Immunoglobulin Gene Sequence as an Inherited and Acquired Risk Factor for Chronic Lymphocytic Leukemia
Abstract
Simple Summary
Abstract
1. Introduction
1.1. CLL Overview
1.2. Classification of CLL—Factors Affecting the CLL Prognosis
1.3. CLL Subsets
2. Importance of Light-Chain Sequence in CLL
3. Factors Affecting CLL-Cell Survival
4. Structural Basis of Autonomous Signaling
5. IGLV3-21R110G Defines a New Category of CLL
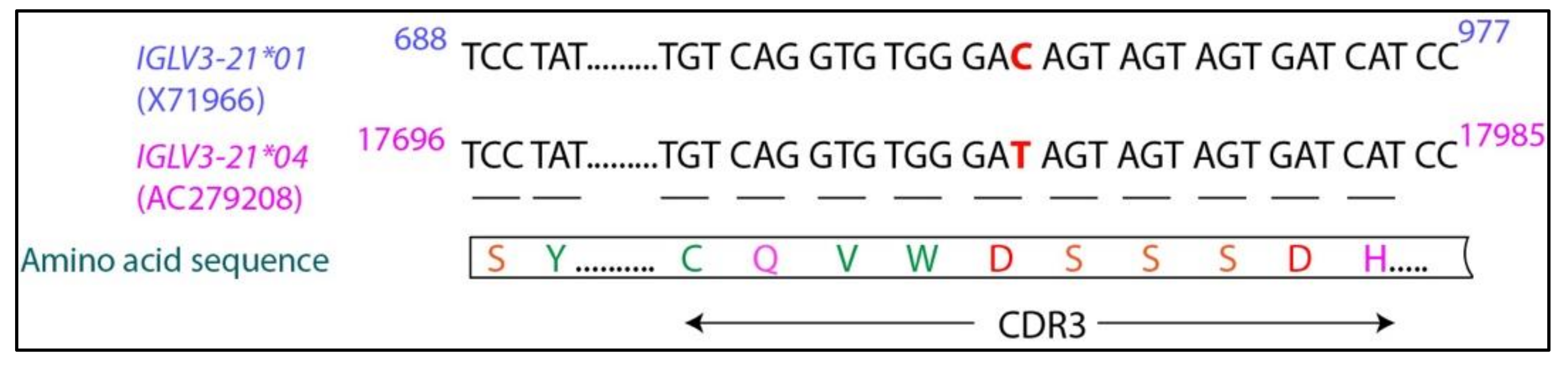
6. IGLV3-21*01/04 Are Potential Risk Alleles for Developing Aggressive CLL
7. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hallek, M. Chronic lymphocytic leukemia: 2015 Update on diagnosis, risk stratification, and treatment. Am. J. Hematol. 2015, 90, 446–460. [Google Scholar] [CrossRef]
- Fabbri, G.; Dalla-Favera, G.F.R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat. Cancer 2016, 16, 145–162. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, F.K.; Forconi, F.; Kipps, T.J. Exploring the pathways to chronic lymphocytic leukemia. Blood 2021, 138, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Chiorazzi, N.; Chen, S.-S.; Rai, K.R. Chronic Lymphocytic Leukemia. Cold Spring Harb. Perspect. Med. 2020, 11, a035220. [Google Scholar] [CrossRef] [PubMed]
- Rozman, C.; Montserrat, E. Chronic Lymphocytic Leukemia. N. Engl. J. Med. 1995, 333, 1052–1057. [Google Scholar] [CrossRef] [PubMed]
- Dameshek, W. Chronic lymphocytic leukemia-an accumulative disease of immunolgically incompetent lymphocytes. Blood 1967, 29, 566–584. [Google Scholar] [CrossRef]
- Chiorazzi, N.; Rai, K.R.; Ferrarini, M. Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2005, 352, 804–815. [Google Scholar] [CrossRef]
- Matutes, E.; Owusu-Ankomah, K.; Morilla, R.; Marco, J.G.; Houlihan, A.; Que, T.H.; Catovsky, D. The immunological profile of B-cell disorders and proposal of a scoring system for the diagnosis of CLL. Leukemia 1994, 8, 1640–1645. [Google Scholar]
- Klein, U.; Tu, Y.; Stolovitzky, G.A.; Mattioli, M.; Cattoretti, G.; Husson, H.; Freedman, A.; Inghirami, G.; Cro, L.M.; Baldini, L.; et al. Gene Expression Profiling of B Cell Chronic Lymphocytic Leukemia Reveals a Homogeneous Phenotype Related to Memory B Cells. J. Exp. Med. 2001, 194, 1625–1638. [Google Scholar] [CrossRef]
- Rosenwald, A.; Alizadeh, A.A.; Widhopf, G.; Simon, R.; Davis, R.E.; Yu, X.; Yang, L.; Pickeral, O.K.; Rassenti, L.Z.; Powell, J.; et al. Relation of Gene Expression Phenotype to Immunoglobulin Mutation Genotype in B Cell Chronic Lymphocytic Leukemia. J. Exp. Med. 2001, 194, 1639–1648. [Google Scholar] [CrossRef]
- Laurenti, L.; Efremov, D. Therapeutic Targets in Chronic Lymphocytic Leukemia. Cancers 2020, 12, 3259. [Google Scholar] [CrossRef]
- Efremov, D.G.; Turkalj, S.; Laurenti, L. Mechanisms of B Cell Receptor Activation and Responses to B Cell Receptor Inhibitors in B Cell Malignancies. Cancers 2020, 12, 1396. [Google Scholar] [CrossRef]
- Hashimoto, S.; Dono, M.; Wakai, M.; Allen, S.; Lichtman, S.M.; Schulman, P.; Vinciguerra, V.P.; Ferrarini, M.; Silver, J.; Chiorazzi, N. Somatic diversification and selection of immunoglobulin heavy and light chain variable region genes in IgG+ CD5+ chronic lymphocytic leukemia B cells. J. Exp. Med. 1995, 181, 1507–1517. [Google Scholar] [CrossRef]
