Simple Summary
The INhibitor of Growth (ING) family of proteins was founded when in 1996 ING1 was identified as a tumour suppressor, a protein that prevents cancer development. Important subsequent genetic and biochemical work in different models, showed that most ING family members are actually required for cellular proliferation, a hallmark of cancer cells. Although several studies suggest that INGs are broadly lost in cancer, some ING family members are amplified and correlate with a bad prognosis. This is especially true in hormone-dependent cancers, such as breast and prostate tumours. Herein, we review these studies and propose that ING proteins are not all tumour suppressors, but can play opposite role. Unquestionably, INGs have various functions, likely many yet to be discovered, and play complex roles during cancer development.
Abstract
The INhibitor of Growth family was defined in the mid-1990s by the identification of a tumour suppressor, ING1, and subsequent expansion of the family based essentially on sequence similarities. However, later work and more recent investigations demonstrate that at least a few ING proteins are actually required for normal proliferation of eukaryotic cells, from yeast to human. ING proteins are also part of a larger family of chromatin-associated factors marked by a plant homeodomain (PHD), which mediates interactions with methylated lysine residues. Herein, we discuss the role of ING proteins and their various roles in chromatin signalling in the context of cancer development and progression.
Keywords:
oncoproteins; plant homeodomain; PHD; INhibitor of Growth; ING; histone mark reader; chromatin; cancer 1. Introduction
Eukaryotic genomes are compacted within the nucleus of the cell. Essentially, the genomic DNA is spooled around histone octamers composed of two copies of each histone—H2A, H2B, H3, and H4—forming the basic repeating unit of chromatin, the nucleosome. Given its role in compacting the genome, the nucleosome obstructs DNA transactions (i.e., transcription, recombination, replication, and repair). To regulate access to genetic information, the cell utilises various molecular mechanisms, including DNA methylation and histone modifications, to elicit access.
The chromodomain of the heterochromatin protein HP1α was identified in 2001 as an adaptor that potently associates with the histone H3 trimethylated on lysine 9 (H3K9me3) [1]. Since then, several other chromatin proteins demonstrated the capacity to distinguish and associate with various modifications on core and linker histones [2]. As such, these proteins are referred to as histone mark readers. These readers regulate access to the genetic information by associating with specific histone marks and stabilizing enzymatic activities to either open up or close the structure of the chromatin fibre.
The INhibitor of Growth (ING) family is composed of five closely related paralogs (Figure 1A). They all associate with enzymatic activities via their amino terminus and bind methylated histones through a plant homeodomain (PHD) at the carboxy terminus [3]. Specifically, ING1 and ING2 associate with the histone deacetylase (HDAC) mSIN3A/HDAC1 complex [4,5], while ING3, ING4, and ING5 associate with TIP60, HBO1, and HBO1 or MOZ/MORF histone acetyltransferases (HAT), respectively [6,7] (Figure 1B). Initially defined as chromatin readers, INGs harbour a PHD domain that mediates protein–protein interaction with the histone H3 trimethylated on lysine 4 (H3K4me3), a histone mark commonly found at the transcriptional start site (TSS) of expressed genes (see [8] for review). By bridging HAT or HDAC activity to H3K4me3 at TSS, INGs can activate [9] or silence [10,11] gene expression, respectively, through gene accessibility/chromatin remodelling [12]. It is evident that ING proteins target their partners to specific regions of the genome through H3K4me3. However, they may also drive specific interactions with methylated nonhistone proteins such as p53, leading to their modification by either HAT or HDAC complexes [13,14]. Moreover, the PHD domains of ING1 and ING2 have also been described as binding important nuclear signalling molecules called phosphatidylinositol phosphates [15,16].
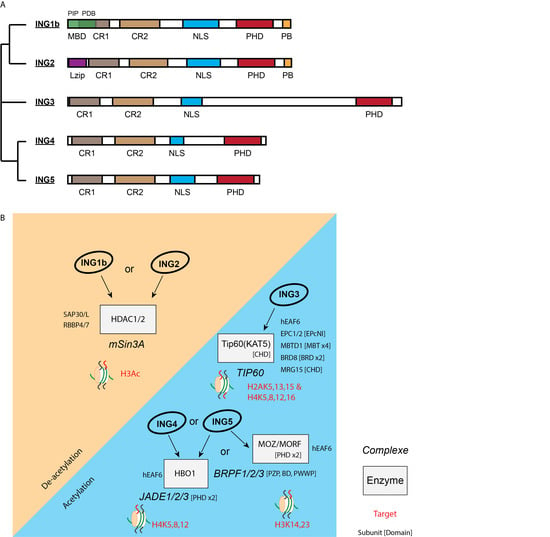
Figure 1.
