng, X.; Liu, H.; Tong, R.; Wu, J.; Li, C.; Yu, H.; Chen, Y.; Cheng, Q.; Chen, J.; et al. High-metastatic cancer cells derived exosomal miR92a-3p promotes epithelial-mesenchymal transition and metastasis of low-metastatic cancer cells by regulating PTEN/Akt pathway in hepatocellular carcinoma. Oncogene 2020, 39, 6529–6543. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Arenaccio, C.; Federico, M. The Multifaceted Functions of Exosomes in Health and Disease: An Overview. Adv. Exp. Med. Biol. 2017, 998, 3–19. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. Discovery of Double-Stranded Genomic DNA in Circulating Exosomes. Cold Spring Harb. Symp. Quant. Biol. 2016, 81, 275–280. [Google Scholar] [CrossRef] [Green Version]
- Castellanos-Rizaldos, E.; Grimm, D.G.; Tadigotla, V.; Hurley, J.; Healy, J.; Neal, P.L.; Sher, M.; Venkatesan, R.; Karlovich, C.; Raponi, M.; et al. Exosome-Based Detection of EGFR T790M in Plasma from Non-Small Cell Lung Cancer Patients. Clin. Cancer Res. 2018, 24, 2944–2950. [Google Scholar] [CrossRef] [Green Version]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borràs, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.K.; Lee, J.; Kim, S.R.; Choi, D.S.; Yoon, Y.J.; Kim, J.H.; Go, G.; Nhung, D.; Hong, K.; Jang, S.C.; et al. EVpedia: A community web portal for extracellular vesicles research. Bioinformatics 2015, 31, 933–939. [Google Scholar] [CrossRef]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Li, Y.; Chen, B.; Zhao, J.; Yu, S.; Tang, Y.; Zheng, Q.; Li, Y.; Wang, P.; He, X.; et al. exoRBase: A database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018, 46, D106–D112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Zhang, Q.; Zhang, J.; Li, C.; Miao, Y.R.; Lei, Q.; Li, Q.; Guo, A.Y. EVmiRNA: A database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. 2019, 47, D89–D93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Ketting, R.F. microRNA Biogenesis and Function: An overview. Adv. Exp. Med. Biol. 2011, 700, 1–14. [Google Scholar] [CrossRef]
- Ardekani, A.M.; Naeini, M.M. The Role of MicroRNAs in Human Diseases. Avicenna J. Med. Biotechnol. 2010, 2, 161–179. [Google Scholar]
- Acunzo, M.; Romano, G.; Wernicke, D.; Croce, C.M. MicroRNA and cancer—A brief overview. Adv. Biol. Regul. 2015, 57, 1–9, Erratum in Adv. Biol. Regul. 2015, 58, 53. [Google Scholar] [CrossRef]
- Santos, R.M.; Moreno, C.; Zhang, W.C. Non-Coding RNAs in Lung Tumor Initiation and Progression. Int. J. Mol. Sci. 2020, 16, 2774. [Google Scholar] [CrossRef]
- Soares, E.; Reis, J.; Rodrigues, M.; Ribeiro, C.F.; Pereira, F.C. Circulating Extracellular Vesicles: The Missing Link between Physical Exercise and Depression Management? Int. J. Mol. Sci. 2021, 22, 542. [Google Scholar] [CrossRef]
- Cheng, G. Circulating miRNAs: Roles in cancer diagnosis, prognosis and therapy. Adv. Drug Deliv. Rev. 2015, 81, 75–93. [Google Scholar] [CrossRef]
- Schwarzenbach, H. Clinical Relevance of Circulating, Cell-Free and Exosomal microRNAs in Plasma and Serum of Breast Cancer Patients. Oncol. Res. Treat. 2017, 40, 423–429. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, W.B.; Zhou, J.; Wei, Y.; Wang, H.M.; Liu, X.D.; Chen, X.C.; Wang, W.; Ye, L.; Yao, L.C.; et al. Circulating exosomal microRNAs as novel potential detection biomarkers in pancreatic cancer. Oncol. Lett. 2020, 20, 1432–1440. [Google Scholar] [CrossRef]
- Jin, X.; Chen, Y.; Chen, H.; Fei, S.; Chen, D.; Cai, X.; Liu, L.; Lin, B.; Su, H.; Zhao, L.; et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin. Cancer Res. 2017, 23, 5311–5319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yagi, Y.; Ohkubo, T.; Kawaji, H.; Machida, A.; Miyata, H.; Goda, S.; Roy, S.; Hayashizaki, Y.; Suzuki, H.; Yokota, T. Next-generation sequencing-based small RNA profiling of cerebrospinal fluid exosomes. Neurosci. Lett. 2017, 636, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Siciliano, V.; Garzilli, I.; Fracassi, C.; Criscuolo, S.; Ventre, S.; di Bernardo, D. MiRNAs confer phenotypic robustness to gene networks by suppressing biological noise. Nat. Commun. 2013, 4, 2364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shore, S.; Henderson, J.M.; Lebedev, A.; Salcedo, M.P.; Zon, G.; McCaffrey, A.P.; Paul, N.; Hogrefe, R.I. Small RNA Library Preparation Method for Next-Generation Sequencing Using Chemical Modifications to Prevent Adapter Dimer Formation. PLoS ONE 2016, 11, e0167009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buschmann, D.; Kirchner, B.; Hermann, S.; Märte, M.; Wurmser, C.; Brandes, F.; Kotschote, S.; Bonin, M.; Steinlein, O.K.; Pfaffl, M.W.; et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J. Extracell. Vesicles 2018, 7, 1481321, Erratum in J. Extracell. Vesicles 2019, 8, 1581487. [Google Scholar] [CrossRef]
- Olivares, D.; Perez-Hernandez, J.; Perez-Gil, D.; Chaves, F.J.; Redon, J.; Cortes, R. Optimization of small RNA library preparation protocol from human urinary exosomes. J. Transl. Med. 2020, 18, 132. [Google Scholar] [CrossRef]
- Loudig, O.; Liu, C.; Rohan, T.; Ben-Dov, I.Z. Retrospective MicroRNA Sequencing: Complementary DNA Library Preparation Protocol Using Formalin-fixed Paraffin-embedded RNA Specimens. J. Vis. Exp. 2018, 135, 57471. [Google Scholar] [CrossRef] [Green Version]
- Loudig, O.; Wang, T.; Ye, K.; Lin, J.; Wang, Y.; Ramnauth, A.; Liu, C.; Stark, A.; Chitale, D.; Greenlee, R.; et al. Evaluation and Adaptation of a Laboratory-Based cDNA Library Preparation Protocol for Retrospective Sequencing of Archived MicroRNAs from up to 35-Year-Old Clinical FFPE Specimens. Int. J. Mol. Sci. 2017, 18, 627. [Google Scholar] [CrossRef] [Green Version]
