Simple Summary
Liquid biopsy is an increasingly used tool for melanoma diagnosis and molecular characterization, but also for monitoring of response to anticancer drugs. The aim of our work is to assess the clinical utility of a real-time quantitative PCR (qPCR)-based platform with a very short turnaround time and identify the best setting for clinical investigation. We investigated the concordance of this technique with tissue analysis in stage III–IV melanoma patients; moreover, we correlated results to clinicopathologic characteristics and outcomes. We found a higher tissue–plasma concordance in melanoma patients with high burden of disease (sum of diameters ≥30 mm, ≥2 metastatic sites, elevated LDH levels), constituting a clinical subgroup worthy of future prospective evaluation; however, the low sensitivity of this technique seems to be not sufficient for predicting relapses in radically resected patients.
Abstract
Background: Liquid biopsy is a potentially useful tool for melanoma patients, also for detecting BRAS/NRAS mutations, even if the tissue analysis remains the current standard. Methods: In this work, we tested ctDNA on plasma samples from 56 BRAF-V600/NRAS mutant stage III/IV melanoma patients using a real-time quantitative PCR (qPCR)-based platform. The study population was divided into two cohorts: the first including 26 patients who had undergone radical resection (resected cohort) and the second including 30 patients who had unresected measurable disease (advanced cohort). Moreover, for 10 patients in the advanced cohort, ctDNA assessment was repeated at specified timepoints after baseline testing. Data were analyzed and correlated to the clinicopathologic characteristics and outcomes. Results: In the baseline cohort, a higher tissue–plasma concordance was seen in patients with high burden of disease (sum of diameters ≥30 mm, ≥2 metastatic sites, elevated LDH levels); furthermore, monitoring of these patients through ctDNA analysis was informative for therapeutic responses. On the other hand, the low sensitivity of this technique did not allow for clinically valuable prediction of relapses in radically resected stage III/IV patients. Conclusions: Overall, our data suggest that qPCR-based ctDNA analysis could be informative in a subset of locally advanced and metastatic melanoma patients with specific clinical–radiological characteristics, supporting further investigations in this setting.
1. Introduction
Cutaneous melanoma is the most lethal type of skin cancer despite treatment advances. In fact, a high percentage of patients relapse after radical resection [1] and less than one patient out of two survive at 5 years from diagnosis of advanced disease [2]. BRAF and NRAS genes are frequently mutated in cutaneous melanoma, approximatively in about 50% and 20% of cases, respectively [3,4]. Guidelines recommend BRAF V600 and NRAS mutation testing on tumor specimens for resectable or unresectable stage III/IV melanoma [5]. However, intra-tumor heterogeneity is a well-established biological characteristic of human malignancies, including melanoma, affecting the predictiveness of tissue BRAF/NRAS mutational testing [6,7].
Liquid biopsy allows isolation of circulating tumor cells (CTCs) or circulating tumor DNA (ctDNA) in human samples [8] to detect tumor traces released from primary tumor and/or metastatic sites and to overcome tumor heterogeneity [9]. In particular, several studies conducted on metastatic melanoma patients have highlighted the utility of liquid biopsy in detecting and monitoring BRAF/NRAS mutations through the use of different technologies [10,11,12,13,14,15,16,17]. Some clinical and radiological characteristics have been correlated to ctDNA levels in metastatic melanoma patients, such as the LDH status (normal/high) and dimensional evaluation of metastatic lesions [15,18], whereas, in radically resected patients, ctDNA detection has prognostic significance, being associated to a higher relapse risk [19,20].