INhibitor of Growth (ING) proteins’ structure and complexes. (A) Phylogenic tree, domains, and conservation of the ING family. MBD: metal-binding domain responsible for zinc binding unique to ING1b. The MBD contains two subdomains: a PIP domain (PCNA-interacting protein motif) and a PDB domain (partial bromodomain); CR1 and 2: conserved regions 1 and 2; Lzip: leucine zipper-like region; NLS: nuclear localisation sequence; PHD: plant homeodomain responsible for binding to H3K4me3; PB: polybasic region unique to ING1 and ING2. ING4 and ING5 are paralogs that may form homo- or heterodimers in the cell [17,18]. ING1b and ING2 are close due to the presence of the PB motif in C-terminus. (B) ING protein association and targets. ING proteins are part of multiprotein complexes. There is a variable complexity of these complexes: from the TIP60 complex, which has around 18 stable subunits, to JADE or BRPF complexes, which harbour 5 subunits. INGs are all associated with an enzymatic activity implicated in the regulation of chromatin acetylation, on one side associated with the MYST (MOZ, Ybf2/Sas3, Sas2, and Tip60) family of acetyltransferases (HBO1 (KAT7), MOZ (KAT6A), MORF (KAT6B), Tip60 (KAT5) for ING3/4/5) but on the other side associated with the deacetylases HDAC1 (RPD3L1) and HDAC2 from the class I family of enzymes for ING1/2. The other subunits play an important targeting role in the ING complexes like the scaffolding subunits JADE (double PHD), BRBF (PZP, BD and PWWP), or EPC1/2 (EPcNI). Subunits having targeting function through chromatin reader domains are mentioned.
Over two decades ago, groundbreaking work identified ING1 as a tumour suppressor [19], and as such most studies of ING proteins have focused on their cancer-protective potential [20]. Unfortunately, almost all investigations merely correlate levels of ING proteins in various cancers without directly linking the functions of ING proteins with tumour suppression [21]. In addition, the evidence that ING proteins could be tumour suppressors is largely based on supraphysiological exogenous overexpression in cancer cell lines put under stress conditions (e.g., [22,23,24,25]). Investigations in transgenic mouse models have so far seemed to confirm that Ing1 and Ing2 are tumour suppressor genes, respectively preventing lymphomas and soft-tissue sarcomas [26,27,28]. However, recent investigations demonstrate that other ING proteins might actually be required for normal proliferation [29,30,31,32]. Thus, this short review will discuss ING proteins’ pleiotropic functions during oncogenesis.
2. Cellular Functions
INGs’ functions can hardly be discriminated from their complex roles. Generally, defects for the different unique subunits show similar or related phenotypes. However, understanding ING-specific functions as part of the different complexes can be difficult [33,34]. Nonetheless, common functions seem to be shared among the ING family (Figure 2).
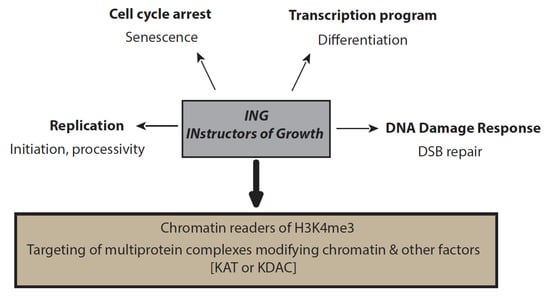
Figure 2.
ING functions. ING proteins are peculiar chromatin readers with a PHD domain. They target their respective complexes to specific regions of the genome, where the different ING-containing complexes are responsible for posttranslational modifications (PTMs) of histones, as well as nonhistone proteins. These PTMs are involved in the regulation of many pathways during transcription, replication, DNA damage, and cell cycle arrest processes.
2.1. ING Proteins and DNA Damage
ING1 and ING2 function within the mSIN3A/HDAC1 complex [6]. This complex seems recruited to DNA damages to induce hypoacetyled H3K56 and the nonhomologous end joining (NHEJ) pathway [35,36]. Acetylation of chromatin is observed to open the damage sites and grant access to the repair machinery [37]. Thus, mSIN3A/HDAC1 might also be engaged in restoring the chromatin landscape after DNA repair [38,39]. Functionally, the PHD domain of ING2 targets the mSIN3A/HDAC1 complex to cell cycle promoters (e.g., CCND1 gene encoding cyclin D1) in response to DNA damages [10]. An intact aromatic cage is also required for ING1 to facilitate DNA repair after UV irradiation and induce cell death [40]. In parallel, ING1 PIP domain (PCNA interacting protein domain) binds PCNA in response to UV radiation [41] (Figure 1A). PCNA acetylation should be tightly regulated, as it is interrelated with other critical posttranslational modifications (PTM), potentially affecting its structure and function during genome replication and repair [42].
Notably, ING3 within the TIP60 complex regulates DNA double-strand break (DSB) repair and influences the choice between NHEJ and homologous recombination (HR) repair pathways [43]. Moreover, ING3 was recently found to directly regulate ATM signalling at DSBs [44], as previously shown for Tip60 [45]. Other ING homologs were found to play a role during the DNA damage response in yeast and nematode [46,47]. Similarly, in response to DNA damages, ING4 appears to associate with H3K4me3 at gene promoters (i.e., SMC4) to regulate their expression and induce apoptosis [9]. Finally, ING5 also seems to play a role during the DNA damage response (DDR). Specifically, an increased level of ING5 is observed upon damages, followed by its translocation into the nucleus [48].
