Fully Automatic Whole-Volume Tumor Segmentation in Cervical Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. MRI Acquisitions
2.2. Inclusion Criteria
2.3. Manual Tumor Segmentation
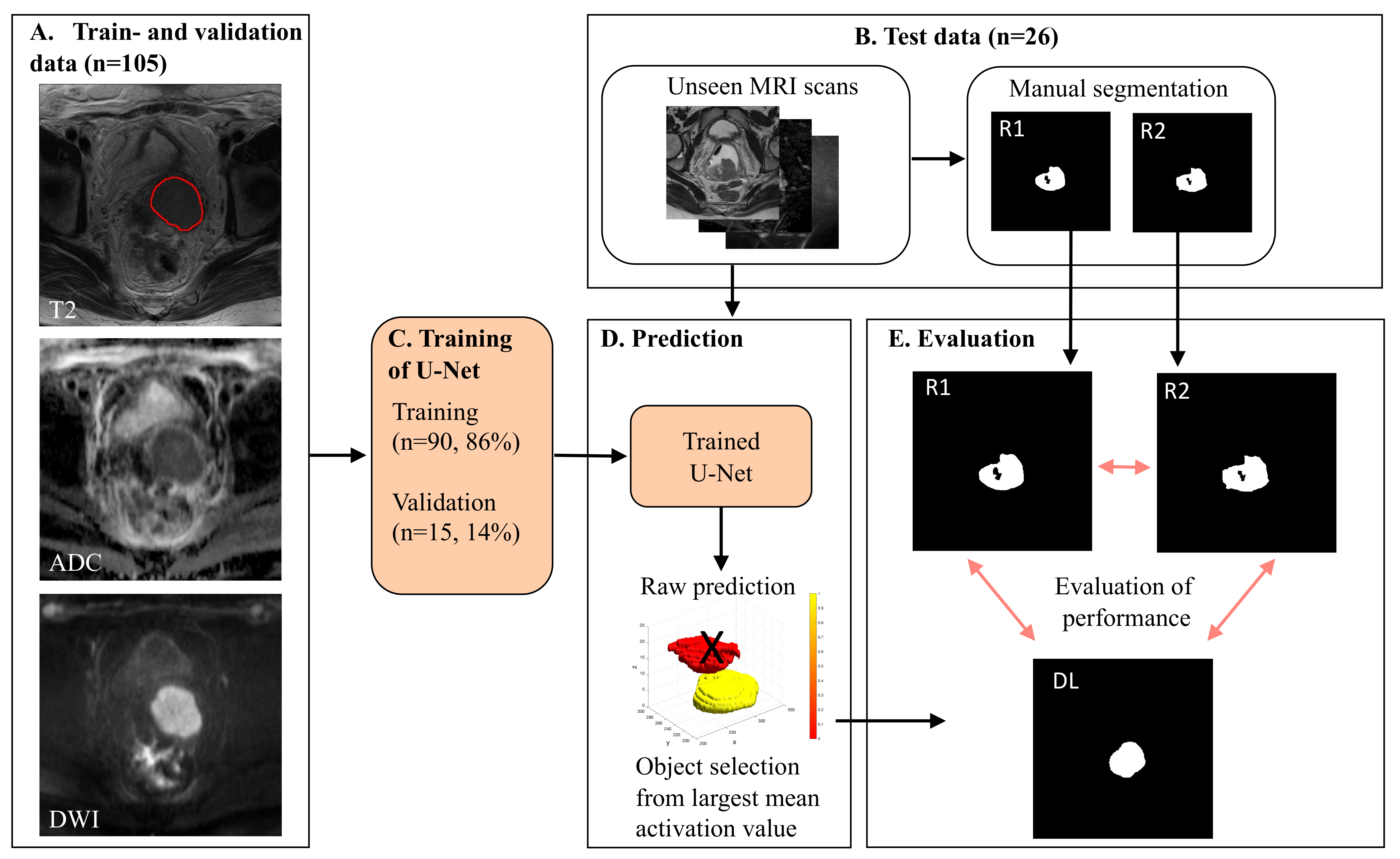
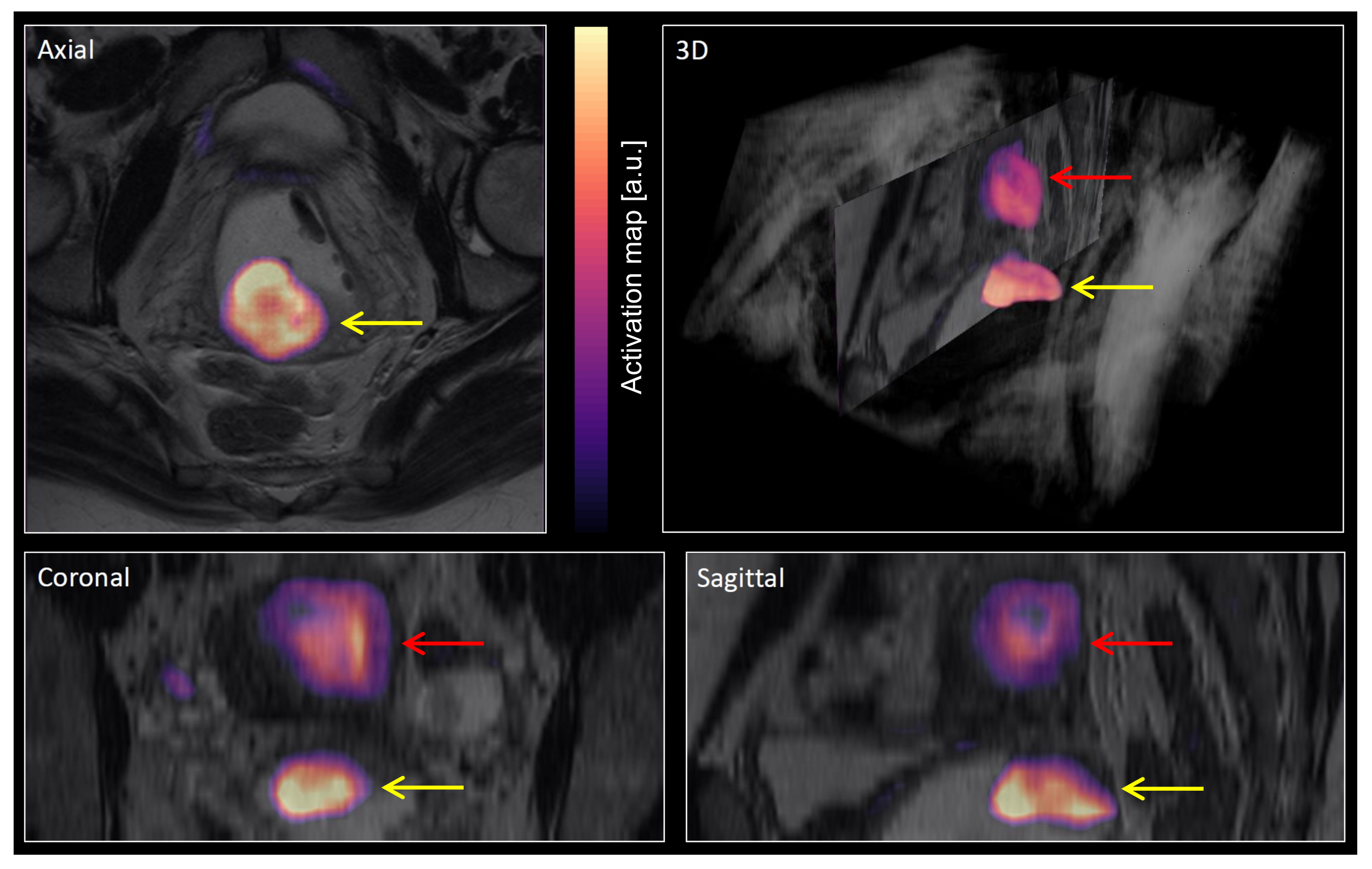
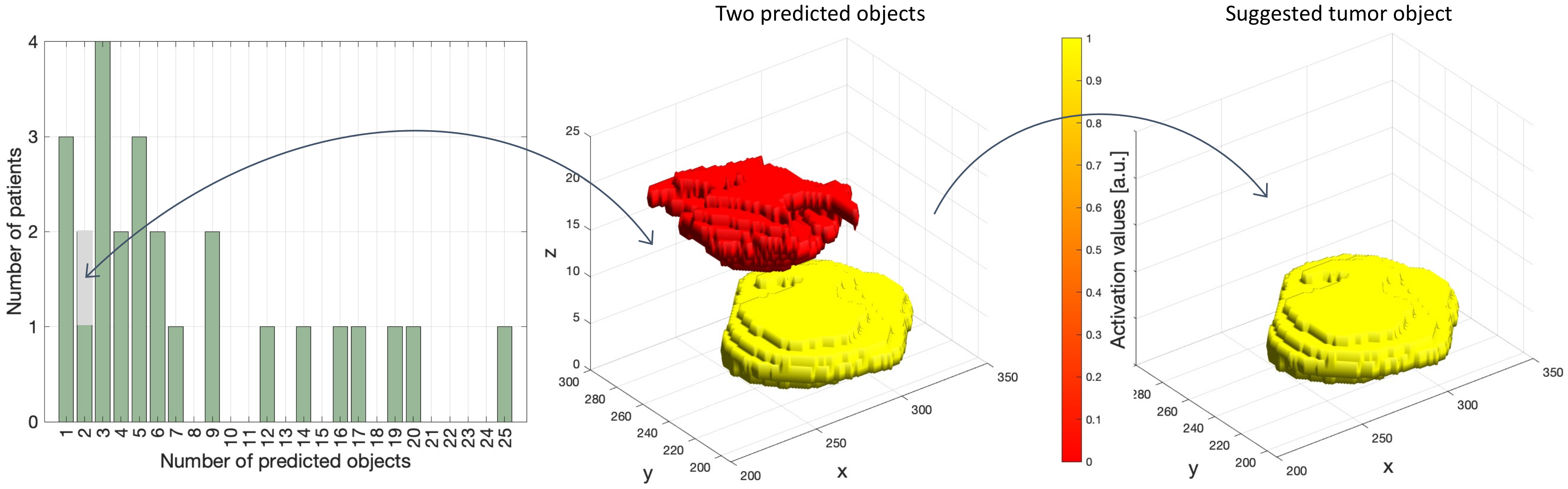
2.4. Major Processing Steps
2.5. Evaluation of Segmentation Performance
2.6. Implementation Details
3. Results
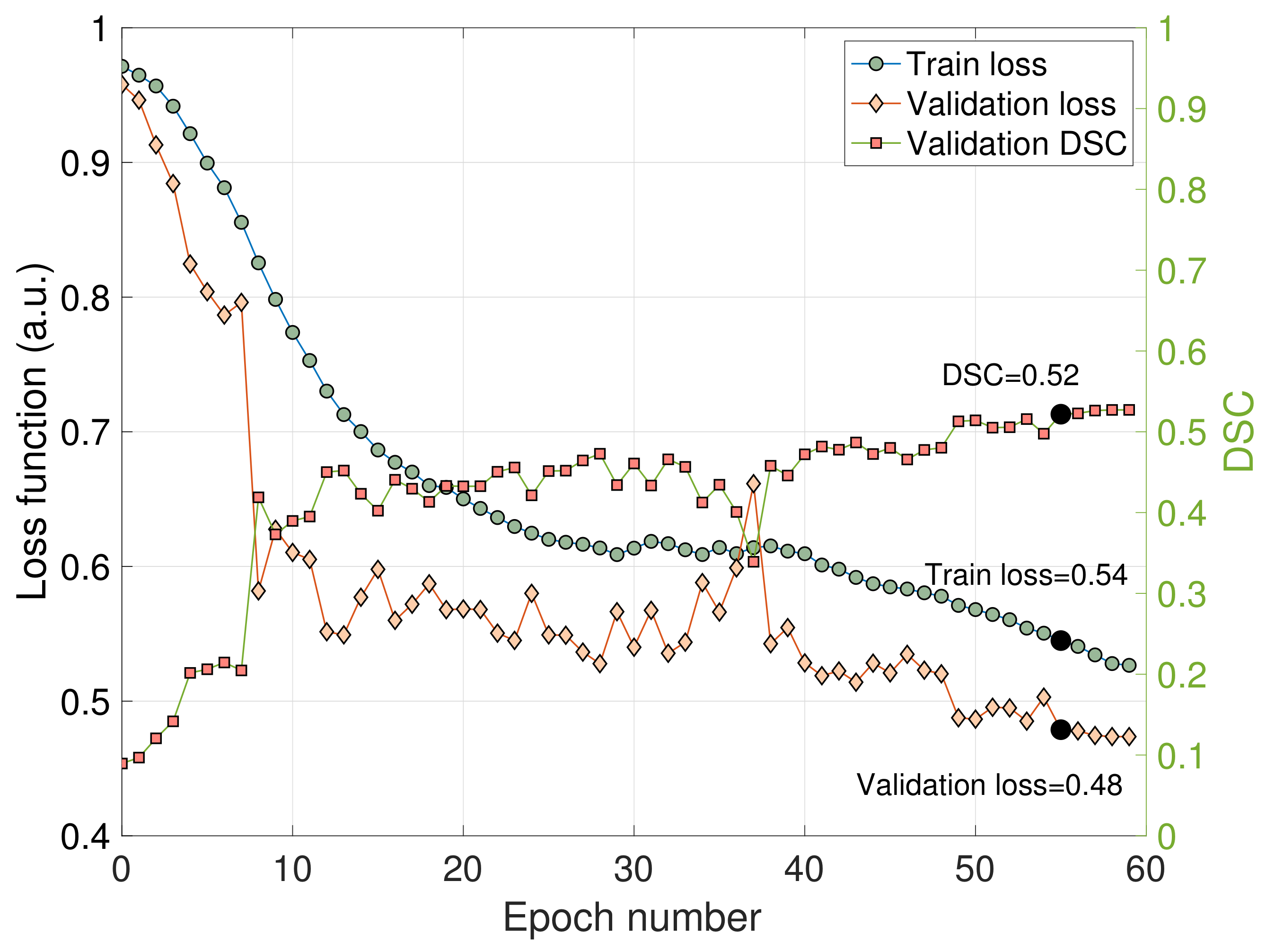
3.1. Train and Validation Metrics
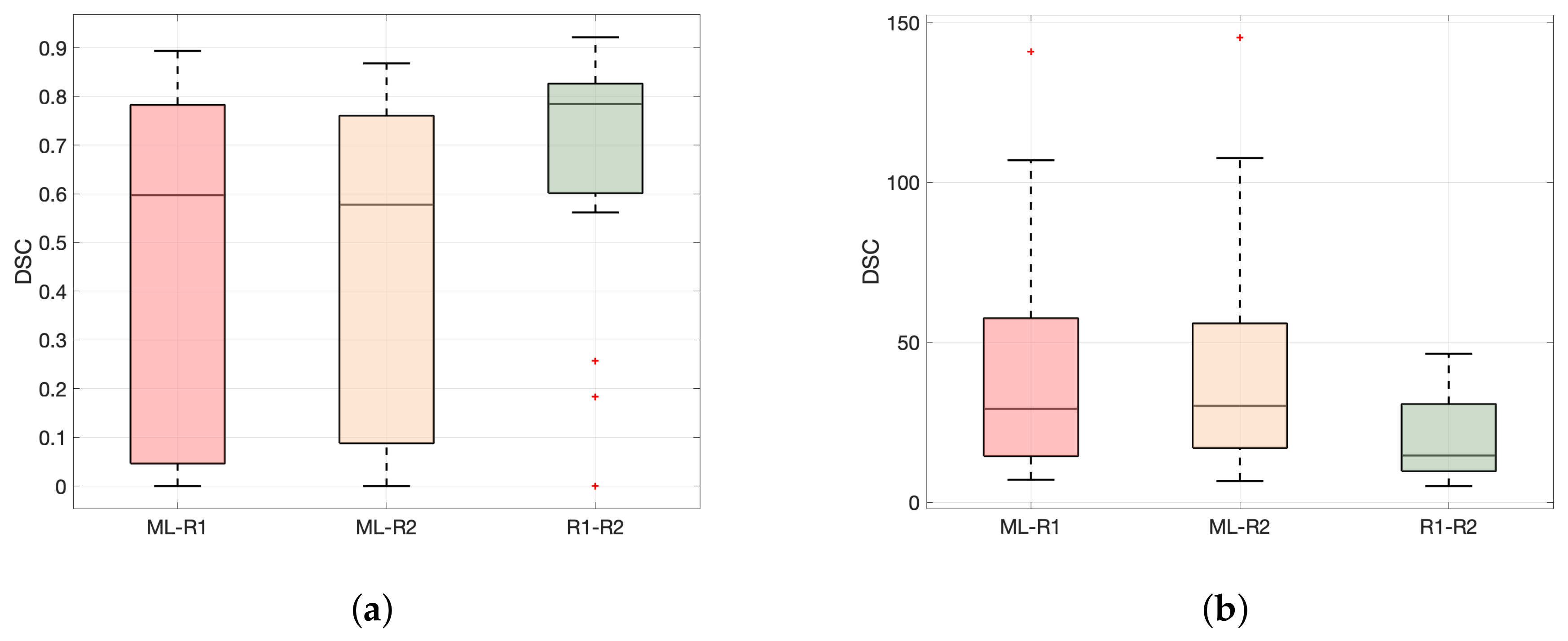
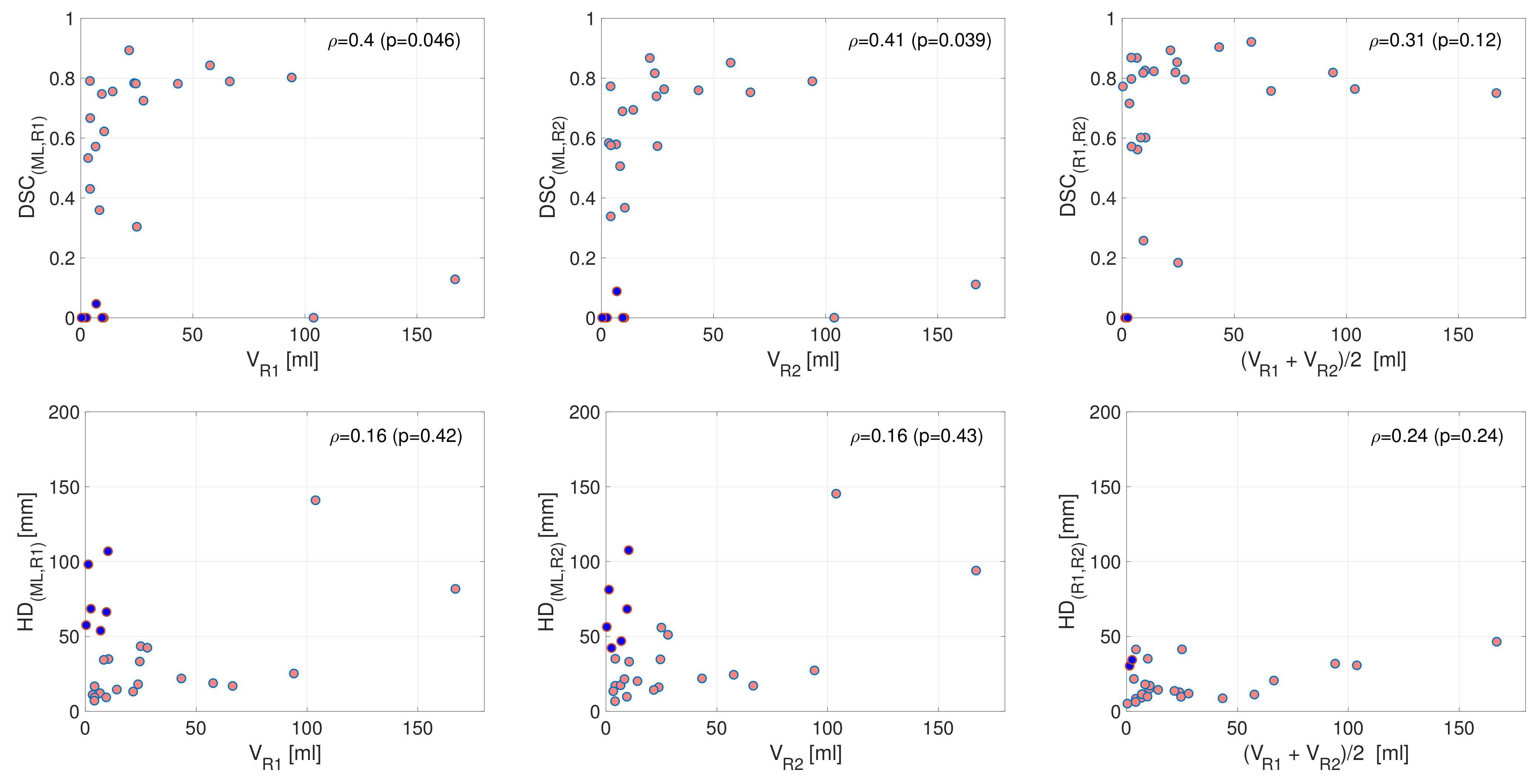
3.2. Performance in Terms of DSC and HD
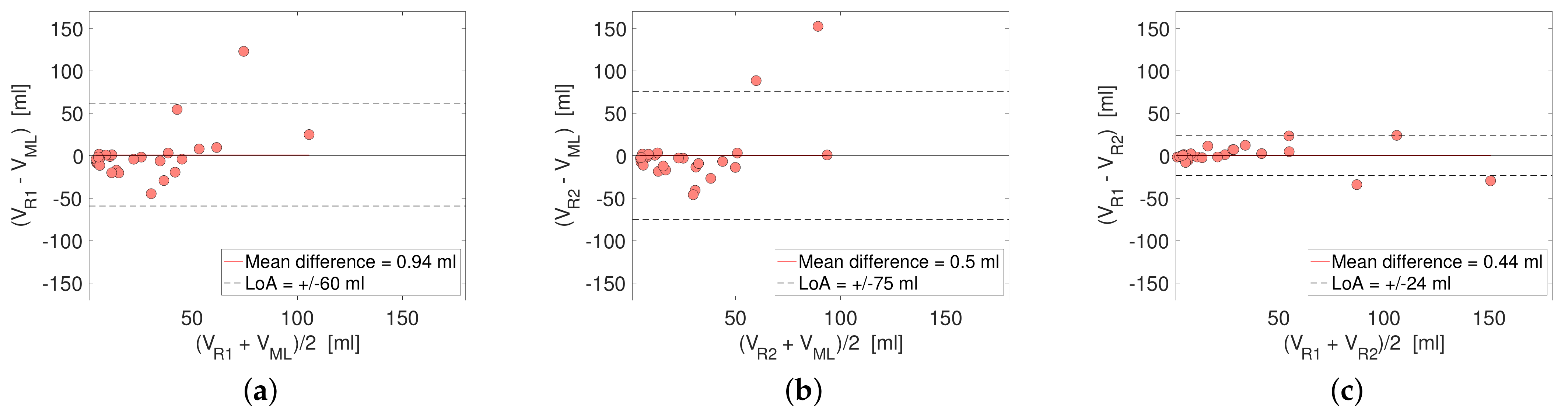
3.3. Performance in Terms of Reported Tumor Volume
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Varghese, B.A.; Cen, S.Y.; Hwang, D.H.; Duddalwar, V.A. Texture analysis of imaging: What radiologists need to know. Am. J. Roentgenol. 2019, 212, 520–528. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yu, X.; Ouyang, H.; Zhang, J.; Chen, S.; Xie, L.; Zhao, X. Whole-tumor texture model based on diffusion kurtosis imaging for assessing cervical cancer: A preliminary study. Eur. Radiol. 2021, 31, 5576–5585. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Ma, F.; Li, Y.; Li, Y.; Li, M.; Zhang, G.; Qiang, J. Multiparametric MRI-based radiomics nomogram for predicting lymph node metastasis in early-stage cervical cancer. J. Magn. Reson. Imaging 2020, 52, 885–896. [Google Scholar] [CrossRef]
- Wang, T.; Gao, T.; Guo, H.; Wang, Y.; Zhou, X.; Tian, J.; Huang, L.; Zhang, M. Preoperative prediction of parametrial invasion in early-stage cervical cancer with MRI-based radiomics nomogram. Eur. Radiol. 2020, 30, 3585–3593. [Google Scholar] [CrossRef]
- Sun, C.; Tian, X.; Liu, Z.; Li, W.; Li, P.; Chen, J.; Zhang, W.; Fang, Z.; Du, P.; Duan, H.; et al. Radiomic analysis for pretreatment prediction of response to neoadjuvant chemotherapy in locally advanced cervical cancer: A multicentre study. EBioMedicine 2019, 46, 160–169. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Gu, H.L.; Zhang, X.L.; Tian, Z.F.; Xu, X.Q.; Tang, W.W. Multiparametric magnetic resonance imaging-derived radiomics for the prediction of disease-free survival in early-stage squamous cervical cancer. Eur. Radiol. 2021, 32, 2540–2551. [Google Scholar] [CrossRef]