In this study, we evaluate the clinical utility of liquid biopsy for testing common BRAF/NRAS mutations using Idylla™ Biocartis, a fully automated real-time quantitative PCR (qPCR)-based platform, in 56 consecutive stage III–IV melanoma patients, divided in two main groups: 30 patients with locally advanced and metastatic melanoma, of which 25 were considered the baseline cohort because the liquid biopsy was collected before any systemic treatment (treatment-naïve), whereas for 5 patients the samples were performed on treatment (non-baseline); the remaining 26 patients had radically resected stage III–IV melanoma and blood samples were collected at baseline (treatment-naïve)
2. Materials and Methods
2.1. Patients’ Cohorts and Clinical Evaluation
In total, 56 consecutive BRAF-V600/NRAS mutant stage III/IV melanoma patients were assessed between September 2018 and April 2020 at the Oncology Unit of University of Campania “Luigi Vanvitelli”. Patients provided informed consent for longitudinal plasma collection and tumor DNA profiling (Protocol n° 59, approved by the Ethics Committee of the University of Campania “Luigi Vanvitelli”, in accordance with the Declaration of Helsinki). Sum of lesion diameters (SoD) was calculated as the sum of the maximum diameters, expressed in millimeters, of all measurable lesions from whole-body CT scans. Tumor response was assessed using Response Evaluation Criteria in Solid Tumours (RECIST) v1.1. Lactate dehydrogenase (LDH) levels for the correlation analysis was expressed as a ratio (LDH value/upper limit of normal LDH as per internal laboratory reference values). Performance status (PS) was assessed using the Eastern Cooperative Oncology Group (ECOG) scale. Data cut-off for survival analysis was 31 October 2021.
2.2. Tissue Analysis
Analyses of the formalin-fixed, paraffin-embedded (FFPE) tissue specimens were all performed in the Pathology Service of University of Campania “Luigi Vanvitelli” using Next Generation Sequencing (NGS) as described in Supplementary Materials and Methods.
2.3. Plasma Collection and qPCR Analysis
Plasma was collected and analyzed immediately after centrifugation or stored at −80 °C until analysis, as described in the Supplementary Materials and Methods. Analyses of plasma were all carried out using the automated Idylla™ qPCR-based platform by Biocartis (Mechelen, Belgium), as previously described (Supplementary Methods) [21,22]. The maximum waiting time from collection to centrifugation of the samples was less than 1 h.
2.4. Statistical Analysis
After checking the assumptions of a normal distribution of the values for the quantitation cycle (Cq), circulating mutational fraction (CMF, %), SoD (mm), and LDH (ratio), using the Anderson–Darling test (normal distribution for Cq and SoD, non-normal distribution for CMF and LDH), a Spearman test was used to assess the pairwise correlation between Cq and both LDH and SoD, and also between CMF and both LDH and SoD. Outlier values were identified using the ROUT (Q = 1%) method [23]. The Kaplan–Meier method was used for survival analysis, and the significance of the split between the survival curves were measured by the log-rank (Mantel–Cox) test. All statistical analyses were performed using GraphPad Prism 8.0.1 software.(GraphPad Software Inc, San Diego, CA, USA)
3. Results
3.1. Patients’ Characteristics
Among the 56 stage III/IV melanoma patients, 30 had unresected disease at the time of first plasma collection (advanced cohort) and 26 had undergone radical resection (confirmed both on pathologic and radiologic assessment) (resected cohort) within 3 months before the first plasma collection. In particular, among the 30 patients in the advanced cohort, 25 were assessed before starting any systemic treatment (baseline cases) and 5 were assessed during systemic treatment (non-baseline cases). Moreover, for 10 out of the 25 baseline patients, the plasma sample was collected also at the time of progression disease or after 6 months in the absence of disease progression (Figure 1).
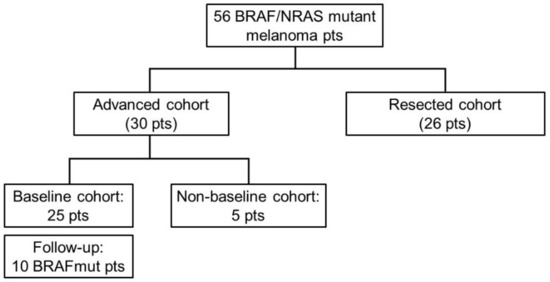
Figure 1.
Flowchart of the enrolled patients. Pts: patients.

Table 1.
Characteristics of the locally advanced/metastatic patients (advanced cohort).

Table 2.
Characteristics of the radically resected stage III/IV patients (resected cohort).
3.2. Baseline Locally Advanced and Metastatic Patients (Baseline Cohort)
Among the 25 consecutive treatment-naive BRAF-V600/NRAS mutant patients with locally advanced or metastatic melanoma who were tested at baseline, the same mutation of the tissues was found in the plasma in 15 patients, with a global concordance of 60%. We therefore analyzed patients’ baseline clinical characteristics and correlate them to qPCR results. In particular, we focused on tumor burden, metastatic sites, LDH levels, and disease-related symptoms (using ECOG PS scale) (Table 3).

