The Role of p53 Signaling in Colorectal Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
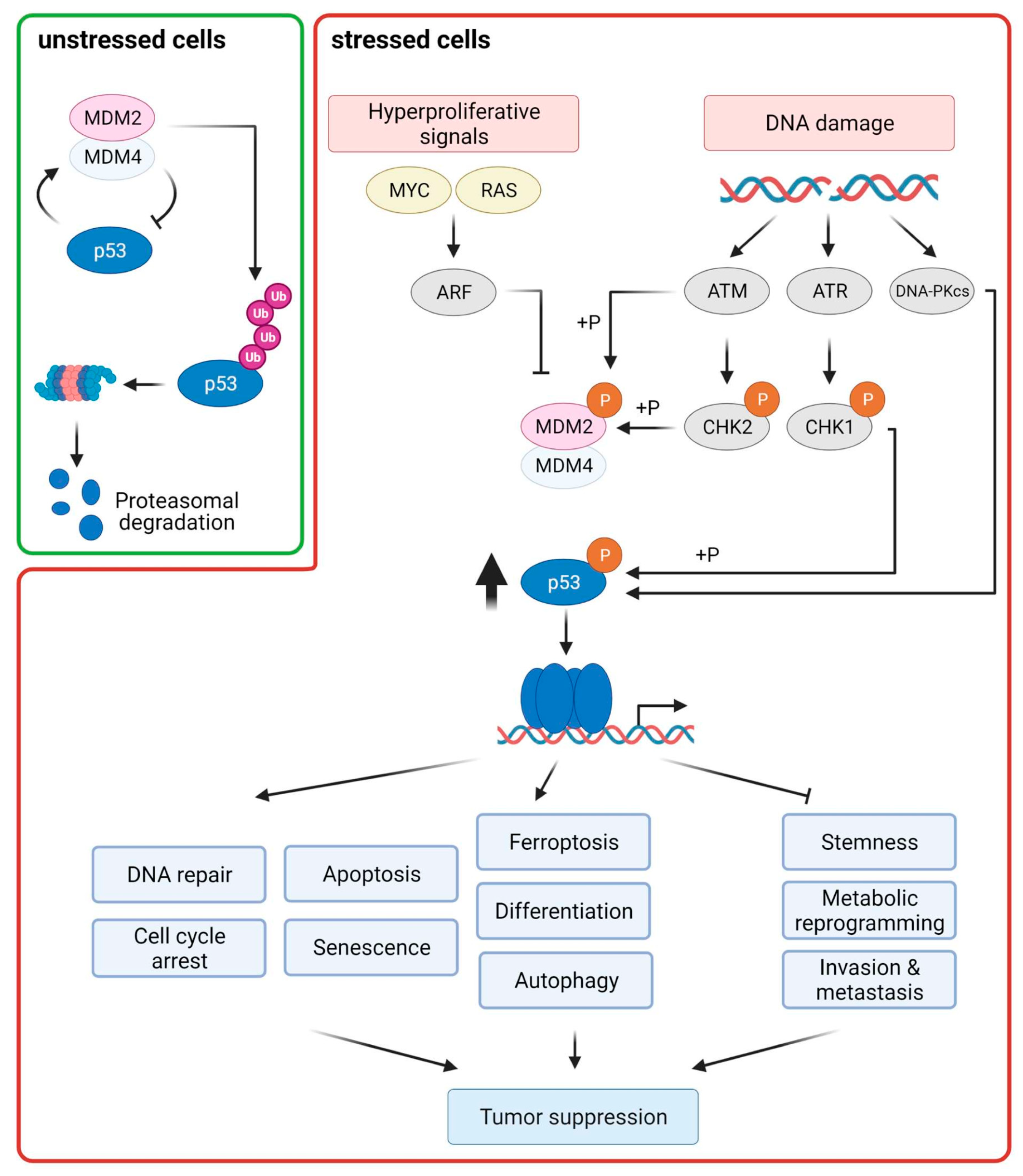
2. The p53 Pathway
2.1. Control of p53 Levels
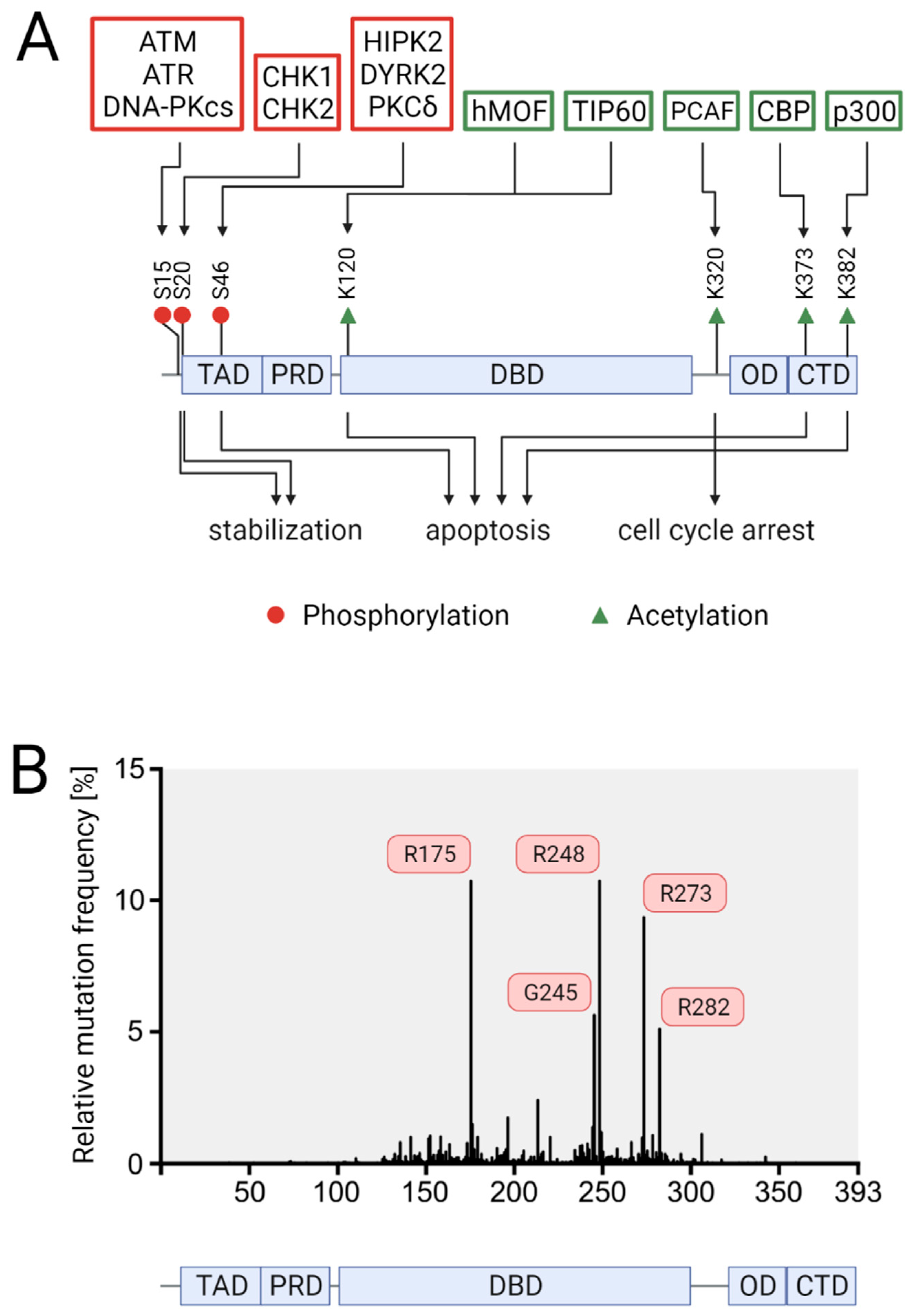
2.2. p53 Downstream Responses
3. p53 Mutations in Colorectal Cancer
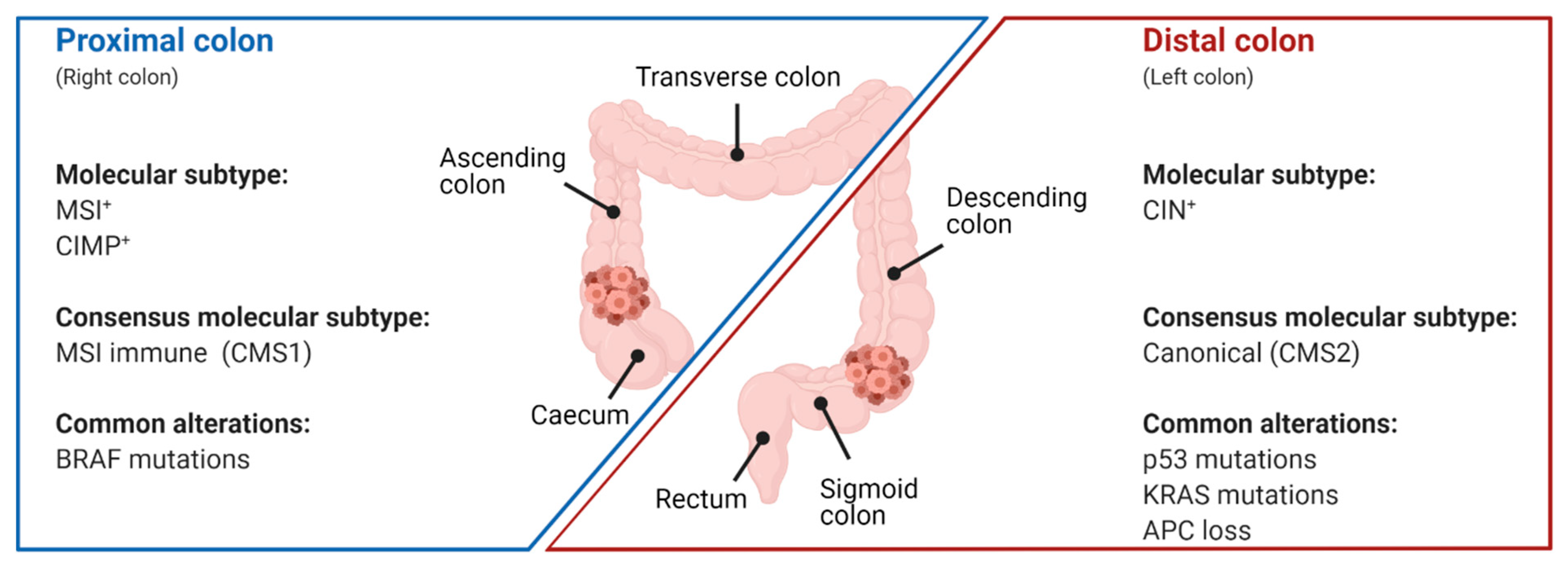
3.1. Prevalence of p53 Mutations
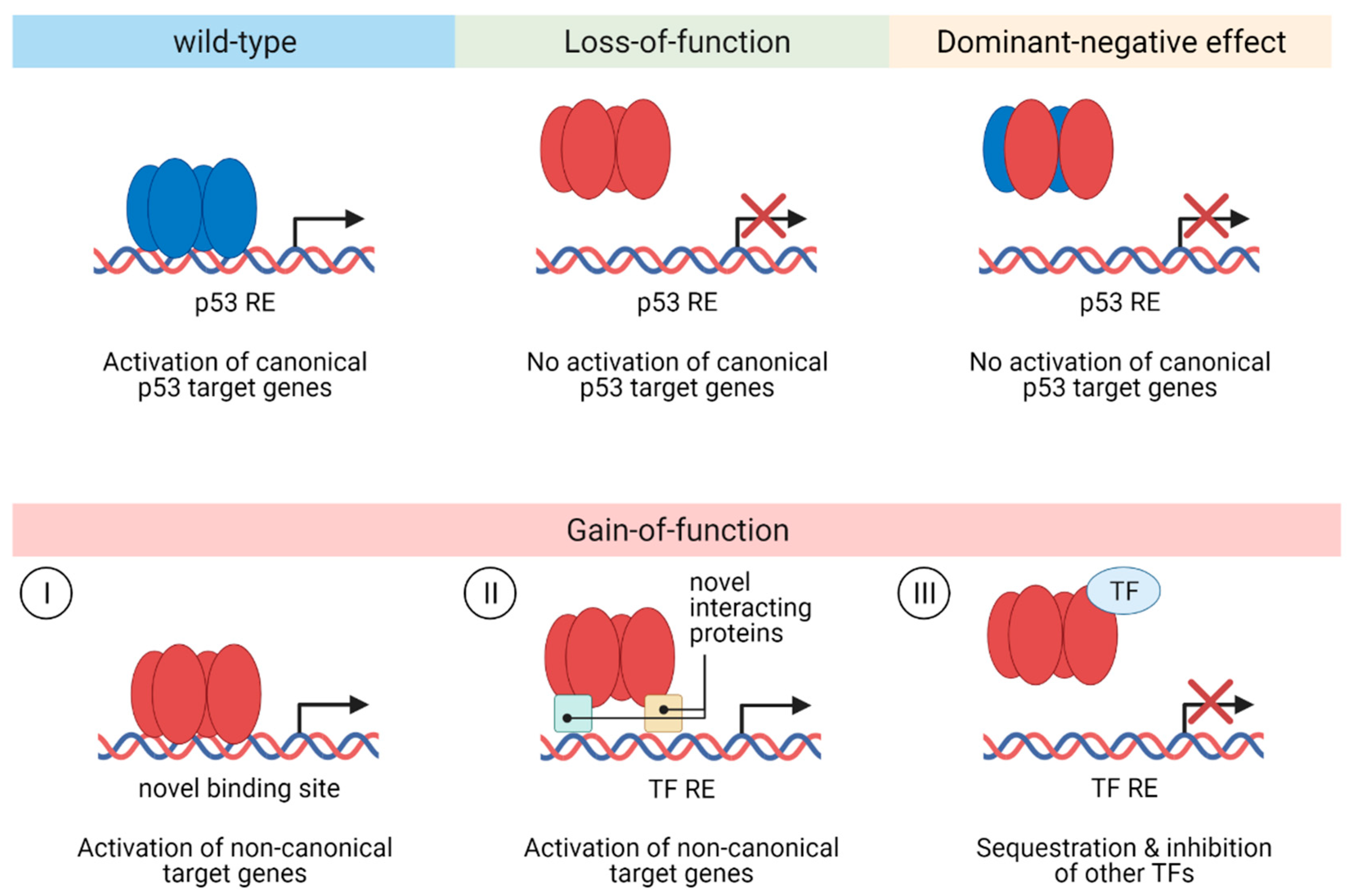
3.2. Functional Effects of p53 Mutations in CRC
3.3. Gain-of-function of p53 Mutants in CRC
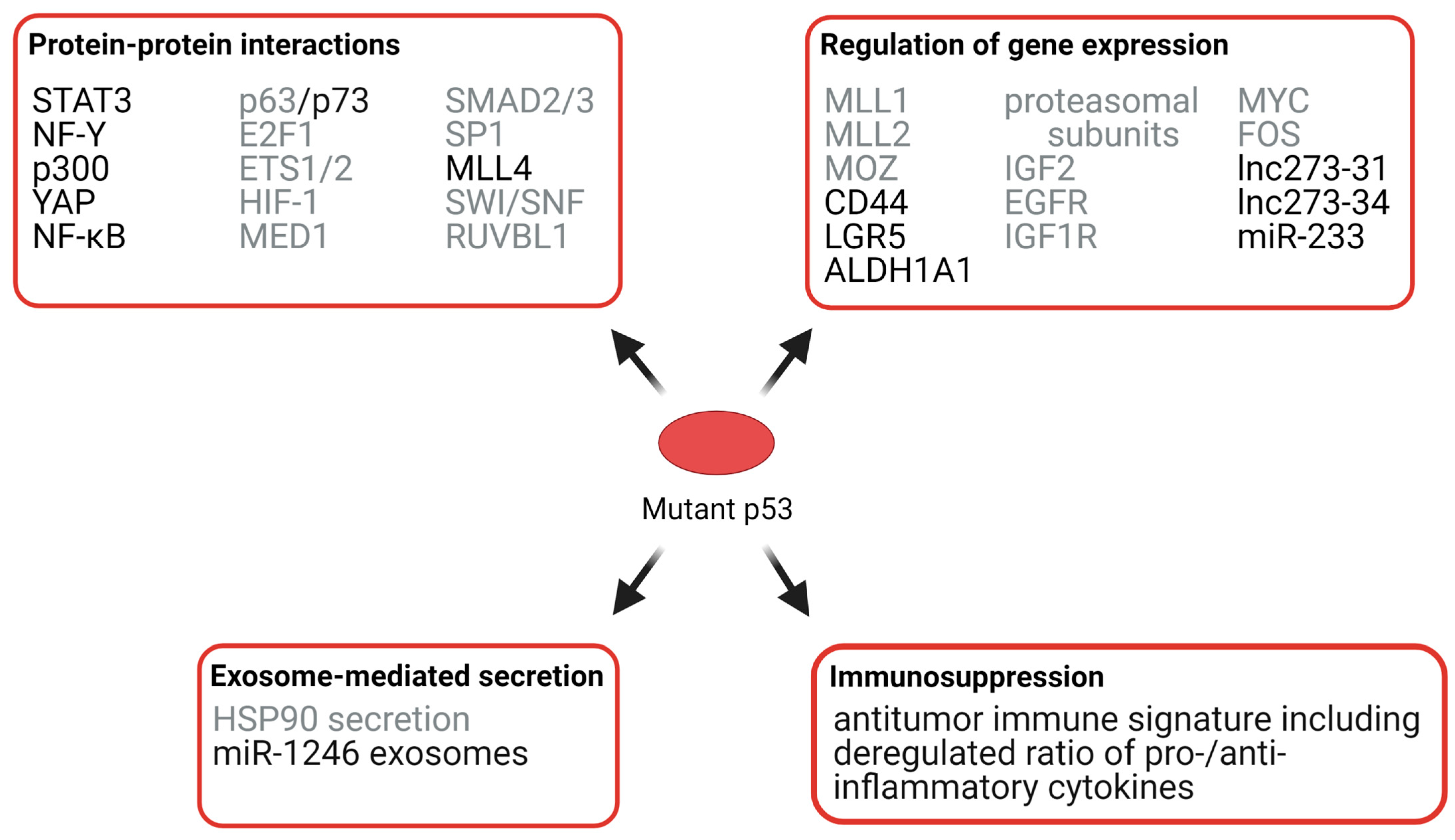
3.3.1. Effects of Mutant p53 on Protein Interactions
3.3.2. Effects of Mutant p53 on Chromatin
3.3.3. Effects of Mutant p53 on RNA Expression, Exosomes, and Immunosuppression
3.4. Effects of p53 Mutations on Therapy Response and Patient Survival
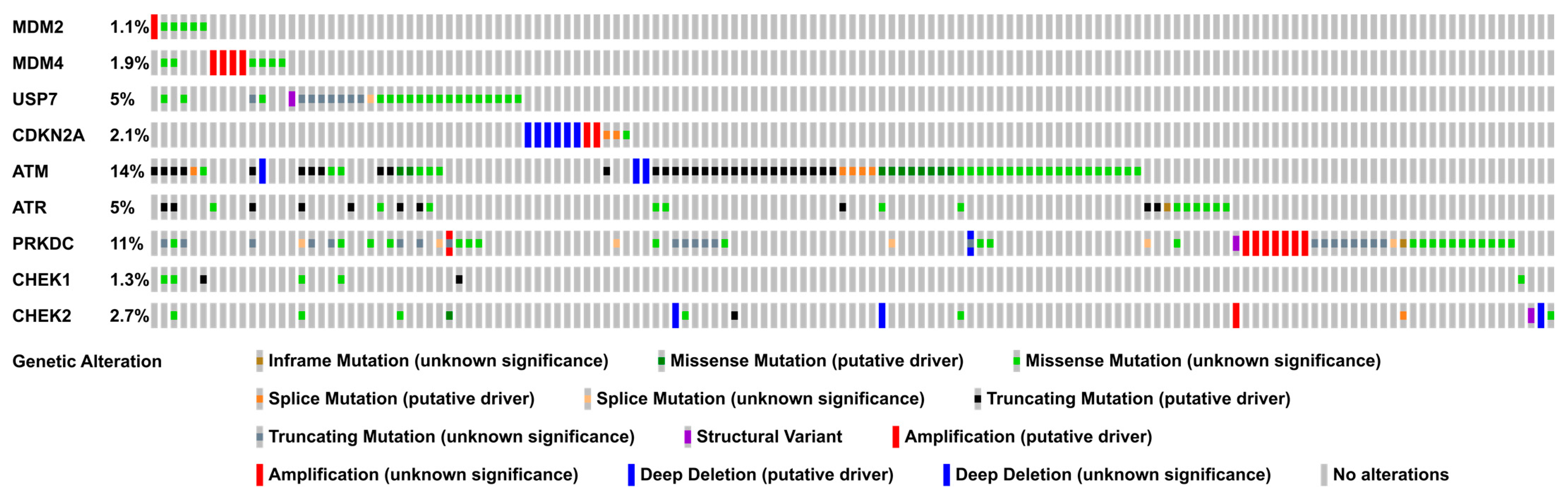
4. Alterations of p53 Regulators in Colorectal Cancer
5. p53 as a Therapeutic Target
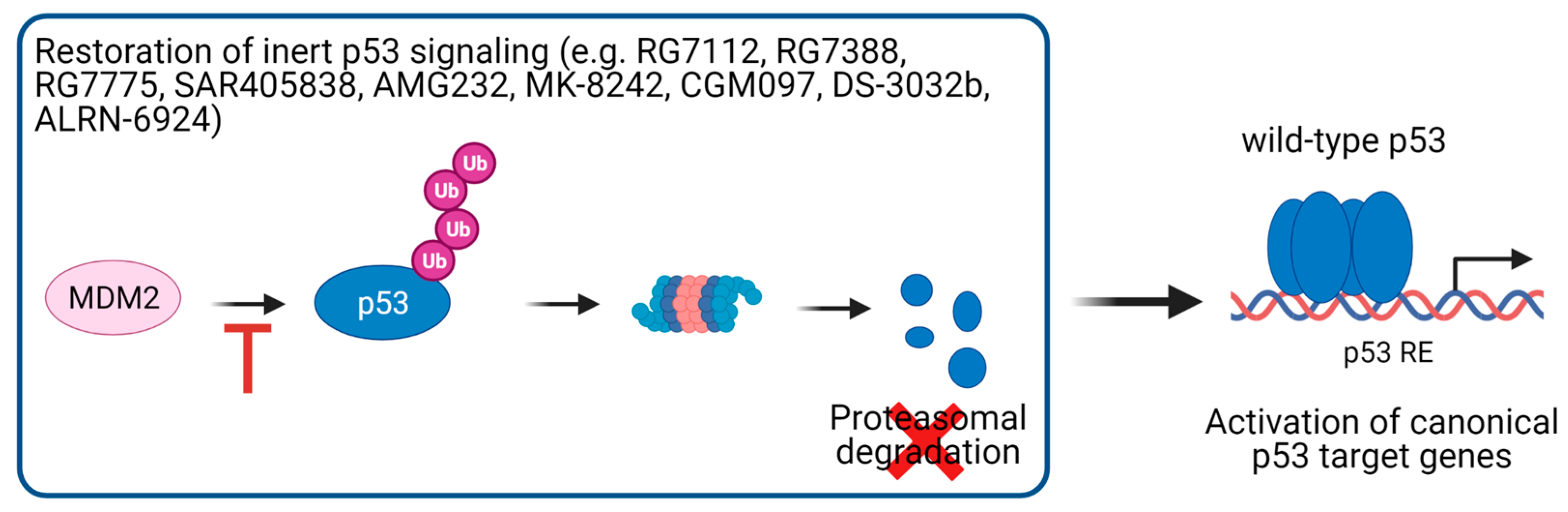
5.1. Activation of p53 Signaling in p53 Wild-Type Cancer Cells
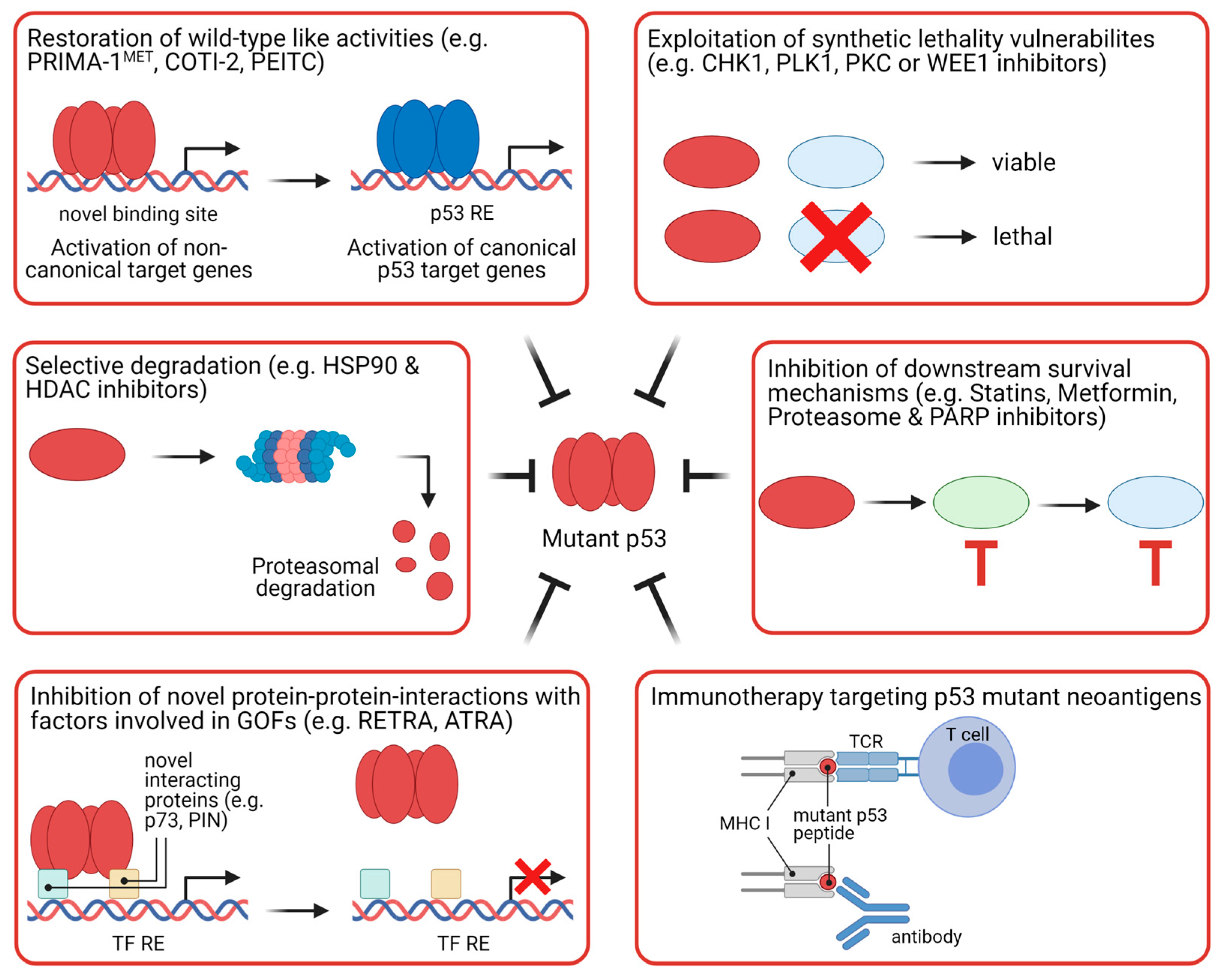
5.2. Strategies to Target Mutant p53
6. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 5-FU | 5-Fluorouracil |
| aa | amino acid |
| ASPP | apoptosis-stimulating of p53 protein |
| CSC | cancer stem cell |
| CIMP | CpG island methylator phenotype |
| CIN | chromosomal instability |
| CMS | Consensus Molecular Subtype |
| COAD | colorectal adenocarcinoma |
| CRC | colorectal cancer |
| CTD | C-terminal domain |
| DBD | DNA-binding domain |
| DN | dominant-negative |
| DNE | dominant-negative effect |
| GOF | gain-of-function |
| HLA | human leukocyte antigen |
| HZF | hematopoietic zinc finger |
| IARC | International Agency for Research on Cancer |
| ICI | immune checkpoint inhibitor |
| IHC | immunohistochemistry |
| lncRNA | long non-coding RNA |
| LOF | loss-of-function |
| MHC I | major histocompatibility complex class I |
| miRNA | microRNA |
| MM | multiple myeloma |
| MSI | microsatellite instability |
| ncRNA | non-coding RNA |
| OD | oligomerization domain |
| PKC | protein kinase C |
| PLK1 | polo-like kinase 1 |
| PRD | proline-rich domain |
| PTM | post-translational modification |
| RE | response element |
| ROS | reactive oxygen species |
| TAD | Transactivation domain |
| TCR | T-cell receptor |
| TF | transcription factor |
References
- Ahmed, M. Colon Cancer: A Clinician’s Perspective in 2019. Gastroenterol. Res. 2020, 13, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mármol, I.; Sánchez-de-Diego, C.; Dieste, A.P.; Cerrada, E.; Yoldi, M.J.R. Colorectal carcinoma: A general overview and future perspectives in colorectal cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Torre, L.A.; Soerjomataram, I.; Hayes, R.B.; Bray, F.; Weber, T.K.; Jemal, A. Global patterns and trends in colorectal cancer incidence in young adults. Gut 2019, 68, 2179–2185. [Google Scholar] [CrossRef] [Green Version]
- Akimoto, N.; Ugai, T.; Zhong, R.; Hamada, T.; Fujiyoshi, K.; Giannakis, M.; Wu, K.; Cao, Y.; Ng, K.; Ogino, S. Rising incidence of early-onset colorectal cancer—A call to action. Nat. Rev. Clin. Oncol. 2020, 1–14. [Google Scholar] [CrossRef]
- Hamilton, S.R.; Aaltonen, L.A. Pathology and Genetics of Tumours of the Digestive System; IARC Press: Lyon, France, 2000. [Google Scholar]
- Remo, A.; Fassan, M.; Vanoli, A.; Bonetti, L.R.; Barresi, V.; Tatangelo, F.; Gafà, R.; Giordano, G.; Pancione, M.; Grillo, F.; et al. Morphology and molecular features of rare colorectal carcinoma histotypes. Cancers 2019, 11, 1036. [Google Scholar] [CrossRef] [Green Version]
- Malki, A.; ElRuz, R.A.; Gupta, I.; Allouch, A.; Vranic, S.; Al Moustafa, A.-E. Molecular Mechanisms of Colon Cancer Progression and Metastasis: Recent Insights and Advancements. Int. J. Mol. Sci. 2020, 22, 130. [Google Scholar] [CrossRef]
- Vousden, K.H.; Lane, D.P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007, 8, 275–283. [Google Scholar] [CrossRef]
- Sabapathy, K.; Lane, D.P. Therapeutic targeting of p53: All mutants are equal, but some mutants are more equal than others. Nat. Rev. Clin. Oncol. 2017, 15, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J.; Oren, M. The first 30 years of p53: Growing ever more complex. Nat. Rev. Cancer 2009, 9, 749–758. [Google Scholar] [CrossRef] [Green Version]
- Joruiz, S.M.; Bourdon, J.C. P53 isoforms: Key regulators of the cell fate decision. Cold Spring Harb. Perspect. Med. 2016, 6. [Google Scholar] [CrossRef] [Green Version]
- Vieler, M.; Sanyal, S. P53 isoforms and their implications in cancer. Cancers 2018, 10, 288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vousden, K.H.; Prives, C. Blinded by the Light: The Growing Complexity of p53. Cell 2009, 137, 413–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laptenko, O.; Shiff, I.; Freed-Pastor, W.; Zupnick, A.; Mattia, M.; Freulich, E.; Shamir, I.; Kadouri, N.; Kahan, T.; Manfredi, J.; et al. The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain. Mol. Cell 2015, 57, 1034–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bode, A.M.; Dong, Z. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer 2004, 4, 793–805. [Google Scholar] [CrossRef] [PubMed]
- Murray-Zmijewski, F.; Slee, E.A.; Lu, X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat. Rev. Mol. Cell Biol. 2008, 9, 702–712. [Google Scholar] [CrossRef] [PubMed]
- Harris, S.L.; Levine, A.J. The p53 pathway: Positive and negative feedback loops. Oncogene 2005, 24, 2899–2908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leng, R.P.; Lin, Y.; Ma, W.; Wu, H.; Lemmers, B.; Chung, S.; Parant, J.M.; Lozano, G.; Hakem, R.; Benchimol, S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 2003, 112, 779–791. [Google Scholar] [CrossRef] [Green Version]