- Geiss, G.K.; Bumgarner, R.E.; Birditt, B.; Dahl, T.; Dowidar, N.; Dunaway, D.L.; Fell, H.P.; Ferree, S.; George, R.D.; Grogan, T.; et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008, 26, 317–325. [Google Scholar] [CrossRef]
- Precazzini, F.; Detassis, S.; Imperatori, A.S.; Denti, M.A.; Campomenosi, P. Measurements Methods for the Development of MicroRNA-Based Tests for Cancer Diagnosis. Int. J. Mol. Sci. 2021, 22, 1176. [Google Scholar] [CrossRef] [PubMed]
- Ryu, K.J.; Lee, J.Y.; Choi, M.E.; Yoon, S.E.; Cho, J.; Ko, Y.H.; Shim, J.H.; Kim, W.S.; Park, C.; Kim, S.J. Serum-Derived Exosomal MicroRNA Profiles Can Predict Poor Survival Outcomes in Patients with Extranodal Natural Killer/T-Cell Lymphoma. Cancers 2020, 12, 3548. [Google Scholar] [CrossRef]
- Sanchez Herrero, J.F.; Pluvinet, R.; Luna de Haro, A.; Sumoy, L. Paired-end small RNA sequencing reveals a possible overestimation in the isomiR sequence repertoire previously reported from conventional single read data analysis. BMC Bioinform. 2021, 22, 215. [Google Scholar] [CrossRef] [PubMed]
- Zelli, V.; Compagnoni, C.; Capelli, R.; Corrente, A.; Cornice, J.; Vecchiotti, D.; Di Padova, M.; Zazzeroni, F.; Alesse, E.; Tessitore, A. Emerging Role of isomiRs in Cancer: State of the Art and Recent Advances. Genes 2021, 12, 1447. [Google Scholar] [CrossRef]
- Giallombardo, M.; Chacártegui Borrás, J.; Castiglia, M.; Van Der Steen, N.; Mertens, I.; Pauwels, P.; Peeters, M.; Rolfo, C. Exosomal miRNA Analysis in Non-small Cell Lung Cancer (NSCLC) Patients’ Plasma Through qPCR: A Feasible Liquid Biopsy Tool. J. Vis. Exp. 2016, 111, 53900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, M.; Bajo-Santos, C.; Hessvik, N.P.; Lorenz, S.; Fromm, B.; Berge, V.; Sandvig, K.; Linē, A.; Llorente, A. Identification of non-invasive miRNAs biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol. Cancer 2017, 16, 156. [Google Scholar] [CrossRef]
- Tang, Y.; Zhao, Y.; Song, X.; Song, X.; Niu, L.; Xie, L. Tumor-derived exosomal miRNA-320d as a biomarker for metastatic colorectal cancer. J. Clin. Lab. Anal. 2019, 33, e23004. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.H.; Choi, B.J.; Nam, S.W.; Park, W.S. Gastric cancer exosomes contribute to the field cancerization of gastric epithelial cells surrounding gastric cancer. Gastric Cancer 2022, 25, 490–502. [Google Scholar] [CrossRef]
- White, T.B.; McCoy, A.M.; Streva, V.A.; Fenrich, J.; Deininger, P.L. A droplet digital PCR detection method for rare L1 insertions in tumors. Mob. DNA 2014, 5, 30. [Google Scholar] [CrossRef] [Green Version]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef]
- Zhu, G.; Ye, X.; Dong, Z.; Lu, Y.C.; Sun, Y.; Liu, Y.; McCormack, R.; Gu, Y.; Liu, X. Highly Sensitive Droplet Digital PCR Method for Detection of EGFR-Activating Mutations in Plasma Cell-Free DNA from Patients with Advanced Non-Small Cell Lung Cancer. J. Mol. Diagn. 2015, 17, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M.; Carnero, A. Cell cycle deregulation: A common motif in cancer. Prog. Cell Cycle Res. 2003, 5, 5–18. [Google Scholar] [PubMed]
- Su, L.; Zhang, J.; Zhang, X.; Zheng, L.; Zhu, Z. Identification of cell cycle as the critical pathway modulated by exosome-derived microRNAs in gallbladder carcinoma. Med. Oncol. 2021, 38, 141. [Google Scholar] [CrossRef]
- Xia, H.; Li, M.; Chen, L.; Leng, W.; Yuan, D.; Pang, X.; Chen, L.; Li, R.; Tang, Q.; Bi, F. Suppression of RND3 activity by AES downregulation promotes cancer cell proliferation and invasion. Int. J. Mol. Med. 2013, 31, 1081–1086. [Google Scholar] [CrossRef] [Green Version]
- Razavipour, S.F.; Harikumar, K.B.; Slingerland, J.M. p27 as a Transcriptional Regulator: New Roles in Development and Cancer. Cancer Res. 2020, 80, 3451–3458. [Google Scholar] [CrossRef]
- Zhang, Z.; Xing, T.; Chen, Y.; Xiao, J. Exosome-mediated miR-200b promotes colorectal cancer proliferation upon TGF-β1 exposure. Biomed. Pharmacother. 2018, 106, 1135–1143. [Google Scholar] [CrossRef]
- Yan, S.; Liu, G.; Jin, C.; Wang, Z.; Duan, Q.; Xu, J.; Xu, D. MicroRNA-6869-5p acts as a tumor suppressor via targeting TLR4/NF-κB signaling pathway in colorectal cancer. J. Cell Physiol. 2018, 233, 6660–6668. [Google Scholar] [CrossRef]
- Yan, S.; Han, B.; Gao, S.; Wang, X.; Wang, Z.; Wang, F.; Zhang, J.; Xu, D.; Sun, B. Exosome-encapsulated microRNAs as circulating biomarkers for colorectal cancer. Oncotarget 2017, 8, 60149–60158. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Yu, F.; Ding, H.; Wang, Y.; Li, P.; Wang, K. Emerging Function and Clinical Values of Exosomal MicroRNAs in Cancer. Mol. Ther. Nucleic Acids 2019, 16, 791–804. [Google Scholar] [CrossRef] [Green Version]
- Avrutsky, M.I.; Troy, C.M. Caspase-9: A Multimodal Therapeutic Target With Diverse Cellular Expression in Human Disease. Front. Pharmacol. 2021, 12, 701301. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Xie, M.; Ding, K.; Xu, T.; Fang, Y.; Ma, P.; Shu, Y. Exosome-transmitted miR-769-5p confers cisplatin resistance and progression in gastric cancer by targeting CASP9 and promoting the ubiquitination degradation of p53. Clin. Transl. Med. 2022, 12, e780. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Li, M.; Cui, S.; Wang, D.; Zhang, C.Y.; Zen, K.; Li, L. Shikonin Inhibits the Proliferation of Human Breast Cancer Cells by Reducing Tumor-Derived Exosomes. Molecules 2016, 21, 777. [Google Scholar] [CrossRef]
- Kwon, Y.; Kim, M.; Kim, Y.; Jung, H.S.; Jeoung, D. Exosomal MicroRNAs as Mediators of Cellular Interactions between Cancer Cells and Macrophages. Front. Immunol. 2020, 11, 1167. [Google Scholar] [CrossRef]
