Comparison of Global DNA Methylation Patterns in Human Melanoma Tissues and Their Derivative Cell Lines
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Generating RRBS Methylomes from Melanoma Tumour Tissue and Matched Cell Lines
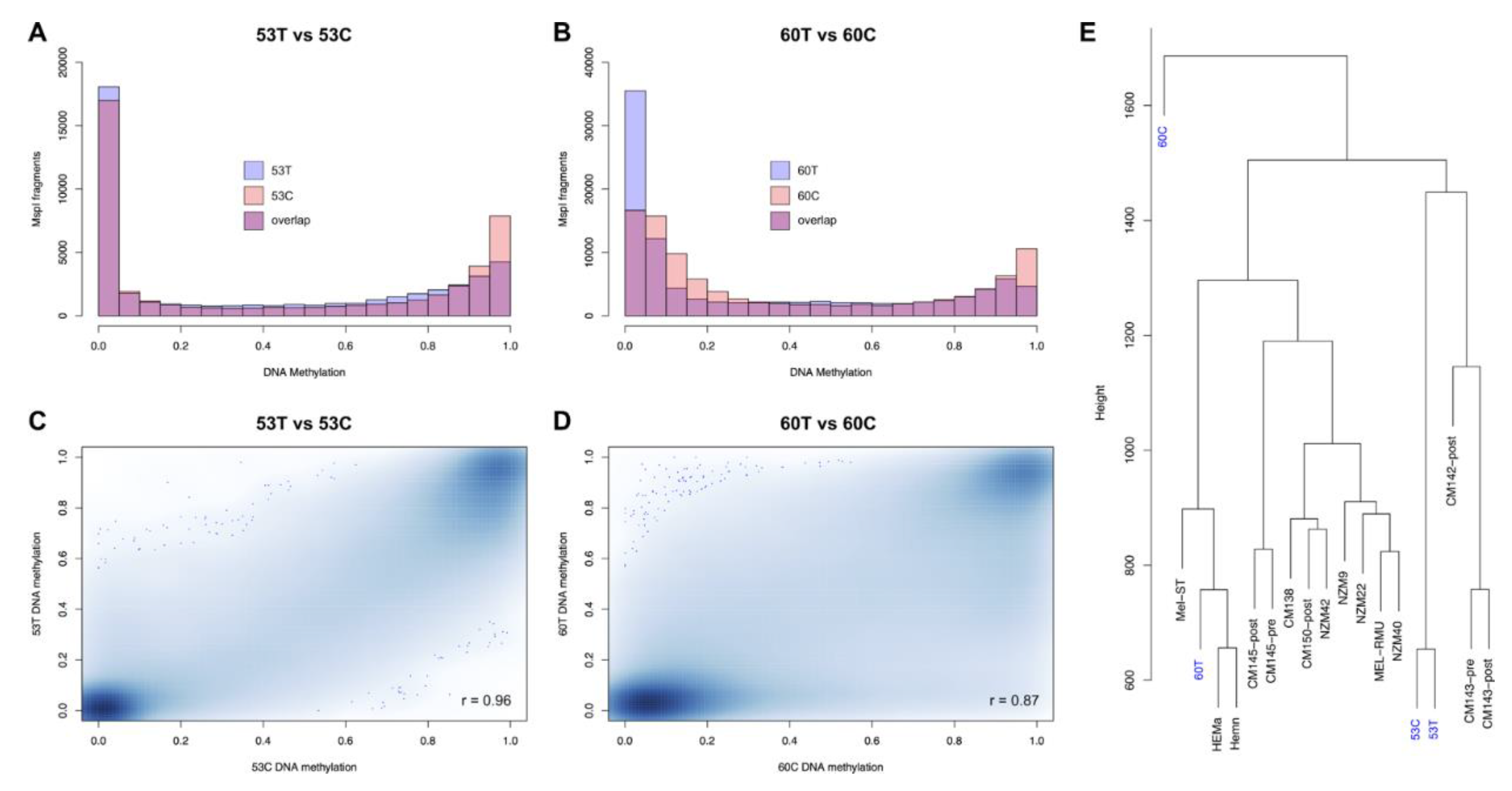
2.2. Metastatic Cell Lines Globally Maintain Tissue Methylation Patterns and Follow Their Origin
2.3. RRBS Methylomes of Melanoma Cell Lines Are Relatively Hypermethylated, When Compared to Their Corresponding Tumour Tissues
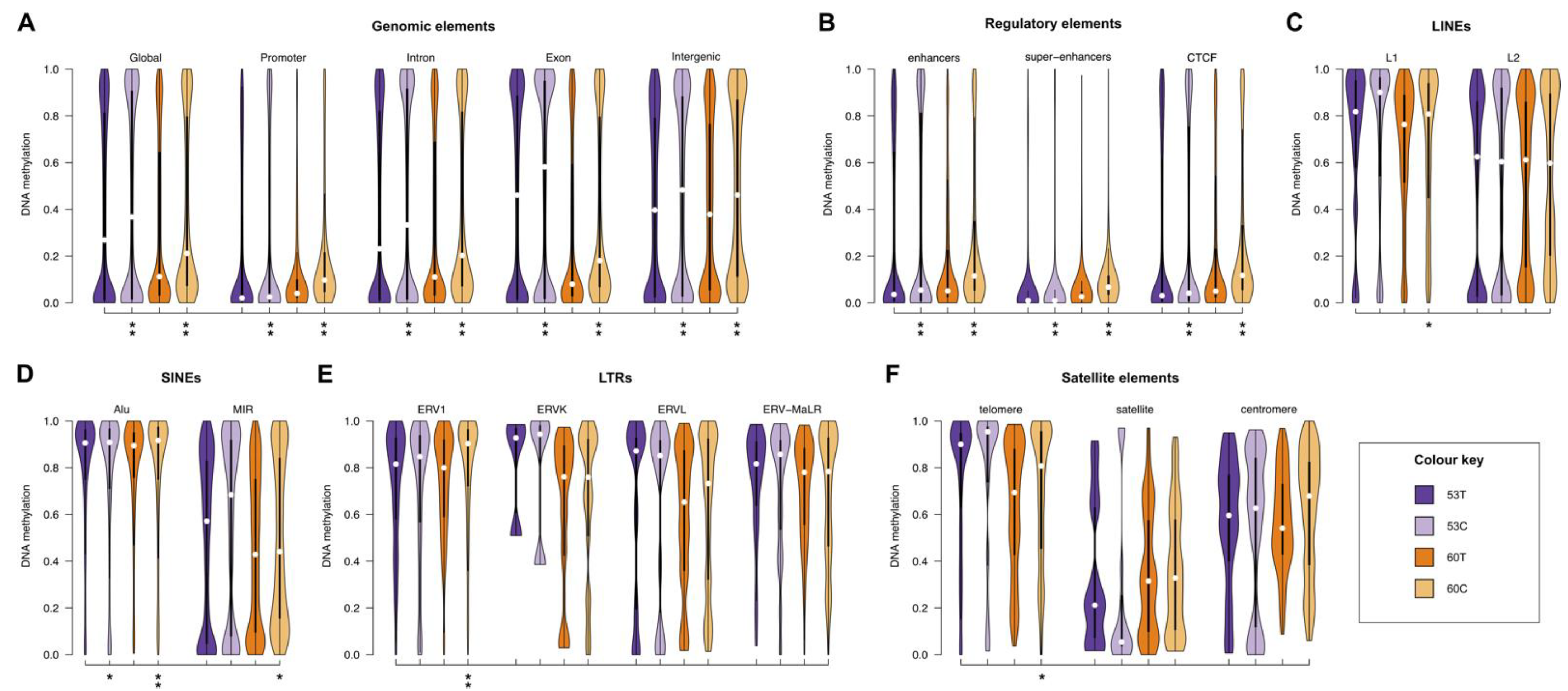
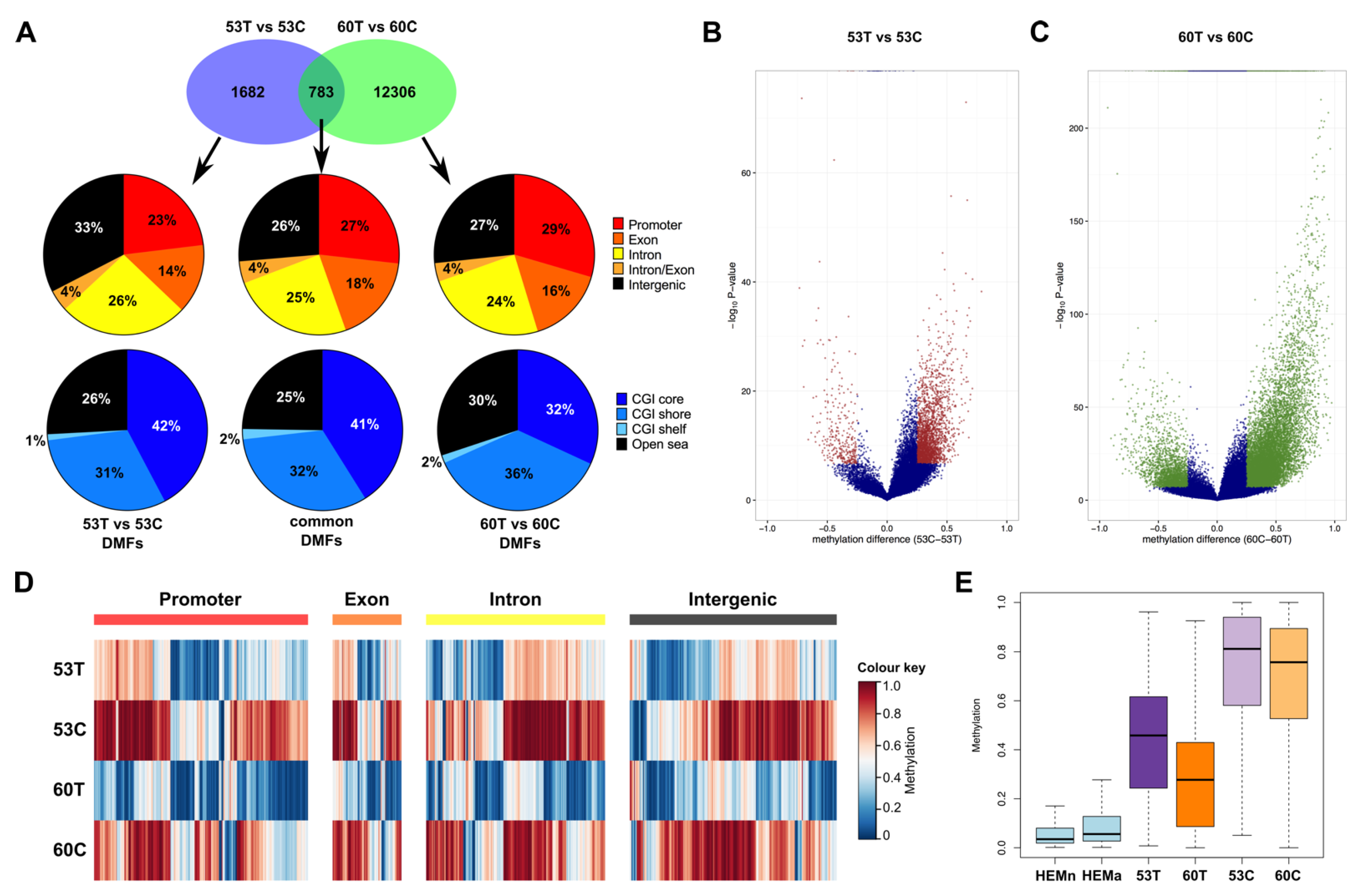
2.4. Melanoma Cell Lines Demonstrate Region-Specific Methylation Differences in Comparison to the Tumour Tissues from Which They Were Derived
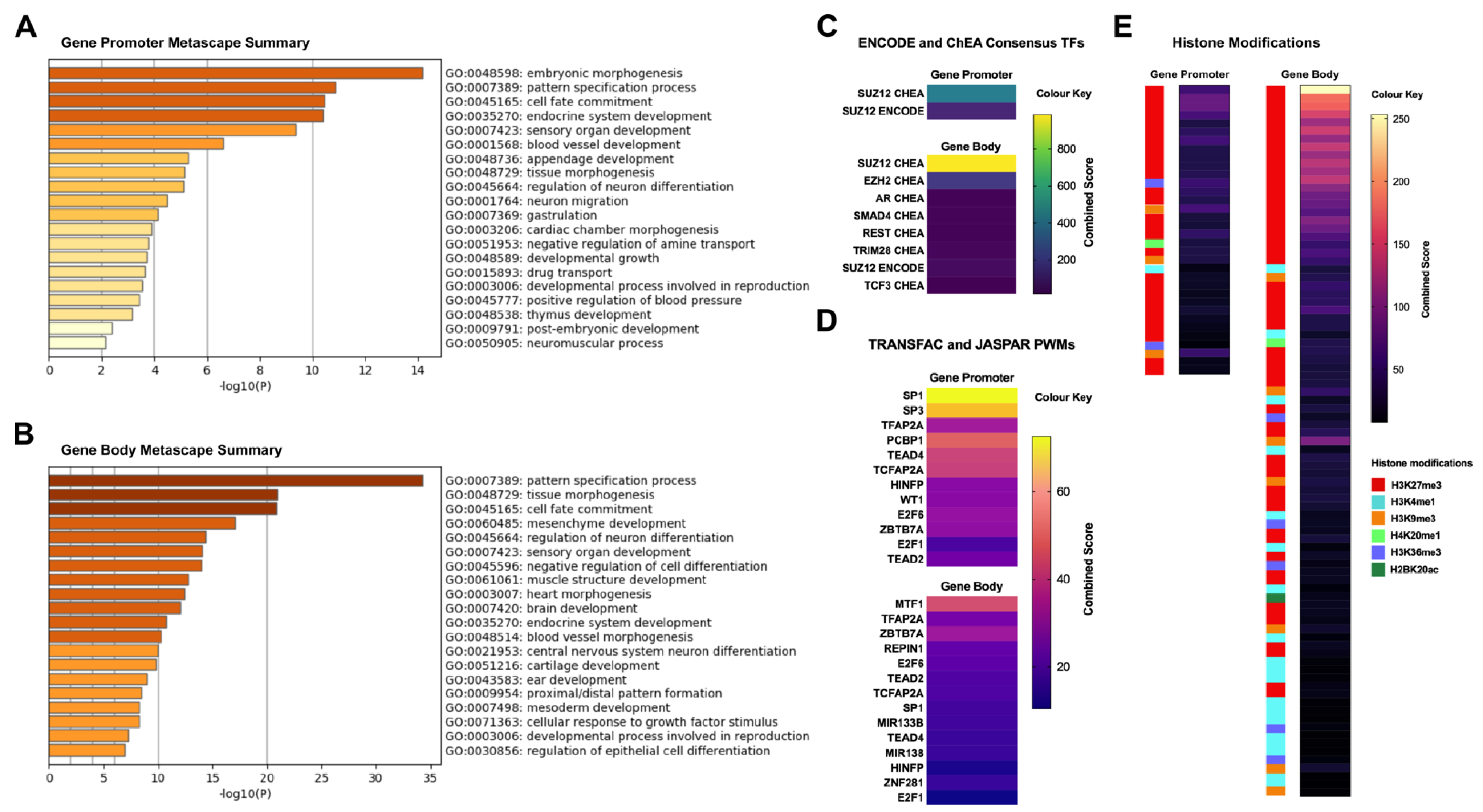
2.5. Differentially Methylated Fragments Shared between Tumour Tissues and Derivative Cell Lines Are Involved in Tissue Morphogenesis and Cellular Differentiation Processes, and Are Enriched for the H3K27me3 Histone Mark and PRC2 Complex Genes
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Molecular Classifications of Cell Lines
4.3. Tissue Sections
4.4. Cell Culture Establishment
4.5. RRBS Library Preparation and Sequencing
4.6. DNA Methylation Data Analysis
4.7. Differential Methylation Analysis
4.8. Functional Enrichment Analyses of the Genomic Features
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reik, W.; Dean, W.; Walter, J. Epigenetic reprogramming in mammalian development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Morgan, H.D.; Santos, F.; Green, K.; Dean, W.; Reik, W. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 2005, 14, R47–R58. [Google Scholar] [CrossRef] [PubMed]
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Rodger, E.J.; Eccles, M.R. Epigenetic drivers of tumourigenesis and cancer metastasis. Semin. Cancer Biol. 2018, 51, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Mirabelli, P.; Coppola, L.; Salvatore, M. Cancer Cell Lines Are Useful Model Systems for Medical Research. Cancers 2019, 11, 1098. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Altering gene expression with 5-azacytidine. Cell 1985, 40, 485–486. [Google Scholar] [CrossRef]
- Wozniak, R.J.; Klimecki, W.T.; Lau, S.S.; Feinstein, Y.; Futscher, B.W. 5-Aza-2′-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene 2007, 26, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Giri, A.K.; Aittokallio, T. DNMT Inhibitors Increase Methylation in the Cancer Genome. Front. Pharmacol. 2019, 10, 385. [Google Scholar] [CrossRef] [PubMed]