- Dornan, D.; Wertz, I.; Shimizu, H.; Arnott, D.; Frantz, G.D.; Dowd, P.; O’ Rourke, K.; Koeppen, H.; Dixit, V.M. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 2004, 429, 86–92. [Google Scholar] [CrossRef]
- Chen, D.; Kon, N.; Li, M.; Zhang, W.; Qin, J.; Gu, W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 2005, 121, 1071–1083. [Google Scholar] [CrossRef] [Green Version]
- Esser, C.; Scheffner, M.; Höhfeld, J. The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation. J. Biol. Chem. 2005, 280, 27443–27448. [Google Scholar] [CrossRef] [Green Version]
- Meek, D.W. The p53 response to DNA damage. DNA Repair 2004, 3, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Kruse, J.P.; Gu, W. Modes of p53 Regulation. Cell 2009, 137, 609–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayo, L.D.; Turchi, J.J.; Berberich, S.J. Mdm-2 phosphorylation by DNA-dependent protein kinase prevents interaction with p53. Cancer Res. 1997, 57, 5013–5016. [Google Scholar] [PubMed]
- Wade, M.; Li, Y.C.; Wahl, G.M. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 2013, 13, 83–96. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Zhao, W.; Chen, Y.; Zhao, Y.; Gu, W. Acetylation Is Indispensable for p53 Activation. Cell 2008, 133, 612–626. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Guan, D.; Dong, M.; Yang, J.; Wei, H.; Liang, Q.; Song, L.; Xu, L.; Bai, J.; Liu, C.; et al. UFMylation maintains tumour suppressor p53 stability by antagonizing its ubiquitination. Nat. Cell Biol. 2020, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bieging, K.T.; Mello, S.S.; Attardi, L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 2014, 14, 359–370. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Kon, N.; Li, T.; Wang, S.-J.; Su, T.; Hibshoosh, H.; Baer, R.; Gu, W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015, 520, 57–62. [Google Scholar] [CrossRef] [Green Version]
- Ho, T.; Tan, B.X.; Lane, D. How the other half lives: What p53 does when it is not being a transcription factor. Int. J. Mol. Sci. 2020, 21, 13. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, K.D.; Galbraith, M.D.; Andrysik, Z.; Espinosa, J.M. Mechanisms of transcriptional regulation by p53. Cell Death Differ. 2017, 25, 133–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, M. Census and evaluation of p53 target genes. Oncogene 2017, 36, 3943–3956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvajal, L.A.; Manfredi, J.J. Another fork in the road-life or death decisions by the tumour suppressor p53. EMBO Rep. 2013, 14, 414–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafner, A.; Bulyk, M.L.; Jambhekar, A.; Lahav, G. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 2019, 20, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, T.G.; Möller, A.; Sirma, H.; Zentgraf, H.; Taya, Y.; Dröge, W.; Will, H.; Schmitz, M.L. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 2002, 4, 1–10. [Google Scholar] [CrossRef]
- D’Orazi, G.; Cecchinelli, B.; Bruno, T.; Manni, I.; Higashimoto, Y.; Saito, S.; Gostissa, M.; Coen, S.; Marchetti, A.; Del Sal, G.; et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 2002, 4, 11–19. [Google Scholar] [CrossRef]
- Taira, N.; Nihira, K.; Yamaguchi, T.; Miki, Y.; Yoshida, K. DYRK2 Is Targeted to the Nucleus and Controls p53 via Ser46 Phosphorylation in the Apoptotic Response to DNA Damage. Mol. Cell 2007, 25, 725–738. [Google Scholar] [CrossRef]
- Chen, S.; Wu, J.; Zhong, S.; Li, Y.; Zhang, P.; Ma, J.; Ren, J.; Tan, Y.; Wang, Y.; Au, K.F.; et al. IASPP mediates p53 selectivity through a modular mechanism fine-tuning DNA recognition. Proc. Natl. Acad. Sci. USA 2019, 116, 17470–17479. [Google Scholar] [CrossRef] [Green Version]
- Liebl, M.C.; Hofmann, T.G. Cell Fate Regulation upon DNA Damage: p53 Serine 46 Kinases Pave the Cell Death Road. BioEssays 2019, 41, 1900127. [Google Scholar] [CrossRef] [Green Version]
- Wulf, G.M.; Liou, Y.-C.; Ryo, A.; Lee, S.W.; Lu, K.P. Role of Pin1 in the regulation of p53 stability and p21 transactivation, and cell cycle checkpoints in response to DNA damage. J. Biol. Chem. 2002, 277, 47976–47979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zacchi, P.; Gostissa, M.; Uchida, T.; Salvagno, C.; Avolio, F.; Volinia, S.; Ronai, Z.; Blandino, G.; Schneider, C.; Sal, G. Del The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature 2002, 419, 853–857. [Google Scholar] [CrossRef]
- Zheng, H.; You, H.; Zhou, X.Z.; Murray, S.A.; Uchida, T.; Wulf, G.; Gu, L.; Tang, X.; Lu, K.P.; Xiao, Z.-X.J. The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature 2002, 419, 849–853. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, F.; Tocco, F.; Girardini, J.; Smith, P.; Gasco, M.; Lu, X.; Crook, T.; Del Sal, G. The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nat. Struct. Mol. Biol. 2007, 14, 912–920. [Google Scholar] [CrossRef]
- Follis, A.V.; Llambi, F.; Merritt, P.; Chipuk, J.E.; Green, D.R.; Kriwacki, R.W. Pin1-Induced Proline Isomerization in Cytosolic p53 Mediates BAX Activation and Apoptosis. Mol. Cell 2015, 59, 677–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sykes, S.M.; Mellert, H.S.; Holbert, M.A.; Li, K.; Marmorstein, R.; Lane, W.S.; McMahon, S.B. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 2006, 24, 841–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.; Luo, J.; Zhang, W.; Gu, W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 2006, 24, 827–839. [Google Scholar] [CrossRef]
- Knights, C.D.; Catania, J.; Di Giovanni, S.; Muratoglu, S.; Perez, R.; Swartzbeck, A.; Quong, A.A.; Zhang, X.; Beerman, T.; Pestell, R.G.; et al. Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J. Cell Biol. 2006, 173, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Samuels-Lev, Y.; O’Connor, D.J.; Bergamaschi, D.; Trigiante, G.; Hsieh, J.K.; Zhong, S.; Campargue, I.; Naumovski, L.; Crook, T.; Lu, X. ASPP proteins specifically stimulate the apoptotic function of p53. Mol. Cell 2001, 8, 781–794. [Google Scholar] [CrossRef]