2.2. Cell Cycle, Cellular Senescence, and Apoptosis
In a simplified view, tumour suppression can function by inducing either apoptosis or a permanent state of exit from the cell cycle called senescence [49]. Interestingly, senescent cells have a marked increase in ING1 levels while silencing of ING1 increases proliferative lifespan [50]. Similarly, ING2 levels are elevated in late-passage human fibroblasts and its overexpression induces senescence, which could be delayed by ING2 silencing [51]. Interestingly, inhibition of the ING5-associated MOZ/MORF acetyltransferases also induces senescence and prevents tumour growth [52].
ING proteins are well-known regulators of the p53 pathway [13,14,25]. They can act at different levels, mostly transcriptionally, but also through direct modification of p53 by acetylation/deacetylation inducing the regulation of the G1/S transition. For example, ING3, as part of TIP60, upregulates the p53 pathway through p21 transcription and induces cell cycle arrest [53,54,55]. The ING3-containing complex TIP60 is also responsible for p53 acetylation leading to the activation of apoptosis [53,56,57]. Overexpression of ING3 in colon cancer cell lines induces apoptosis and inhibits tumour growth [25], whereas downregulation of ING3 is associated with a poor prognosis in cutaneous melanoma [58].
Of note, ING4 expression in normal fibroblasts induces proliferation arrest and a senescence-associated secretory phenotype (SASP) [59]. In agreement with previous work showing that SASP stimulates tumour progression in the tissue microenvironment [60], these ING4-induced senescent cells promote tumour growth in mice [59].
Finally, ING5 can interact with the replication machinery. The HBO1 (histone acetyltransferase binding to ORC1) complex is recruited at replication origins for hyperacetylation of H3 [61,62]. Similarly, ING5 has been described as interacting with the minichromosome maintenance helicases (MCM) complex and is critical for normal progression through the S phase [6].
2.3. Other Functions of the INGs
ING4 interacts with the NFκB pathway to suppress angiogenesis in glioma, colorectal, and breast cancers [63,64,65]. ING4 also represses HIF (hypoxia-inducible factors) and regulates its transcriptional activity as well [66]. Hence, ING4 plays a key antitumorigenic function. ING1 seems to play a similar role through the regulation of angiopoietins [67].
3. ING Family Roles in Cancer
Expression of ING Proteins in Cancer
The levels of ING genes and proteins have been reported to be lost in several studies involving a restricted number of cases (reviewed in [3,19]). However, contrarily to what would be expected from a family of genes called INhibitor of Growth, ING genes are, according to cBioPortal for Cancer Genomics [68,69], broadly altered, i.e., amplified, deleted, or mutated in cancers, but at moderate levels (usually below a frequency of 10%).
Although previous investigations have reported a loss of ING3 expression in small sample number studies, we found, using the cBioPortal, that INGs are broadly amplified in a variety of cancers [32]. Using a high-throughput tissue microarray approach consisting of benign prostate hyperplasia and prostate cancer samples, we have validated that ING3 protein levels are elevated in prostate cancer, corresponding to cBioPortal genomic data [32]. Moreover, although altered in non-small-cell lung cancers, ING2 seems to be required for cancer proliferation [70]. In other words, the expression of ING proteins in cancer does not always correlate with cell proliferation.
4. ING Proteins in Eukaryotic Model Organisms
4.1. Yeast Deletion Models
ING proteins are conserved in unicellular eukaryotes, such as Saccharomyces cerevisiae, which has three ING-like proteins called Yng1, Yng2, and Pho23 [46]. The closest relatives of Yng1 are ING4/5, while Yng2 is related to human ING3. Since Pho23 is associated with Rpd3 [71], it would be related to ING1 and ING2. Yng2 associates with Tra1 (TRRAP in mammals) [46] through a direct interaction with Epl1, further supporting that Yng2 and ING3 are related as ING3 associates with the TIP60/TRRAP complex [6]. Notably, the three yeast ING homologs (i.e., Pho23, Yng1, and Yng2) were purified as stable components of Sin3/Rpd3 deacetylase complex or NuA3 and NuA4 acetyltransferase complexes, respectively [72,73,74].
Counterintuitively, ΔYng2 S. cerevisiae cells grow slower than wild-type cells (~5 h doubling time versus ~3 h normally) and are multibudded, while ΔYng1 and ΔPho23 grow normally, but combined ΔYng1/ΔYng2/ΔPho23 deletion leads to further growth impairment (~10 h doubling time) [46]. Moreover, both ΔYng1 and ΔYng2 cells fail to grow from nonfermentable carbon sources (e.g., galactose, glycerol) [46]. Mild growth defects were revealed in the ΔPho23 strain grown on galactose or glycerol, but also in response to heat stress [46]. Importantly, the expression of human ING1 or Schizosaccharomyces pombe Png1 can rescue the ΔYng2 growth defect phenotypes [46]. These experiments demonstrate that the functions of ING proteins are conserved between unicellular eukaryotes and humans but also highlight that, contrary to what their names suggest, at least some ING family members are actually required for growth [74].