- Lucia, F.; Visvikis, D.; Desseroit, M.C.; Miranda, O.; Malhaire, J.P.; Robin, P.; Pradier, O.; Hatt, M.; Schick, U. Prediction of outcome using pretreatment 18 F-FDG PET/CT and MRI radiomics in locally advanced cervical cancer treated with chemoradiotherapy. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 768–786. [Google Scholar] [CrossRef] [Green Version]
- Lucia, F.; Visvikis, D.; Vallières, M.; Desseroit, M.C.; Miranda, O.; Robin, P.; Bonaffini, P.A.; Alfieri, J.; Masson, I.; Mervoyer, A.; et al. External validation of a combined PET and MRI radiomics model for prediction of recurrence in cervical cancer patients treated with chemoradiotherapy. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 864–877. [Google Scholar] [CrossRef]
- Torheim, T.; Malinen, E.; Hole, K.H.; Lund, K.V.; Indahl, U.G.; Lyng, H.; Kvaal, K.; Futsaether, C.M. Autodelineation of cervical cancers using multiparametric magnetic resonance imaging and machine learning. Acta Oncol. 2017, 56, 806–812. [Google Scholar] [CrossRef]
- Kano, Y.; Ikushima, H.; Sasaki, M.; Haga, A. Automatic contour segmentation of cervical cancer using artificial intelligence. J. Radiat. Res. 2021, 62, 934–944. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Lin, C.H.; Lu, H.Y.; Chiang, H.J.; Wang, H.K.; Huang, Y.T.; Ng, S.H.; Hong, J.H.; Yen, T.C.; Lai, C.H.; et al. Deep learning for fully automated tumor segmentation and extraction of magnetic resonance radiomics features in cervical cancer. Eur. Radiol. 