Context-Dependent Effects Explain Divergent Prognostic Roles of Tregs in Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials & Methods
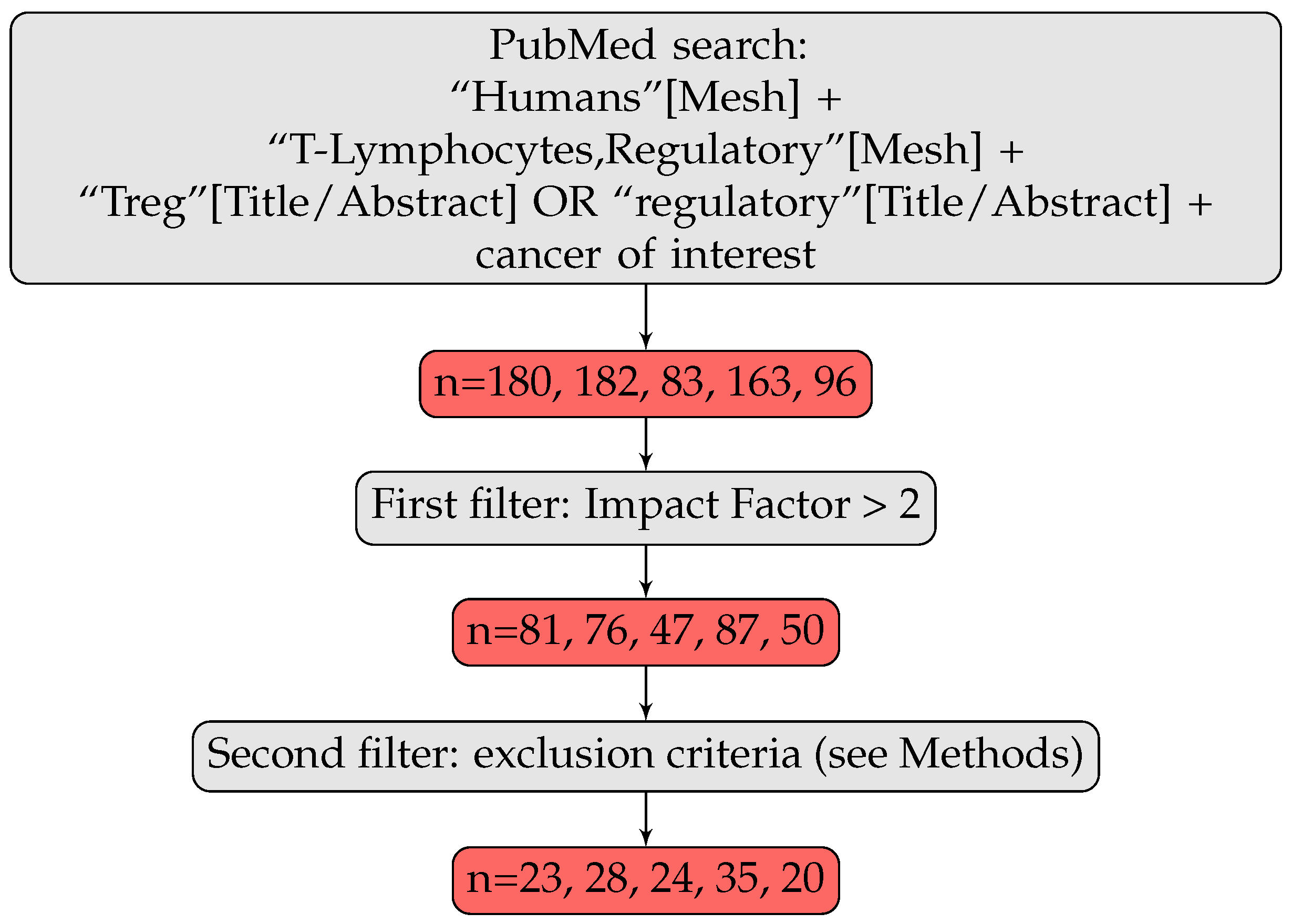
2.1. Articles Selection
2.2. Features Retrieval
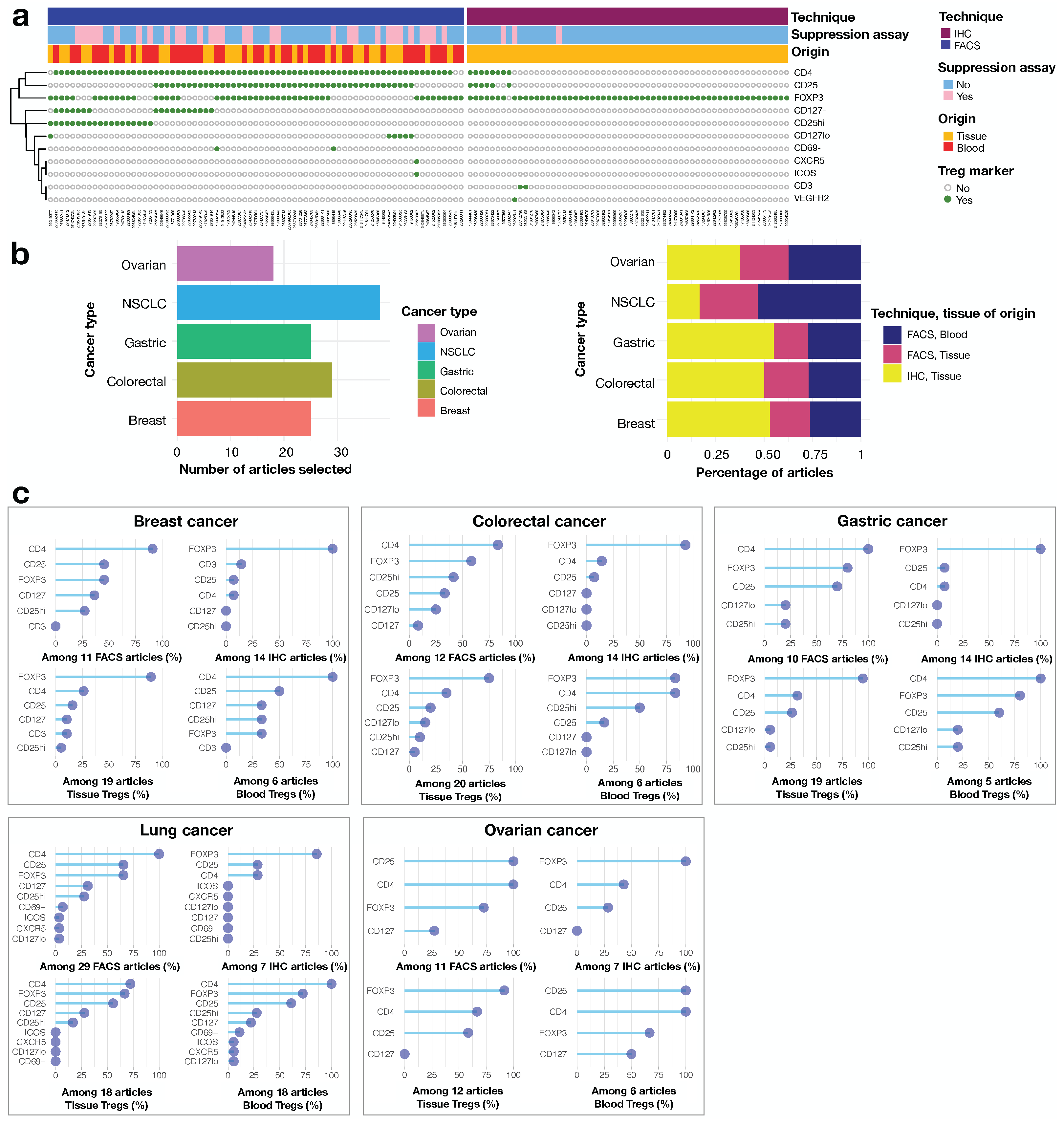
2.3. Regulatory T Cells Definition
2.4. Evaluation of the Degree of Agreement
3. Results
3.1. The Definition of Tregs Is Fuzzy in Human Cancer Literature
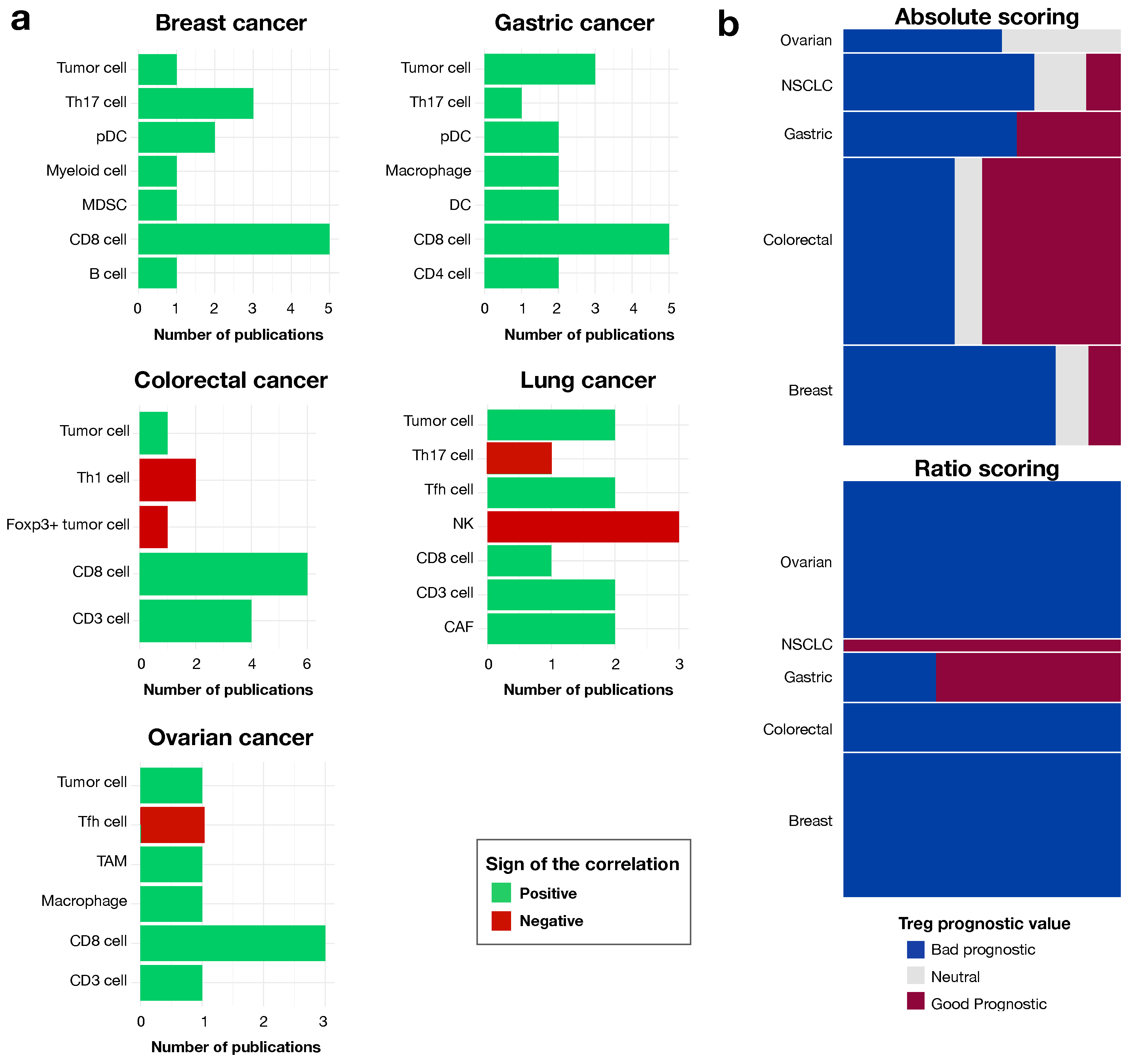
3.2. The Contribution of Tregs to Prognosis Depends on the Treg Population
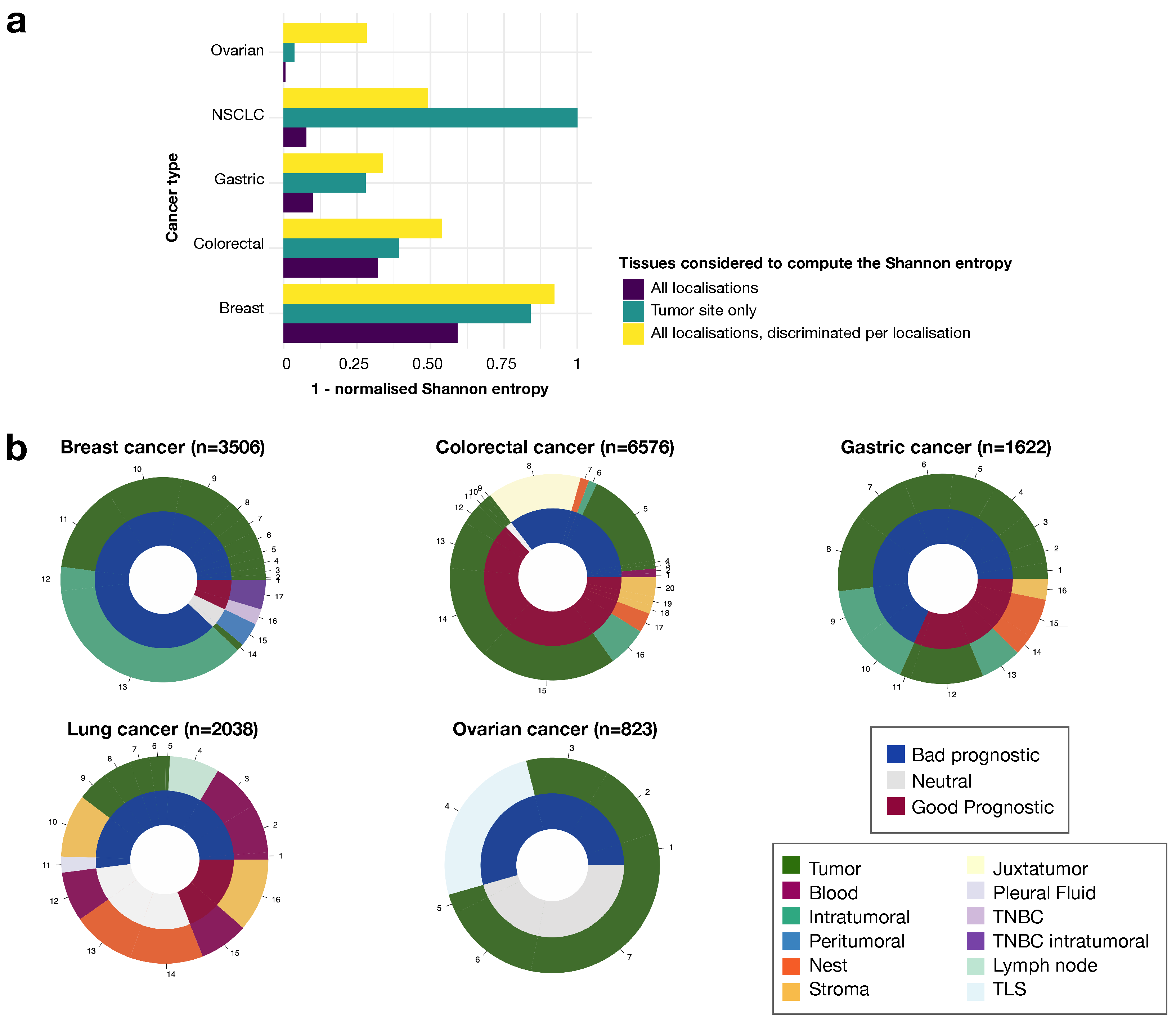
3.3. The Prognostic Value of Tregs Is Context-Dependent, as Shown by Analyses of Ratios to Other Cells in the Local Environment
3.4. Tumor Tissue Tregs Have a Greater Impact on Prognosis Than Blood Tregs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Bissell, M.; Hines, W. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med. 2011, 17, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Hsu, D.; Kim, M.; Balakumaran, B.; Acharya, C.; Anders, C.; Clay, T.; Lyerly, H.K.; Drake, C.; Morse, M.; Febbo, P. Immune Signatures Predict Prognosis in Localized Cancer. Cancer Investig. 2010, 28, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Tosolini, M.; Kirilovsky, A.; Mlecnik, B.; Fredriksen, T.; Mauger, S.; Bindea, G.; Berger, A.; Bruneval, P.; Fridman, W.H.; Pagès, F.; et al. Clinical Impact of Different Classes of Infiltrating T Cytotoxic and Helper Cells (Th1, Th2, Treg, Th17) in Patients with Colorectal Cancer. Cancer Res. 2011, 71, 1263–1271. [Google Scholar] [CrossRef]