Table 3.
Baseline locally advanced and metastatic patients.
Interestingly, plasma analysis was positive in all BRAF-V600 mutant patients, with SoD ≥ 30 mm (13/13); extrapulmonary visceral, excluding exclusive brain metastases (8/8), with ≥2 metastatic sites (8/8), with liver metastases (6/6), with elevated baseline LDH levels (6/6), or with symptomatic disease (4/4). Conversely, ctDNA analysis did not reveal blood mutations in all three patients with locally advanced disease and in the two patients with CNS-limited disease. M1a stage patients were found positive in 7 out of 13 patients (53.8%), but, in case of nodal metastases and SoD ≥ 30 mm, the positivity rate was higher (87.5%). The miss rate (or false negative rate, FNR) was calculated for the baseline cohort, this being 0.4. The characteristics most closely related to false negative results were low SoD (<30 mm) and cutaneous or nodal disease only.
After a median follow-up of 18.7 months (range 3.5–39.7), 17 (68%) patients had progressed to first-line treatment and 14 (56%) had died. The median progression-free survival (PFS) and overall survival (OS) was 13.3 and 18.7 months, respectively.
PFS was calculated for baseline patients from the time of first plasma collection to time of disease progression or death of any cause, whichever occurred first; similarly, OS was calculated for baseline patients from the time of first plasma collection to death of any cause. For both PFS and OS analysis, we divided the baseline patients in positive and negative groups according to their ctDNA result (Figure 2).
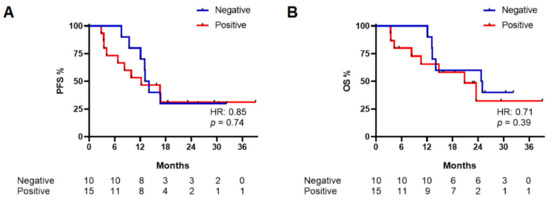
Figure 2.
Progression-free survival (PFS) (A) and overall survival (OS) (B) in baseline patients according to their qPCR result.
A non-statistically significant difference between the negative and positive groups was observed for both PFS (median PFS: 13.8 vs. 12.4 months, respectively; HR: 0.85, 95%CI: 0.33–2.2, p = 0.74) and OS (median OS: 25.2 vs. 21.1 months, respectively; HR: 0.71, 95%CI: 0.25–2.02, p = 0.39) (Figure 2).
3.3. Non-Baseline Locally Advanced and Metastatic Patients
Four BRAF-V600/NRAS mutant pre-treated patients with locally advanced or metastatic melanoma and one patient (#A30) with de novo metastases during adjuvant therapy for radically resected BRAF-V600/NRAS wild-type melanoma were tested. Only 1 case out of 4 was ctDNA positive for the BRAF-V600/NRAS mutation (Table 4).

Table 4.
Non-baseline locally advanced and metastatic patients.
Notably, patient #A30 was in treatment with anti-PD-1 adjuvant therapy for radically resected stage IIIC melanoma; the BRAF-V600/NRAS analysis was performed on the metastatic sentinel node, and no mutation was found on this tissue. However, for rapid development of symptomatic metastases (bone, liver, and lungs, SoD: 70 mm, elevated LDH level) at 6 months after starting adjuvant treatment, we tested a plasma sample and, surprisingly, a BRAF-V600 mutation was found. The patient underwent liver biopsy, which confirmed the presence of the BRAF-V600 mutation, allowing for the initiation of BRAF and MEK inhibitors.
3.4. Monitoring of Locally Advanced and Metastatic BRAF-V600 Patients
Ten BRAF-V600 mutant locally advanced and metastatic patients repeated plasma sample collection after 6 months, if their plasma sample was positive at baseline, or at evidence of progressive disease (PD), in case their plasma sample was negative at baseline (Table 5).

Table 5.
Monitoring of BRAF-V600 mutant locally advanced and metastatic patients after 6 months of treatment.