- Rőszer, T. Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediat. Inflamm. 2015, 2015, 816460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owen, J.L.; Mohamadzadeh, M. Macrophages and chemokines as mediators of angiogenesis. Front. Physiol. 2013, 4, 159. [Google Scholar] [CrossRef] [Green Version]
- Ying, X.; Wu, Q.; Wu, X.; Zhu, Q.; Wang, X.; Jiang, L.; Chen, X.; Wang, X. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget 2016, 7, 43076–43087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Ying, X.; Wang, X.; Wu, X.; Zhu, Q.; Wang, X. Exosomes derived from hypoxic epithelial ovarian cancer deliver microRNA-940 to induce macrophage M2 polarization. Oncol. Rep. 2017, 38, 522–528. [Google Scholar] [CrossRef] [Green Version]
- Deregowska, A.; Wnuk, M. RAP1/TERF2IP-A Multifunctional Player in Cancer Development. Cancers 2021, 13, 5970. [Google Scholar] [CrossRef]
- Medzhitov, R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001, 1, 135–145. [Google Scholar] [CrossRef]
- Martin-Gayo, E.; Yu, X.G. Role of Dendritic Cells in Natural Immune Control of HIV-1 Infection. Front. Immunol. 2019, 10, 1306. [Google Scholar] [CrossRef] [PubMed]
- Veglia, F.; Gabrilovich, D.I. Dendritic cells in cancer: The role revisited. Curr. Opin. Immunol. 2017, 45, 43–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romano, R.; Picca, A.; Eusebi, L.H.U.; Marzetti, E.; Calvani, R.; Moro, L.; Bucci, C.; Guerra, F. Extracellular Vesicles and Pancreatic Cancer: Insights on the Roles of miRNA, lncRNA, and Protein Cargos in Cancer Progression. Cells 2021, 10, 1361. [Google Scholar] [CrossRef]
- Zhou, M.; Chen, J.; Zhou, L.; Chen, W.; Ding, G.; Cao, L. Pancreatic cancer derived exosomes regulate the expression of TLR4 in dendritic cells via miR-203. Cell Immunol. 2014, 292, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Lundholm, M.; Schröder, M.; Nagaeva, O.; Baranov, V.; Widmark, A.; Mincheva-Nilsson, L.; Wikström, P. Prostate tumor-derived exosomes down-regulate NKG2D expression on natural killer cells and CD8+ T cells: Mechanism of immune evasion. PLoS ONE 2014, 9, e108925. [Google Scholar] [CrossRef]
- Childs, R.W.; Carlsten, M. Therapeutic approaches to enhance natural killer cell cytotoxicity against cancer: The force awakens. Nat. Rev. Drug Discov. 2015, 14, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Thompson, T.W.; Kim, A.B.; Li, P.J.; Wang, J.; Jackson, B.T.; Huang, K.T.H.; Zhang, L.; Raulet, D.H. Endothelial cells express NKG2D ligands and desensitize antitumor NK responses. Elife 2017, 6, e30881. [Google Scholar] [CrossRef]
- Briand, J.; Garnier, D.; Nadaradjane, A.; Clément-Colmou, K.; Potiron, V.; Supiot, S.; Bougras-Cartron, G.; Frenel, J.S.; Heymann, D.; Vallette, F.M.; et al. Radiotherapy-induced overexpression of exosomal miRNA-378a-3p in cancer cells limits natural killer cells cytotoxicity. Epigenomics 2020, 12, 397–408. [Google Scholar] [CrossRef]
- Duan, S.; Guo, W.; Xu, Z.; He, Y.; Liang, C.; Mo, Y.; Wang, Y.; Xiong, F.; Guo, C.; Li, Y.; et al. Natural killer group 2D receptor and its ligands in cancer immune escape. Mol. Cancer 2019, 18, 29. [Google Scholar] [CrossRef]
- Berchem, G.; Noman, M.Z.; Bosseler, M.; Paggetti, J.; Baconnais, S.; Le Cam, E.; Nanbakhsh, A.; Moussay, E.; Mami-Chouaib, F.; Janji, B.; et al. Hypoxic tumor-derived microvesicles negatively regulate NK cell function by a mechanism involving TGF-β and miR23a transfer. Oncoimmunology 2015, 5, e1062968. [Google Scholar] [CrossRef] [Green Version]
- Alečković, M.; Kang, Y. Regulation of cancer metastasis by cell-free miRNAs. Biochim. Biophys. Acta 2015, 1855, 24–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, X.; Qian, N.; Ling, S.; Li, Y.; Sun, W.; Li, J.; Du, R.; Zhong, G.; Liu, C.; Yu, G.; et al. Breast cancer exosomes contribute to pre-metastatic niche formation and promote bone metastasis of tumor cells. Theranostics 2021, 11, 1429–1445. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Zhu, A.; Gong, L. Exosomes of glioma cells deliver miR-148a to promote proliferation and metastasis of glioblastoma via targeting CADM1. Bull. Cancer 2018, 105, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef] [Green Version]
- Felicetti, F.; De Feo, A.; Coscia, C.; Puglisi, R.; Pedini, F.; Pasquini, L.; Bellenghi, M.; Errico, M.C.; Pagani, E.; Carè, A. Exosome-mediated transfer of miR-222 is sufficient to increase tumor malignancy in melanoma. J. Transl. Med. 2016, 14, 56. [Google Scholar] [CrossRef] [Green Version]
- Sai, B.; Dai, Y.; Fan, S.; Wang, F.; Wang, L.; Li, Z.; Tang, J.; Wang, L.; Zhang, X.; Zheng, L.; et al. Cancer-educated mesenchymal stem cells promote the survival of cancer cells at primary and distant metastatic sites via the expansion of bone marrow-derived-PMN-MDSCs. Cell Death Dis. 2019, 10, 941. [Google Scholar] [CrossRef]
- Zhang, X.; Sai, B.; Wang, F.; Wang, L.; Wang, Y.; Zheng, L.; Li, G.; Tang, J.; Xiang, J. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol. Cancer 2019, 18, 40. [Google Scholar] [CrossRef] [Green Version]
- Théry, C.; Regnault, A.; Garin, J.; Wolfers, J.; Zitvogel, L.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Molecular Characterization of Dendritic Cell-Derived Exosomes. Selective Accumulation of the Heat Shock Protein Hsc73. J. Cell Biol. 1999, 147, 599–610. [Google Scholar] [CrossRef] [Green Version]
- Théry, C.; Boussac, M.; Véron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic Analysis of Dendritic Cell-Derived Exosomes: A Secreted Subcellular Compartment Distinct from Apoptotic Vesicles. J. Immunol. 2001, 166, 7309–7318. [Google Scholar] [CrossRef] [Green Version]