- Olkhanud, P.; Damdinsuren, B.; Bodogai, M.; Gress, R.; Sen, R.; Wejksza, K.; Malchinkhuu, E.; Wersto, R.; Biragyn, A. Tumor-Evoked Regulatory B Cells Promote Breast Cancer Metastasis by Converting Resting CD4+ T Cells to T-Regulatory Cells. Cancer Res. 2011, 71, 3505–3515. [Google Scholar] [CrossRef]
- Toh, B.; Wang, X.; Keeble, J.; Sim, W.J.; Khoo, K.; Wong, W.C.; Kato, M.; Prevost-Blondel, A.; Thiery, J.P.; Abastado, J.P. Mesenchymal Transition and Dissemination of Cancer Cells Is Driven by Myeloid-Derived Suppressor Cells Infiltrating the Primary Tumor. PLoS Biol. 2011, 9, e1001162. [Google Scholar] [CrossRef]
- Binnewies, M.; Roberts, E.; Kersten, K.; Chan, V.; Fearon, D.; Merad, M.; Coussens, L.; Gabrilovich, D.; Ostrand-Rosenberg, S.; Hedrick, C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef]
- Galon, J.; Pagès, F.; Marincola, F.; Angell, H.; Thurin, M.; Lugli, A.; Zlobec, I.; Berger, A.; Bifulco, C.; Botti, G.; et al. Cancer classification using the Immunoscore: A worldwide task force. J. Transl. Med. 2012, 10, 205. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.; Brown, S.; Wolf, D.; Bortone, D.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.; Plaisier, C.; Eddy, J.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830. [Google Scholar] [CrossRef]
- Fridman, W.; Pagès, F.; Sautès-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef]
- Fridman, W.; Zitvogel, L.; Sautès-Fridman, C.; Kroemer, G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017, 14, 717–734. [Google Scholar] [CrossRef]
- Ahrends, T.; Borst, J. The opposing roles of CD4 + T cells in anti-tumour immunity. Immunology 2018, 154, 582–592. [Google Scholar] [CrossRef]