- Bergamaschi, D.; Samuels, Y.; O’Neil, N.J.; Trigiante, G.; Crook, T.; Hsieh, J.-K.; O’Connor, D.J.; Zhong, S.; Campargue, I.; Tomlinson, M.L.; et al. iASPP oncoprotein is a key inhibitor of p53 conserved from worm to human. Nat. Genet. 2003, 33, 162–167. [Google Scholar] [CrossRef]
- Das, S.; Raj, L.; Zhao, B.; Kimura, Y.; Bernstein, A.; Aaronson, S.A.; Lee, S.W. Hzf Determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell 2007, 130, 624–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liebl, M.C.; Moehlenbrink, J.; Becker, H.; Raddatz, G.; Abdeen, S.K.; Aqeilan, R.I.; Lyko, F.; Hofmann, T.G. DAZAP2 acts as specifier of the p53 response to DNA damage. Nucleic Acids Res. 2021, 49, 2759–2776. [Google Scholar] [CrossRef] [PubMed]
- Paek, A.L.; Liu, J.C.; Loewer, A.; Forrester, W.C.; Lahav, G. Cell-to-Cell Variation in p53 Dynamics Leads to Fractional Killing. Cell 2016, 165, 631–642. [Google Scholar] [CrossRef] [Green Version]
- Purvis, J.E.; Karhohs, K.W.; Mock, C.; Batchelor, E.; Loewer, A.; Lahav, G. p53 dynamics control cell fate. Science 2012, 336, 1440–1444. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Chen, J.; Gan, S.; Guan, H.; Zhou, Y.; Ouyang, Q.; Shi, J. DNA damage strength modulates a bimodal switch of p53 dynamics for cell-fate control. BMC Biol. 2013, 11, 73. [Google Scholar] [CrossRef] [Green Version]
- Tsabar, M.; Mock, C.S.; Venkatachalam, V.; Reyes, J.; Karhohs, K.W.; Oliver, T.G.; Regev, A.; Jambhekar, A.; Lahav, G. A Switch in p53 Dynamics Marks Cells That Escape from DSB-Induced Cell Cycle Arrest. Cell Rep. 2020, 33, 108392. [Google Scholar] [CrossRef]
- Porter, J.R.; Fisher, B.E.; Batchelor, E. p53 Pulses Diversify Target Gene Expression Dynamics in an mRNA Half-Life-Dependent Manner and Delineate Co-regulated Target Gene Subnetworks. Cell Syst. 2016, 2, 272–282. [Google Scholar] [CrossRef] [Green Version]
- Hafner, A.; Stewart-Ornstein, J.; Purvis, J.E.; Forrester, W.C.; Bulyk, M.L.; Lahav, G. P53 pulses lead to distinct patterns of gene expression albeit similar DNA-binding dynamics. Nat. Struct. Mol. Biol. 2017, 24, 840–847. [Google Scholar] [CrossRef]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef] [Green Version]
- Baugh, E.H.; Ke, H.; Levine, A.J.; Bonneau, R.A.; Chan, C.S. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 2018, 25, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.A.; Vousden, K.H. p53 mutations in cancer. Nat. Cell Biol. 2013, 15, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Curtin, N.J. DNA repair dysregulation from cancer driver to therapeutic target. Nat. Rev. Cancer 2012, 12, 801–817. [Google Scholar] [CrossRef]
- Basak, D.; Uddin, M.N.; Hancock, J. The Role of Oxidative Stress and Its Counteractive Utility in Colorectal Cancer (CRC). Cancers 2020, 12, 3336. [Google Scholar] [CrossRef]
- Donehower, L.A.; Soussi, T.; Korkut, A.; Liu, Y.; Schultz, A.; Cardenas, M.; Li, X.; Babur, O.; Hsu, T.-K.; Lichtarge, O.; et al. Integrated Analysis of TP53 Gene and Pathway Alterations in The Cancer Genome Atlas. Cell Rep. 2019, 28, 1370–1384.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frum, R.A.; Grossman, S.R. Mechanisms of mutant p53 stabilization in cancer. Subcell. Biochem. 2014, 85, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Lieu, C.H.; Golemis, E.A.; Serebriiskii, I.G.; Newberg, J.; Hemmerich, A.; Connelly, C.; Messersmith, W.A.; Eng, C.; Gail Eckhardt, S.; Frampton, G.; et al. Comprehensive genomic landscapes in early and later onset colorectal cancer. Clin. Cancer Res. 2019, 25, 5852–5858. [Google Scholar] [CrossRef] [Green Version]
- Russo, A.; Bazan, V.; Iacopetta, B.; Kerr, D.; Soussi, T.; Gebbia, N. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: Influence of tumor site, type of mutation, and adjuvant treatment. J. Clin. Oncol. 2005, 23, 7518–7528. [Google Scholar] [CrossRef]
- Kadosh, E.; Snir-Alkalay, I.; Venkatachalam, A.; May, S.; Lasry, A.; Elyada, E.; Zinger, A.; Shaham, M.; Vaalani, G.; Mernberger, M.; et al. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature 2020, 586, 133–138. [Google Scholar] [CrossRef]
- Muzny, D.M.; Bainbridge, M.N.; Chang, K.; Dinh, H.H.; Drummond, J.A.; Fowler, G.; Kovar, C.L.; Lewis, L.R.; Morgan, M.B.; Newsham, I.F.; et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [Green Version]
- Zaidi, S.H.; Harrison, T.A.; Phipps, A.I.; Steinfelder, R.; Trinh, Q.M.; Qu, C.; Banbury, B.L.; Georgeson, P.; Grasso, C.S.; Giannakis, M.; et al. Landscape of somatic single nucleotide variants and indels in colorectal cancer and impact on survival. Nat. Commun. 2020, 11, 3644. [Google Scholar] [CrossRef]
- Grady, W.M.; Carethers, J.M. Genomic and Epigenetic Instability in Colorectal Cancer Pathogenesis. Gastroenterology 2008, 135, 1079–1099. [Google Scholar] [CrossRef] [Green Version]
- Keum, N.N.; Giovannucci, E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Rajagopalan, H.; Nowak, M.A.; Vogelstein, B.; Lengauer, C. The significance of unstable chromosomes in colorectal cancer. Nat. Rev. Cancer 2003, 3, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Guinney, J.; Dienstmann, R.; Wang, X.; De Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Giacomelli, A.O.; Yang, X.; Lintner, R.E.; McFarland, J.M.; Duby, M.; Kim, J.; Howard, T.P.; Takeda, D.Y.; Ly, S.H.; Kim, E.; et al. Mutational processes shape the landscape of TP53 mutations in human cancer. Nat. Genet. 2018, 50, 1381–1387. [Google Scholar] [CrossRef] [PubMed]