Of eight patients with a mutation detected in their baseline plasma sample, only one patient was still positive after 6 months (#A11); notably, it was also the only patient with concurrent progressive disease at CT scan. Of the two patients with a negative baseline plasma sample, a new analysis was performed at the time of PD: patient #A7, with de-novo liver metastases (SoD: 15 mm) after 10 months of targeted therapy, was found positive (same mutation of primary tumor), but patient #A10, despite evidence of nodal PD (SoD: 50 mm) after 12 months of targeted therapy, was still negative; intriguingly, a new molecular analysis on nodal biopsy at time of PD in patient #A10 was performed, and no BRAF-V600/NRAS mutation was found.
3.5. Cq and CMF in Advanced Patients’ Cohort
Cq values are reported in Table S1, showing a normal distribution (Figure S1); the median Cq value in baseline cohort was 49.46 (range: 36.94–53.4). Therefore, we obtained PFS and OS curves dividing patients in two groups according to their Cq values (< or ≥49.46), including negative patients in the group with higher Cq values—starting from the assumption that higher Cq values mean a lower quantity of ctDNA and vice versa (Figure 2). PFS analysis showed a non-statistically significant difference between the Cq low and Cq high groups (median PFS: 15.5 vs. 6.9 months, respectively; HR: 0.46, 95%CI: 0.14–1.51, p = 0.12) while a difference in OS was observed (median OS: 25.3 vs. 10.7 months, respectively; HR: 0.32, 95%CI: 0.09–1.21, p = 0.027) (Figure 3).
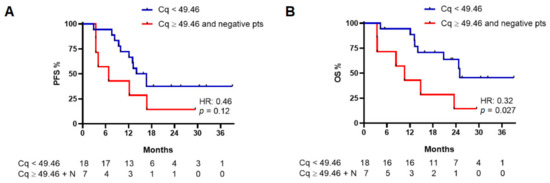
Figure 3.
Progression-free survival (PFS) (A) and overall survival (OS) (B) in baseline patients with a Cq value lower than 49.46 and patients with a Cq value equal to or higher than 49.46 (including in this group patients with no mutation detected by the instrument; see text for details). N, patients with no detectable mutation at baseline.
CMF values are reported in Table S1; a non-normal distribution of all CMF values was observed (Figure S1). In a similar way to Cq, we obtained PFS and OS curves according to the CMF values (< or ≥0.011%, which is the median value of CMF in the baseline cohort excluding the two outlier values), including negative patients in the group with lower CMF values (Figure 2). The result was not significant for the correlation of CMF with both PFS and OS (Figure S2). Moreover, we analyzed the potential correlation between the Cq and CMF values with clinical factors such as the SoD and LDH ratios, finding no correlation (Figure S3).
3.6. Baseline Radically Resected Stage III–IV Patients (Resected Cohort)
Among the 26 consecutive radically resected stage III–IV melanoma patients who were tested before starting adjuvant treatment, the BRAF V600E mutation was found on ctDNA only in one patient. At the data cut-off time, after a median follow up of 26.7 months (range: 14.3–38.9), 9 out of 26 patients (34.6%) had disease relapse, with a median disease-free survival (DFS, defined as the time from randomization to recurrence of tumor or death whichever occurred first) of 20.6 months (range: 11.6–38.9) (Table 6).

Table 6.
Radically resected stage III–IV patients.
Noteworthy, among the relapsed patients, there was the only one of this cohort with a positive ctDNA result at baseline (#B6, see Table 6), who developed CNS metastases at the end of the year of adjuvant treatment with targeted therapy.
Accuracy in relapse detection by qPCR on ctDNA was therefore calculated in this cohort: the positive predictive value (PPV) was 100% and negative predictive value (NPV) was 68%, with an FNR of 0.875, reflecting an extremely low power for identifying patients at higher risk of relapse.
4. Discussion
The present work has evaluated the overall clinical performance of Idylla™ Biocartis in the characterization of BRAF/NRAS-mutated melanoma patients, either in radically operated stage III–IV or in locally advanced/metastatic ones.