- Curotto de Lafaille, M.A.; Lafaille, J.J. Natural and adaptive foxp3+ regulatory T cells: More of the same or a division of labor? Immunity 2009, 30, 626–635. [Google Scholar] [CrossRef]
- Mohr, A.; Malhotra, R.; Mayer, G.; Gorochov, G.; Miyara, M. Human FOXP3+ T regulatory cell heterogeneity. Clin. Transl. Immunol. 2018, 7, e1005. [Google Scholar] [CrossRef]
- Xydia, M.; Rahbari, R.; Ruggiero, E.; Macaulay, I.; Tarabichi, M.; Lohmayer, R.; Wilkening, S.; Michels, T.; Brown, D.; Vanuytven, S.; et al. Common clonal origin of conventional T cells and induced regulatory T cells in breast cancer patients. Nat. Commun. 2021, 12, 1119. [Google Scholar] [CrossRef]
- Grossman, W.J.; Verbsky, J.W.; Barchet, W.; Colonna, M.; Atkinson, J.P.; Ley, T.J. Human T regulatory cells can use the perforin pathway to cause autologous target cell death. Immunity 2004, 21, 589–601. [Google Scholar] [CrossRef]
- Liu, F.; Tong, F.; He, Y.; Liu, H. Detectable expression of IL-35 in CD4+ T cells from peripheral blood of chronic Hepatitis B patients. Clin. Immunol. 2011, 139, 1–5. [Google Scholar] [CrossRef]
- Nakamura, K.; Kitani, A.; Strober, W. Cell contact-dependent immunosuppression by CD4(+)CD25(+) regulatory T cells is mediated by cell surface-bound transforming growth factor beta. J. Exp. Med. 2001, 194, 629–644. [Google Scholar] [CrossRef]
- Frydrychowicz, M.; Boruczkowski, M.; Kolecka-Bednarczyk, A.; Dworacki, G. The Dual Role of Treg in Cancer. Scand. J. Immunol. 2017, 86, 436–443. [Google Scholar] [CrossRef]
- Whiteside, T. Regulatory T cell subsets in human cancer: Are they regulating for or against tumor progression? Cancer Immunol. Immunother CII 2014, 63, 67–72. [Google Scholar] [CrossRef]