- Boettcher, S.; Miller, P.G.; Sharma, R.; McConkey, M.; Leventhal, M.; Krivtsov, A.V.; Giacomelli, A.O.; Wong, W.; Kim, J.; Chao, S.; et al. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science 2019, 365, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Bougeard, G.; Sesboüé, R.; Baert-Desurmont, S.; Vasseur, S.; Martin, C.; Tinat, J.; Brugières, L.; Chompret, A.; Bressac-de Paillerets, B.; Stoppa-Lyonnet, D.; et al. Molecular basis of the Li-Fraumeni syndrome: An update from the French LFS families. J. Med. Genet. 2008, 45, 535–538. [Google Scholar] [CrossRef]
- Xu, J.; Qian, J.; Hu, Y.; Wang, J.; Zhou, X.; Chen, H.; Fang, J.Y. Heterogeneity of Li-Fraumeni Syndrome links to unequal gain-of-function effects of p53 mutations. Sci. Rep. 2014, 4, 4223. [Google Scholar] [CrossRef] [Green Version]
- Alexandrova, E.M.; Yallowitz, A.R.; Li, D.; Xu, S.; Schulz, R.; Proia, D.A.; Lozano, G.; Dobbelstein, M.; Moll, U.M. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature 2015, 523, 352–356. [Google Scholar] [CrossRef] [Green Version]
- Olive, K.P.; Tuveson, D.A.; Ruhe, Z.C.; Yin, B.; Willis, N.A.; Bronson, R.T.; Crowley, D.; Jacks, T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 2004, 119, 847–860. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.; Feng, Y.; Kuick, R.; Green, M.; Green, M.; Sakamoto, N.; Kurosu, Y.; Lin, J.; Cho, K.R.; Fearon, E.R. Trp53 null and R270H mutant alleles have comparable effects in regulating invasion, metastasis, and gene expression in mouse colon tumorigenesis. Lab. Investig. 2019, 99, 1454–1469. [Google Scholar] [CrossRef] [PubMed]
- Pfister, N.T.; Prives, C. Transcriptional regulation by wild-type and cancer-related mutant forms of p53. Cold Spring Harb. Perspect. Med. 2017, 7, a026054. [Google Scholar] [CrossRef] [Green Version]
- Schulz-Heddergott, R.; Stark, N.; Edmunds, S.J.; Li, J.; Conradi, L.C.; Bohnenberger, H.; Ceteci, F.; Greten, F.R.; Dobbelstein, M.; Moll, U.M. Therapeutic Ablation of Gain-of-Function Mutant p53 in Colorectal Cancer Inhibits Stat3-Mediated Tumor Growth and Invasion. Cancer Cell 2018, 34, 298–314.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Agostino, S.; Strano, S.; Emiliozzi, V.; Zerbini, V.; Mottolese, M.; Sacchi, A.; Blandino, G.; Piaggio, G. Gain of function of mutant p53: The mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell 2006, 10, 191–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, K.; Ling, S.; Lin, W.-C. TopBP1 Mediates Mutant p53 Gain of Function through NF-Y and p63/p73. Mol. Cell. Biol. 2011, 31, 4464–4481. [Google Scholar] [CrossRef] [Green Version]
- Di Agostino, S.; Sorrentino, G.; Ingallina, E.; Valenti, F.; Ferraiuolo, M.; Bicciato, S.; Piazza, S.; Strano, S.; Del Sal, G.; Blandino, G. YAP enhances the pro-proliferative transcriptional activity of mutant p53 proteins. EMBO Rep. 2016, 17, 188–201. [Google Scholar] [CrossRef]
- Alam, S.K.; Yadav, V.K.; Bajaj, S.; Datta, A.; Dutta, S.K.; Bhattacharyya, M.; Bhattacharya, S.; Debnath, S.; Roy, S.; Boardman, L.A.; et al. DNA damage-induced ephrin-B2 reverse signaling promotes chemoresistance and drives EMT in colorectal carcinoma harboring mutant p53. Cell Death Differ. 2016, 23, 707–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooks, T.; Pateras, I.S.; Tarcic, O.; Solomon, H.; Schetter, A.J.; Wilder, S.; Lozano, G.; Pikarsky, E.; Forshew, T.; Rozenfeld, N.; et al. Mutant p53 Prolongs NF-κB Activation and Promotes Chronic Inflammation and Inflammation-Associated Colorectal Cancer. Cancer Cell 2013, 23, 634–646. [Google Scholar] [CrossRef] [Green Version]
- Gaiddon, C.; Lokshin, M.; Ahn, J.; Zhang, T.; Prives, C. A Subset of Tumor-Derived Mutant Forms of p53 Down-Regulate p63 and p73 through a Direct Interaction with the p53 Core Domain. Mol. Cell. Biol. 2001, 21, 1874–1887. [Google Scholar] [CrossRef] [Green Version]
- Adorno, M.; Cordenonsi, M.; Montagner, M.; Dupont, S.; Wong, C.; Hann, B.; Solari, A.; Bobisse, S.; Rondina, M.B.; Guzzardo, V.; et al. A Mutant-p53/Smad Complex Opposes p63 to Empower TGFβ-Induced Metastasis. Cell 2009, 137, 87–98. [Google Scholar] [CrossRef]
- Fontemaggi, G.; Dell’Orso, S.; Trisciuoglio, D.; Shay, T.; Melucci, E.; Fazi, F.; Terrenato, I.; Mottolese, M.; Muti, P.; Domany, E.; et al. The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis. Nat. Struct. Mol. Biol. 2009, 16, 1086–1093. [Google Scholar] [CrossRef]
- Do, P.M.; Varanasi, L.; Fan, S.; Li, C.; Kubacka, I.; Newman, V.; Chauhan, K.; Daniels, S.R.; Boccetta, M.; Garrett, M.R.; et al. Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev. 2012, 26, 830–845. [Google Scholar] [CrossRef] [Green Version]
- Ji, L.; Xu, J.; Liu, J.; Amjad, A.; Zhang, K.; Liu, Q.; Zhou, L.; Xiao, J.; Li, X. Mutant p53 promotes tumor cell malignancy by both positive and negative regulation of the transforming growth factor β (TGF-β) pathway. J. Biol. Chem. 2015, 290, 11729–11740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amelio, I.; Mancini, M.; Petrova, V.; Cairns, R.A.; Vikhreva, P.; Nicolai, S.; Marini, A.; Antonov, A.A.; Le Quesne, J.; Baena Acevedo, J.D.; et al. p53 mutants cooperate with HIF-1 in transcriptional regulation of extracellular matrix components to promote tumor progression. Proc. Natl. Acad. Sci. USA 2018, 115, E10869–E10878. [Google Scholar] [CrossRef] [Green Version]