Concerning locally advanced and metastatic melanoma patients (advanced cohort), we obtained an overall plasma–tissue concordance of 60%, despite a lower sensitivity compared to other similar methods. The same technology had been already investigated in metastatic melanoma patients in two previous works [13,14], reporting baseline plasma–tissue concordance for the BRAF mutation of 47% and 64.2%, respectively, whilst an overall agreement of 84% was shown in the work by Long-Mira et al. [11]. Moreover, we investigated the correlation between ctDNA quantity and clinical–radiological tumor parameters, in our baseline cohort. In particular, a high rate of circulating mutation identification was obtained in patients with high burden of disease, high LDH levels, and/or symptomatic disease; all these characteristics are in fact associated with the highest probability of finding relevant ctDNA concentration in plasma samples, detecting BRAF-V600/NRAS mutations virtually in all cases. On the contrary, we did not find any mutation in plasma samples from metastatic patients with the brain as the unique site of metastasis; this finding, in line with previous reports [13,24,25,26], is probably linked to a lower ctDNA quantity released into the circulation by the blood–brain barrier; similarly, patients with locally advanced disease likely have ctDNA levels lower than the sensibility threshold of this technique.
With respect to the prognostic value of ctDNA, previous works highlighted a correlation between ctDNA levels and survival [13,15,16,25]. In our work, we used Cq values to investigate their potential prognostic significance, finding a non-significant trend towards a better PFS and a statistically significant improvement in OS in those patients without detectable mutations and with a Cq value ≥49.46 (median value in test-positive patients). Similarly, Rutkowsky and colleagues failed to demonstrate a correlation between Cq values and PFS, and also between Cq and duration of response (DoR) [14].
In line with previous reports [11,13,25], our results support the potential use of liquid biopsy to monitor the response to treatment together with radiological and/or clinical assessment in locally advanced or metastatic melanoma patients with a BRAF-V600/NRAS mutation already identified by baseline liquid biopsy. The interesting cases of patient #A10, in which ctDNA remained negative despite disease progression to iliac lymph nodes—a result which was later confirmed to be truly BRAF WT at tissue analysis—highlight a good clinical correlation that better recapitulate melanoma biology in a clinical scenario. Moreover, in patient #A30, BRAF-V600 ctDNA positivity constituted a de novo event during anti-PD1 adjuvant treatment and, after confirmation on metastatic tissue analysis, allowed the patient to access targeted therapy. In this circumstance, ctDNA analysis was used to better capture tumor heterogeneity and allowed the patient to access alternative treatment strategies in a short period of time.
Finally, concerning radically resected stage III/IV melanoma patients, our results indicate that qPCR-based plasma analysis could not be used in predicting disease relapses. In fact, despite the high PPV, the low sensitivity of the test in this scenario translates into a high FNR. In this setting, more sensitive techniques, such as ddPCR, better correlate with relapse, as previously described [20].
Taken all together, our data suggest that qPCR-based ctDNA analysis on plasma samples using Idylla™ Biocartis (Mechelen, Belgium) could be used to achieve a better understanding of melanoma biology and provides valuable clinical information in patients with specific clinical–radiological characteristics, in addition to the current gold-standard tissue-based mutational analysis [26]. In particular, the performance of this technique in disease monitoring for advanced disease is worth further investigation and validation, while its potential for the identification of relapsing patients is not clinically reliable to differentiate patients at higher risk of relapse, though the presence of detectable ctDNA is strongly associated with relapse before imaging detection.
Among the advantages of this method, it must be underlined that the time required to obtain the analysis is approximatively 120 min after sampling, possibly allowing to anticipate access to targeted patients, if our results are prospectively confirmed in larger cohorts, especially for symptomatic patients with a high disease burden who could benefit from the rapid effect of targeted therapy without delay [27]. Limitations of our work are the inclusion of a low number of patients with a known BRAF-V600/NRAS mutation in tissues and the impossibility of excluding confounding factors that limit our analysis, particularly for the Cq and CMF values.
Finally, with regard to cost-effectiveness assessments that are strictly dependent on the healthcare system of reference, a definitive estimation cannot be accurately derived from the present study. However, the identification of a target population with higher diagnostic accuracy of the test achieved by this study can strongly enhance its feasibility and effectiveness by refining patients’ selection, and this technique was recently shown to be the cheapest in centers with a low sample throughput per year [28].