- Whiteside, T. Human regulatory T cells (Treg) and their response to cancer. Expert Rev. Precis. Med. Drug Dev. 2019, 4, 1–14. [Google Scholar] [CrossRef]
- Shang, B.; Liu, Y.; Jiang, S.j.; Liu, Y. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: A systematic review and meta-analysis. Sci. Rep. 2015, 5, 15179. [Google Scholar] [CrossRef]
- Allan, S.; Crome, S.; Crellin, N.; Passerini, L.; Steiner, T.; Bacchetta, R.; Roncarolo, M.; Levings, M. Activation-induced FOXP3 in human T effector cells does not suppress proliferation or cytokine production. Int. Immunol. 2007, 19, 345–354. [Google Scholar] [CrossRef]
- Sharma, M.D.; Huang, L.; Choi, J.H.; Lee, E.J.; Wilson, J.M.; Lemos, H.; Pan, F.; Blazar, B.R.; Pardoll, D.M.; Mellor, A.L.; et al. An inherently bi-functional subset of Foxp3+ T helper cells is controlled by the transcription factor Eos. Immunity 2013, 38, 998–1012. [Google Scholar] [CrossRef]
- Wang, J.; Ioan-Facsinay, A.; van der Voort, E.I.H.; Huizinga, T.W.J.; Toes, R.E.M. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur. J. Immunol. 2007, 37, 129–138. [Google Scholar] [CrossRef]
- Otsubo, K.; Kanegane, H.; Kamachi, Y.; Kobayashi, I.; Tsuge, I.; Imaizumi, M.; Sasahara, Y.; Hayakawa, A.; Nozu, K.; Iijima, K.; et al. Identification of FOXP3-negative regulatory T-like (CD4+CD25+CD127low) cells in patients with immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome. Clin. Immunol. 2011, 141, 111–120. [Google Scholar] [CrossRef]