- Rahnamoun, H.; Hong, J.; Sun, Z.; Lee, J.; Lu, H.; Lauberth, S.M. Mutant p53 regulates enhancer-associated H3K4 monomethylation through interactions with the methyltransferase MLL4. J. Biol. Chem. 2018, 293, 13234–13246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfister, N.T.; Fomin, V.; Regunath, K.; Zhou, J.Y.; Zhou, W.; Silwal-Pandit, L.; Freed-Pastor, W.A.; Laptenko, O.; Neo, S.P.; Bargonetti, J.; et al. Mutant p53 cooperates with the SWI/SNF chromatin remodeling complex to regulate VEGFR2 in breast cancer cells. Genes Dev. 2015, 29, 1298–1315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Zhang, C.; Yue, X.; Li, X.; Liu, J.; Yu, H.; Belyi, V.A.; Yang, Q.; Feng, Z.; Hu, W. Pontin, a new mutant p53-binding protein, promotes gain-of-function of mutant p53. Cell Death Differ. 2015, 22, 1824–1836. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Sammons, M.A.; Donahue, G.; Dou, Z.; Vedadi, M.; Getlik, M.; Barsyte-Lovejoy, D.; Al-awar, R.; Katona, B.W.; Shilatifard, A.; et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature 2015, 525, 206–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solomon, H.; Dinowitz, N.; Pateras, I.S.; Cooks, T.; Shetzer, Y.; Molchadsky, A.; Charni, M.; Rabani, S.; Koifman, G.; Tarcic, O.; et al. Mutant p53 gain of function underlies high expression levels of colorectal cancer stem cells markers. Oncogene 2018, 37, 1669–1684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walerych, D.; Lisek, K.; Sommaggio, R.; Piazza, S.; Ciani, Y.; Dalla, E.; Rajkowska, K.; Gaweda-Walerych, K.; Ingallina, E.; Tonelli, C.; et al. Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer. Nat. Cell Biol. 2016, 18, 897–909. [Google Scholar] [CrossRef]
- Weisz, L.; Oren, M.; Rotter, V. Transcription regulation by mutant p53. Oncogene 2007, 26, 2202–2211. [Google Scholar] [CrossRef] [Green Version]
- Di Agostino, S. The impact of mutant p53 in the non-coding RNA world. Biomolecules 2020, 10, 472. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Li, Y.; Sheng, J.; Wu, F.; Li, K.; Huang, R.; Wang, X.; Jiao, T.; Guan, X.; Lu, Y.; et al. P53-R273H mutation enhances colorectal cancer stemness through regulating specific lncRNAs. J. Exp. Clin. Cancer Res. 2019, 38, 379. [Google Scholar] [CrossRef] [Green Version]
- Masciarelli, S.; Fontemaggi, G.; Di Agostino, S.; Donzelli, S.; Carcarino, E.; Strano, S.; Blandino, G. Gain-of-function mutant p53 downregulates miR-223 contributing to chemoresistance of cultured tumor cells. Oncogene 2014, 33, 1601–1608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Wang, C.; Ma, B.; Xu, M.; Xu, S.; Liu, J.; Tian, Y.; Fu, Y.; Luo, Y. Mutant p53 Drives Cancer Metastasis via RCP-Mediated Hsp90α Secretion. Cell Rep. 2020, 32, 107879. [Google Scholar] [CrossRef] [PubMed]
- Cooks, T.; Pateras, I.S.; Jenkins, L.M.; Patel, K.M.; Robles, A.I.; Morris, J.; Forshew, T.; Appella, E.; Gorgoulis, V.G.; Harris, C.C. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat. Commun. 2018, 9, 771. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Li, M.; Wang, X. Cancer type-dependent correlations between TP53 mutations and antitumor immunity. DNA Repair 2020, 88, 102785. [Google Scholar] [CrossRef]
- Hientz, K.; Mohr, A.; Bhakta-Guha, D.; Efferth, T. The role of p53 in cancer drug resistance and targeted chemotherapy. Oncotarget 2017, 8, 8921–8946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elsaleh, H.; Powell, B.; Mccaul, K.; Grieu, F.; Grant, R.; Joseph, D.; Iacopetta, B. P53 Alteration and Microsatellite Instability Have Predictive Value for Survival Benefit from Chemotherapy in Stage III Colorectal Carcinoma 1. Clin. Cancer Res. 2001, 7, 1343–1349. [Google Scholar] [PubMed]
- Bunz, F.; Hwang, P.M.; Torrance, C.; Waldman, T.; Zhang, Y.; Dillehay, L.; Williams, J.; Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J. Clin. Investig. 1999, 104, 263–269. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Liu, N.; Liu, J.; Liu, Y.; Zhang, C.; Long, S.; Luo, G.; Zhang, L.; Zhang, Y. Mutant p53 drives cancer chemotherapy resistance due to loss of function on activating transcription of PUMA. Cell Cycle 2019, 18, 3442–3455. [Google Scholar] [CrossRef]
- Wang, X.; Sun, Q. TP53 mutations, expression and interaction networks in human cancers. Oncotarget 2017, 8, 624–643. [Google Scholar] [CrossRef] [Green Version]
- Chun, Y.S.; Passot, G.; Yamashita, S.; Nusrat, M.; Katsonis, P.; Loree, J.M.; Conrad, C.; Tzeng, C.W.D.; Xiao, L.; Aloia, T.A.; et al. Deleterious Effect of RAS and Evolutionary High-risk TP53 Double Mutation in Colorectal Liver Metastases. Ann. Surg. 2019, 269, 917–923. [Google Scholar] [CrossRef] [PubMed]
- Sclafani, F.; Wilson, S.H.; Cunningham, D.; Gonzalez De Castro, D.; Kalaitzaki, E.; Begum, R.; Wotherspoon, A.; Capdevila, J.; Glimelius, B.; Roselló, S.; et al. Analysis of KRAS, NRAS, BRAF, PIK3CA and TP53 mutations in a large prospective series of locally advanced rectal cancer patients. Int. J. Cancer 2020, 146, 94–102. [Google Scholar] [CrossRef]
- Forslund, A.; Zeng, Z.; Qin, L.X.; Rosenberg, S.; Ndubuisi, M.; Pincas, H.; Gerald, W.; Notterman, D.A.; Barany, F.; Paty, P.B. MDM2 gene amplification is correlated to tumor progression but not to the presence of SNP309 or TP53 mutational status in primary colorectal cancers. Mol. Cancer Res. 2008, 6, 205–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilkes, D.M.; Pan, Y.; Coppola, D.; Yeatman, T.; Reuther, G.W.; Chen, J. Regulation of MDMX Expression by Mitogenic Signaling. Mol. Cell. Biol. 2008, 28, 1999–2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Chen, D.; Shiloh, A.; Luo, J.; Nikolaev, A.Y.; Qin, J.; Gu, W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 2002, 416, 648–653. [Google Scholar] [CrossRef]
- Cummins, J.M.; Rago, C.; Kohli, M.; Kinzler, K.W.; Lengauer, C.; Vogelstein, B. Disruption of HAUSP gene stabilizes p53. Nature 2004, 428, 1–2. [Google Scholar] [CrossRef]
- Li, M.; Brooks, C.L.; Kon, N.; Gu, W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell 2004, 13, 879–886. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Tan, C.; Yue, X.; Zhao, Y.; Peng, J.; Wang, X.; Laddha, S.V.; Chan, C.S.; Zheng, S.; et al. microRNA-1827 represses MDM2 to positively regulate tumor suppressor p53 and suppress tumorigenesis. Oncotarget 2016, 7, 8783–8796. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Cai, G.; Liao, Z.; Lin, K.; Li, G.; Li, Y. miRNA-766 induces apoptosis of human colon cancer cells through the p53/Bax signaling pathway by MDM4. Exp. Ther. Med. 2019, 17, 4100–4108. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.J.; Lee, J.H.; Jin, S.; Kim, J.H.; Kim, S.H. Primate-specific miR-944 activates p53-dependent tumor suppression in human colorectal cancers. Cancer Lett. 2019, 440–441, 168–179. [Google Scholar] [CrossRef]