- Oscier, D.G.; Thompsett, A.; Zhu, D.; Stevenson, F. Differential rates of somatic hypermutation in V(H) genes among subsets of chronic lymphocytic leukemia defined by chromosomal abnormalities. Blood 1997, 89, 4153–4160. [Google Scholar] [CrossRef]
- Fais, F.; Ghiotto, F.; Hashimoto, S.; Sellars, B.; Valetto, A.; Allen, S.; Schulman, P.; Vinciguerra, V.P.; Rai, K.; Rassenti, L.Z.; et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J. Clin. Investig. 1998, 102, 1515–1525. [Google Scholar] [CrossRef]
- Damle, R.N.; Wasil, T.; Fais, F.; Ghiotto, F.; Valetto, A.; Allen, S.L.; Buchbinder, A.; Budman, D.; Dittmar, K.; Kolitz, J.; et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 1999, 94, 1840–1847. [Google Scholar] [CrossRef]
- Hamblin, T.J.; Davis, Z.; Gardiner, A.; Oscier, D.G.; Stevenson, F. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 1999, 94, 1848–1854. [Google Scholar] [CrossRef]
- Raponi, S.; Ilari, C.; Della Starza, I.; Cappelli, L.V.; Cafforio, L.; Piciocchi, A.; Arena, V.; Mariglia, P.; Mauro, F.R.; Gentile, M.; et al. Redefining the prognostic likelihood of chronic lymphocytic leukaemia patients with borderline percentage of immunoglobulin variable heavy chain region mutations. Br. J. Haematol. 2020, 189, 853–859. [Google Scholar] [CrossRef]
- D’Arena, G.; Nunziata, G.; Coppola, G.; Vigliotti, M.L.; Tartarone, A.; Carpinelli, N.; Matera, R.; Bisogno, R.C.; Pistolese, G.; Di Renzo, N. CD38 expression does not change in B-cell chronic lymphocytic leukemia. Blood 2002, 100, 3052. [Google Scholar] [CrossRef]
- Chevallier, P.; Penther, D.; Avet-Loiseau, H.; Robillard, N.; Ifrah, N.; Mahé, B.; Hamidou, M.; Maisonneuve, H.; Moreau, P.; Jardel, H.; et al. CD38 expression and secondary 17p deletion are important prognostic factors in chronic lymphocytic leukaemia. Br. J. Haematol. 2002, 116, 142–150. [Google Scholar] [CrossRef]
- Ghia, P.; Guida, G.; Scielzo, C.; Geuna, M.; Caligaris-Cappio, F. CD38 modifications in chronic lymphocytic leukemia: Are they relevant? Leukemia 2004, 18, 1733–1735. [Google Scholar] [CrossRef]
- Crespo, M.; Bosch, F.; Villamor, N.; Bellosillo, B.; Colomer, D.; Rozman, M.; Marcé, S.; López-Guillermo, A.; Campo, E.; Montserrat, E. ZAP-70 Expression as a Surrogate for Immunoglobulin-Variable-Region Mutations in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2003, 348, 1764–1775. [Google Scholar] [CrossRef]
- Shanafelt, T.D.; Geyer, S.M.; Bone, N.D.; Tschumper, R.C.; Witzig, T.E.; Nowakowski, G.S.; Zent, C.S.; Call, T.G.; LaPlant, B.; Dewald, G.W.; et al. CD49d expression is an independent predictor of overall survival in patients with chronic lymphocytic leukaemia: A prognostic parameter with therapeutic potential. Br. J. Haematol. 2008, 140, 537–546. [Google Scholar] [CrossRef]
