Prognostic Value of KRAS Mutations in Colorectal Cancer Patients
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Enrolled Patients
2.2. DNA Extraction and Molecular Analysis
3. Results
3.1. Patient’s Characteristics
3.2. RAS Mutations
3.3. KRAS and NRAS Mutations: Prognostic Value Evaluation
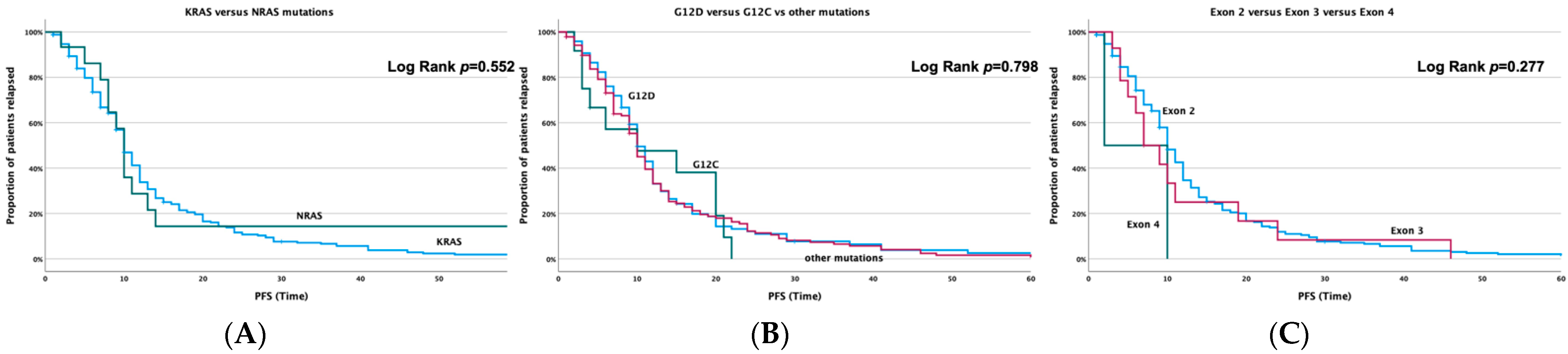
3.3.1. Correlation with PFS
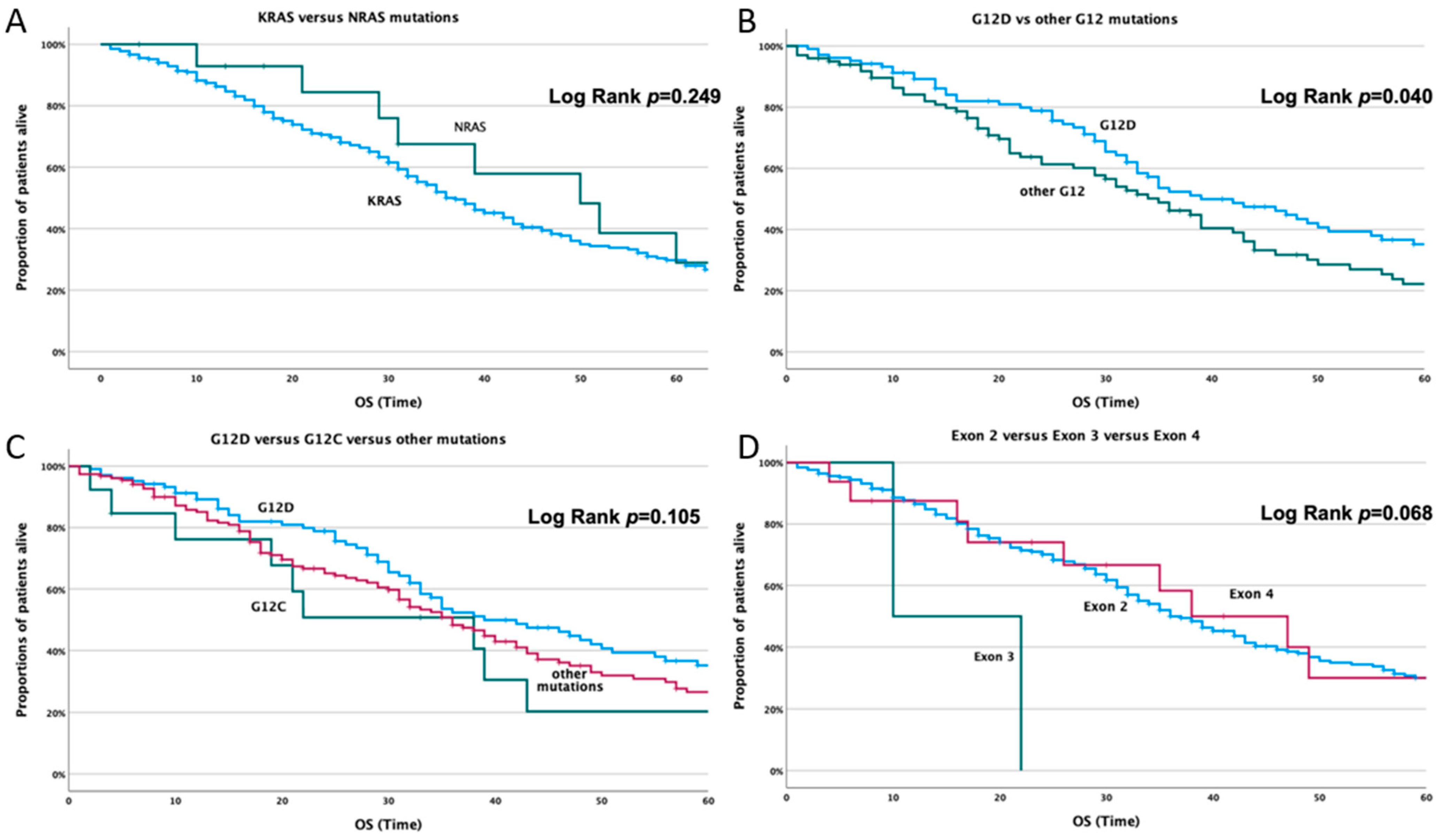
3.3.2. Correlation with Overall Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ucar, G.; Ergun, Y.; Aktürk Esen, S.; Acikgoz, Y.; Dirikoc, M.; Esen, İ.; Bal, Ö.; Uncu, D. Prognostic and Predictive Value of KRAS Mutation Number in Metastatic Colorectal Cancer. Medicine 2020, 99, e22407. [Google Scholar] [CrossRef] [PubMed]
- Saeed, O.; Lopez-Beltran, A.; Fisher, K.W.; Scarpelli, M.; Montironi, R.; Cimadamore, A.; Massari, F.; Santoni, M.; Cheng, L. RAS Genes in Colorectal Carcinoma: Pathogenesis, Testing Guidelines and Treatment Implications. J. Clin. Pathol. 2019, 72, 135–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tariq, K.; Ghias, K. Colorectal Cancer Carcinogenesis: A Review of Mechanisms. Cancer Biol. Med. 2016, 13, 120–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pino, M.S.; Chung, D.C. The Chromosomal Instability Pathway in Colon Cancer. Gastroenterology 2010, 138, 2059–2072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, D.; Wang, Y.; Zhu, L.; Jin, H.; Wang, X. Prognostic Value of KRAS Mutation Status in Colorectal Cancer Patients: A Population-Based Competing Risk Analysis. PeerJ 2020, 8, e9149. [Google Scholar] [CrossRef] [PubMed]
- Cefalì, M.; Epistolio, S.; Palmarocchi, M.C.; Frattini, M.; De Dosso, S. Research Progress on KRAS Mutations in Colorectal Cancer. J. Cancer Metastasis Treat 2021, 7, 26. [Google Scholar] [CrossRef]
- Sepulveda, A.R.; Hamilton, S.R.; Allegra, C.J.; Grody, W.; Cushman-Vokoun, A.M.; Funkhouser, W.K.; Kopetz, S.E.; Lieu, C.; Lindor, N.M.; Minsky, B.D.; et al. Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and the American Society of Clinical Oncology. J. Clin. Oncol. 2017, 35, 1453–1486. [Google Scholar] [CrossRef]