A number of potential mechanisms have been suggested for the growth defects observed in yeast models. The Yng2 deletion impairs progression through mitosis and meiosis [75] but is also required to overcome DNA damage-induced S phase arrest [76]. As in humans, Yng2 (ING3) and Yaf9 (YEATS4/GAS41) associate with the acetyltransferase Esa1 (Tip60/KAT5). Interestingly, both ΔYaf9 and ΔYng2 have growth defects that are enhanced by spindle stress [77], supporting their role during chromosome segregation in M phase.
In addition, the ΔPho23 strain displays elevated levels of autophagy gene expression, along with corresponding ATG proteins, as well as increased autophagy activity upon nitrogen starvation [78].
Finally, genetic deletion of the S. pombe ING ortholog Png1 also results in slower proliferation [46,79].
4.2. Ing1−/− Mouse Model
Interestingly, deletion of all Ing1 isoforms in CJ-7 mouse strain predisposes animals to lymphomas [27], suggesting that Ing1 is indeed a tumour suppressor. Moreover, disruption of the p37Ing1 isoform in 129/SVJ strain, while leaving the p31Ing1 isoform intact, was sufficient to increase the incidence of follicular B-cell lymphomas [26]. Since p31Ing1 remains intact in the p37Ing1-null animals, this suggests that p31Ing1 does not compensate for the loss of p37Ing1 even though the levels of p31Ing1 are elevated in p37Ing1-null tissues [26].
Although CJ-7 Ing1−/− mouse embryonic fibroblasts (MEF) have a normal replicative lifespan and appear to have normal response to growth-inhibitory treatments, they display mild cell cycle profile alterations in response to various genotoxic insults with a reduced G2 population [27]. Comparably, 129/SVJ p37Ing1-null MEFs had increased growth rates [26]. Interestingly, the reduced body weight reported for CJ-7 Ing1−/− animals is in agreement with growth defects observed in the ΔYng2 S. cerevisiae strain.
Interestingly, knockout of all Ing1 isoforms in TBV-2 strain results in MEFs that are resistant to RasV12-induced senescence [80]. Oncogene-induced senescence (OIS) is triggered by increased Ras/Raf/MAPK signalling through mutations (e.g., RasV12), upstream cascade activation (e.g., EGFR), or downstream loss of pRB function and increased E2F1 activity. It remains unknown how loss of Ing1 prevents RASV12-induced senescence and if or how and where it regulates the signalling cascade. Since Ing1 is required for OIS, it is likely acting downstream of Ras.
4.3. Ing2−/− Mouse Model
Ing2 mRNA is highly expressed in testes, followed by lung and spleen [28]. In agreement with a role in testes biology, male Ing2−/− 129/SV mice had smaller testes, lower sperm counts, and impaired sperm motility, with abnormal morphology, which resulted in male infertility [28].
Although the authors did not demonstrate that Ing2 regulates directly the genes that they have identified by microarray, they have found that a number of sperm (e.g., Prss21, Sly, Spef2) and chromatin (e.g., Setdb2, Suv39h2, Ing3) genes were downregulated in Ing2−/− testes [28]. Since ING3 is required for cellular proliferation [32], decreased Ing3 levels in Ing2−/− testes may hypothetically explain smaller testes in Ing2−/− males. In addition, p53 and p53-regulated apoptotic factor PUMA were upregulated in Ing2−/− testes, which correlated with increased apoptotic cells, while senescence appeared unaffected [28].
Importantly, Ing2−/− mice were afflicted by increased soft-tissue sarcoma [28], demonstrating that loss of Ing2 is sufficient to increase tumorigenicity and that Ing2 is likely a bona fide tumour suppressor.
4.4. Ing3−/− Mouse Model
So far, there is one recent report of Ing3 KO mice [81]. In this study, as expected, Ing3−/− mice were embryonic lethal at stage E10.5. Thus, they display very strong growth defects as well as severe developmental disorders of the nervous system, revealing a lack of ectodermal differentiation. Tip60 KO causes early embryonic lethality near blastocyst stage and embryo death before implantation [82,83]. However, because of the embryonic lethal phenotype, it could not be concluded that ING3 functions as an oncoprotein or a tumour suppressor.
4.5. Ing4−/− Mouse Model
Lipopolysaccharides (LPSs) are membrane components of Gram-negative bacteria and trigger a potent immune-inflammatory response by activating the NFκB transcription factor. The ING protein ING4 was found to regulate NFκB-dependent transcription and thus is hypothesised to be involved in inflammatory responses. Basically, LPS binds to the toll-like TLR4 receptor, activating a signalling cascade that results in the phosphorylation of the NFκB inhibitor IκB by IKK1/2, phospho-dependent degradation of IκB, and expression of NFκB-responsive genes [84].