5. Conclusions
Our work shows the potential clinical utility of a ready-to-use diagnostic tool in stage III–IV melanoma patients, from molecular diagnosis to response monitoring, whose results could be integrated with the currently used clinical–radiological factors. Results from our work, if prospectively validated using a wider cohort of patients, could therefore improve the outcomes of melanoma patients. In fact, the automated qPCR-based ctDNA analysis using this platform could provide useful information in a very short timeframe and help decision making for the treating clinicians.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers14133053/s1, Files S1: Supplementary Method; Figure S1. Normality distribution; Figure S2: Progression free survival (PFS) and overall survival (OS) according to CMF values; Figure S3: Spearman correlation test; Table S1: All positive qPCR results, in relation to time of plasma collection.
Author Contributions
Conceptualization, E.F.G., V.D.F., P.P.V. and T.T.; methodology, E.F.G., V.D.F., P.P.V., G.A., E.M., F.C., S.N. and T.T.; software, E.F.G. and P.P.V.; validation, S.N.; formal analysis, E.F.G., V.D.F., P.P.V., R.F. and M.C.; investigation, E.F.G., V.D.F., P.P.V., G.A. and T.T.; resources, L.P.G., G.S., R.N., A.P., G.A., R.F. and M.C.; data curation, E.F.G., L.P.G. and M.C.; writing—original draft preparation, E.F.G., V.D.F. and P.P.V.; writing—review and editing, E.F.G., V.D.F., P.P.V., S.N. and T.T.; visualization, E.M., D.C. and F.C.; supervision, F.C., S.N. and T.T.; project administration, E.F.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by Ethics Committee of the University of Campania “Luigi Vanvitelli”, (Protocol n° 59).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available in this article (and Supplementary Materials).
Acknowledgments
I-CURE research project: “Identification, characterization and mining of Colorectal tumorigenesis: cause, prevention & cure (ICURE)”. Roche Foundation Medicine: Roche S.p.A., Monza, Italy.
Conflicts of Interest
E.F.G. reports personal fees from Novartis. P.P.V. reports to be consultant of Biocartis and speaker for Merck. E.M. reports advisory boards for Bayer, Merck, Pierre Fabre, Roche, Servier and giving expert opinion for ESMO. D.C. reports travel grant from Sanofi. F.C. reports advisory boards for Amgen, Bayer, BMS, Cellgene, Lilly, Merck, Pfizer, Roche, Sanofi, Servier and institutional research grants for Amgen, AstraZeneca, Bayer, Merck, Roche, Takeda. T.T. reports institutional research grants from Bayer, Merck, Novartis, Roche, Sanofi, Servier. The other authors declare no conflict of interest.
References
- Troiani, T.; De Falco, V.; Napolitano, S.; Trojaniello, C.; Ascierto, P.A. How we treat locoregional melanoma. ESMO Open 2021, 6, 100136. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Rutkowski, P.; Lao, C.D.; Cowey, C.L.; Schadendorf, D.; Wagstaff, J.; Dummer, R.; et al. Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2019, 381, 1535–1546. [Google Scholar] [CrossRef] [PubMed]
- Rutkowski, P.; Gos, A.; Jurkowska, M.; Switaj, T.; Dziewirski, W.; Zdzienicki, M.; Ptaszyński, K.; Michej, W.; Tysarowski, A.; Siedlecki, J.A. Molecular alterations in clinical stage III cutaneous melanoma: Correlation with clinicopathological features and patient outcome. Oncol. Lett. 2014, 8, 47–54. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fedorenko, I.V.; Gibney, G.T.; Smalley, K.S.M. NRAS mutant melanoma: Biological behavior and future strategies for therapeutic management. Oncogene 2013, 32, 3009–3018. [Google Scholar] [CrossRef] [PubMed]