- van Niel, G.; Raposo, G.; Candalh, C.; Boussac, M.; Hershberg, R.; Cerf-Bensussan, N.; Heyman, M. Intestinal Epithelial Cells Secrete Exosome-like Vesicles. Gastroenterology 2001, 121, 337–349. [Google Scholar] [CrossRef]
- Skokos, D.; Botros, H.G.; Demeure, C.; Morin, J.; Peronet, R.; Birkenmeier, G.; Boudaly, S.; Mécheri, S. Mast Cell-Derived Exosomes Induce Phenotypic and Functional Maturation of Dendritic Cells and Elicit Specific Immune Responses In Vivo. J. Immunol. Baltim. Md. 2003, 170, 3037–3045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- György, B.; Szabó, T.G.; Pásztói, M.; Pál, Z.; Misják, P.; Aradi, B.; László, V.; Pállinger, E.; Pap, E.; Kittel, A.; et al. Membrane Vesicles, Current State-of-the-Art: Emerging Role of Extracellular Vesicles. Cell. Mol. Life Sci. 2011, 68, 2667–2688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological Properties of Extracellular Vesicles and Their Physiological Functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef] [Green Version]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-to-Cell Communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Teng, F.; Fussenegger, M. Shedding Light on Extracellular Vesicle Biogenesis and Bioengineering. Adv. Sci. 2021, 8, 2003505. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Marar, C.; Starich, B.; Wirtz, D. Extracellular Vesicles in Immunomodulation and Tumor Progression. Nat. Immunol. 2021, 22, 560–570. [Google Scholar] [CrossRef]
- Cheng, L.; Hill, A.F. Therapeutically Harnessing Extracellular Vesicles. Nat. Rev. Drug Discov. 2022, 21, 379–399. [Google Scholar] [CrossRef] [PubMed]
- Østergaard, O.; Nielsen, C.T.; Iversen, L.V.; Jacobsen, S.; Tanassi, J.T.; Heegaard, N.H.H. Quantitative Proteome Profiling of Normal Human Circulating Microparticles. J. Proteome Res. 2012, 11, 2154–2163. [Google Scholar] [CrossRef] [PubMed]
- Baranyai, T.; Herczeg, K.; Onódi, Z.; Voszka, I.; Módos, K.; Marton, N.; Nagy, G.; Mäger, I.; Wood, M.J.; El Andaloussi, S.; et al. Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PLoS ONE 2015, 10, e0145686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brennan, K.; Martin, K.; FitzGerald, S.P.; O’Sullivan, J.; Wu, Y.; Blanco, A.; Richardson, C.; Mc Gee, M.M. A Comparison of Methods for the Isolation and Separation of Extracellular Vesicles from Protein and Lipid Particles in Human Serum. Sci. Rep. 2020, 10, 1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Askeland, A.; Borup, A.; Østergaard, O.; Olsen, J.V.; Lund, S.M.; Christiansen, G.; Kristensen, S.R.; Heegaard, N.H.H.; Pedersen, S. Mass-Spectrometry Based Proteome Comparison of Extracellular Vesicle Isolation Methods: Comparison of ME-Kit, Size-Exclusion Chromatography, and High-Speed Centrifugation. Biomedicines 2020, 8, 246. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic Comparison Defines Novel Markers to Characterize Heterogeneous Populations of Extracellular Vesicle Subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [Green Version]
- Kasprzyk, J.; Stępień, E.; Piekoszewski, W. Application of Nano-LC-MALDI-TOF/TOF-MS for Proteomic Analysis of Microvesicles. Clin. Biochem. 2017, 50, 241–243. [Google Scholar] [CrossRef]
- Fel, A.; Lewandowska, A.E.; Petrides, P.E.; Wiśniewski, J.R. Comparison of Proteome Composition of Serum Enriched in Extracellular Vesicles Isolated from Polycythemia Vera Patients and Healthy Controls. Proteomes 2019, 7, 20. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.C.; Morgan, C.P.; Adrian Leu, N.; Shetty, A.; Cisse, Y.M.; Nugent, B.M.; Morrison, K.E.; Jašarević, E.; Huang, W.; Kanyuch, N.; et al. Reproductive Tract Extracellular Vesicles Are Sufficient to Transmit Intergenerational Stress and Program Neurodevelopment. Nat. Commun. 2020, 11, 1499. [Google Scholar] [CrossRef] [Green Version]
- Zougman, A.; Selby, P.J.; Banks, R.E. Suspension Trapping (STrap) Sample Preparation Method for Bottom-up Proteomics Analysis. Proteomics 2014, 14, 1006–1000. [Google Scholar] [CrossRef]
- Wu, C.; Zhou, S.; Mitchell, M.I.; Hou, C.; Byers, S.; Loudig, O.; Ma, J. Coupling Suspension Trapping–Based Sample Preparation and Data-Independent Acquisition Mass Spectrometry for Sensitive Exosomal Proteomic Analysis. Anal. Bioanal. Chem. 2022, 414, 2585–2595. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.J.; Lim, J.W.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic Insights and Diagnostic Potential. Expert Rev. Proteom. 2009, 6, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Schey, K.L.; Luther, J.M.; Rose, K.L. Proteomics Characterization of Exosome Cargo. Methods 2015, 87, 75–82. [Google Scholar] [CrossRef] [Green Version]
- Kreimer, S.; Belov, A.M.; Ghiran, I.; Murthy, S.K.; Frank, D.A.; Ivanov, A.R. Mass-Spectrometry-Based Molecular Characterization of Extracellular Vesicles: Lipidomics and Proteomics. J. Proteome Res. 2015, 14, 2367–2384. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.-S.; Kim, D.-K.; Kim, Y.-K.; Gho, Y.S. Proteomics of Extracellular Vesicles: Exosomes and Ectosomes. Mass Spectrom. Rev. 2015, 34, 474–490. [Google Scholar] [CrossRef]
- Rosa-Fernandes, L.; Rocha, V.B.; Carregari, V.C.; Urbani, A.; Palmisano, G. A Perspective on Extracellular Vesicles Proteomics. Front. Chem. 2017, 5, 102. [Google Scholar] [CrossRef] [Green Version]
- Geis-Asteggiante, L.; Dhabaria, A.; Edwards, N.; Ostrand-Rosenberg, S.; Fenselau, C. Top–down Analysis of Low Mass Proteins in Exosomes Shed by Murine Myeloid-Derived Suppressor Cells. Int. J. Mass Spectrom. 2015, 378, 264–269. [Google Scholar] [CrossRef] [Green Version]