2020, 30, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Bnouni, N.; Rekik, I.; Rhim, M.S.; Amara, N.E.B. Context-Aware Synergetic Multiplex Network for Multi-organ Segmentation of Cervical Cancer MRI. In Proceedings of the International Workshop on Predictive Intelligence in Medicine, Lima, Peru, 8 October 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 1–11. [Google Scholar]
- Renard, F.; Guedria, S.; Palma, N.D.; Vuillerme, N. Variability and reproducibility in deep learning for medical image segmentation. Sci. Rep. 2020, 10, 13724. [Google Scholar] [CrossRef]
- Almeida, G.; Tavares, J.M.R. Deep learning in radiation oncology treatment planning for prostate cancer: A systematic review. J. Med. Syst. 2020, 44, 1–15. [Google Scholar] [CrossRef]
- Lundervold, A.S.; Lundervold, A. An overview of deep learning in medical imaging focusing on MRI. Z. Med. Phys. 2019, 29, 102–127. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Ruan, S.; Canu, S. A review: Deep learning for medical image segmentation using multi-modality fusion. Array 2019, 3, 100004. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Kerfoot, E.; Clough, J.; Oksuz, I.; Lee, J.; King, A.P.; Schnabel, J.A. Left-ventricle quantification using residual U-Net. In Proceedings of the International Workshop on Statistical Atlases and Computational Models of the Heart, Granada, Spain, 16 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 371–380. [Google Scholar]
- Yushkevich, P.A.; Piven, J.; Cody Hazlett, H.; Gimpel Smith, R.; Ho, S.; Gee, J.C.; Gerig, G. User-Guided 3D Active Contour Segmentation of Anatomical Structures: Significantly Improved Efficiency and Reliability. Neuroimage 2006, 31, 1116–1128. [Google Scholar] [CrossRef] [Green Version]
- Cox, R.; Ashburner, J.; Breman, H.; Fissell, K.; Haselgrove, C.; Holmes, C.; Lancaster, J.; Rex, D.; Smith, S.; Woodward, J.; et al. A (sort of) new image data format standard: NiFTI-1. Presented at the 10th Annual Meeting of the Organization for Human Brain Mapping, Budapest, Hungary, 13–17 June 2004. [Google Scholar]
- Zhang, Y.; Chen, W.; Chen, Y.; Tang, X. A post-processing method to improve the white matter hyperintensity segmentation accuracy for randomly-initialized U-net. In Proceedings of the 2018 IEEE 23rd International Conference on Digital Signal Processing (DSP), Shanghai, China, 19–21 November 2018; pp. 1–5. [Google Scholar]
- Kikinis, R.; Pieper, S.D.; Vosburgh, K.G. 3D Slicer: A platform for subject-specific image analysis, visualization, and clinical support. In Intraoperative Imaging and Image-Guided Therapy; Springer: Berlin/Heidelberg, Germany, 2014; pp. 277–289. [Google Scholar]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Hausdorff, F. Grundzüge der Mengenlehre. In SSVM; Leipzig Viet: Leipzig, Germany, 1949. [Google Scholar]
- Andersen, E. Imagedata: A Python library to handle medical image data in NumPy array subclass Series. J. Open Source Softw. 2022, submitted.
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- Howard, J.; Gugger, S. Fastai: A layered API for deep learning. Information 2020, 11, 108. [Google Scholar] [CrossRef] [Green Version]
- Kaliyugarasan, S.K.; Lundervold, A.; Lundervold, A.S. Pulmonary Nodule Classification in Lung Cancer from 3D Thoracic CT Scans Using fastai and MONAI. Int. J. Interact. Multimed. Artif. Intell. 2021, 6, 83–89. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Wright, L. Ranger—A Synergistic Optimizer. 2019. Available online: https://github.com/lessw2020/Ranger-Deep-Learning-Optimizer (accessed on 16 December 2021).