- Michielin, O.; van Akkooi, A.C.J.; Ascierto, P.A.; Dummer, R.; Keilholz, U. Cutaneous melanoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2019, 30, 1884–1901. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.A.; Wiggins, J.M.; Corless, B.C.; Syeda, M.M.; Tadepalli, J.S.; Blake, S.; Fleming, N.; Darvishian, F.; Pavlick, A.; Berman, R.S.; et al. TERT, BRAF, and NRAS Mutational Heterogeneity between Paired Primary and Metastatic Melanoma Tumors. J. Investig. Dermatol. 2020, 140, 1609–1618.e7. [Google Scholar] [CrossRef] [PubMed]
- Grzywa, T.M.; Paskal, W.; Wlodarski, P.K. Intratumor and Intertumor Heterogeneity in Melanoma. Transl. Oncol. 2017, 10, 956–975. [Google Scholar] [CrossRef]
- Poulet, G.; Massias, J.; Taly, V. Liquid Biopsy: General Concepts. Acta Cytol. 2019, 63, 449–455. [Google Scholar] [CrossRef]
- Venesio, T.; Siravegna, G.; Bardelli, A.; Sapino, A. Liquid Biopsies for Monitoring Temporal Genomic Heterogeneity in Breast and Colon Cancers. Pathobiology 2018, 85, 146–154. [Google Scholar] [CrossRef]
- Burjanivova, T.; Malicherova, B.; Grendar, M.; Minarikova, E.; Dusenka, R.; Vanova, B.; Bobrovska, M.; Pecova, T.; Homola, I.; Lasabova, Z.; et al. Detection of BRAFV600E Mutation in Melanoma Patients by Digital PCR of Circulating DNA. Genet. Test. Mol. Biomark. 2019, 23, 241–245. [Google Scholar] [CrossRef]
- Long-Mira, E.; Ilie, M.; Chamorey, E.; Leduff-Blanc, F.; Montaudié, H.; Tanga, V.; Allégra, M.; Lespinet-Fabre, V.; Bordone, O.; Bonnetaud, C.; et al. Monitoring BRAF and NRAS mutations with cell-free circulating tumor DNA from metastatic melanoma patients. Oncotarget 2018, 9, 36238–36249. [Google Scholar] [CrossRef] [PubMed]
- Alrabadi, N.; Haddad, R.; Alomari, A.K. Detection of Gene Mutations in Liquid Biopsy of Melanoma Patients: Overview and Future Perspectives. Curr. Treat. Options Oncol. 2020, 21, 19. [Google Scholar] [CrossRef] [PubMed]
- Seremet, T.; Jansen, Y.; Planken, S.; Njimi, H.; Delaunoy, M.; El Housni, H.; Awada, G.; Schwarze, J.K.; Keyaerts, M.; Everaert, H.; et al. Undetectable circulating tumor DNA (ctDNA) levels correlate with favorable outcome in metastatic melanoma patients treated with anti-PD1 therapy. J. Transl. Med. 2019, 17, 303. [Google Scholar] [CrossRef] [PubMed]
- Rutkowski, P.; Pauwels, P.; Kerger, J.; Jacobs, B.; Maertens, G.; Gadeyne, V.; Thielemans, A.; de Backer, K.; Neyns, B. Characterization and Clinical Utility of BRAFV600 Mutation Detection Using Cell-Free DNA in Patients with Advanced Melanoma. Cancers 2021, 13, 3591. [Google Scholar] [CrossRef]
- Tolmeijer, S.H.; Koornstra, R.H.T.; de Groot, J.W.B.; Geerlings, M.J.; van Rens, D.H.; Boers-Sonderen, M.J.; Schalken, J.A.; Gerritsen, W.R.; Ligtenberg, M.J.L.; Mehra, N. Plasma BRAF Mutation Detection for the Diagnostic and Monitoring Trajectory of Patients with LDH-High Stage IV Melanoma. Cancers 2021, 13, 3913. [Google Scholar] [CrossRef]
- Marczynski, G.T.; Laus, A.C.; Dos Reis, M.B.; Reis, R.M.; Vazquez, V.L. Circulating tumor DNA (ctDNA) detection is associated with shorter progression-free survival in advanced melanoma patients. Sci. Rep. 2020, 10, 18682. [Google Scholar] [CrossRef]