- Urbano, A.; Smith, J.; Weeks, R.J.; Chatterjee, A. Gene-Specific Targeting of DNA Methylation in the Mammalian Genome. Cancers 2019, 11, 1515. [Google Scholar] [CrossRef] [PubMed]
- Sung, C.K.; Yim, H. CRISPR-mediated promoter de/methylation technologies for gene regulation. Arch. Pharm. Res. 2020, 43, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S.R.; Cui, Y.; Lubecka, K.; Stefanska, B.; Irudayaraj, J. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget 2016, 7, 46545–46556. [Google Scholar] [CrossRef] [PubMed]
- Vojta, A.; Dobrinic, P.; Tadic, V.; Bockor, L.; Korac, P.; Julg, B.; Klasic, M.; Zoldos, V. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016, 44, 5615–5628. [Google Scholar] [CrossRef] [PubMed]
- Moure, C.J.; Diplas, B.H.; Chen, L.H.; Yang, R.; Pirozzi, C.J.; Wang, Z.; Spasojevic, I.; Waitkus, M.S.; He, Y.; Yan, H. CRISPR Editing of Mutant IDH1 R132H Induces a CpG Methylation-Low State in Patient-Derived Glioma Models of G-CIMP. Mol. Cancer Res. 2019, 17, 2042–2050. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Noor, D.A.M.; Jeyapalan, J.N.; Alhazmi, S.; Carr, M.; Squibb, B.; Wallace, C.; Tan, C.; Cusack, M.; Hughes, J.; Reader, T. Genome-wide methylation analysis identifies genes silenced in non-seminoma cell lines. NPJ Genom. Med. 2016, 1, 1–13. [Google Scholar] [CrossRef]
- Katt, M.E.; Placone, A.L.; Wong, A.D.; Xu, Z.S.; Searson, P.C. In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Front. Bioeng. Biotechnol. 2016, 4, 12. [Google Scholar] [CrossRef]
- Ferreira, D.; Adega, F.; Chaves, R. The importance of cancer cell lines as in vitro models in cancer methylome analysis and anticancer drugs testing. In Oncogenomics and Cancer Proteomics-Novel Approaches in Biomarkers Discovery and Therapeutic Targets in Cancer; IntechOpen: London, UK, 2013; pp. 139–166. [Google Scholar]
- Garnett, M.J.; McDermott, U. The evolving role of cancer cell line-based screens to define the impact of cancer genomes on drug response. Curr. Opin. Genet. Dev. 2014, 24, 114–119. [Google Scholar] [CrossRef][Green Version]
- Futscher, B.W. Epigenetic changes during cell transformation. Adv. Exp. Med. Biol. 2013, 754, 179–194. [Google Scholar] [CrossRef]