Interestingly, the Ing4−/− mouse model is viable; the mice developed normally and remained free of tumours over the 20-month period investigated [85]. However, Ing4−/− mice expressed high levels of inflammatory cytokines (e.g., IL6) and died within 24 h of exposure to LPS.
In agreement with the Ing4−/− model, small interfering RNA-mediated silencing of ING4 prevents normal cell cycle progression of human breast cancer MCF7 cells, marked with an accumulation in the G2/M phase of the cell cycle [6]. Together, these results suggest that, although ING4 is involved to some degree in the regulation of the cell cycle, its principal function is not to suppress tumour development.
4.6. Ing5−/− Mouse Model
To our knowledge there are no published Ing5−/− knockout mouse models yet, thus limiting the interpretation of ING5′s role as a candidate tumour suppressor.
4.7. Cell-Based Gene Silencing
Silencing of either HBO1 (ING4 complex subunit) or TIP60 (ING3 complex subunit) acetyltransferases by shRNA reduced proliferation of human embryonic kidney cells (HEK293T), which accumulated in G2/M [6], a phenotype also observed in yeast [7]. In agreement, knockout of ING4 in myelogenous leukaemia HAP1 cells led to reduced proliferation [86]. Silencing of HBO1 in human breast cancer cells (MCF7) also decreased BrdU incorporation, suggesting a role in DNA replication [6]. In contrast, silencing of ING4 only modestly reduced BrdU incorporation in MCF7 cells but led to an increase in G2/M population. However, silencing of ING5, which also associates with HBO1, led to the disappearance of the G2/M phase population and entirely blocked DNA synthesis [6]. Interestingly, ING5-silenced human osteosarcoma cells (U2OS) are partially resistant to DNA damage-induced apoptosis [48]. Moreover, two independent shRNA targeting ING5 inhibited the growth of HCT116, HCT116-p53−/−, U2OS, and HeLa cell lines, possibly by inducing apoptosis [87]. In agreement, silencing of ING5 in primary human keratinocytes reduced clone formation in an organotypic skin reconstitution assay due to terminal differentiation [88].
In agreement with the requirement of TIP60 for cellular proliferation [6], we observed that ING3 was also essential for the proliferation of human breast (MCF7) and prostate (LNCaP, PC3) cancer cell lines [32], suggesting a central role for the ING3/TIP60 complex in cell cycle progression. Indeed, ING3 regulates the expression of cell cycle genes by interacting with H3K4me3 at transcriptional start sites [32]. In addition, silencing of ING3 induces cellular differentiation and promotes adipogenesis using a human mesenchymal progenitor cell-derived osteoblast cellular model [44], again highlighting the requirement of ING3 for sustained cellular proliferation in at least these instances. Interestingly, cell-based silencing experiments recently demonstrated that ING3 is necessary for proper nonhomologous end joining (NHEJ) and homologous recombination (HR) DNA damage repair pathways [44], potentially leading to cell cycle arrest, in agreement with previous work [32,46].
4.8. Other Models
INGs have been studied in other models. In Caenorhabditis elegans, ING3 activates apoptosis induced by DNA damages through a p53-dependent pathway [47]. In Xenopus laevis, several splice variants were identified for ING1 (seven variants so far), all playing a positive role in apoptosis [89].
5. Reclassification of ING Proteins
There is increasing evidence that ING proteins have different, sometimes opposite, functions. Animal models clearly define ING1 and ING2 as bona fide tumour suppressors. However, yeast models suggested early on that ING proteins are required for cellular proliferation under some environmental conditions. In support of this hypothesis, we have recently found that ING3 (which is, based on sequence similarity, the closest relative to yeast Yng2, an ING member required for growth in a nonfermentable carbon-source environment) is also required for cellular proliferation of human breast and prostate cancer cells [32].
In agreement with yeast models, we found that ING3 is required for proliferation of breast, ovarian, and prostate cancer cells [32], suggesting that it may function as an oncoprotein instead of a tumour suppressor. Indeed, increased ING3 levels in prostate cancer patient samples correlate with poor survival. Moreover, ING3 expression in normal human cells was sufficient for transformation [32].
Thus, ING proteins should more accurately be called INstructors of Growth, to retain the gene symbols and refer to cell cycle regulators instead of candidate tumour suppressors, to better reflect the broad pleiotropic functions of the family members.
6. Conclusions
In light of limited previous observations and recent systematic approaches, it appears that ING3 is not a tumour suppressor, at least in some cancers [29,32]. Interestingly, ING4 and ING5 are also required for proliferation of breast cancer cells, supporting the hypothesis that ING proteins play pleiotropic functions in eukaryotic cells.