- Guo, T.; Wu, Y.; Tan, C.; Jin, Y.; Sheng, W.; Cai, S.; Liu, F.; Xu, Y. Clinicopathologic Features and Prognostic Value of KRAS, NRAS and BRAF Mutations and DNA Mismatch Repair Status: A Single-center Retrospective Study of 1,834 Chinese Patients with Stage I–IV Colorectal Cancer. Int. J. Cancer 2019, 145, 1625–1634. [Google Scholar] [CrossRef] [Green Version]
- Bellio, H.; Fumet, J.D.; Ghiringhelli, F. Targeting BRAF and RAS in Colorectal Cancer. Cancers 2021, 13, 2201. [Google Scholar] [CrossRef]
- Souglakos, J.; Philips, J.; Wang, R.; Marwah, S.; Silver, M.; Tzardi, M.; Silver, J.; Ogino, S.; Hooshmand, S.; Kwak, E.; et al. Prognostic and Predictive Value of Common Mutations for Treatment Response and Survival in Patients with Metastatic Colorectal Cancer. Br. J. Cancer 2009, 101, 465–472. [Google Scholar] [CrossRef]
- Brenner, H.; Chen, C. The Colorectal Cancer Epidemic: Challenges and Opportunities for Primary, Secondary and Tertiary Prevention. Br. J. Cancer 2018, 119, 785–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Cancer Genome Atlas Network. Comprehensive Molecular Characterization of Human Colon and Rectal Cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obuch, J.C.; Ahnen, D.J. Colorectal Cancer. Gastroenterol. Clin. N. Am. 2016, 45, 459–476. [Google Scholar] [CrossRef]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A.; Kinzler, K.W. Cancer Genome Landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- Eklöf, V.; Wikberg, M.L.; Edin, S.; Dahlin, A.M.; Jonsson, B.-A.; Öberg, Å.; Rutegård, J.; Palmqvist, R. The Prognostic Role of KRAS, BRAF, PIK3CA and PTEN in Colorectal Cancer. Br. J. Cancer 2013, 108, 2153–2163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, X.; Wang, X.; Duanmu, J.; Li, T.; Jiang, Q. KRAS Mutations Are Negatively Correlated with Immunity in Colon Cancer. Aging 2021, 13, 750–768. [Google Scholar] [CrossRef]
- Andreyev, H.J.N.; Norman, A.R.; Cunningham, D.; Oates, J.; Dix, B.R.; Iacopetta, B.J.; Young, J.; Walsh, T.; Ward, R.; Hawkins, N.; et al. Kirsten Ras Mutations in Patients with Colorectal Cancer: The ‘RASCAL II’ Study. Br. J. Cancer 2001, 85, 692–696. [Google Scholar] [CrossRef] [PubMed]