- Smith, L.N.; Topin, N. Super-convergence: Very fast training of neural networks using large learning rates. In Artificial Intelligence and Machine Learning for Multi-Domain Operations Applications; International Society for Optics and Photonics: Bellingham, WA, USA, 2019; Volume 11006, p. 1100612. [Google Scholar]
- Cawley, G.C.; Talbot, N.L. On over-fitting in model selection and subsequent selection bias in performance evaluation. J. Mach. Learn. Res. 2010, 11, 2079–2107. [Google Scholar]
- Lai, C.C.; Wang, H.K.; Wang, F.N.; Peng, Y.C.; Lin, T.P.; Peng, H.H.; Shen, S.H. Autosegmentation of Prostate Zones and Cancer Regions from Biparametric Magnetic Resonance Images by Using Deep-Learning-Based Neural Networks. Sensors 2021, 21, 2709. [Google Scholar] [CrossRef]
- Hodneland, E.; Dybvik, J.A.; Wagner-Larsen, K.S.; Šoltészová, V.; Munthe-Kaas, A.Z.; Fasmer, K.E.; Krakstad, C.; Lundervold, A.; Lundervold, A.S.; Salvesen, Ø.; et al. Automated segmentation of endometrial cancer on MR images using deep learning. Sci. Rep. 2021, 11, 179. [Google Scholar] [CrossRef]
- Kurata, Y.; Nishio, M.; Moribata, Y.; Kido, A.; Himoto, Y.; Otani, S.; Fujimoto, K.; Yakami, M.; Minamiguchi, S.; Mandai, M.; et al. Automatic segmentation of uterine endometrial cancer on multi-sequence MRI using a convolutional neural network. Sci. Rep. 2021, 11, 14440. [Google Scholar] [CrossRef]
- Trebeschi, S.; van Griethuysen, J.J.; Lambregts, D.M.; Lahaye, M.J.; Parmar, C.; Bakers, F.C.; Peters, N.H.; Beets-Tan, R.G.; Aerts, H.J. Deep learning for fully-automated localization and segmentation of rectal cancer on multiparametric MR. Sci. Rep. 2017, 7, 5301. [Google Scholar] [CrossRef]
- Zhu, H.T.; Zhang, X.Y.; Shi, Y.J.; Li, X.T.; Sun, Y.S. Automatic segmentation of rectal tumor on diffusion-weighted images by deep learning with U-Net. J. Appl. Clin. Med. Phys. 2021, 22, 324–331. [Google Scholar] [CrossRef]
- Liechti, M.R.; Muehlematter, U.J.; Schneider, A.F.; Eberli, D.; Rupp, N.J.; Hötker, A.M.; Donati, O.F.; Becker, A.S. Manual prostate cancer segmentation in MRI: Interreader agreement and volumetric correlation with transperineal template core needle biopsy. Eur. Radiol. 2020, 30, 4806–4815. [Google Scholar] [CrossRef]
- Ji, W.; Yu, S.; Wu, J.; Ma, K.; Bian, C.; Bi, Q.; Li, J.; Liu, H.; Cheng, L.; Zheng, Y. Learning calibrated medical image segmentation via multi-rater agreement modeling. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 12341–12351. [Google Scholar]
- Warfield, S.K.; Zou, K.H.; Wells, W.M. Simultaneous truth and performance level estimation (STAPLE): An algorithm for the validation of image segmentation. IEEE Trans. Med. Imaging 2004, 23, 903–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.; Whitehead, T.D.; Quirk, J.D.; Salter, A.; Ademuyiwa, F.O.; Li, S.; An, H.; Shoghi, K.I. Optimal co-clinical radiomics: Sensitivity of radiomic features to tumour volume, image noise and resolution in co-clinical T1-weighted and T2-weighted magnetic resonance imaging. EBioMedicine 2020, 59, 102963. [Google Scholar] [CrossRef] [PubMed]
- Bento, M.; Fantini, I.; Park, J.; Rittner, L.; Frayne, R. Deep Learning in Large and Multi-Site Structural Brain MR Imaging Datasets. Front. Neuroinformatics 2021, 15, 805669. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Fang, B.; Liu, Y.; Gao, M.; Zheng, S.; Wang, Y. Liver vessels segmentation based on 3D residual U-NET. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; pp. 250–254. [Google Scholar]
- Tashk, A.; Herp, J.; Nadimi, E. Fully automatic polyp detection based on a novel U-Net architecture and morphological post-process. In Proceedings of the 2019 International Conference on Control, Artificial Intelligence, Robotics & Optimization (ICCAIRO), Athens, Greece, 8–10 December 2019; pp. 37–41. [Google Scholar]
- Ngo, D.K.; Tran, M.T.; Kim, S.H.; Yang, H.J.; Lee, G.S. Multi-task learning for small brain tumor segmentation from MRI. Appl. Sci. 2020, 10, 7790. [Google Scholar] [CrossRef]
| Parameter | Siemens 1.5T | GE 1.5T | Philips 1.5 | Siemens 3T | Philips 3T | |
|---|---|---|---|---|---|---|
| T2 | Pixel spacing [mm] (inplane) | (0.39, 0.39) | (0.35, 0.35) | (0.40, 0.40) | (0.52, 0.52) | (0.35, 0.35) |
| Matrix (x, y) | (512, 512) | (512, 512) | (512, 512) | (384, 384) | (512, 512) | |