- Nestor, C.E.; Ottaviano, R.; Reinhardt, D.; Cruickshanks, H.A.; Mjoseng, H.K.; McPherson, R.C.; Lentini, A.; Thomson, J.P.; Dunican, D.S.; Pennings, S.; et al. Rapid reprogramming of epigenetic and transcriptional profiles in mammalian culture systems. Genome Biol. 2015, 16, 11. [Google Scholar] [CrossRef]
- Yang, X.; Shao, X.; Gao, L.; Zhang, S. Systematic DNA methylation analysis of multiple cell lines reveals common and specific patterns within and across tissues of origin. Hum. Mol. Genet. 2015, 24, 4374–4384. [Google Scholar] [CrossRef]
- Varley, K.E.; Gertz, J.; Bowling, K.M.; Parker, S.L.; Reddy, T.E.; Pauli-Behn, F.; Cross, M.K.; Williams, B.A.; Stamatoyannopoulos, J.A.; Crawford, G.E.; et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013, 23, 555–567. [Google Scholar] [CrossRef]
- Massie, C.E.; Mills, I.G.; Lynch, A.G. The importance of DNA methylation in prostate cancer development. J. Steroid Biochem. Mol. Biol. 2017, 166, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, D.; Eide, P.W.; Eilertsen, I.A.; Danielsen, S.A.; Eknaes, M.; Hektoen, M.; Lind, G.E.; Lothe, R.A. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2013, 2, e71. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.; Sayols, S.; Moran, S.; Guillaumet-Adkins, A.; Schroeder, M.P.; Royo, R.; Orozco, M.; Gut, M.; Gut, I.; Lopez-Bigas, N.; et al. A DNA methylation map of human cancer at single base-pair resolution. Oncogene 2017, 36, 5648–5657. [Google Scholar] [CrossRef] [PubMed]
- Meissner, A.; Gnirke, A.; Bell, G.W.; Ramsahoye, B.; Lander, E.S.; Jaenisch, R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005, 33, 5868–5877. [Google Scholar] [CrossRef]
- Chatterjee, A.; Macaulay, E.C.; Ahn, A.; Ludgate, J.L.; Stockwell, P.A.; Weeks, R.J.; Parry, M.F.; Foster, T.J.; Knarston, I.M.; Eccles, M.R.; et al. Comparative assessment of DNA methylation patterns between reduced representation bisulfite sequencing and Sequenom EpiTyper methylation analysis. Epigenomics 2017, 9, 823–832. [Google Scholar] [CrossRef]
- Meissner, A.; Mikkelsen, T.S.; Gu, H.; Wernig, M.; Hanna, J.; Sivachenko, A.; Zhang, X.; Bernstein, B.E.; Nusbaum, C.; Jaffe, D.B.; et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008, 454, 766–770. [Google Scholar] [CrossRef]
- Chatterjee, A.; Stockwell, P.A.; Rodger, E.J.; Morison, I.M. Comparison of alignment software for genome-wide bisulphite sequence data. Nucleic Acids Res. 2012, 40, e79. [Google Scholar] [CrossRef]
- Stockwell, P.A.; Chatterjee, A.; Rodger, E.J.; Morison, I.M. DMAP: Differential methylation analysis package for RRBS and WGBS data. Bioinformatics 2014, 30, 1814–1822. [Google Scholar] [CrossRef]