- Fujita, K.; Kume, H.; Matsuzaki, K.; Kawashima, A.; Ujike, T.; Nagahara, A.; Uemura, M.; Miyagawa, Y.; Tomonaga, T.; Nonomura, N. Proteomic Analysis of Urinary Extracellular Vesicles from High Gleason Score Prostate Cancer. Sci. Rep. 2017, 7, 42961. [Google Scholar] [CrossRef] [Green Version]
- Fricke, F.; Michalak, M.; Warnken, U.; Hausser, I.; Schnölzer, M.; Kopitz, J.; Gebert, J. SILAC-Based Quantification of TGFBR2-Regulated Protein Expression in Extracellular Vesicles of Microsatellite Unstable Colorectal Cancers. Int. J. Mol. Sci. 2019, 20, E4162. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, S.; Koller, A.; Mani, K.M.; Wen, R.; Alfieri, A.; Saha, S.; Wang, J.; Patel, P.; Bandeira, N.; Guha, C.; et al. Identifying Urinary and Serum Exosome Biomarkers For Radiation Exposure Using A DDA and SWATH-MS Combined Workflow. Int. J. Radiat. Oncol. Biol. Phys. 2016, 96, 566–577. [Google Scholar] [CrossRef] [Green Version]
- Ole, N.; Jensen, O.N. Interpreting the protein language using proteomics. Nat. Rev. Mol. Cell Biol. 2006, 7, 391–403. [Google Scholar]
- Muller, M. Post-Translational Modifications of Protein Backbones: Unique Functions, Mechanisms, and Challenges. Biochemistry 2018, 57, 177–185. [Google Scholar] [CrossRef]
- Gonzales, P.A.; Pisitkun, T.; Hoffert, J.D.; Tchapyjnikov, D.; Star, R.A.; Kleta, R.; Wang, N.S.; Knepper, M.A. Large-Scale Proteomics and Phosphoproteomics of Urinary Exosomes. J. Am. Soc. Nephrol. 2009, 20, 363–379. [Google Scholar] [CrossRef] [Green Version]
- Saraswat, M.; Joenväära, S.; Musante, L.; Peltoniemi, H.; Holthofer, H.; Renkonen, R. N-Linked (N-) Glycoproteomics of Urinary Exosomes. Mol. Cell. Proteom. 2015, 14, 2298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, C.J.; Gaunitz, S.; Wang, Z.; Strindelius, L.; Jacobson, S.C.; Clemmer, D.E.; Trinidad, J.C.; Novotny, M.V. Glycoproteomic Analysis of Human Urinary Exosomes. Anal. Chem. 2020, 92, 14357–14365. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.-H.; Xue, L.; Hsu, C.-C.; Paez, J.S.P.; Pan, L.; Andaluz, H.; Wendt, M.K.; Iliuk, A.B.; Zhu, J.-K.; Tao, W.A. Phosphoproteins in Extracellular Vesicles as Candidate Markers for Breast Cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 3175–3180. [Google Scholar] [CrossRef] [Green Version]
- Rontogianni, S.; Synadaki, E.; Li, B.; Liefaard, M.C.; Lips, E.H.; Wesseling, J.; Wu, W.; Altelaar, M. Proteomic Profiling of Extracellular Vesicles Allows for Human Breast Cancer Subtyping. Commun. Biol. 2019, 2, 325. [Google Scholar] [CrossRef] [Green Version]
- Lo Cicero, A.; Stahl, P.D.; Raposo, G. Extracellular Vesicles Shuffling Intercellular Messages: For Good or for Bad. Curr. Opin. Cell Biol. 2015, 35, 69–77. [Google Scholar] [CrossRef]
- Yu, R.K.; Tsai, Y.-T.; Ariga, T. Functional Roles of Gangliosides in Neurodevelopment: An Overview of Recent Advances. Neurochem. Res. 2012, 37, 1230–1244. [Google Scholar] [CrossRef] [Green Version]
- Sunshine, H.; Iruela-Arispe, M.L. Membrane lipids and cell signaling. Curr. Opin. Lipidol. 2017, 28, 408–413. [Google Scholar] [CrossRef]
- Butler, L.M.; Perone, Y.; Dehairs, J.; Lupien, L.E.; de Laat, V.; Talebi, A.; Loda, M.; Kinlaw, W.B.; Swinnen, J.V. Lipids and cancer: Emerging roles in pathogenesis, diagnosis and therapeutic intervention. Adv. Drug Deliv. Rev. 2020, 159, 245–293. [Google Scholar] [CrossRef] [PubMed]
- Gould, S.B. Membranes and evolution. Curr. Biol. 2018, 28, R381–R385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singer, S.J.; Nicolson, G.L. The fluid mosaic model of the structure of cell membranes. Science 1972, 175, 720–731. [Google Scholar] [CrossRef]
- Jackson Catherine, L.; Walch, L.; Verbavatz, J.-M. Lipids and Their Trafficking: An Integral Part of Cellular Organization. Dev. Cell 2016, 39, 139–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, J.X.; Wan, J.B.; He, C. Concise Review: Regulation of Stem Cell Proliferation and Differentiation by Essential Fatty Acids and Their Metabolites. Stem Cells 2014, 32, 1092–1098. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Song, Y.H.; Tang, Z.; Wang, Z.P.; Xu, Q.; Bao, N. Effects of ganglioside GM1 and neural growth factor on neural stem cell proliferation and differentiation. Genet. Mol. Res. 2016, 15, 1–10. [Google Scholar] [CrossRef]
- Newton, J.; Lima, S.; Maceyka, M.; Spiegel, S. Revisiting the sphingolipid rheostat: Evolving concepts in cancer therapy. Exp. Cell Res. 2015, 333, 195–200. [Google Scholar] [CrossRef] [Green Version]
- Houten, S.M.; Wanders, R.J.A. A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation. J. Inherit. Metab. Dis. 2010, 33, 469–477. [Google Scholar] [CrossRef] [Green Version]
- Abuhusain, H.J.; Matin, A.; Qiao, Q.; Shen, H.; Kain, N.; Day, B.W.; Stringer, B.W.; Daniels, B.; Laaksonen, M.A.; Teo, C.; et al. A metabolic shift favoring sphingosine 1-phosphate at the expense of ceramide controls glioblastoma angiogenesis. J. Biol. Chem. 2013, 288, 37355–37364. [Google Scholar] [CrossRef] [Green Version]
- Koizumi, A.; Narita, S.; Nakanishi, H.; Ishikawa, M.; Eguchi, S.; Kimura, H.; Takasuga, S.; Huang, M.; Inoue, T.; Sasaki, J.; et al. Increased fatty acyl saturation of phosphatidylinositol phosphates in prostate cancer progression. Sci. Rep. 2019, 9, 13257. [Google Scholar] [CrossRef] [Green Version]
- Greenlee, J.D.; Subramanian, T.; Liu, K.; King, M.R. Rafting Down the Metastatic Cascade: The Role of Lipid Rafts in Cancer Metastasis, Cell Death, and Clinical Outcomes. Cancer Res. 2021, 81, 5. [Google Scholar] [CrossRef] [PubMed]