- Busser, B.; Lupo, J.; Sancey, L.; Mouret, S.; Faure, P.; Plumas, J. Plasma Circulating Tumor DNA Levels for the Monitoring of Melanoma Patients: Landscape of Available Technologies and Clinical Applications. Biomed. Res. Int. 2017, 2017, 5986129. [Google Scholar] [CrossRef]
- Valpione, S.; Gremel, G.; Mundra, P.; Middlehurst, P.; Galvani, E.; Girotti, M.R.; Lee, R.J.; Garner, G.; Dhomen, N.; Lorigan, P.C.; et al. Plasma total cell-free DNA (cfDNA) is a surrogate biomarker for tumour burden and a prognostic biomarker for survival in metastatic melanoma patients. Eur. J. Cancer 2018, 88, 1–9. [Google Scholar] [CrossRef]
- Lee, R.J.; Gremel, G.; Marshall, A.; Myers, K.A.; Fisher, N.; Dunn, J.A.; Dhomen, N.; Corrie, P.G.; Middleton, M.R.; Lorigan, P.; et al. Circulating tumor DNA predicts survival in patients with resected high-risk stage II/III melanoma. Ann. Oncol. 2018, 29, 490–496. [Google Scholar] [CrossRef]
- Tan, L.; Sandhu, S.; Lee, R.J.; Li, J.; Callahan, J.; Ftouni, S.; Dhomen, N.; Middlehurst, P.; Wallace, A.; Raleigh, J.; et al. Prediction and monitoring of relapse in stage III melanoma using circulating tumor DNA. Ann. Oncol. 2019, 30, 804–814. [Google Scholar] [CrossRef]
- Vitiello, P.P.; De Falco, V.; Giunta, E.F.; Ciardiello, D.; Cardone, C.; Vitale, P.; Zanaletti, N.; Borrelli, C.; Poliero, L.; Terminiello, M.; et al. Clinical Practice Use of Liquid Biopsy to Identify RAS/BRAF Mutations in Patients with Metastatic Colorectal Cancer (mCRC): A Single Institution Experience. Cancers 2019, 11, 1504. [Google Scholar] [CrossRef] [PubMed]
- ctNRAS-BRAF Cartridge Technical Sheet. Available online: https://media.biocartis.com/biocartis/documents/Tech_Sheet-ctNRAS-BRAF-IVD-A4_WEB.pdf (accessed on 27 December 2021).
- Motulsky, H.J.; Brown, R.E. Detecting outliers when fitting data with nonlinear regression—A new method based on robust nonlinear regression and the false discovery rate. BMC Bioinform. 2006, 7, 123. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Menzies, A.M.; Carlino, M.S.; McEvoy, A.C.; Sandhu, S.; Weppler, A.M.; Diefenbach, R.J.; Dawson, S.J.; Kefford, R.F.; Millward, M.J.; et al. Longitudinal Monitoring of ctDNA in Patients with Melanoma and Brain Metastases Treated with Immune Checkpoint Inhibitors. Clin. Cancer Res. 2020, 26, 4064–4071. [Google Scholar] [CrossRef] [PubMed]
- Syeda, M.M.; Wiggins, J.M.; Corless, B.C.; Long, G.V.; Flaherty, K.T.; Schadendorf, D.; Nathan, P.D.; Robert, C.; Ribas, A.; Davies, M.A. Circulating tumour DNA in patients with advanced melanoma treated with dabrafenib or dabrafenib plus trametinib: A clinical validation study. Lancet Oncol. 2021, 22, 370–380. [Google Scholar] [CrossRef]
- Vanni, I.; Tanda, E.T.; Spagnolo, F.; Andreotti, V.; Bruno, W.; Ghiorzo, P. The Current State of Molecular Testing in the BRAF-Mutated Melanoma Landscape. Front. Mol. Biosci. 2020, 7, 113. [Google Scholar] [CrossRef] [PubMed]
- Giunta, E.F.; De Falco, V.; Napolitano, S.; Argenziano, G.; Brancaccio, G.; Moscarella, E.; Ciardiello, D.; Ciardiello, F.; Troiani, T. Optimal treatment strategy for metastatic melanoma patients harboring BRAF-V600 mutations. Ther. Adv. Med. Oncol. 2020, 12, 1–15. [Google Scholar] [CrossRef]
- Vessies, D.C.L.; Greuter, M.J.E.; van Rooijen, K.L.; Linders, T.C.; Lanfermeijer, M.; Ramkisoensing, K.L.; Meijer, G.A.; Koopman, M.; Coupé, V.M.H.; Vink, G.R.; et al. Performance of four platforms for KRAS mutation detection in plasma cell-free DNA: ddPCR, Idylla, COBAS z480 and BEAMing. Sci. Rep. 2020, 10, 8122. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).