- Samanta, M.; Mondal, R.; Ray, S.; Sabui, T.K.; Kundu, C.K.; Hazra, A.; Chatterjee, K.; Sarkar, D. Blood pressure variation with gestational age and birth weight in Indian newborn. J. Trop. Pediatrics 2015, 61, 197–205. [Google Scholar] [CrossRef]
- Chatterjee, A.; Rodger, E.J.; Ahn, A.; Stockwell, P.A.; Parry, M.; Motwani, J.; Gallagher, S.J.; Shklovskaya, E.; Tiffen, J.; Eccles, M.R.; et al. Marked Global DNA Hypomethylation Is Associated with Constitutive PD-L1 Expression in Melanoma. iScience 2018, 4, 312–325. [Google Scholar] [CrossRef]
- Chatterjee, A.; Stockwell, P.A.; Ahn, A.; Rodger, E.J.; Leichter, A.L.; Eccles, M.R. Genome-wide methylation sequencing of paired primary and metastatic cell lines identifies common DNA methylation changes and a role for EBF3 as a candidate epigenetic driver of melanoma metastasis. Oncotarget 2017, 8, 6085–6101. [Google Scholar] [CrossRef]
- Bork, S.; Pfister, S.; Witt, H.; Horn, P.; Korn, B.; Ho, A.D.; Wagner, W. DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells. Aging Cell 2010, 9, 54–63. [Google Scholar] [CrossRef]
- Kolat, D.; Kaluzinska, Z.; Bednarek, A.K.; Pluciennik, E. The biological characteristics of transcription factors AP-2alpha and AP-2gamma and their importance in various types of cancers. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Chen, H.Z.; Tsai, S.Y.; Leone, G. Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat. Rev. Cancer 2009, 9, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Archer, M.C. Role of sp transcription factors in the regulation of cancer cell metabolism. Genes Cancer 2011, 2, 712–719. [Google Scholar] [CrossRef]
- Vizoso, M.; Ferreira, H.J.; Lopez-Serra, P.; Carmona, F.J.; Martinez-Cardus, A.; Girotti, M.R.; Villanueva, A.; Guil, S.; Moutinho, C.; Liz, J.; et al. Epigenetic activation of a cryptic TBC1D16 transcript enhances melanoma progression by targeting EGFR. Nat. Med. 2015. [Google Scholar] [CrossRef]
- Smiraglia, D.J.; Rush, L.J.; Fruhwald, M.C.; Dai, Z.; Held, W.A.; Costello, J.F.; Lang, J.C.; Eng, C.; Li, B.; Wright, F.A.; et al. Excessive CpG island hypermethylation in cancer cell lines versus primary human malignancies. Hum. Mol. Genet. 2001, 10, 1413–1419. [Google Scholar] [CrossRef] [PubMed]