- Boldt, A.; Kentouche, K.; Fricke, S.; Borte, S.; Kahlenberg, F.; Sack, U. Differences in FOXP3 and CD127 expression in Treg-like cells in patients with IPEX syndrome. Clin. Immunol. 2014, 153, 109–111. [Google Scholar] [CrossRef] [PubMed]
- Miyara, M.; Yoshioka, Y.; Kitoh, A.; Shima, T.; Wing, K.; Niwa, A.; Parizot, C.; Taflin, C.; Heike, T.; Valeyre, D.; et al. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity 2009, 30, 899–911. [Google Scholar] [CrossRef]
- Endharti, A.T.; Rifa, M.; Shi, Z.; Fukuoka, Y.; Nakahara, Y.; Kawamoto, Y.; Takeda, K.; Isobe, K.i.; Suzuki, H. Cutting Edge: CD8+CD122+ Regulatory T Cells Produce IL-10 to Suppress IFN-γ Production and Proliferation of CD8+ T Cells. J. Immunol. 2005, 175, 7093–7097. [Google Scholar] [CrossRef]
- Chaput, N.; Louafi, S.; Bardier, A.; Charlotte, F.; Vaillant, J.C.; Ménégaux, F.; Rosenzwajg, M.; Lemoine, F.; Klatzmann, D.; Taieb, J. Identification of CD8+CD25+Foxp3+ suppressive T cells in colorectal cancer tissue. Gut 2009, 58, 520–529. [Google Scholar] [CrossRef]
- Arruvito, L.; Payaslián, F.; Baz, P.; Podhorzer, A.; Billordo, A.; Pandolfi, J.; Semeniuk, G.; Arribalzaga, E.; Fainboim, L. Identification and clinical relevance of naturally occurring human CD8+HLA-DR+ regulatory T cells. J. Immunol. 2014, 193, 4469–4476. [Google Scholar] [CrossRef] [PubMed]
- Waniczek, D.; Lorenc, Z.; Śnietura, M.; Wesecki, M.; Kopec, A.; Muc-Wierzgoń, M. Tumor-Associated Macrophages and Regulatory T Cells Infiltration and the Clinical Outcome in Colorectal Cancer. Arch. Immunol. Ther. Exp. 2017, 65, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Panda, A.K.; Bose, S.; Roy, D.; Kajal, K.; Guha, D.; Sa, G. Transcriptional regulation of FOXP3 requires integrated activation of both promoter and CNS regions in tumor-induced CD8+ Treg cells. Sci. Rep. 2017, 7, 1628. [Google Scholar] [CrossRef] [PubMed]