Alternative splicing might be a mechanism that modulates ING proteins’ functions, as for ING1 [90,91]. Most probably, posttranslational modifications (PTMs) of the ING proteins should be considered as a way to switch/modulate their functions ON and OFF. Among those modifications, acetylation is central because we already know that HAT complexes can autoacetylate, not only on the catalytic subunit but also on the other subunits [92]. Furthermore, PTM of the PHD in particular is already documented for ING4 and ING5 [93]. Those PTMs could regulate their half-life (i.e., ubiquitylation), subcellular localization (i.e., PTM in NLS), and interaction with partner subunits or interactors (through phosphorylation and others) (for review, see [94]). For example, ING4 was reported to be phosphorylated on threonine 197 (T197) and tyrosine 198 (Y198) [95]. The latter is one of the residues composing the H3K4me3-binding aromatic cage [9]. Based on mutational analyses of other readers, phosphorylation of ING4-Y198 would likely impact its interaction with H3K4me3 [96].
We thus propose to rename the ING proteins the INstructors of Growth family of histone mark readers to retain the gene symbol by which they are known, while highlighting the inhibiting and stimulating impact on proliferation of ING1–2 and ING3–5, respectively. Much work remains to definitively characterise the expanding cellular roles of the ING family of epigenetic regulators in normal and pathological states.
Funding
This research received no external funding.
Acknowledgments
We apologise for omitted references due to space limitation.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lachner, M.; O’Carroll, D.; Rea, S.; Mechtler, K.; Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 2001, 410, 116–120. [Google Scholar] [CrossRef]
- Musselman, C.A.; Lalonde, M.E.; Côté, J.; Kutateladze, T.G. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012, 19, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Bua, D.J.; Binda, O. The return of the INGs, histone mark sensors and phospholipid signaling effectors. Curr. Drug Targets 2009, 10, 418–431. [Google Scholar] [CrossRef]
- Kuzmichev, A.; Zhang, Y.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Role of the SIN3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol. Cell Biol. 2002, 22, 835–848. [Google Scholar] [CrossRef]
- Skowyra, D.; Zeremski, M.; Neznanov, N.; Li, M.; Choi, Y.; Uesugi, M.; Hauser, C.A.; Gu, W.; Gudkov, A.V.; Qin, J. Differential association of products of alternative transcripts of the candidate tumor suppressor ing1 with the mSIN3/HDAC1 transcriptional corepressor complex. J. Biol. Chem. 2001, 276, 8734–8739. [Google Scholar] [CrossRef]
- Doyon, Y.; Cayrou, C.; Ullah, M.; Landry, A.J.; Côté, V.; Selleck, W.; Lane, W.S.; Tan, S.; Yang, X.J.; Côté, J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell. 2006, 21, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Doyon, Y.; Selleck, W.; Lane, W.S.; Tan, S.; Côté, J. Structural and functional conservation of the nua4 histone acetyltransferase complex from yeast to humans. Mol. Cell Biol. 2004, 24, 1884–1896. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, G.W.; Kwon, S.H.; Lee, J.S. Broad domains of histone H3 lysine 4 trimethylation in transcriptional regulation and disease. FEBS J. 2020, 287, 2891–2902. [Google Scholar] [CrossRef] [PubMed]
- Hung, T.; Binda, O.; Champagne, K.S.; Kuo, A.J.; Johnson, K.; Chang, H.Y.; Simon, M.D.; Kutateladze, T.G.; Gozani, O. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol. Cell 2009, 33, 248–256. [Google Scholar] [CrossRef]
- Shi, X.; Hong, T.; Walter, K.L.; Ewalt, M.; Michishita, E.; Hung, T.; Carney, D.; Peña, P.; Lan, F.; Kaadige, M.R.; et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 2006, 442, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Peña, P.V.; Davrazou, F.; Shi, X.; Walter, K.L.; Verkhusha, V.V.; Gozani, O.; Zhao, R.; Kutateladze, T.G. Molecular mechanism of histone h3k4me3 recognition by plant homeodomain of ING2. Nature 2006, 442, 100–103. [Google Scholar] [CrossRef]