- Gal-Yam, E.N.; Egger, G.; Iniguez, L.; Holster, H.; Einarsson, S.; Zhang, X.; Lin, J.C.; Liang, G.; Jones, P.A.; Tanay, A. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc. Natl. Acad. Sci. USA 2008, 105, 12979–12984. [Google Scholar] [CrossRef]
- Rada-Iglesias, A.; Enroth, S.; Andersson, R.; Wanders, A.; Pahlman, L.; Komorowski, J.; Wadelius, C. Histone H3 lysine 27 trimethylation in adult differentiated colon associated to cancer DNA hypermethylation. Epigenetics 2009, 4, 107–113. [Google Scholar] [CrossRef]
- Widschwendter, M.; Fiegl, H.; Egle, D.; Mueller-Holzner, E.; Spizzo, G.; Marth, C.; Weisenberger, D.J.; Campan, M.; Young, J.; Jacobs, I.; et al. Epigenetic stem cell signature in cancer. Nat. Genet. 2007, 39, 157–158. [Google Scholar] [CrossRef]
- Antequera, F.; Boyes, J.; Bird, A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell 1990, 62, 503–514. [Google Scholar] [CrossRef]
- Weissbein, U.; Plotnik, O.; Vershkov, D.; Benvenisty, N. Culture-induced recurrent epigenetic aberrations in human pluripotent stem cells. PLoS Genet. 2017, 13, e1006979. [Google Scholar] [CrossRef] [PubMed]
- Aref-Eshghi, E.; Biswas, S.; Chen, C.; Sadikovic, B.; Chakrabarti, S. Glucose-induced, duration-dependent genome-wide DNA methylation changes in human endothelial cells. Am. J. Physiol. Cell Physiol. 2020, 319, C268–C276. [Google Scholar] [CrossRef]
- Hamadneh, L.; Al-Majawleh, M.; Jarrar, Y.; Shraim, S.; Hasan, M.; Abu-Irmaileh, B. Culturing conditions highly affect DNA methylation and gene expression levels in MCF7 breast cancer cell line. In Vitro Cell Dev. Biol. Anim. 2018, 54, 331–334. [Google Scholar] [CrossRef]
- Allegrucci, C.; Wu, Y.Z.; Thurston, A.; Denning, C.N.; Priddle, H.; Mummery, C.L.; Ward-van Oostwaard, D.; Andrews, P.W.; Stojkovic, M.; Smith, N.; et al. Restriction landmark genome scanning identifies culture-induced DNA methylation instability in the human embryonic stem cell epigenome. Hum. Mol. Genet. 2007, 16, 1253–1268. [Google Scholar] [CrossRef] [PubMed]
- Novak, P.; Jensen, T.J.; Garbe, J.C.; Stampfer, M.R.; Futscher, B.W. Stepwise DNA methylation changes are linked to escape from defined proliferation barriers and mammary epithelial cell immortalization. Cancer Res. 2009, 69, 5251–5258. [Google Scholar] [CrossRef]