- Sriram, V.; Cho, S.; Li, P.; O’Donnell Patrick, W.; Dunn, C.; Hayakawa, K.; Blum Janice, S.; Brutkiewicz Randy, R. Inhibition of glycolipid shedding rescues recognition of a CD1+ T cell lymphoma by natural killer T (NKT) cells. Proc. Natl. Acad. Sci. USA 2002, 99, 8197–8202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skotland, T.; Hessvik, N.P.; Sandvig, K.; Llorente, A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. 2019, 60, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017, 66, 30–41. [Google Scholar] [CrossRef]
- Wang, W.; Zhu, N.; Yan, T.; Shi, Y.-N.; Chen, J.; Zhang, C.-J.; Xie, X.-J.; Liao, D.-F.; Qin, L. The crosstalk: Exosomes and lipid metabolism. Cell Commun. Signal. 2020, 18, 119. [Google Scholar] [CrossRef]
- Donoso-Quezada, J.; Ayala-Mar, S.; González-Valdez, J. The role of lipids in exosome biology and intercellular communication: Function, analytics and applications. Traffic 2021, 22, 204–220. [Google Scholar] [CrossRef]
- Skotland, T.; Sagini, K.; Sandvig, K.; Llorente, A. An emerging focus on lipids in extracellular vesicles. Adv. Drug Deliv. Rev. 2020, 159, 308–321. [Google Scholar] [CrossRef]
- Zebrowska, A.; Skowronek, A.; Wojakowska, A.; Widlak, P.; Pietrowska, M. Metabolome of Exosomes: Focus on Vesicles Released by Cancer Cells and Present in Human Body Fluids. Int. J. Mol. Sci. 2019, 20, 3461. [Google Scholar] [CrossRef] [Green Version]
- Llorente, A.; Skotland, T.; Sylvänne, T.; Kauhanen, D.; Róg, T.; Orłowski, A.; Vattulainen, I.; Ekroos, K.; Sandvig, K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim. Biophys. Acta 2013, 1831, 1302–1309. [Google Scholar] [CrossRef]
- Yi, X.; Li, Y.; Hu, X.; Wang, F.; Liu, T. Changes in phospholipid metabolism in exosomes of hormone-sensitive and hormone-resistant prostate cancer cells. J. Cancer 2021, 12, 2893–2902. [Google Scholar] [CrossRef]
- Lydic, T.A.; Townsend, S.; Adda, C.G.; Collins, C.; Mathivanan, S.; Reid, G.E. Rapid and comprehensive ‘shotgun’ lipidome profiling of colorectal cancer cell derived exosomes. Methods 2015, 87, 83–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haraszti, R.A.; Didiot, M.-C.; Sapp, E.; Leszyk, J.; Shaffer, S.A.; Rockwell, H.E.; Gao, F.; Narain, N.R.; DiFiglia, M.; Kiebish, M.A.; et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles 2016, 5, 32570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fahy, E.; Cotter, D.; Sud, M.; Subramaniam, S. Lipid classification, structures and tools. Biochim. Biophys. Acta 2011, 1811, 637–647. [Google Scholar] [CrossRef] [Green Version]
- Fahy, E.; Subramaniam, S.; Murphy, R.C.; Nishijima, M.; Raetz, C.R.H.; Shimizu, T.; Spener, F.; van Meer, G.; Wakelam, M.J.O.; Dennis, E.A. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009, 50, S9–S14. [Google Scholar] [CrossRef] [Green Version]
- Lobasso, S.; Tanzarella, P.; Mannavola, F.; Tucci, M.; Silvestris, F.; Felici, C.; Ingrosso, C.; Corcelli, A.; Lopalco, P. A Lipidomic Approach to Identify Potential Biomarkers in Exosomes from Melanoma Cells with Different Metastatic Potential. Front. Physiol. 2021, 12, 748895. [Google Scholar] [CrossRef]
- Peterka, O.; Jirásko, R.; Chocholoušková, M.; Kuchař, L.; Wolrab, D.; Hájek, R.; Vrána, D.; Strouhal, O.; Melichar, B.; Holčapek, M. Lipidomic characterization of exosomes isolated from human plasma using various mass spectrometry techniques. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158634. [Google Scholar] [CrossRef] [PubMed]
- Altadill, T.; Campoy, I.; Lanau, L.; Gill, K.; Rigau, M.; Gil-Moreno, A.; Reventos, J.; Byers, S.; Colas, E.; Cheema, A.K. Enabling Metabolomics Based Biomarker Discovery Studies Using Molecular Phenotyping of Exosome-Like Vesicles. PLoS ONE 2016, 11, e0151339. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Ye, M.; Xu, D.; Li, X.; Pan, C.; Zou, H. Matrix with high salt tolerance for the analysis of peptide and protein samples by desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 2006, 78, 2593–2599. [Google Scholar] [CrossRef]
- Tran, A.; Wan, L.; Xu, Z.; Haro, J.M.; Li, B.; Jones, J.W. Lithium Hydroxide Hydrolysis Combined with MALDI TOF Mass Spectrometry for Rapid Sphingolipid Detection. J. Am. Soc. Mass. Spectrom. 2021, 32, 289–300. [Google Scholar] [CrossRef]
- Paglia, G.; Kliman, M.; Claude, E.; Geromanos, S.; Astarita, G. Applications of ion-mobility mass spectrometry for lipid analysis. Anal. Bioanal. Chem. 2015, 407, 4995–5007. [Google Scholar] [CrossRef]
- Bowman, A.P.; Blakney, G.T.; Hendrickson, C.L.; Ellis, S.R.; Heeren, R.M.A.; Smith, D.F. Ultra-High Mass Resolving Power, Mass Accuracy, and Dynamic Range MALDI Mass Spectrometry Imaging by 21-T FT-ICR MS. Anal. Chem. 2020, 92, 3133–3142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodds, J.N.; Baker, E.S. Ion Mobility Spectrometry: Fundamental Concepts, Instrumentation, Applications, and the Road Ahead. J. Am. Soc. Mass Spectrom. 2019, 30, 2185–2195. [Google Scholar] [CrossRef] [PubMed]
- Giles, K.; Ujma, J.; Wildgoose, J.; Pringle, S.; Richardson, K.; Langridge, D.; Green, M. A Cyclic Ion Mobility-Mass Spectrometry System. Anal. Chem. 2019, 91, 8564–8573. [Google Scholar] [CrossRef] [Green Version]
- Claude, E.; Jones, E.A.; Pringle, S.D. DESI Mass Spectrometry Imaging (MSI). Methods Mol. Biol. 2017, 1618, 65–75. [Google Scholar] [PubMed]
- Han, X.; Gross, R.W. Shotgun lipidomics: Electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass. Spectrom. Rev. 2005, 24, 367–412. [Google Scholar] [CrossRef] [PubMed]