- De Roock, W.; Claes, B.; Bernasconi, D.; De Schutter, J.; Biesmans, B.; Fountzilas, G.; Kalogeras, K.T.; Kotoula, V.; Papamichael, D.; Laurent-Puig, P.; et al. Effects of KRAS, BRAF, NRAS, and PIK3CA Mutations on the Efficacy of Cetuximab plus Chemotherapy in Chemotherapy-Refractory Metastatic Colorectal Cancer: A Retrospective Consortium Analysis. Lancet Oncol. 2010, 11, 753–762. [Google Scholar] [CrossRef]
- Hayama, T.; Hashiguchi, Y.; Okamoto, K.; Okada, Y.; Ono, K.; Shimada, R.; Ozawa, T.; Toyoda, T.; Tsuchiya, T.; Iinuma, H.; et al. G12V and G12C Mutations in the Gene KRAS Are Associated with a Poorer Prognosis in Primary Colorectal Cancer. Int. J. Colorectal Dis. 2019, 34, 1491–1496. [Google Scholar] [CrossRef]
- Bai, B.; Shan, L.; Xie, B.; Huang, X.; Mao, W.; Wang, X.; Wang, D.; Zhu, H. Mutations in KRAS Codon 12 Predict Poor Survival in Chinese Patients with Metastatic Colorectal Cancer. Oncol. Lett. 2018, 15, 3161–3166. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.-L.; Liau, J.-Y.; Yu, S.-C.; Ou, D.-L.; Lin, L.-I.; Tseng, L.-H.; Chang, Y.-L.; Yeh, K.-H.; Cheng, A.-L. KRAS Mutation Is a Predictor of Oxaliplatin Sensitivity in Colon Cancer Cells. PLoS ONE 2012, 7, e50701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddiqui, A.D.; Piperdi, B. KRAS Mutation in Colon Cancer: A Marker of Resistance to EGFR-I Therapy. Ann. Surg. Oncol. 2010, 17, 1168–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nieder, C.; Hintz, M.; Grosu, A.L. Colorectal Cancer Metastatic to the Brain: Analysis of Prognostic Factors and Impact of KRAS Mutations on Presentation and Outcome. Clin. Transl. Oncol. 2016, 18, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Carbonero, N.; Martinez-Useros, J.; Li, W.; Orta, A.; Perez, N.; Carames, C.; Hernandez, T.; Moreno, I.; Serrano, G.; Garcia-Foncillas, J. KRAS and BRAF Mutations as Prognostic and Predictive Biomarkers for Standard Chemotherapy Response in Metastatic Colorectal Cancer: A Single Institutional Study. Cells 2020, 9, 219. [Google Scholar] [CrossRef] [Green Version]