- Habibi, E.; Brinkman, A.B.; Arand, J.; Kroeze, L.I.; Kerstens, H.H.; Matarese, F.; Lepikhov, K.; Gut, M.; Brun-Heath, I.; Hubner, N.C.; et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell 2013, 13, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Peng, Y.; Gao, A.; Du, C.; Herman, J.G. Epigenetic heterogeneity in cancer. Biomark Res. 2019, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Roesch, A.; Fukunaga-Kalabis, M.; Schmidt, E.C.; Zabierowski, S.E.; Brafford, P.A.; Vultur, A.; Basu, D.; Gimotty, P.; Vogt, T.; Herlyn, M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 2010, 141, 583–594. [Google Scholar] [CrossRef]
- Sharma, S.V.; Lee, D.Y.; Li, B.; Quinlan, M.P.; Takahashi, F.; Maheswaran, S.; McDermott, U.; Azizian, N.; Zou, L.; Fischbach, M.A.; et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010, 141, 69–80. [Google Scholar] [CrossRef]
- Ben-David, U.; Siranosian, B.; Ha, G.; Tang, H.; Oren, Y.; Hinohara, K.; Strathdee, C.A.; Dempster, J.; Lyons, N.J.; Burns, R.; et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 2018, 560, 325–330. [Google Scholar] [CrossRef]
- Wagner, D.E.; Klein, A.M. Lineage tracing meets single-cell omics: Opportunities and challenges. Nat. Rev. Genet. 2020, 21, 410–427. [Google Scholar] [CrossRef]
- Quinn, J.J.; Jones, M.G.; Okimoto, R.A.; Nanjo, S.; Chan, M.M.; Yosef, N.; Bivona, T.G.; Weissman, J.S. Single-cell lineages reveal the rates, routes, and drivers of metastasis in cancer xenografts. Science 2021, 371. [Google Scholar] [CrossRef] [PubMed]
- Arneson, D.; Yang, X.; Wang, K. MethylResolver-a method for deconvoluting bulk DNA methylation profiles into known and unknown cell contents. Commun. Biol. 2020, 3, 422. [Google Scholar] [CrossRef]
- Muller, J. Transcriptional silencing by the Polycomb protein in Drosophila embryos. EMBO J. 1995, 14, 1209–1220. [Google Scholar] [CrossRef] [PubMed]
- Laugesen, A.; Hojfeldt, J.W.; Helin, K. Role of the Polycomb Repressive Complex 2 (PRC2) in Transcriptional Regulation and Cancer. Cold Spring Harb. Perspect. Med. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Ji, G.; Gao, Z.; Han, X.; Ye, M.; Yuan, Z.; Luo, H.; Huang, X.; Natarajan, K.; Wang, J.; et al. Direct ChIP-bisulfite sequencing reveals a role of H3K27me3 mediating aberrant hypermethylation of promoter CpG islands in cancer cells. Genomics 2014, 103, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Pinello, L.; Xu, J.; Orkin, S.H.; Yuan, G.C. Analysis of chromatin-state plasticity identifies cell-type-specific regulators of H3K27me3 patterns. Proc. Natl. Acad. Sci. USA 2014, 111, E344–E353. [Google Scholar] [CrossRef] [PubMed]