- Deng, X.; Li, S.; Kong, F.; Ruan, H.; Xu, X.; Zhang, X.; Wu, Z.; Zhang, L.; Xu, Y.; Yuan, H.; et al. Long noncoding RNA PiHL regulates p53 protein stability through GRWD1/RPL11/MDM2 axis in colorectal cancer. Theranostics 2020, 10, 65–280. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, Y.; Kong, P.; Han, M.; Li, B. Expression of circZNF609 is down-regulated in colorectal cancer tissue and promotes apoptosis in colorectal cancer cells by upregulating p53. Med. Sci. Monit. 2019, 25, 5977–5985. [Google Scholar] [CrossRef]
- Wei, L.J.; Bai, D.M.; Wang, Z.Y.; Liu, B.C. Upregulated lncRNA CACNA1G-AS1 aggravates the progression of colorectal cancer by downregulating p53. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 130–136. [Google Scholar] [CrossRef]
- Chandrasekaran, K.S.; Sathyanarayanan, A.; Karunagaran, D. MicroRNA-214 suppresses growth, migration and invasion through a novel target, high mobility group AT-hook 1, in human cervical and colorectal cancer cells. Br. J. Cancer 2016, 115, 741–751. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekaran, K.S.; Sathyanarayanan, A.; Karunagaran, D. miR-214 activates TP53 but suppresses the expression of RELA, CTNNB1, and STAT3 in human cervical and colorectal cancer cells. Cell Biochem. Funct. 2017, 35, 464–471. [Google Scholar] [CrossRef]
- Dong, Y.X.; Pang, Z.G.; Zhang, J.C.; Hu, J.Q.; Wang, L.Y. Long non-coding RNA GClnc1 promotes progression of colorectal cancer by inhibiting p53 signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5705–5713. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bykov, V.J.N.; Eriksson, S.E.; Bianchi, J.; Wiman, K.G. Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer 2017, 18, 89–102. [Google Scholar] [CrossRef]
- Cheok, C.F.; Lane, D.P. Exploiting the p53 pathway for therapy. Cold Spring Harb. Perspect. Med. 2017, 7, a026310. [Google Scholar] [CrossRef]
- Li, H.; Zhang, J.; Tong, J.H.M.; Chan, A.W.H.; Yu, J.; Kang, W.; To, K.F. Targeting the Oncogenic p53 Mutants in Colorectal Cancer and Other Solid Tumors. Int. J. Mol. Sci. 2019, 20, 5999. [Google Scholar] [CrossRef] [Green Version]
- Duffy, M.J.; Synnott, N.C.; O’Grady, S.; Crown, J. Targeting p53 for the treatment of cancer. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef]
- Vassilev, L.T.; Vu, B.T.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In Vivo Activation of the p53 Pathway by Small-Molecule Antagonists of MDM2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef] [Green Version]
- A Study of RO5045337 [RG7112] in Patients with Advanced Solid Tumors. Available online: https://www.clinicaltrials.gov/ct2/show/NCT00559533 (accessed on 17 March 2021).
- A Study to Determine the Excretion Balance, Pharmacokinetics, Metabolism and Absolute Oral Bioavailability of a Single Oral Dose of [14C]-Labeled Idasanutlin and an Intravenous Tracer Dose of [13C]-Labeled Idasanutlin in a Single Cohort of Participants With Solid Tumors (Malignancies). Available online: https://clinicaltrials.gov/ct2/show/NCT02828930 (accessed on 17 March 2021).
- A Study to Investigate the Bioequivalence or Relative Bioavailability of Three New Idasanutlin Tablet Variants Following Oral Administration in Participants With Solid Tumors. Available online: https://clinicaltrials.gov/ct2/show/NCT03362723 (accessed on 17 March 2021).
- A Study Evaluating the Safety, Tolerability, Pharmacokinetics, and Preliminary Activity of Idasanutlin in Combination With Either Chemotherapy or Venetoclax in the Treatment of Pediatric and Young Adult Participants With Relapsed/Refractory Acute Leukemias or Solid Tumors. Available online: https://clinicaltrials.gov/ct2/show/NCT04029688 (accessed on 17 March 2021).
- Tumor-Agnostic Precision Immuno-Oncology and Somatic Targeting Rational for You (TAPISTRY) Platform Study. Available online: https://clinicaltrials.gov/ct2/show/NCT04589845 (accessed on 17 March 2021).
- A Study of the Safety and Pharmacokinetics of RO6839921, an MDM2 Antagonist, in Patients with Advanced Cancers, Including Acute Myeloid Leukemia. Available online: https://clinicaltrials.gov/ct2/show/NCT02098967 (accessed on 17 March 2021).
- Phase 1 Safety Testing of SAR405838. Available online: https://clinicaltrials.gov/ct2/show/NCT01636479 (accessed on 17 March 2021).
- A Safety and Efficacy Study of SAR405838 and Pimasertib in Cancer Patients. Available online: https://clinicaltrials.gov/ct2/show/NCT01985191 (accessed on 17 March 2021).
- A Phase 1 Study Evaluating AMG 232 in Advanced Solid Tumors or Multiple Myeloma. Available online: https://clinicaltrials.gov/ct2/show/NCT01723020 (accessed on 17 March 2021).
- Study of Safety and Pharmacokinetics of MK-8242 in Participants with Advanced Solid Tumors (P07650). Available online: https://clinicaltrials.gov/ct2/show/NCT01463696 (accessed on 17 March 2021).
- A Phase I Dose Escalation Study of CGM097 in Adult Patients With Selected Advanced Solid Tumors. Available online: https://clinicaltrials.gov/ct2/show/NCT01760525 (accessed on 17 March 2021).
- APG-115 in Patients with Advanced Solid Tumors or Lymphomas. Available online: https://clinicaltrials.gov/ct2/show/NCT02935907 (accessed on 20 April 2021).
- A Study of APG-115 in Combination with Pembrolizumab in Patients with Metastatic Melanomas or Advanced Solid Tumors. Available online: https://www.clinicaltrials.gov/ct2/show/NCT03611868 (accessed on 20 April 2021).