- Liang, L.; Tian, J.; Yu, Y.; Wang, Z.; Peng, K.; Liu, R.; Wang, Y.; Xu, X.; Li, H.; Zhuang, R.; et al. An Analysis of Relationship between RAS Mutations and Prognosis of Primary Tumour Resection for Metastatic Colorectal Cancer Patients. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 50, 768–782. [Google Scholar] [CrossRef]
- He, K.; Wang, Y.; Zhong, Y.; Pan, X.; Si, L.; Lu, J. KRAS Codon 12 Mutation Is Associated with More Aggressive Invasiveness in Synchronous Metastatic Colorectal Cancer (MCRC): Retrospective Research. OncoTargets Ther. 2020, 13, 12601–12613. [Google Scholar] [CrossRef]
- Liao, W.; Overman, M.J.; Boutin, A.T.; Shang, X.; Zhao, D.; Dey, P.; Li, J.; Wang, G.; Lan, Z.; Li, J.; et al. KRAS-IRF2 Axis Drives Immune Suppression and Immune Therapy Resistance in Colorectal Cancer. Cancer Cell 2019, 35, 559–572.e7. [Google Scholar] [CrossRef] [Green Version]
- Skoulidis, F.; Heymach, J.V. Co-Occurring Genomic Alterations in Non-Small-Cell Lung Cancer Biology and Therapy. Nat. Rev. Cancer 2019, 19, 495–509. [Google Scholar] [CrossRef]
- Sfakianaki, M.; Papadaki, C.; Tzardi, M.; Trypaki, M.; Alam, S.; Lagoudaki, E.D.; Messaritakis, I.; Zoras, O.; Mavroudis, D.; Georgoulias, V.; et al. Loss of LKB1 Protein Expression Correlates with Increased Risk of Recurrence and Death in Patients with Resected, Stage II or III Colon Cancer. Cancer Res. Treat. 2019, 51, 1518–1526. [Google Scholar] [CrossRef]
- Wang, C.; Fakih, M. Targeting KRAS in Colorectal Cancer. Curr. Oncol. Rep. 2021, 23, 28. [Google Scholar] [CrossRef]
- Scott, A.; Goffredo, P.; Ginader, T.; Hrabe, J.; Gribovskaja-Rupp, I.; Kapadia, M.R.; Weigel, R.J.; Hassan, I. The Impact of KRAS Mutation on the Presentation and Prognosis of Non-Metastatic Colon Cancer: An Analysis from the National Cancer Database. J. Gastrointest. Surg. 2020, 24, 1402–1410. [Google Scholar] [CrossRef] [PubMed]
| KRAS | ||||
|---|---|---|---|---|
| KRAS12_F | 5’ | ACTGGTGGAGTATTTGATAGTGTAT | 3’ | exon 2 |
| KRAS12_EXR | 5’ | TGTATCAAAGAATGGTCCTGCAC | 3’ | |
| KRAS12_INR | 5’ | GGTCCTGCACCAGTAATATGC | 3’ | |
| KRAS61_EXF | 5’ | AGGTGCACTGTAATAATCCAGACT | 3’ | exon 3 |
| KRAS61_INF | 5’ | TCCAGACTGTGTTTCTCCCT | 3’ | |
| KRAS61_R | 5’ | AACCCACCTATAATGGTGAATATCT | 3’ | |