- Enroth, S.; Rada-Iglesisas, A.; Andersson, R.; Wallerman, O.; Wanders, A.; Pahlman, L.; Komorowski, J.; Wadelius, C. Cancer associated epigenetic transitions identified by genome-wide histone methylation binding profiles in human colorectal cancer samples and paired normal mucosa. BMC Cancer 2011, 11, 450. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Shi, J.; Li, W.; Tanaka, K.; Allton, K.L.; Richardson, D.; Li, J.; Franco, H.L.; Nagari, A.; Malladi, V.S.; et al. Histone modification profiling in breast cancer cell lines highlights commonalities and differences among subtypes. BMC Genom. 2018, 19, 150. [Google Scholar] [CrossRef]
- Wiles, E.T.; Selker, E.U. H3K27 methylation: A promiscuous repressive chromatin mark. Curr. Opin. Genet. Dev. 2017, 43, 31–37. [Google Scholar] [CrossRef]
- Tran, K.B.; Gimenez, G.; Tsai, P.; Kolekar, S.; Rodger, E.J.; Chatterjee, A.; Jabed, A.; Shih, J.H.; Joseph, W.R.; Marshall, E.S. Genomic and signalling pathway characterization of the NZM panel of melanoma cell lines: A valuable model for studying the impact of genetic diversity in melanoma. Pigment Cell Melanoma Res. 2020, 34, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Jeffs, A.R.; Glover, A.C.; Slobbe, L.J.; Wang, L.; He, S.; Hazlett, J.A.; Awasthi, A.; Woolley, A.G.; Marshall, E.S.; Joseph, W.R. A gene expression signature of invasive potential in metastatic melanoma cells. PLoS ONE 2009, 4, e8461. [Google Scholar] [CrossRef]
- Liu, Y.; Siegmund, K.D.; Laird, P.W.; Berman, B.P. Bis-SNP: Combined DNA methylation and SNP calling for Bisulfite-seq data. Genome Biol. 2012, 13, R61. [Google Scholar] [CrossRef]
- Ludgate, J.L.; Wright, J.; Stockwell, P.A.; Morison, I.M.; Eccles, M.R.; Chatterjee, A. A streamlined method for analysing genome-wide DNA methylation patterns from low amounts of FFPE DNA. BMC Med. Genom. 2017, 10, 54. [Google Scholar] [CrossRef] [PubMed]
- Marshall, E.S.; Matthews, J.H.; Shaw, J.H.; Nixon, J.; Tumewu, P.; Finlay, G.J.; Holdaway, K.M.; Baguley, B.C. Radiosensitivity of new and established human melanoma cell lines: Comparison of [3H]thymidine incorporation and soft agar clonogenic assays. Eur. J. Cancer 1994, 30A, 1370–1376. [Google Scholar] [CrossRef]
- Chatterjee, A.; Rodger, E.J.; Stockwell, P.A.; Weeks, R.J.; Morison, I.M. Technical considerations for reduced representation bisulfite sequencing with multiplexed libraries. J. Biomed. Biotechnol. 2012, 2012, 741542. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Rodger, E.J.; Stockwell, P.A.; Le Mee, G.; Morison, I.M. Generating Multiple Base-Resolution DNA Methylomes Using Reduced Representation Bisulfite Sequencing. Methods Mol. Biol. 2017, 1537, 279–298. [Google Scholar] [CrossRef]