- Hsu, F.-F. Mass spectrometry-based shotgun lipidomics—A critical review from the technical point of view. Anal. Bioanal. Chem. 2018, 410, 6387–6409. [Google Scholar] [CrossRef] [PubMed]
- Su, B.; Bettcher, L.F.; Hsieh, W.-Y.; Hornburg, D.; Pearson, M.J.; Blomberg, N.; Giera, M.; Snyder, M.P.; Raftery, D.; Bensinger, S.J.; et al. A DMS Shotgun Lipidomics Workflow Application to Facilitate High-Throughput, Comprehensive Lipidomics. J. Am. Soc. Mass Spectrom. 2021, 32, 2655–2663. [Google Scholar] [CrossRef]
- Wojakowska, A.; Zebrowska, A.; Skowronek, A.; Rutkowski, T.; Polanski, K.; Widlak, P.; Marczak, L.; Pietrowska, M. Metabolic Profiles of Whole Serum and Serum-Derived Exosomes Are Different in Head and Neck Cancer Patients Treated by Radiotherapy. J. Pers. Med. 2020, 10, 229. [Google Scholar] [CrossRef]
- Fuchs, B.; Süß, R.; Teuber, K.; Eibisch, M.; Schiller, J. Lipid analysis by thin-layer chromatography—A review of the current state. J. Chromatogr. A 2011, 1218, 2754–2774. [Google Scholar] [CrossRef]
- Engel, K.M.; Prabutzki, P.; Leopold, J.; Nimptsch, A.; Lemmnitzer, K.; Vos, D.R.N.; Hopf, C.; Schiller, J. A new update of MALDI-TOF mass spectrometry in lipid research. Prog. Lipid Res. 2022, 86, 101145. [Google Scholar] [CrossRef]
- Cajka, T.; Fiehn, O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Anal. Chem. TRAC 2014, 61, 192–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holčapek, M.; Liebisch, G.; Ekroos, K. Lipidomic Analysis. Anal. Chem. 2018, 90, 4249–4257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lange, M.; Fedorova, M. Evaluation of lipid quantification accuracy using HILIC and RPLC MS on the example of NIST® SRM® 1950 metabolites in human plasma. Anal. Bioanal. Chem. 2020, 412, 3573–3584. [Google Scholar] [CrossRef] [Green Version]
- Rampler, E.; Criscuolo, A.; Zeller, M.; El Abiead, Y.; Schoeny, H.; Hermann, G.; Sokol, E.; Cook, K.; Peake, D.A.; Delanghe, B.; et al. A Novel Lipidomics Workflow for Improved Human Plasma Identification and Quantification Using RPLC-MSn Methods and Isotope Dilution Strategies. Anal. Chem. 2018, 90, 6494–6501. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Lee, M.Y.; Choi, D.Y.; Lee, J.W.; You, S.; Lee, K.Y.; Kim, J.; Kim, K.P. Phospholipids of tumor extracellular vesicles stratify gefitinib-resistant nonsmall cell lung cancer cells from gefitinib-sensitive cells. Proteomics 2015, 15, 824–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aiello, N.M.; Brabletz, T.; Kang, Y.; Nieto, M.A.; Weinberg, R.A.; Stanger, B.Z. Upholding a role for EMT in pancreatic cancer metastasis. Nature 2017, 547, E7–E8. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Li, B.; Liu, F.; Zhang, M.; Wang, Q.; Liu, Y.; Yao, Y.; Li, D. The epithelial to mesenchymal transition (EMT) and cancer stem cells: Implication for treatment resistance in pancreatic cancer. Mol. Cancer 2017, 16, 52. [Google Scholar] [CrossRef] [Green Version]
- Brzozowski, J.S.; Jankowski, H.; Bond, D.R.; McCague, S.B.; Munro, B.R.; Predebon, M.J.; Scarlett, C.J.; Skelding, K.A.; Weidenhofer, J. Lipidomic profiling of extracellular vesicles derived from prostate and prostate cancer cell lines. Lipids Health Dis. 2018, 17, 211. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Zhang, K.; Qing, Y.; Li, D.; Cui, M.; Jin, P.; Xu, T. Proteomic and lipidomic analysis of exosomes derived from ovarian cancer cells and ovarian surface epithelial cells. J. Ovarian Res. 2020, 13, 9. [Google Scholar] [CrossRef]
- Kuc, N.; Doermann, A.; Shirey, C.; Lee, D.D.; Lowe, C.-W.; Awasthi, N.; Schwarz, R.E.; Stahelin, R.V.; Schwarz, M.A. Pancreatic ductal adenocarcinoma cell secreted extracellular vesicles containing ceramide-1-phosphate promote pancreatic cancer stem cell motility. Biochem. Pharmacol. 2018, 156, 458–466. [Google Scholar] [CrossRef]
- Fan, T.W.M.; Zhang, X.; Wang, C.; Yang, Y.; Kang, W.-Y.; Arnold, S.; Higashi, R.M.; Liu, J.; Lane, A.N. Exosomal lipids for classifying early and late stage non-small cell lung cancer. Anal. Chim. Acta 2018, 1037, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Zhou, J.; Yuan, C.; Zhang, L.; Li, D.; Si, D.; Xiu, D.; Zhong, L. Metabolomics identifies serum and exosomes metabolite markers of pancreatic cancer. Metabolomics 2019, 15, 86. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Ekroos, K.; Kauhanen, D.; Simolin, H.; Seierstad, T.; Berge, V.; Sandvig, K.; Llorente, A. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur. J. Cancer 2017, 70, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-T.; Purcell, E.; Palacios-Rolston, C.; Lo, T.-W.; Ramnath, N.; Jolly, S.; Nagrath, S. Isolation and Profiling of Circulating Tumor-Associated Exosomes Using Extracellular Vesicular Lipid-Protein Binding Affinity Based Microfluidic Device. Small 2019, 15, e1903600. [Google Scholar] [CrossRef]
- Matsumura, S.; Minamisawa, T.; Suga, K.; Kishita, H.; Akagi, T.; Ichiki, T.; Ichikawa, Y.; Shiba, K. Subtypes of tumour cell-derived small extracellular vesicles having differently externalized phosphatidylserine. J. Extracell. Vesicles 2019, 8, 1579541. [Google Scholar] [CrossRef] [Green Version]
- Sharma, R.; Huang, X.; Brekken, R.A.; Schroit, A.J. Detection of phosphatidylserine-positive exosomes for the diagnosis of early-stage malignancies. Br. J. Cancer 2017, 117, 545–552. [Google Scholar] [CrossRef] [Green Version]
- Nakai, W.; Yoshida, T.; Diez, D.; Miyatake, Y.; Nishibu, T.; Imawaka, N.; Naruse, K.; Sadamura, Y.; Hanayama, R. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci. Rep. 2016, 6, 33935. [Google Scholar] [CrossRef] [Green Version]