- Sarrabayrouse, G.; Bossard, C.; Chauvin, J.M.; Jarry, A.; Meurette, G.; Quévrain, E.; Bridonneau, C.; Preisser, L.; Asehnoune, K.; Labarrière, N.; et al. CD4CD8αα lymphocytes, a novel human regulatory T cell subset induced by colonic bacteria and deficient in patients with inflammatory bowel disease. PLoS Biol. 2014, 12, e1001833. [Google Scholar] [CrossRef]
- Kim, M.; Grimmig, T.; Grimm, M.; Lazariotou, M.; Meier, E.; Rosenwald, A.; Tsaur, I.; Blaheta, R.; Heemann, U.; Germer, C.T.; et al. Expression of Foxp3 in colorectal cancer but not in Treg cells correlates with disease progression in patients with colorectal cancer. PLoS ONE 2013, 8, e53630. [Google Scholar] [CrossRef]
- Saito, T.; Nishikawa, H.; Wada, H.; Nagano, Y.; Sugiyama, D.; Atarashi, K.; Maeda, Y.; Hamaguchi, M.; Ohkura, N.; Sato, E.; et al. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat. Med. 2016, 22, 679–684. [Google Scholar] [CrossRef]
- Kotsakis, A.; Koinis, F.; Katsarou, A.; Gioulbasani, M.; Aggouraki, D.; Kentepozidis, N.; Georgoulias, V.; Vetsika, E.K. Prognostic value of circulating regulatory T cell subsets in untreated non-small cell lung cancer patients. Sci. Rep. 2016, 6, 39247. [Google Scholar] [CrossRef]
- Plitas, G.; Konopacki, C.; Wu, K.; Bos, P.; Morrow, M.; Putintseva, E.V.; Chudakov, D.M.; Rudensky, A.Y. Regulatory T cells exhibit distinct features in human breast cancer. Immunity 2016, 45, 1122–1134. [Google Scholar] [CrossRef]