- Bulian, P.; Shanafelt, T.D.; Fegan, C.; Zucchetto, A.; Cro, L.M.; Nückel, H.; Baldini, L.; Kurtova, A.V.; Ferrajoli, A.; Burger, J.A.; et al. CD49d Is the Strongest Flow Cytometry–Based Predictor of Overall Survival in Chronic Lymphocytic Leukemia. J. Clin. Oncol. 2014, 32, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Gattei, V.; Bulian, P.; del Principe, M.I.; Zucchetto, A.; Maurillo, L.; Buccisano, F.; Bomben, R.; Bo, M.D.; Luciano, F.; Rossi, F.M.; et al. Relevance of CD49d protein expression as overall survival and progressive disease prognosticator in chronic lymphocytic leukemia. Blood 2008, 111, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Tissino, E.; Pozzo, F.; Benedetti, D.; Caldana, C.; Bittolo, T.; Rossi, F.M.; Bomben, R.; Nanni, P.; Chivilò, H.; Cattarossi, I.; et al. CD49d promotes disease progression in chronic lymphocytic leukemia: New insights from CD49d bimodal expression. Blood 2020, 135, 1244–1254. [Google Scholar] [CrossRef]
- Yun, X.; Zhang, Y.; Wang, X. Recent progress of prognostic biomarkers and risk scoring systems in chronic lymphocytic leukemia. Biomark. Res. 2020, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Puiggros, A.; Blanco, G.; Espinet, B. Genetic Abnormalities in Chronic Lymphocytic Leukemia: Where We Are and Where We Go. BioMed Res. Int. 2014, 2014, 435983. [Google Scholar] [CrossRef] [PubMed]
- Rosati, E.; Baldoni, S.; De Falco, F.; Del Papa, B.; Dorillo, E.; Rompietti, C.; Albi, E.; Falzetti, F.; Di Ianni, M.; Sportoletti, P. NOTCH1 Aberrations in Chronic Lymphocytic Leukemia. Front. Oncol. 2018, 8, 229. [Google Scholar] [CrossRef] [PubMed]
- Di Ianni, M.; Baldoni, S.; Rosati, E.; Ciurnelli, R.; Cavalli, L.; Martelli, M.F.; Marconi, P.; Screpanti, I.; Falzetti, F. A new genetic lesion in B-CLL: A NOTCH1 PEST domain mutation. Br. J. Haematol. 2009, 146, 689–691. [Google Scholar] [CrossRef]
- Wan, Y.; Wu, C.J. SF3B1 mutations in chronic lymphocytic leukemia. Blood 2013, 121, 4627–4634. [Google Scholar] [CrossRef]
- Rossi, D.; Bruscaggin, A.; Spina, V.; Rasi, S.; Khiabanian, H.; Messina, M.; Fangazio, M.; Vaisitti, T.; Monti, S.; Chiaretti, S.; et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: Association with progression and fludarabine-refractoriness. Blood 2011, 118, 6904–6908. [Google Scholar] [CrossRef]
- Kikushige, Y.; Ishikawa, F.; Miyamoto, T.; Shima, T.; Urata, S.; Yoshimoto, G.; Mori, Y.; Iino, T.; Yamauchi, T.; Eto, T.; et al. Self-Renewing Hematopoietic Stem Cell Is the Primary Target in Pathogenesis of Human Chronic Lymphocytic Leukemia. Cancer Cell 2011, 20, 246–259. [Google Scholar] [CrossRef]
- Damm, F.; Mylonas, E.; Cosson, A.; Yoshida, K.; Della Valle, V.; Mouly, E.; Diop, M.; Scourzic, L.; Shiraishi, Y.; Chiba, K.; et al. Acquired Initiating Mutations in Early Hematopoietic Cells of CLL Patients. Cancer Discov. 2014, 4, 1088–1101. [Google Scholar] [CrossRef]
- Nguyen-Khac, F. “Double-Hit” Chronic Lymphocytic Leukemia, Involving the TP53 and MYC Genes. Front. Oncol. 2022, 11, 826245. [Google Scholar] [CrossRef]
- Kanduri, M.; Cahill, N.; Göransson, H.; Enström, C.; Ryan, F.; Isaksson, A.; Rosenquist, R. Differential genome-wide array–based methylation profiles in prognostic subsets of chronic lymphocytic leukemia. Blood 2010, 115, 296–305. [Google Scholar] [CrossRef]