- Wysocka, J.; Swigut, T.; Xiao, H.; Milne, T.A.; Kwon, S.Y.; Landry, J.; Kauer, M.; Tackett, A.J.; Chait, B.T.; Badenhorst, P.; et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 2006, 442, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, M.; Shiseki, M.; Miura, K.; Hagiwara, K.; Linke, S.P.; Pedeux, R.; Wang, X.W.; Yokota, J.; Riabowol, K.; Harris, C.C. DNA damage-inducible gene p33ING2 negatively regulates cell proliferation through acetylation of p53. Proc. Natl. Acad. Sci. USA 2001, 98, 9671–9676. [Google Scholar] [CrossRef]
- Shiseki, M.; Nagashima, M.; Pedeux, R.M.; Kitahama-Shiseki, M.; Miura, K.; Okamura, S.; Onogi, H.; Higashimoto, Y.; Appella, E.; Yokota, J.; et al. P29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 2003, 63, 2373–2378. [Google Scholar]
- Gozani, O.; Karuman, P.; Jones, D.R.; Ivanov, D.; Cha, J.; Lugovskoy, A.A.; Baird, C.L.; Zhu, H.; Field, S.J.; Lessnick, S.L.; et al. The phd finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 2003, 114, 99–111. [Google Scholar] [CrossRef]
- Bua, D.J.; Martin, G.M.; Binda, O.; Gozani, O. Nuclear phosphatidylinositol-5-phosphate regulates ING2 stability at discrete chromatin targets in response to DNA damage. Sci. Rep. 2013, 3, 2137. [Google Scholar] [CrossRef]
- Culurgioni, S.; Munoz, I.G.; Moreno, A.; Palacios, A.; Villate, M.; Palmero, I.; Montoya, G.; Blanco, F.J. Crystal structure of inhibitor of growth 4 (ING4) dimerization domain reveals functional organization of ING family of chromatin-binding proteins. J. Biol. Chem. 2012, 287, 10876–10884. [Google Scholar] [CrossRef]
- Ormaza, G.; Rodriguez, J.A.; Ibanez de Opakua, A.; Merino, N.; Villate, M.; Gorrono, I.; Rabano, M.; Palmero, I.; Vilaseca, M.; Kypta, R.; et al. The tumor suppressor ING5 is a dimeric, bivalent recognition molecule of the histone H3K4me3 mark. J. Mol. Biol. 2019, 431, 2298–2319. [Google Scholar] [CrossRef]
- Garkavtsev, I.; Kazarov, A.; Gudkov, A.; Riabowol, K. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat. Genet. 1996, 14, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Campos, E.I.; Chin, M.Y.; Kuo, W.H.; Li, G. Biological functions of the ing family tumor suppressors. Cell. Mol. Life Sci. CMLS 2004, 61, 2597–2613. [Google Scholar] [CrossRef]
- Guérillon, C.; Bigot, N.; Pedeux, R. The ING tumor suppressor genes: Status in human tumors. Cancer Lett. 2014, 345, 1–16. [Google Scholar] [CrossRef]
- Garkavtsev, I.; Grigorian, I.A.; Ossovskaya, V.S.; Chernov, M.V.; Chumakov, P.M.; Gudkov, A.V. The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control. Nature 1998, 391, 295–298. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, L.S.; Wang, Z.Q.; Wang, K.S.; Li, N.; Cheng, Z.H.; Huang, S.Z.; Wei, D.Z.; Han, Z.G. ING4 induces G2/M cell cycle arrest and enhances the chemosensitivity to DNA-damage agents in HepG2 cells. FEBS Lett. 2004, 570, 7–12. [Google Scholar] [CrossRef]
- Kim, S.; Chin, K.; Gray, J.W.; Bishop, J.M. A screen for genes that suppress loss of contact inhibition: Identification of ING4 as a candidate tumor suppressor gene in human cancer. Proc. Natl. Acad. Sci. USA 2004, 101, 16251–16256. [Google Scholar] [CrossRef]
- Nagashima, M.; Shiseki, M.; Pedeux, R.M.; Okamura, S.; Kitahama-Shiseki, M.; Miura, K.; Yokota, J.; Harris, C.C. A novel PHD-finger motif protein, p47ING3, modulates p53-mediated transcription, cell cycle control, and apoptosis. Oncogene 2003, 22, 343–350. [Google Scholar] [CrossRef]
- Coles, A.H.; Liang, H.; Zhu, Z.; Marfella, C.G.; Kang, J.; Imbalzano, A.N.; Jones, S.N. Deletion of p37Ing1 in mice reveals a p53-independent role for Ing1 in the suppression of cell proliferation, apoptosis, and tumorigenesis. Cancer Res. 2007, 67, 2054–2061. [Google Scholar] [CrossRef] [PubMed]
- Kichina, J.V.; Zeremski, M.; Aris, L.; Gurova, K.V.; Walker, E.; Franks, R.; Nikitin, A.Y.; Kiyokawa, H.; Gudkov, A.V. Targeted disruption of the mouse Ing1 locus results in reduced body size, hypersensitivity to radiation and elevated incidence of lymphomas. Oncogene 2006, 25, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Kumamoto, K.; Robles, A.I.; Horikawa, I.; Furusato, B.; Okamura, S.; Goto, A.; Yamashita, T.; Nagashima, M.; Lee, T.L.; et al. Targeted disruption of Ing2 results in defective spermatogenesis and development of soft-tissue sarcomas. PLoS ONE 2010, 5, e15541. [Google Scholar] [CrossRef] [PubMed]
- Nabbi, A.; McClurg, U.L.; Thalappilly, S.; Almami, A.; Mobahat, M.; Bismar, T.A.; Binda, O.; Riabowol, K.T. ING3 promotes prostate cancer growth by activating the androgen receptor. BMC Med. 2017, 15, 103. [Google Scholar] [CrossRef] [PubMed]