| KRAS146_EXF | 5’ | CTCTGAAGATGTACCTATGGTCCT | 3’ | exon 4 |
| KRAS146_INF | 5’ | AGACACAAAACAGGCTCAGGA | 3’ | |
| KRAS146_R | 5’ | GCCCTCTCAAGAGACAAAAACAT | 3’ | |
| NRAS | ||||
| NRAS12_F | 5’ | GGCTCGCCAATTAACCCTGA | 3’ | exon 2 |
| NRAS12_EXR | 5’ | CACTGGGCCTCACCTCTATG | 3’ | |
| NRAS12_INR | 5’ | GCCTCACCTCTATGGTGGGAT | 3’ | |
| NRAS61_F | 5’ | ATTGAACTTCCCTCCCTCCCT | 3’ | exon 3 |
| NRAS61_EXR | 5’ | ACCTGTAGAGGTTAATATCCGCAAA | 3’ | |
| NRAS61_INR | 5’ | ATTGATGGCAAATACACAGAGGA | 3’ | |
| NRAS146_EXF | 5’ | AGCGAGTAAAAGACTCGGATGA | 3’ | exon 4 |
| NRAS146_INF | 5’ | TCGGATGATGTACCTATGGTGC | 3’ | |
| NRAS146_R | 5’ | TGGATCACATCTCTACCAGAGTTA | 3’ | |
| Characteristics | Frequency (N = 578) | % |
|---|---|---|
| Age median | 66 (28–88 years) | |
| <70 | 354 | 61.4 |
| ≥70 | 224 | 38.9 |
| Gender | 578 | |
| Male | 347 | 60 |
| Female | 231 | 40 |
| Performance status | ||
| 0–1 | 547 | 94.6 |
| ≥2 | 31 | 5.4 |
| Stage at diagnosis | ||
| I | 5 | 0.9 |
| II | 62 | 10.7 |
| III | 227 | 39.3 |
| IV | 284 | 49.1 |
| Location | ||
| Cecum | 77 | 13.4 |
| Ascending | 63 | 10.9 |
| Transverse | 24 | 4.2 |
| Descending | 34 | 5.8 |
| Sigmoid | 210 | 36.3 |
| Rectum | 169 | 29.3 |
| Right/Left | ||
| Right | 165 | 28.6 |
| Left | 413 | 71.4 |
| Adjuvant Treatment | ||
| Yes | 274 | 47.4 |
| No | 304 | 52.6 |
| Adjuvant Regimen | ||
| None | 304 | 52.6 |
| 5FU-like | 101 | 17.6 |
| LOHP-based | 175 | 29.8 |
| First Line Regimen | ||
| Irinotecan-based | 308 | 53.3 |
| LOHP-based * | 246 | 42.5 |
| 5FU-based * | 24 | 4.2 |
| Metastasectomy | ||
| Yes | 95 | 16.5 |
| No | 483 | 83.5 |
| KRAS mut | 539 | 93.2 |
| NRAS mut | 39 | 6.8 |
| Characteristics | Frequency (N = 28) | % |
|---|---|---|
| Age | 64 (28–83 years) | |
| <70 | 19 | 67.9 |
| ≥70 | 9 | 32.1 |
| Gender | ||
| Male | 19 | 67.9 |
| Female | 9 | 32.1 |
| Performance status | ||
| 0–1 | 27 | 96.4 |
| ≥2 | 1 | 3.6 |
| Stage at diagnosis | ||
| I–III | 11 | 39.3 |
| IV | 17 | 60.7 |
| Right/Left | ||
| Right | 6 | 21.4 |
| Left | 22 | 78.6 |
| Metastasectomy | ||
| Yes | 10 | 37.7 |
| No | 18 | 64.3 |
| KRAS Mutation | Frequency (N = 539) | % |
|---|---|---|
| G12D | 190 | 33.1 |