- Kulis, M.; Heath, S.; Bibikova, M.; Queirós, A.C.; Navarro, A.; Clot, G.; Martínez-Trillos, A.; Castellano, G.; Brun-Heath, I.; Pinyol, M.; et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat. Genet. 2012, 44, 1236–1242. [Google Scholar] [CrossRef]
- Queiros, A.; Villamor, N.; Clot, G.; Martineztrillos, A.; Kulis, M.; Navarro, A.; Penas, E.M.M.; Jayne, S.; Majid, A.M.S.A.; Richter, J.A.; et al. A B-cell epigenetic signature defines three biologic subgroups of chronic lymphocytic leukemia with clinical impact. Leukemia 2014, 29, 598–605. [Google Scholar] [CrossRef]
- Stamatopoulos, K.; Agathangelidis, A.; Rosenquist, R.; Ghia, P. Antigen receptor stereotypy in chronic lymphocytic leukemia. Leukemia 2016, 31, 282–291. [Google Scholar] [CrossRef]
- Marcatili, P.; Ghiotto, F.; Tenca, C.; Chailyan, A.; Mazzarello, A.N.; Yan, X.-J.; Colombo, M.; Albesiano, E.; Bagnara, D.; Cutrona, G.; et al. Igs Expressed by Chronic Lymphocytic Leukemia B Cells Show Limited Binding-Site Structure Variability. J. Immunol. 2013, 190, 5771–5778. [Google Scholar] [CrossRef]
- Gerousi, M.; Laidou, S.; Gemenetzi, K.; Stamatopoulos, K.; Chatzidimitriou, A. Distinctive Signaling Profiles with Distinct Biological and Clinical Implications in Aggressive CLL Subsets with Stereotyped B-Cell Receptor Immunoglobulin. Front. Oncol. 2021, 11, 771454. [Google Scholar] [CrossRef] [PubMed]
- Dühren-Von Minden, M.; Übelhart, R.; Schneider, D.; Wossning, T.; Bach, M.P.; Buchner, M.; Hofmann, D.; Surova, E.; Follo, M.; Köhler, F.; et al. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature 2012, 489, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Minici, C.; Gounari, M.; Übelhart, R.; Scarfo’, L.; Minden, M.D.-V.; Schneider, D.; Tasdogan, A.; Alkhatib, A.; Agathangelidis, A.; Ntoufa, S.; et al. Distinct homotypic B-cell receptor interactions shape the outcome of chronic lymphocytic leukaemia. Nat. Commun. 2017, 8, 15746. [Google Scholar] [CrossRef]
- Agathangelidis, A.; Chatzidimitriou, A.; Gemenetzi, K.; Giudicelli, V.; Karypidou, M.; Plevova, K.; Davis, Z.; Yan, X.-J.; Jeromin, S.; Schneider, C.; et al. Higher-order connections between stereotyped subsets: Implications for improved patient classification in CLL. Blood 2021, 137, 1365–1376. [Google Scholar] [CrossRef] [PubMed]
- Tobin, G.; Thunberg, U.; Johnson, A.; Eriksson, I.; Söderberg, O.; Karlsson, K.; Merup, M.; Juliusson, G.; Vilpo, J.; Enblad, G.; et al. Chronic lymphocytic leukemias utilizing the VH3-21 gene display highly restricted Vλ2-14 gene use and homologous CDR3s: Implicating recognition of a common antigen epitope. Blood 2003, 101, 4952–4957. [Google Scholar] [CrossRef] [PubMed]
- Stamatopoulos, K.; Belessi, C.; Hadzidimitriou, A.; Smilevska, T.; Kalagiakou, E.; Hatzi, K.; Stavroyianni, N.; Athanasiadou, A.; Tsompanakou, A.; Papadaki, T.; et al. Immunoglobulin light chain repertoire in chronic lymphocytic leukemia. Blood 2005, 106, 3575–3583. [Google Scholar] [CrossRef] [PubMed]
- Ghia, P.; Stamatopoulos, K.; Belessi, C.; Moreno, C.; Stella, S.; Guida, G.; Michel, A.; Crespo, M.; Laoutaris, N.; Montserrat, E.; et al. Geographic patterns and pathogenetic implications of IGHV gene usage in chronic lymphocytic leukemia: The lesson of the IGHV3-21 gene. Blood 2005, 105, 1678–1685. [Google Scholar] [CrossRef]