- APG-115 in Combination with PD-1 Inhibitor in Patients with Advanced Liposarcoma or Advanced Solid Tumors. Available online: https://clinicaltrials.gov/ct2/show/NCT04785196 (accessed on 20 April 2021).
- This Study Aims to Find the Best Dose of BI 907828 in Patients with Different Types of Advanced Cancer (Solid Tumors). Available online: https://clinicaltrials.gov/ct2/show/NCT03449381 (accessed on 20 April 2021).
- A Study in Patients with Different Types of Advanced Cancer (Solid Tumors) to Test Different Doses of BI 907828 in Combination with BI 754091 (Ezabenlimab) and 754111 or BI 907828 in Combination with BI 754091 (Ezabenlimab). Available online: https://clinicaltrials.gov/ct2/show/NCT03964233 (accessed on 20 April 2021).
- Study to Determine and Evaluate a Safe and Tolerated Dose of HDM201 in Patients with Selected Advanced Tumors That Are TP53wt. Available online: https://clinicaltrials.gov/ct2/show/NCT02143635 (accessed on 20 April 2021).
- A Study of PDR001 in Combination with LCL161, Everolimus or Panobinostat. Available online: https://www.clinicaltrials.gov/ct2/show/NCT02890069 (accessed on 20 April 2021).
- Trametinib + HDM201 in CRC Patients with RAS/RAF Mutant and TP53 Wild-type Advanced/Metastatic Colorectal Cancer Mutant and TP53 Wild-type. Available online: https://clinicaltrials.gov/ct2/show/NCT03714958 (accessed on 20 April 2021).
- A Study Evaluating the Activity of Anti-cancer Treatments Targeting Tumor Molecular Alterations/Characteristics in Advanced/Metastatic Tumors. Available online: https://clinicaltrials.gov/ct2/show/NCT04116541 (accessed on 20 April 2021).
- A Phase 1 Multiple Ascending Dose Study of Milademetan in Subjects with Advanced Solid Tumors or Lymphomas. Available online: https://clinicaltrials.gov/ct2/show/NCT01877382 (accessed on 17 March 2021).
- ALRN-6924 and Paclitaxel in Treating Patients with Advanced, Metastatic, or Unresectable Solid Tumors. Available online: https://clinicaltrials.gov/ct2/show/NCT03725436 (accessed on 17 March 2021).
- Phase 1 Study of the Dual MDM2/MDMX Inhibitor ALRN-6924 in Pediatric Cancer. Available online: https://clinicaltrials.gov/ct2/show/NCT03654716 (accessed on 17 March 2021).
- ALRN-6924 in Patients with Advanced Solid Tumors or Lymphomas. Available online: https://clinicaltrials.gov/ct2/show/NCT02264613 (accessed on 17 March 2021).
- Jeay, S.; Gaulis, S.; Ferretti, S.; Bitter, H.; Ito, M.; Valat, T.; Murakami, M.; Ruetz, S.; Guthy, D.A.; Rynn, C.; et al. A distinct p53 target gene set predicts for response to the selective p53- HDM2 inhibitor NVP-CGM097. Elife 2015, 4. [Google Scholar] [CrossRef]
- Sonkin, D. Expression signature based on TP53 target genes doesn’t predict response to TP53-MDM2 inhibitor in wild type TP53 tumors. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Chen, G.; Jukofsky, L.; Geho, D.; Han, S.W.; Birzele, F.; Bader, S.; Himmelein, L.; Cai, J.; Albertyn, Z.; et al. MDM2 antagonist clinical response association with a gene expression signature in acute myeloid leukaemia. Br. J. Haematol. 2015, 171, 432–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishizawa, J.; Nakamaru, K.; Seki, T.; Tazaki, K.; Kojima, K.; Chachad, D.; Zhao, R.; Heese, L.; Ma, W.; Ma, M.C.J.; et al. Predictive gene signatures determine tumor sensitivity to MDM2 inhibition. Cancer Res. 2018, 78, 2721–2731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Webster, M.R.; Fane, M.E.; Alicea, G.M.; Basu, S.; Kossenkov, A.V.; Marino, G.E.; Douglass, S.M.; Kaur, A.; Ecker, B.L.; Gnanapradeepan, K.; et al. Paradoxical Role for Wild-Type p53 in Driving Therapy Resistance in Melanoma. Mol. Cell 2020, 77, 633–644.e5. [Google Scholar] [CrossRef]
- Soussi, T.; Wiman, K.G. Shaping Genetic Alterations in Human Cancer: The p53 Mutation Paradigm. Cancer Cell 2007, 12, 303–312. [Google Scholar] [CrossRef] [Green Version]
- Martins, C.P.; Brown-Swigart, L.; Evan, G.I. Modeling the Therapeutic Efficacy of p53 Restoration in Tumors. Cell 2006, 127, 1323–1334. [Google Scholar] [CrossRef] [Green Version]
- Ventura, A.; Kirsch, D.G.; McLaughlin, M.E.; Tuveson, D.A.; Grimm, J.; Lintault, L.; Newman, J.; Reczek, E.E.; Weissleder, R.; Jacks, T. Restoration of p53 function leads to tumour regression in vivo. Nature 2007, 445, 661–665. [Google Scholar] [CrossRef]
- Xue, W.; Zender, L.; Miething, C.; Dickins, R.A.; Hernando, E.; Krizhanovsky, V.; Cordon-Cardo, C.; Lowe, S.W. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007, 445, 656–660. [Google Scholar] [CrossRef] [Green Version]
- Phase 1/2 Study of APR-246 in Combination with Pembrolizumab in Subjects With Solid Tumor Malignancies. Available online: https://clinicaltrials.gov/ct2/show/NCT04383938 (accessed on 17 March 2021).
- Lambert, J.M.R.; Gorzov, P.; Veprintsev, D.B.; Söderqvist, M.; Segerbäck, D.; Bergman, J.; Fersht, A.R.; Hainaut, P.; Wiman, K.G.; Bykov, V.J.N. PRIMA-1 Reactivates Mutant p53 by Covalent Binding to the Core Domain. Cancer Cell 2009, 15, 376–388. [Google Scholar] [CrossRef] [Green Version]
- Study of COTI-2 as Monotherapy or Combination Therapy for the Treatment of Malignancies. Available online: https://www.clinicaltrials.gov/ct2/show/NCT02433626 (accessed on 17 March 2021).
- Aggarwal, M.; Saxena, R.; Sinclair, E.; Fu, Y.; Jacobs, A.; Dyba, M.; Wang, X.; Cruz, I.; Berry, D.; Kallakury, B.; et al. Reactivation of mutant p53 by a dietary-related compound phenethyl isothiocyanate inhibits tumor growth. Cell Death Differ. 2016, 23, 1615–1627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Marchenko, N.D.; Schulz, R.; Fischer, V.; Velasco-Hernandez, T.; Talos, F.; Moll, U.M. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol. Cancer Res. 2011, 9, 577–588. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Marchenko, N.D.; Moll, U.M. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ. 2011, 18, 1904–1913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrales, A.; Iwakuma, T. Targeting oncogenic mutant p53 for cancer therapy. Front. Oncol. 2015, 5, 288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kravchenko, J.E.; Ilyinskaya, G.V.; Komarov, P.G.; Agapova, L.S.; Kochetkov, D.V.; Strom, E.; Frolova, E.I.; Kovriga, I.; Gudkov, A.V.; Feinstein, E.; et al. Small-molecule RETRA suppresses mutant p53-bearing cancer cells through a p73-dependent salvage pathway. Proc. Natl. Acad. Sci. USA 2008, 105, 6302–6307. [Google Scholar] [CrossRef] [Green Version]
- Girardini, J.E.; Napoli, M.; Piazza, S.; Rustighi, A.; Marotta, C.; Radaelli, E.; Capaci, V.; Jordan, L.; Quinlan, P.; Thompson, A.; et al. A Pin1/Mutant p53 Axis Promotes Aggressiveness in Breast Cancer. Cancer Cell 2011, 20, 79–91. [Google Scholar] [CrossRef]
- Liao, P.; Zeng, S.X.; Zhou, X.; Chen, T.; Zhou, F.; Cao, B.; Jung, J.H.; Del Sal, G.; Luo, S.; Lu, H. Mutant p53 Gains Its Function via c-Myc Activation upon CDK4 Phosphorylation at Serine 249 and Consequent PIN1 Binding. Mol. Cell 2017, 68, 1134–1146.e6. [Google Scholar] [CrossRef] [Green Version]
- Wei, S.; Kozono, S.; Kats, L.; Nechama, M.; Li, W.; Guarnerio, J.; Luo, M.; You, M.H.; Yao, Y.; Kondo, A.; et al. Active Pin1 is a key target of all-trans retinoic acid in acute promyelocytic leukemia and breast cancer. Nat. Med. 2015, 21, 457–466. [Google Scholar] [CrossRef]
- Bitomsky, N.; Conrad, E.; Moritz, C.; Polonio-Vallon, T.; Sombroek, D.; Schultheiss, K.; Glas, C.; Greiner, V.; Herbel, C.; Mantovani, F.; et al. Autophosphorylation and Pin1 binding coordinate DNA damage-induced HIPK2 activation and cell death. Proc. Natl. Acad. Sci. USA 2013, 110, E4203-12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofmann, T.G.; Stollberg, N.; Schmitz, M.L.; Will, H. HIPK2 regulates transforming growth factor-beta-induced c-Jun NH(2)-terminal kinase activation and apoptosis in human hepatoma cells. Cancer Res. 2003, 63, 8271–8277. [Google Scholar] [PubMed]
- Zhang, Q.; Yoshimatsu, Y.; Hildebrand, J.; Frisch, S.M.; Goodman, R.H. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell 2003, 115, 177–186. [Google Scholar] [CrossRef] [Green Version]