| G12V | 121 | 21.2 |
| G13D | 96 | 16.7 |
| G12C | 28 | 4.8 |
| G12S | 27 | 4.7 |
| G12A | 21 | 3.6 |
| A146T | 15 | 2.6 |
| A146A | 6 | 1 |
| A146V | 6 | 1 |
| Q61H | 5 | 0.9 |
| G12R | 3 | 0.5 |
| G13R | 3 | 0.5 |
| A59T | 2 | 0.3 |
| G13_V14 > D | 2 | 0.3 |
| G13C | 2 | 0.3 |
| K117N | 2 | 0.3 |
| Q61K | 2 | 0.3 |
| Q61L | 2 | 0.3 |
| Q61R | 2 | 0.3 |
| A146X | 1 | 0.2 |
| A59E | 1 | 0.2 |
| E62Q | 1 | 0.2 |
| G12S, G12V | 1 | 0.2 |
| NRAS Mutations | Frequency (N = 39) | % |
|---|---|---|
| Q61R | 9 | 1.6 |
| G12D | 8 | 1.4 |
| Q61K | 7 | 1.2 |
| G12A | 3 | 0.5 |
| G12V | 2 | 0.3 |
| G13R | 2 | 0.3 |
| Q61L | 2 | 0.3 |
| S145L | 1 | 0.2 |
| G12S | 1 | 0.2 |
| G13D, A59T | 1 | 0.2 |
| G13V | 1 | 0.2 |
| K117K | 1 | 0.2 |
| Q61H | 1 | 0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koulouridi, A.; Karagianni, M.; Messaritakis, I.; Sfakianaki, M.; Voutsina, A.; Trypaki, M.; Bachlitzanaki, M.; Koustas, E.; Karamouzis, M.V.; Ntavatzikos, A.; et al. Prognostic Value of KRAS Mutations in Colorectal Cancer Patients. Cancers 2022, 14, 3320. https://doi.org/10.3390/cancers14143320
Koulouridi A, Karagianni M, Messaritakis I, Sfakianaki M, Voutsina A, Trypaki M, Bachlitzanaki M, Koustas E, Karamouzis MV, Ntavatzikos A, et al. Prognostic Value of KRAS Mutations in Colorectal Cancer Patients. Cancers. 2022; 14(14):3320. https://doi.org/10.3390/cancers14143320
Chicago/Turabian StyleKoulouridi, Asimina, Michaela Karagianni, Ippokratis Messaritakis, Maria Sfakianaki, Alexandra Voutsina, Maria Trypaki, Maria Bachlitzanaki, Evangelos Koustas, Michalis V. Karamouzis, Anastasios Ntavatzikos, and et al. 2022. "Prognostic Value of KRAS Mutations in Colorectal Cancer Patients" Cancers 14, no. 14: 3320. https://doi.org/10.3390/cancers14143320
APA StyleKoulouridi, A., Karagianni, M., Messaritakis, I., Sfakianaki, M., Voutsina, A., Trypaki, M., Bachlitzanaki, M., Koustas, E., Karamouzis, M. V., Ntavatzikos, A., Koumarianou, A., Androulakis, N., Mavroudis, D., Tzardi, M., & Souglakos, J. (2022). Prognostic Value of KRAS Mutations in Colorectal Cancer Patients. Cancers, 14(14), 3320. https://doi.org/10.3390/cancers14143320