- Shih, C.-L.; Chong, K.-Y.; Hsu, S.-C.; Chien, H.-J.; Ma, C.-T.; Chang, J.W.-C.; Yu, C.-J.; Chiou, C.-C. Development of a magnetic bead-based method for the collection of circulating extracellular vesicles. New Biotechnol. 2016, 33, 116–122. [Google Scholar] [CrossRef]
- Vallabhapurapu, S.D.; Blanco, V.M.; Sulaiman, M.K.; Vallabhapurapu, S.L.; Chu, Z.; Franco, R.S.; Qi, X. Variation in human cancer cell external phosphatidylserine is regulated by flippase activity and intracellular calcium. Oncotarget 2015, 6, 34375–34388. [Google Scholar] [CrossRef] [Green Version]
- Utsugi, T.; Schroit, A.J.; Connor, J.; Bucana, C.D.; Fidler, I.J. Elevated Expression of Phosphatidylserine in the Outer Membrane Leaflet of Human Tumor Cells and Recognition by Activated Human Blood Monocytes. Cancer Res. 1991, 51, 3062–3066. [Google Scholar]
- Riedl, S.; Rinner, B.; Asslaber, M.; Schaider, H.; Walzer, S.; Novak, A.; Lohner, K.; Zweytick, D. In search of a novel target—Phosphatidylserine exposed by non-apoptotic tumor cells and metastases of malignancies with poor treatment efficacy. Biochim. Biophys. Acta 2011, 1808, 2638–2645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, H.P.; Holth, A.; Kleinberg, L.; Ruud, M.G.; Elstrand, M.B.; Tropé, C.G.; Davidson, B.; Risberg, B. Evaluation of cell surface expression of phosphatidylserine in ovarian carcinoma effusions using the annexin-V/7-AAD assay: Clinical relevance and comparison with other apoptosis parameters. Am. J. Clin. Pathol. 2009, 132, 756–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelleher, R.J., Jr.; Balu-Iyer, S.; Loyall, J.; Sacca, A.J.; Shenoy, G.N.; Peng, P.; Iyer, V.; Fathallah, A.M.; Berenson, C.S.; Wallace, P.K.; et al. Extracellular Vesicles Present in Human Ovarian Tumor Microenvironments Induce a Phosphatidylserine-Dependent Arrest in the T-cell Signaling Cascade. Cancer Immunol. Res. 2015, 3, 1269–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lea, J.; Sharma, R.; Yang, F.; Zhu, H.; Ward, E.S.; Schroit, A.J. Detection of phosphatidylserine-positive exosomes as a diagnostic marker for ovarian malignancies: A proof of concept study. Oncotarget 2017, 8, 14395–14407. [Google Scholar] [CrossRef] [Green Version]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
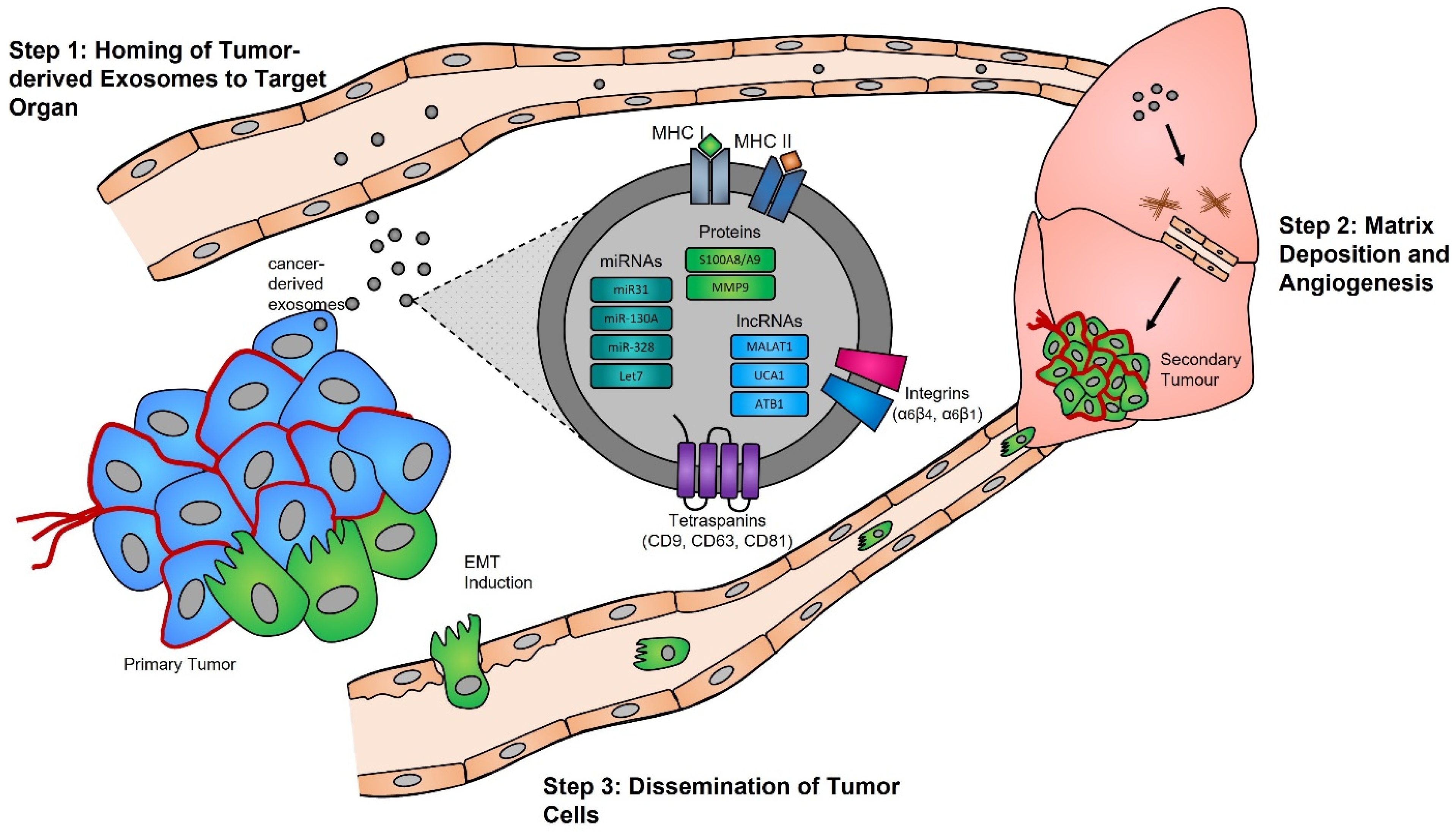




| Cancer Type | Differentially Expressed Between Healthy and Cancer | Reference | |
|---|---|---|---|
| miRNA Biomarkers | |||
| Colorectal Cancer | ↑ | miR-224-5p, miR-548d-5p, miR-200a-3p, miR-320d, miR-200b-3p, miR-1246 | Tang et al., 2019 [310] |
| ↓ | novel_246, novel_301, miR-27a-5p | ||
| miR-135a-5p, miR-204-5p | Sun et al., 2021 [104] | ||
| ↓ | miR-6869-5p | Yan et al., 2018 [321] | |
| ↑ | miR-486-5p, miR-3180-5p | Yan et al., 2017 [322] | |
| ↓ | miR-638, miR-5787, miR-8075, miR-6869-5p, miR-548c-5p | ||
| Ovarian Cancer | ↑ | miR-21, miR-141, miR-200a, miR-200b, miR-200c, miR-203, miR-205, miR-214, miR-215 | Taylor and Taylor, 2008 [89] |
| ↑ | miR-940 | Chen et al., 2017 [331] | |
| ↑ | miR-222-3p | Ying et al., 2016 [330] | |
| Glioblastoma | ↑ | let-7a, miR-15b, miR-16, miR-19b, miR-21, miR-26a, miR-27a, miR-92, miR-93, miR-320, miR-20 | Skog et al., 2008 [91] |
| ↑ | miR-148a | Cai et al., 2018 [346] | |
| Liver Cancer | ↑ | miR-17, miR-18a, miR-19a, miR-19b, miR-20a, miR-92a-3p | Yang et al., 2020 [276] |
| ↑ | miR-193a-3p, miR-210-3p, miR-5100 | Zhang et al., 2019 [350] | |
| Pancreatic Cancer | ↑ | miR-21, miR-210 | Wu et al., 2020 [294] |
| ↑ | miR-193a-3p, miR-210-3p, miR-5100 | Zhang et al., 2019 [350] | |
| Lung Cancer | ↑ | miR-132-3p, miR-181b-5p, miR-27a-3p, miR-27b-3p, miR-320a, miR-361-5p, let-7b-5p, miR-24-3p, miR-3184-5p, miR-486-5p, miR-486-3p, miR-320b | Jin et al., 2017 [295] |
| ↓ | let-7a-5p, let-7d-5p, let-7f-5p, miR-26b-5p, miR-30a-3p, miR-30e-3p, miR-744-5p, miR-744-5p, let-7e-5p, miR-191-5p, miR-191-5p, miR-206, miR-21-5p, miR-23a-5p, miR-23b-5p, miR-10b-5p, miR-15b-5p | ||
| miR-30b, miR-30c, miR-103, miR-122, miR-195, miR-203, miR-221, miR-222 | Giallombardo et al., 2016 [308] | ||
| ↑ | miR-193a-3p, miR-210-3p, miR-5100 | Zhang et al., 2019 [350] | |
| Extranodal Natural Killer/T-Cell Lymphoma | ↑ | miR-320e, miR-4454, miR-4516, miR-630, miR-122-5p, miR-574-5p, miR-22-3p, miR-486-3p, miR-1915-5p, miR-1972, miR-1285-5p, miR-222-3p, miR-1305, miR-891b, miR-4455, miR-21-5p, miR-1258, let-7b-5p, miR-25-3p, miR-1268a | Ryu et al., 2020 [305] |
| ↓ | miR-564, miR-196a-5p, miR-520c-3p, let-7d-5p, let-7i-5p, miR-212-3p, miR-29a-3p, miR-608, miR-503-5p, miR-587, miR-548g-3p, miR-765, miR-34c-3p, miR-770-5p, miR-301a-5p, miR-526a, miR-340-5p, miR-325, miR-199a-3p+miR-199b-3p, miR-423-3p | ||
| Prostate Cancer | ↓ | miR-196a-5p, miR-34a-5p, miR-501-3p, miR-92a-1-5p | Rodríguez et al., 2017 [309] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitchell, M.I.; Ma, J.; Carter, C.L.; Loudig, O. Circulating Exosome Cargoes Contain Functionally Diverse Cancer Biomarkers: From Biogenesis and Function to Purification and Potential Translational Utility. Cancers 2022, 14, 3350. https://doi.org/10.3390/cancers14143350
Mitchell MI, Ma J, Carter CL, Loudig O. Circulating Exosome Cargoes Contain Functionally Diverse Cancer Biomarkers: From Biogenesis and Function to Purification and Potential Translational Utility. Cancers. 2022; 14(14):3350. https://doi.org/10.3390/cancers14143350
Chicago/Turabian StyleMitchell, Megan I., Junfeng Ma, Claire L. Carter, and Olivier Loudig. 2022. "Circulating Exosome Cargoes Contain Functionally Diverse Cancer Biomarkers: From Biogenesis and Function to Purification and Potential Translational Utility" Cancers 14, no. 14: 3350. https://doi.org/10.3390/cancers14143350