- Freed-Pastor, W.A.; Mizuno, H.; Zhao, X.; Langerød, A.; Moon, S.H.; Rodriguez-Barrueco, R.; Barsotti, A.; Chicas, A.; Li, W.; Polotskaia, A.; et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 2012, 148, 244–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, S.H.; Huang, C.H.; Houlihan, S.L.; Regunath, K.; Freed-Pastor, W.A.; Morris, J.P.; Tschaharganeh, D.F.; Kastenhuber, E.R.; Barsotti, A.M.; Culp-Hill, R.; et al. p53 Represses the Mevalonate Pathway to Mediate Tumor Suppression. Cell 2019, 176, 564–580.e19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrales, A.; Ranjan, A.; Iyer, S.V.; Padhye, S.; Weir, S.J.; Roy, A.; Iwakuma, T. DNAJA1 controls the fate of misfolded mutant p53 through the mevalonate pathway. Nat. Cell Biol. 2016, 18, 1233–1243. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, J.; Liang, Y.; Wu, R.; Zhao, Y.; Hong, X.; Lin, M.; Yu, H.; Liu, L.; Levine, A.J.; et al. Tumour-associated mutant p53 drives the Warburg effect. Nat. Commun. 2013, 4, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polotskaia, A.; Xiao, G.; Reynoso, K.; Martin, C.; Qiu, W.G.; Hendrickson, R.C.; Bargonettia, J. Proteome-wide analysis of mutant p53 targets in breast cancer identifies new levels of gain-of-function that influence PARP, PCNA, and MCM4. Proc. Natl. Acad. Sci. USA 2015, 112, E1220–E1229. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Fan, S.; Eastman, A.; Worland, P.J.; Sausville, E.A.; O’Connor, P.M. UCN-01: A potent abrogator of G2 checkpoint function in cancer cells with disrupted p53. J. Natl. Cancer Inst. 1996, 88, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.; Booher, R.N.; Kraker, A.; Lawrence, T.; Leopold, W.R.; Sun, Y. Radiosensitization of p53 mutant cells by PD0166285, a novel G2 checkpoint abrogator. Cancer Res. 2001, 61, 8211–8217. [Google Scholar] [PubMed]
- Sur, S.; Pagliarini, R.; Bunz, F.; Rago, C.; Diaz, L.A.; Kinzler, K.W.; Vogelstein, B.; Papadopoulos, N. A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc. Natl. Acad. Sci. USA 2009, 106, 3964–3969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.X.; Cai, S.; Li, S.; Ryan, C.E.; Guo, Z.; Schaiff, W.T.; Lin, L.; Hoog, J.; Goiffon, R.J.; Prat, A.; et al. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J. Clin. Investig. 2012, 122, 1541–1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houbiers, J.G.A.; van der Burg, S.H.; van de Watering, L.M.G.; Tollenaar, R.A.E.M.; Brand, A.; van de Velde, C.J.H.; Melief, C.J.M. Antibodies against p53 are associated with poor prognosis of colorectal cancer. Br. J. Cancer 1995, 72, 637–641. [Google Scholar] [CrossRef] [Green Version]
- Angelopoulou, K.; Stratis, M.; Diamandis, E.P. Humoral immune response against p53 protein in patients with colorectal carcinoma. Int. J. Cancer 1997, 70, 46–51. [Google Scholar] [CrossRef]
- Vermeij, R.; Leffers, N.; Van Der Burg, S.H.; Melief, C.J.; Daemen, T.; Nijman, H.W. Immunological and clinical effects of vaccines targeting p53-overexpressing malignancies. J. Biomed. Biotechnol. 2011, 2011. [Google Scholar] [CrossRef]
- Lo, W.; Parkhurst, M.; Robbins, P.F.; Tran, E.; Lu, Y.C.; Jia, L.; Gartner, J.J.; Pasetto, A.; Deniger, D.; Malekzadeh, P.; et al. Immunologic recognition of a shared p53 mutated neoantigen in a patient with metastatic colorectal cancer. Cancer Immunol. Res. 2019, 7, 534–543. [Google Scholar] [CrossRef]
- Hsiue, E.H.-C.; Wright, K.M.; Douglass, J.; Hwang, M.S.; Mog, B.J.; Pearlman, A.H.; Paul, S.; DiNapoli, S.R.; Konig, M.F.; Wang, Q.; et al. Targeting a neoantigen derived from a common TP53 mutation. Science 2021, 371, eabc8697. [Google Scholar] [CrossRef]
- Matt, S.; Hofmann, T.G. The DNA damage-induced cell death response: A roadmap to kill cancer cells. Cell. Mol. Life Sci. 2016, 73, 2829–2850. [Google Scholar] [CrossRef] [PubMed]
- Frappart, P.O.; Hofmann, T.G. Pancreatic ductal adenocarcinoma (Pdac) organoids: The shining light at the end of the tunnel for drug response prediction and personalized medicine. Cancers 2020, 12, 2750. [Google Scholar] [CrossRef] [PubMed]
- Janic, A.; Valente, L.J.; Wakefield, M.J.; Di Stefano, L.; Milla, L.; Wilcox, S.; Yang, H.; Tai, L.; Vandenberg, C.J.; Kueh, A.J.; et al. DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat. Med. 2018, 1. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, A.M.; Attardi, L.D. Deconstructing networks of p53-mediated tumor suppression in vivo. Cell Death Differ. 2017. [Google Scholar] [CrossRef] [Green Version]
| Compounds Targeting p53 Wild-Type | ||||||
|---|---|---|---|---|---|---|
| Compound | Combination Therapy | Disease | Phase | ClinicalTrials.Gov ID (accessed on 1 April 2021) | Inclusion Criteria Regarding p53 Status | (Possible) Mechanism of Action |
| AMG232 | — | Solid tumor, MM | 1 | NCT01723020 | p53 wild-type | MDM2 inhibition |
| APG-115 | — | Solid tumor, lymphoma | 1 | NCT02935907 | p53 mutation analysis | MDM2 inhibition |
| Pembrolizumab | Solid tumor, melanoma | 1, 2 | NCT03611868 | p53 wild-type | MDM2 inhibition + ICI | |
| Toripalimab | Solid tumor, liposarcoma | 1, 2 | NCT04785196 | —1 | MDM2 inhibition + ICI | |
| BI 907828 | — | Solid tumor | 1 | NCT03449381 | p53 wild-type or unknown | MDM2 inhibition |
| alone or plus BI 754,091 ± BI 754,111 | Solid tumor | 1 | NCT03964233 | p53 wild-type or unknown | MDM2 inhibition ± ICI | |
| CGM097 | — | Solid tumor | 1 | NCT01760525 | p53 wild-type | MDM2 inhibition |
| HDM201 | Alone or plus ancillary treatment | Solid or hematological tumors | 1 | NCT02143635 | p53 wild-type | MDM2 inhibition |
| PDR0012 | CRC, other solid tumors | 1 | NCT02890069 | — | MDM2 inhibition ± ICI | |
| Trametinib | CRC | 1 | NCT03714958 | p53 wild-type | MDM2 + MEK inhibition | |
| Ribociclib | Solid tumor | 2 | NCT04116541 | p53 wild-type | MDM2 + CDK4/6 inhibition | |
| Idasanutlin (RG7388/ RO5503781) | — | Solid tumor | 1 | NCT02828930 | — | MDM2 inhibition |
| — | Solid tumor | 1 | NCT03362723 | — | MDM2 inhibition | |
| — | Neoplasm (except leukemia) | 1 | NCT01462175 | p53 mutation analysis | MDM2 inhibition | |
| Posaconazole | Solid tumor | 1 | NCT01901172 | — | MDM2 inhibition + anti-fungal drug | |
| —1 | Solid tumor, leukemia | 1, 2 | NCT04029688 | —1 | MDM2 inhibition | |
| Atezolizumab | CRC | 1, 2 | NCT03555149 | — | MDM2 inhibition + ICI | |
| — | Solid tumor | 2 | NCT04589845 | p53-wildtype | MDM2 inhibition | |
| Milademetan (DS-3032b) | — | Solid tumor, lymphoma | 1 | NCT01877382 | No known p53 mutation, p53 mutation analysis | MDM2 inhibition |
| MK-8242 | — | Solid tumor | 1 | NCT01463696 | p53 mutation analysis | MDM2 inhibition |
| RO5045337 (RG7112) | — | Solid tumor | 1 | NCT00559533 | — | MDM2 inhibition |
| — | Solid tumor | 1 | NCT01164033 | — | MDM2 inhibition | |
| — | Neoplasm | 1 | NCT01677780 | — | MDM2 inhibition | |
| RO6839921 (RG7775) | — | Neoplasm | 1 | NCT02098967 | — | MDM2 inhibition |
| SAR405838 (MI-77031) | — | Neoplasm | 1 | NCT01636479 | — | MDM2 inhibition |
| Pimasertib | Neoplasm | 1 | NCT01985191 | — | MDM2 + MEK1/2 inhibition | |
| ALRN-6924 | —3 | Pediatric cancer | 1 | NCT03654716 | p53 wild-type | MDM2/4 inhibition |
| Paclitaxel | Solid tumor | 1 | NCT03725436 | p53 wild-type | MDM2/4 inhibition + chemotherapy | |
| — | Solid tumor, lymphoma | 1, 2 | NCT02264613 | p53 wild-type | MDM2/4 inhibition | |
| Ad-p53 (Gendicine) | — | CRC, other solid tumors | 1 | NCT01191684 | >10% of p53-positive cells in IHC | Gene therapy to deliver p53 wild-type |
| Pembrolizumab | CRC, other solid tumors | 1 | NCT02432963 | p53 mutation or ≥10% of p53-positive cells in IHC | Gene therapy to deliver p53 wild-type + ICI | |
| Xeloda or Keytruda or Opdivo | Solid tumor | 1, 2 | NCT02842125 | p53 wild-type or < 20% of p53-positive cells in IHC | Gene therapy to deliver p53 wild-type + chemotherapy or ICI | |
| Approved ICI | Solid tumor, lymphoma | 2 | NCT03544723 | p53 wild-type or < 20% of p53-positive cells in IHC | Gene therapy to deliver p53 wild-type | |
| Compounds Targeting p53 Mutant | ||||||
| Compound | Combination Therapy | Disease | Phase | ClinicalTrials.govID (accessed on 1 April 2021) | Inclusion Criteria regarding p53 Status | (Possible) Mechanism of Action |
| PRIMA-1MET (APR-246) | Pembrolizumab | Solid tumor | 1, 2 | NCT04383938 | p53 mutation | Restoration of wild-type functions + ICI |
| COTI-2 | Alone or plus Cisplatin | CRC, other solid tumors | 1 | NCT02433626 | —2 | Restoration of wild-type functions + chemotherapy |
| Kevetrin (thioureidobutyronitrile) | — | Solid tumor | 1 | NCT01664000 | — | Degradation of mutant p53 (activation of wild-type p53) |
| Atrovastatin | — | Ulcerative colitis | 2 | NCT04767984 | DN missense p53 mutation | Degradation of mutant p53 + Inhibition of the melanovate pathway |
| Tanespimycin (17-AAG) | Irinotecan | Solid tumor | 1 | NCT00119236 | — | Degradation of mutant p53 by HSP90 inhibition + chemotherapy (Investigation if response depends on p53 mutational status?) |
| Mutant p53 peptide pulsed dendritic cell vaccine | Colorectal cancer, other solid tumors | 2 | NCT00019084 | p53 mutation | Mutant p53 vaccine | |
| ALT-801 | — | Neoplasm | 1 | NCT00496860 | HLA-A2.1/p53 positive | IL-2 fused to TCR recognizing p53 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liebl, M.C.; Hofmann, T.G. The Role of p53 Signaling in Colorectal Cancer. Cancers 2021, 13, 2125. https://doi.org/10.3390/cancers13092125
Liebl MC, Hofmann TG. The Role of p53 Signaling in Colorectal Cancer. Cancers. 2021; 13(9):2125. https://doi.org/10.3390/cancers13092125
Chicago/Turabian StyleLiebl, Magdalena C., and Thomas G. Hofmann. 2021. "The Role of p53 Signaling in Colorectal Cancer" Cancers 13, no. 9: 2125. https://doi.org/10.3390/cancers13092125
APA StyleLiebl, M. C., & Hofmann, T. G. (2021). The Role of p53 Signaling in Colorectal Cancer. Cancers, 13(9), 2125. https://doi.org/10.3390/cancers13092125