- Thorsélius, M.; Kröber, A.; Murray, F.; Thunberg, U.; Tobin, G.; Bühler, A.; Kienle, D.; Albesiano, E.; Maffei, R.; Dao-Ung, L.-P.; et al. Strikingly homologous immunoglobulin gene rearrangements and poor outcome in VH3-21-using chronic lymphocytic leukemia patients independent of geographic origin and mutational status. Blood 2006, 107, 2889–2894. [Google Scholar] [CrossRef]
- Hadzidimitriou, A.; Darzentas, N.; Murray, F.; Smilevska, T.; Arvaniti, E.; Tresoldi, C.; Tsaftaris, A.; Laoutaris, N.; Anagnostopoulos, A.; Davi, F.; et al. Evidence for the significant role of immunoglobulin light chains in antigen recognition and selection in chronic lymphocytic leukemia. Blood 2009, 113, 403–411. [Google Scholar] [CrossRef]
- Stamatopoulos, B.; Smith, T.; Crompot, E.; Pieters, K.; Clifford, R.; Mraz, M.; Robbe, P.; Burns, A.; Timbs, A.; Bruce, D.; et al. The Light Chain IgLV3-21 Defines a New Poor Prognostic Subgroup in Chronic Lymphocytic Leukemia: Results of a Multicenter Study. Clin. Cancer Res. 2018, 24, 5048–5057. [Google Scholar] [CrossRef]
- Maity, P.C.; Bilal, M.; Koning, M.T.; Young, M.; van Bergen, C.A.M.; Renna, V.; Nicolò, A.; Datta, M.; Gentner-Göbel, E.; Barendse, R.S.; et al. IGLV3-21 * 01 is an inherited risk factor for CLL through the acquisition of a single-point mutation enabling autonomous BCR signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 4320–4327. [Google Scholar] [CrossRef]
- Nadeu, F.; Royo, R.; Clot, G.; Duran-Ferrer, M.; Navarro, A.; Martin, S.; Lu, J.; Zenz, T.; Baumann, T.S.; Jares, P.; et al. IGLV3-21R110 identifies an aggressive biological subtype of chronic lymphocytic leukemia with intermediate epigenetics. Blood 2020, 137, 2935–2946. [Google Scholar] [CrossRef]
- Messmer, B.T.; Albesiano, E.; Efremov, D.; Ghiotto, F.; Allen, S.; Kolitz, J.; Foa, R.; Damle, R.N.; Fais, F.; Messmer, D.; et al. Multiple Distinct Sets of Stereotyped Antigen Receptors Indicate a Role for Antigen in Promoting Chronic Lymphocytic Leukemia. J. Exp. Med. 2004, 200, 519–525. [Google Scholar] [CrossRef]
- Iype, J.; Datta, M.; Khadour, A.; Übelhart, R.; Nicolò, A.; Rollenske, T.; Minden, M.D.-V.; Wardemann, H.; Maity, P.C.; Jumaa, H. Differences in Self-Recognition between Secreted Antibody and Membrane-Bound B Cell Antigen Receptor. J. Immunol. 2019, 202, 1417–1427. [Google Scholar] [CrossRef]
- Calissano, C.; Damle, R.N.; Marsilio, S.; Yan, X.-J.; Yancopoulos, S.; Hayes, G.; Emson, C.; Murphy, E.J.; Hellerstein, M.K.; Sison, C.; et al. Intraclonal Complexity in Chronic Lymphocytic Leukemia: Fractions Enriched in Recently Born/Divided and Older/Quiescent Cells. Mol. Med. 2011, 17, 1374–1382. [Google Scholar] [CrossRef]
- Herndon, T.M.; Chen, S.-S.; Saba, N.S.; Valdez, J.; Emson, C.; Gatmaitan, M.; Tian, X.; Hughes, T.E.; Sun, C.; Arthur, D.C.; et al. Direct in vivo evidence for increased proliferation of CLL cells in lymph nodes compared to bone marrow and peripheral blood. Leukemia 2017, 31, 1340–1347. [Google Scholar] [CrossRef]