- Chatterjee, A.; Rodger, E.J.; Morison, I.M.; Eccles, M.R.; Stockwell, P.A. Tools and Strategies for Analysis of Genome-Wide and Gene-Specific DNA Methylation Patterns. Methods Mol. Biol. 2017, 1537, 249–277. [Google Scholar] [CrossRef]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef]
- Rodger, E.J.; Chatterjee, A.; Stockwell, P.A.; Eccles, M.R. Characterisation of DNA methylation changes in EBF3 and TBC1D16 associated with tumour progression and metastasis in multiple cancer types. Clin. Epigenetics 2019, 11, 114. [Google Scholar] [CrossRef] [PubMed]
- Bowden, S.A.; Stockwell, P.A.; Rodger, E.J.; Parry, M.F.; Eccles, M.R.; Stayner, C.; Chatterjee, A. Extensive Inter-Cyst DNA Methylation Variation in Autosomal Dominant Polycystic Kidney Disease Revealed by Genome Scale Sequencing. Front. Genet. 2020, 11, 348. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Stockwell, P.A.; Rodger, E.J.; Morison, I.M. Genome-scale DNA methylome and transcriptome profiling of human neutrophils. Sci. Data 2016, 3, 160019. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodger, E.J.; Almomani, S.N.; Ludgate, J.L.; Stockwell, P.A.; Baguley, B.C.; Eccles, M.R.; Chatterjee, A. Comparison of Global DNA Methylation Patterns in Human Melanoma Tissues and Their Derivative Cell Lines. Cancers 2021, 13, 2123. https://doi.org/10.3390/cancers13092123
Rodger EJ, Almomani SN, Ludgate JL, Stockwell PA, Baguley BC, Eccles MR, Chatterjee A. Comparison of Global DNA Methylation Patterns in Human Melanoma Tissues and Their Derivative Cell Lines. Cancers. 2021; 13(9):2123. https://doi.org/10.3390/cancers13092123
Chicago/Turabian StyleRodger, Euan J., Suzan N. Almomani, Jackie L. Ludgate, Peter A. Stockwell, Bruce C. Baguley, Michael R. Eccles, and Aniruddha Chatterjee. 2021. "Comparison of Global DNA Methylation Patterns in Human Melanoma Tissues and Their Derivative Cell Lines" Cancers 13, no. 9: 2123. https://doi.org/10.3390/cancers13092123
APA StyleRodger, E. J., Almomani, S. N., Ludgate, J. L., Stockwell, P. A., Baguley, B. C., Eccles, M. R., & Chatterjee, A. (2021). Comparison of Global DNA Methylation Patterns in Human Melanoma Tissues and Their Derivative Cell Lines. Cancers, 13(9), 2123. https://doi.org/10.3390/cancers13092123