- De Simone, M.; Arrigoni, A.; Rossetti, G.; Gruarin, P.; Ranzani, V.; Politano, C.; Bonnal, R.J.P.; Provasi, E.; Sarnicola, M.L.; Panzeri, I.; et al. Transcriptional Landscape of Human Tissue Lymphocytes Unveils Uniqueness of Tumor-Infiltrating T Regulatory Cells. Immunity 2016, 45, 1135–1147. [Google Scholar] [CrossRef]
- Bailur, J.K.; Gueckel, B.; Derhovanessian, E.; Pawelec, G. Presence of circulating Her2-reactive CD8+ T-cells is associated with lower frequencies of myeloid-derived suppressor cells and regulatory T cells, and better survival in older breast cancer patients. Breast Cancer Res. BCR 2015, 17, 34. [Google Scholar] [CrossRef]
- Liu, F.; Li, Y.; Ren, M.; Zhang, X.; Guo, X.; Lang, R.; Gu, F.; Fu, L. Peritumoral FOXP3+ regulatory T cell is sensitive to chemotherapy while intratumoral FOXP3+ regulatory T cell is prognostic predictor of breast cancer patients. Breast Cancer Res. Treat. 2012, 135, 459–467. [Google Scholar] [CrossRef]
- Blatner, N.R.; Gounari, F.; Khazaie, K. The two faces of regulatory T cells in cancer. Oncoimmunology 2013, 2, e23852. [Google Scholar] [CrossRef][Green Version]
- Mao, Y.; Qu, Q.; Chen, X.; Huang, O.; Wu, J.; Shen, K. The Prognostic Value of Tumor-Infiltrating Lymphocytes in Breast Cancer: A Systematic Review and Meta-Analysis. PLoS ONE 2016, 11, e0152500. [Google Scholar] [CrossRef]
- Whiteside, T.L. The role of regulatory T cells in cancer immunology. Immunotargets Ther. 2015, 4, 159–171. [Google Scholar] [CrossRef]
- Jiménez-Sánchez, A.; Cybulska, P.; Mager, K.L.; Koplev, S.; Cast, O.; Couturier, D.L.; Memon, D.; Selenica, P.; Nikolovski, I.; Mazaheri, Y.; et al. Unraveling tumor–immune heterogeneity in advanced ovarian cancer uncovers immunogenic effect of chemotherapy. Nat. Genet. 2020, 52, 582–593. [Google Scholar] [CrossRef]