| FOV [mm] (x, y) | (180, 180) | (180, 180) | (205, 205) | (200, 200) | (180, 180) | |
| TR [ms] | 4790 | 3157 | 5362 | 4610 | 4074 | |
| TE [ms] | 100 | 81 | 100 | 94 | 110 | |
| FA [degrees] | 150 | 160 | 90 | 148 | 90 | |
| Slice thickness [mm] | 3.00 | 3.00 | 3.00 | 3.00 | 2.50 | |
| Number of averages | 2 | 2 | 6 | 2 | 2 | |
| Interslice gap [mm] | 0.50 | 0.00 | 0.30 | 0.30 | 0.25 | |
| Number of slices | 25 | 30 | 26 | 24 | 35 | |
| DWI | Pixel spacing [mm] (x, y) | (1.56, 1.56) | (1.37, 1.37) | (1.46, 1.46) | (1.43, 1.43) | (0.80, 0.80) |
| Matrix (x, y) | (144, 144) | (256, 256) | (256, 256) | (144, 144) | (352, 352) | |
| FOV [mm] (x, y) | (250, 250) | (350, 350) | (375, 375) | (200, 200) | (280, 280) | |
| TR [ms] | 3200 | 4000 | 1716.30 | 5640 | 3280 | |
| TE [ms] | 82 | 52 | 69.18 | 63 | 85 | |
| FA [degrees] | 90 | 90 | 90 | 180 | 90 | |
| Slice thickness [mm] | 4.00 | 5.00 | 5.00 | 3.00 | 4.00 | |
| Number of averages | 10 | 2 | 3 | 2 | 2 | |
| Interslice gap [mm] | 0.60 | 0.50 | 1.00 | 0.40 | 0.40 | |
| Number of slices | 22 | 25 | 30 | 25 | 33 | |
| b-values [s/mm] | [0/50, 800/1000] | NA * | [0, 1000] | [0/50, 800/1000] | NA * | |
| N | Number of patients | 51 | 9 | 27 | 27 | 9 |
| Variable | Train (n = 90) and Validation (n = 15) Data | Test Data (n = 26) | p |
|---|---|---|---|
| Age (yrs.) | 0.73 | ||
| Median (IQR) | 48 (37–60) | 49 (41–59) | |
| FIGO (2009) stage | 0.21 | ||
| I | 52 (49%) | 14 (54%) | |
| II | 27 (26%) | 6 (23%) | |
| III | 18 (17%) | 5 (19%) | |
| IV | 8 (8%) | 1 (4%) | |
| MRI-assessed maximum tumor size (cm) | 0.24 | ||
| Median (IQR) | 4.6 (3.0–5.6) | 3.9 (2.5–5.1) | |
| Primary treatment | 0.21 | ||
| Surgery only | 26 (25%) | 9 (34%) | |
| Surgery and adjuvant therapy | 63 (60%) | 15 (58%) | |
| Primary radiotherapy w/o chemotherapy | 12 (11%) | 2 (8%) | |
| Palliative treatment | 4 (4%) | 0 | |
| Histologic subtype | 0.19 | ||
| Squamous cell carcinoma | 82 (78%) | 21 (81%) | |
| Adenocarcinoma | 18 (17%) | 3 (11%) | |
| Other * | 5 (5%) | 2 (8%) | |
| Histologic grade | 0.76 | ||
| Low/medium | 80 (82%) | 22 (88%) | |
| High | 17 (18%) | 3 (12%) |
| Measure | Median Value of Estimate (IQR) | Absolute Difference (p-Value) | ||||
|---|---|---|---|---|---|---|
| . (DL, R1) | . (DL, R2) | . (R1, R2) | (p) | (p) | ||
| I. Unadjusted | DSC | 0.60 (0.05, 0.78) | 0.58 (0.09, 0.76) | 0.78 (0.60, 0.83) | 0.19 (0.01 *) | 0.21 (0.005 *) |
| HD [mm] | 29.2 (14.5, 57.5) | 30.2 (17.1, 55.9) | 14.6 (9.80, 30.7) | 14.6 (0.01 *) | 15.5 (0.003 *) | |
| II. Adjusted for R1-R2 disagreement | DSC | 0.81 | 0.79 | 1 (ref.) | - | - |
| HD [mm] | 3.73 | 9.10 | 0 (ref.) | - | - | |
| Estimate | SE | p | |
|---|---|---|---|
| (Intercept) | 0.08 | 0.36 | 0.82 |
| Field strength | −0.06 | 0.15 | 0.71 |
| Anisotropy T2 | 0.04 | 0.04 | 0.33 |
| FOV T2 | 1.77 | 23.69 | 0.94 |
| Anisotropy DWI | 0.04 | 0.07 | 0.54 |
| FOV DWI | 4.10 | 6.11 | 0.51 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hodneland, E.; Kaliyugarasan, S.; Wagner-Larsen, K.S.; Lura, N.; Andersen, E.; Bartsch, H.; Smit, N.; Halle, M.K.; Krakstad, C.; Lundervold, A.S.; et al. Fully Automatic Whole-Volume Tumor Segmentation in Cervical Cancer. Cancers 2022, 14, 2372. https://doi.org/10.3390/cancers14102372
Hodneland E, Kaliyugarasan S, Wagner-Larsen KS, Lura N, Andersen E, Bartsch H, Smit N, Halle MK, Krakstad C, Lundervold AS, et al. Fully Automatic Whole-Volume Tumor Segmentation in Cervical Cancer. Cancers. 2022; 14(10):2372. https://doi.org/10.3390/cancers14102372
Chicago/Turabian StyleHodneland, Erlend, Satheshkumar Kaliyugarasan, Kari Strøno Wagner-Larsen, Njål Lura, Erling Andersen, Hauke Bartsch, Noeska Smit, Mari Kyllesø Halle, Camilla Krakstad, Alexander Selvikvåg Lundervold, and et al. 2022. "Fully Automatic Whole-Volume Tumor Segmentation in Cervical Cancer" Cancers 14, no. 10: 2372. https://doi.org/10.3390/cancers14102372
APA StyleHodneland, E., Kaliyugarasan, S., Wagner-Larsen, K. S., Lura, N., Andersen, E., Bartsch, H., Smit, N., Halle, M. K., Krakstad, C., Lundervold, A. S., & Haldorsen, I. S. (2022). Fully Automatic Whole-Volume Tumor Segmentation in Cervical Cancer. Cancers, 14(10), 2372. https://doi.org/10.3390/cancers14102372