- Haselager, M.V.; Kater, A.P.; Eldering, E. Proliferative Signals in Chronic Lymphocytic Leukemia; What Are We Missing? Front. Oncol. 2020, 10, 592205. [Google Scholar] [CrossRef]
- Burger, J.A. Nurture versus Nature: The Microenvironment in Chronic Lymphocytic Leukemia. Hematol. Am. Soc. Hematol. Educ. Program 2011, 2011, 96–103. [Google Scholar] [CrossRef]
- De Totero, D.; Meazza, R.; Zupo, S.; Cutrona, G.; Matis, S.; Colombo, M.; Balleari, E.; Pierri, I.; Fabbi, M.; Capaia, M.; et al. Interleukin-21 receptor (IL-21R) is up-regulated by CD40 triggering and mediates proapoptotic signals in chronic lymphocytic leukemia B cells. Blood 2006, 107, 3708–3715. [Google Scholar] [CrossRef]
- Chapman, A.E.; Oates, M.; Mohammad, I.S.; Davies, B.R.; Stockman, P.; Zhuang, J.; Pettitt, A.R. Delineating the distinct role of AKT in mediating cell survival and proliferation induced by CD154 and IL-4/IL-21 in chronic lymphocytic leukemia. Oncotarget 2017, 8, 102948–102964. [Google Scholar] [CrossRef][Green Version]
- Ghia, P.; Strola, G.; Granziero, L.; Geuna, M.; Guida, G.; Sallusto, F.; Ruffing, N.; Montagna, L.; Piccoli, P.; Chilosi, M.; et al. Chronic lymphocytic leukemia B cells are endowed with the capacity to attract CD4+, CD40L+ T cells by producing CCL22. Eur. J. Immunol. 2002, 32, 1403–1413. [Google Scholar] [CrossRef]
- Efremov, D.G.; Bomben, R.; Gobessi, S.; Gattei, V. TLR9 signaling defines distinct prognostic subsets in CLL. Front. Biosci. 2013, 18, 371–386. [Google Scholar] [CrossRef][Green Version]
- Liang, X.; Moseman, E.A.; Farrar, M.; Bachanova, V.; Weisdorf, D.J.; Blazar, B.R.; Chen, W. Toll-like receptor 9 signaling by CpG-B oligodeoxynucleotides induces an apoptotic pathway in human chronic lymphocytic leukemia B cells. Blood 2010, 115, 5041–5052. [Google Scholar] [CrossRef]
- Greaves, M. Clonal expansion in B-CLL: Fungal drivers or self-service? J. Exp. Med. 2013, 210, 1–3. [Google Scholar] [CrossRef][Green Version]
- Meixlsperger, S.; Köhler, F.; Wossning, T.; Reppel, M.; Müschen, M.; Jumaa, H. Conventional Light Chains Inhibit the Autonomous Signaling Capacity of the B Cell Receptor. Immunity 2007, 26, 323–333. [Google Scholar] [CrossRef]
- Köhler, F.; Hug, E.; Eschbach, C.; Meixlsperger, S.; Hobeika, E.; Kofer, J.; Wardemann, H.; Jumaa, H. Autoreactive B Cell Receptors Mimic Autonomous Pre-B Cell Receptor Signaling and Induce Proliferation of Early B Cells. Immunity 2008, 29, 912–921. [Google Scholar] [CrossRef]
- Binder, M.; Müller, F.; Jackst, A.; Léchenne, B.; Pantic, M.; Bacher, U.; Zu Eulenburg, C.; Veelken, H.; Mertelsmann, R.; Pasqualini, R.; et al. B-cell receptor epitope recognition correlates with the clinical course of chronic lymphocytic leukemia. Cancer 2010, 117, 1891–1900. [Google Scholar] [CrossRef] [PubMed]