- Hamy, A.S.; Bonsang-Kitzis, H.; De Croze, D.; Laas, E.; Darrigues, L.; Topciu, L.; Menet, E.; Vincent-Salomon, A.; Lerebours, F.; Pierga, J.Y.; et al. Interaction between Molecular Subtypes and Stromal Immune Infiltration before and after Treatment in Breast Cancer Patients Treated with Neoadjuvant Chemotherapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 6731–6741. [Google Scholar] [CrossRef]
- Pesenacker, A.M.; Wang, A.; Singh, A.; Gillies, J.; Kim, Y.; Piccirillo, C.; Nguyen, D.; Haining, N.; Tebbutt, S.; Panagiotopoulos, C.; et al. A Regulatory T-Cell Gene Signature Is a Specific and Sensitive Biomarker to Identify Children With New-Onset Type 1 Diabetes. Diabetes 2016, 65, 1031–1039. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amblard, E.; Soumelis, V. Context-Dependent Effects Explain Divergent Prognostic Roles of Tregs in Cancer. Cancers 2022, 14, 2991. https://doi.org/10.3390/cancers14122991
Amblard E, Soumelis V. Context-Dependent Effects Explain Divergent Prognostic Roles of Tregs in Cancer. Cancers. 2022; 14(12):2991. https://doi.org/10.3390/cancers14122991
Chicago/Turabian StyleAmblard, Elise, and Vassili Soumelis. 2022. "Context-Dependent Effects Explain Divergent Prognostic Roles of Tregs in Cancer" Cancers 14, no. 12: 2991. https://doi.org/10.3390/cancers14122991
APA StyleAmblard, E., & Soumelis, V. (2022). Context-Dependent Effects Explain Divergent Prognostic Roles of Tregs in Cancer. Cancers, 14(12), 2991. https://doi.org/10.3390/cancers14122991