- Agathangelidis, A.; Chatzidimitriou, A.; Chatzikonstantinou, T.; Tresoldi, C.; Davis, Z.; Giudicelli, V.; Kossida, S.; Belessi, C.; Rosenquist, R.; Ghia, P.; et al. Immunoglobulin gene sequence analysis in chronic lymphocytic leukemia: The 2022 update of the recommendations by ERIC, the European Research Initiative on CLL. Leukemia 2022, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kolijn, P.M.; Muggen, A.F.; Ljungström, V.; Agathangelidis, A.; Wolvers-Tettero, I.L.M.; Beverloo, H.B.; Pál, K.; Hengeveld, P.J.; Darzentas, N.; Hendriks, R.W.; et al. Consistent B Cell Receptor Immunoglobulin Features Between Siblings in Familial Chronic Lymphocytic Leukemia. Front. Oncol. 2021, 11, 740083. [Google Scholar] [CrossRef] [PubMed]
- Janovska, P.; Poppova, L.; Plevova, K.; Plesingerova, H.; Behal, M.; Kaucka, M.; Ovesna, P.; Hlozkova, M.; Borsky, M.; Stehlikova, O.; et al. Autocrine Signaling by Wnt-5a Deregulates Chemotaxis of Leukemic Cells and Predicts Clinical Outcome in Chronic Lymphocytic Leukemia. Clin. Cancer Res. 2016, 22, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Mangolini, M.; Götte, F.; Moore, A.; Ammon, T.; Oelsner, M.; Lutzny-Geier, G.; Klein-Hitpass, L.; Williamson, J.C.; Lehner, P.J.; Dürig, J.; et al. Notch2 controls non-autonomous Wnt-signalling in chronic lymphocytic leukaemia. Nat. Commun. 2018, 9, 3839. [Google Scholar] [CrossRef]
- Oppezzo, P.; Navarrete, M.; Chiorazzi, N. AID in Chronic Lymphocytic Leukemia: Induction and Action During Disease Progression. Front. Oncol. 2021, 11, 634383. [Google Scholar] [CrossRef]
- Yuan, C.; Chu, C.C.; Yan, X.-J.; Bagnara, D.; Chiorazzi, N.; MacCarthy, T. The Number of Overlapping AID Hotspots in Germline IGHV Genes Is Inversely Correlated with Mutation Frequency in Chronic Lymphocytic Leukemia. PLoS ONE 2017, 12, e0167602. [Google Scholar] [CrossRef]
- Paschold, L.; Simnica, D.; Brito, R.B.; Zhang, T.; Schultheiß, C.; Dierks, C.; Binder, M. Subclonal heterogeneity sheds light on the transformation trajectory in IGLV3-21R110 chronic lymphocytic leukemia. Blood Cancer J. 2022, 12, 49. [Google Scholar] [CrossRef]
- Gemenetzi, K.; Psomopoulos, F.; Carriles, A.A.; Gounari, M.; Minici, C.; Plevova, K.; Sutton, L.-A.; Tsagiopoulou, M.; Baliakas, P.; Pasentsis, K.; et al. Higher-order immunoglobulin repertoire restrictions in CLL: The illustrative case of stereotyped subsets 2 and 169. Blood 2021, 137, 1895–1904. [Google Scholar] [CrossRef]
- Nicolò, A.; Linder, A.T.; Jumaa, H.; Maity, P.C. The Determinants of B Cell Receptor Signaling as Prototype Molecular Biomarkers of Leukemia. Front. Oncol. 2021, 11, 771669. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Datta, M.; Jumaa, H. Immunoglobulin Gene Sequence as an Inherited and Acquired Risk Factor for Chronic Lymphocytic Leukemia. Cancers 2022, 14, 3045. https://doi.org/10.3390/cancers14133045
Datta M, Jumaa H. Immunoglobulin Gene Sequence as an Inherited and Acquired Risk Factor for Chronic Lymphocytic Leukemia. Cancers. 2022; 14(13):3045. https://doi.org/10.3390/cancers14133045
Chicago/Turabian StyleDatta, Moumita, and Hassan Jumaa. 2022. "Immunoglobulin Gene Sequence as an Inherited and Acquired Risk Factor for Chronic Lymphocytic Leukemia" Cancers 14, no. 13: 3045. https://doi.org/10.3390/cancers14133045
APA StyleDatta, M., & Jumaa, H. (2022). Immunoglobulin Gene Sequence as an Inherited and Acquired Risk Factor for Chronic Lymphocytic Leukemia. Cancers, 14(13), 3045. https://doi.org/10.3390/cancers14133045