- Almami, A.; Hegazy, S.A.; Nabbi, A.; Alshalalfa, M.; Salman, A.; Abou-Ouf, H.; Riabowol, K.; Bismar, T.A. ING3 is associated with increased cell invasion and lethal outcome in erg-negative prostate cancer patients. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2016, 37, 9731–9738. [Google Scholar] [CrossRef]
- Nabbi, A.; Almami, A.; Thakur, S.; Suzuki, K.; Boland, D.; Bismar, T.A.; Riabowol, K. ING3 protein expression profiling in normal human tissues suggest its role in cellular growth and self-renewal. Eur. J. Cell Biol. 2015, 94, 214–222. [Google Scholar] [CrossRef] [PubMed]
- McClurg, U.L.; Nabbi, A.; Ricordel, C.; Korolchuk, S.; McCracken, S.; Heer, R.; Wilson, L.; Butler, L.M.; Irving-Hooper, B.K.; Pedeux, R.; et al. Human ex vivo prostate tissue model system identifies ING3 as an oncoprotein. Br. J. Cancer 2018, 118, 713–726. [Google Scholar] [CrossRef] [PubMed]
- Lalonde, M.E.; Avvakumov, N.; Glass, K.C.; Joncas, F.H.; Saksouk, N.; Holliday, M.; Paquet, E.; Yan, K.; Tong, Q.; Klein, B.J.; et al. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 2013, 27, 2009–2024. [Google Scholar] [CrossRef]
- Lalonde, M.E.; Cheng, X.; Cote, J. Histone target selection within chromatin: An exemplary case of teamwork. Genes Dev. 2014, 28, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.M.; Tjeertes, J.V.; Coates, J.; Legube, G.; Polo, S.E.; Britton, S.; Jackson, S.P. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 2010, 17, 1144–1151. [Google Scholar] [CrossRef]
- Zhu, Q.; Battu, A.; Ray, A.; Wani, G.; Qian, J.; He, J.; Wang, Q.E.; Wani, A.A. Damaged DNA-binding protein down-regulates epigenetic mark H3K56ac through histone deacetylase 1 and 2. Mutat. Res. 2015, 776, 16–23. [Google Scholar] [CrossRef]
- Rossetto, D.; Truman, A.W.; Kron, S.J.; Côté, J. Epigenetic modifications in double-strand break DNA damage signaling and repair. Clin. Cancer Res. 2010, 16, 4543–4552. [Google Scholar] [CrossRef]
- Jazayeri, A.; McAinsh, A.D.; Jackson, S.P. Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 2004, 101, 1644–1649. [Google Scholar] [CrossRef]
- Tamburini, B.A.; Tyler, J.K. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol. Cell Biol. 2005, 25, 4903–4913. [Google Scholar] [CrossRef]
- Peña, P.V.; Hom, R.A.; Hung, T.; Lin, H.; Kuo, A.J.; Wong, R.P.; Subach, O.M.; Champagne, K.S.; Zhao, R.; Verkhusha, V.V.; et al. Histone H3K4me3 binding is required for the DNA repair and apoptotic activities of ING1 tumor suppressor. J. Mol. Biol. 2008, 380, 303–312. [Google Scholar] [CrossRef]
- Scott, M.; Bonnefin, P.; Vieyra, D.; Boisvert, F.M.; Young, D.; Bazett-Jones, D.P.; Riabowol, K. Uv-induced binding of ING1 to PCNA regulates the induction of apoptosis. J. Cell Sci. 2001, 114, 3455–3462. [Google Scholar] [CrossRef]
- Billon, P.; Li, J.; Lambert, J.P.; Chen, Y.; Tremblay, V.; Brunzelle, J.S.; Gingras, A.C.; Verreault, A.; Sugiyama, T.; Couture, J.F.; et al. Acetylation of PCNA sliding surface by ECO1 promotes genome stability through homologous recombination. Mol. Cell 2017, 65, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Jacquet, K.; Fradet-Turcotte, A.; Avvakumov, N.; Lambert, J.P.; Roques, C.; Pandita, R.K.; Paquet, E.; Herst, P.; Gingras, A.C.; Pandita, T.K.; et al. The TIP60 complex regulates bivalent chromatin recognition by 53BP1 through direct H4K20me binding and h2ak15 acetylation. Mol. Cell 2016, 62, 409–421. [Google Scholar] [CrossRef]
- Mouche, A.; Archambeau, J.; Ricordel, C.; Chaillot, L.; Bigot, N.; Guillaudeux, T.; Grenon, M.; Pedeux, R. IIN3 is required for ATM signaling and DNA repair in response to DNA double strand breaks. Cell Death Differ. 2019, 26, 2344–2357. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Jiang, X.; Chen, S.; Fernandes, N.; Price, B.D. A role for the TIP60 histone acetyltransferase in the acetylation and activation of atm. Proc. Natl. Acad. Sci. USA 2005, 102, 13182–13187. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Meijer, M.; Lees-Miller, S.P.; Riabowol, K.; Young, D. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol. Cell Biol. 2000, 20, 3807–3816. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Shah, S.; Riabowol, K.; Mains, P.E. The caenorhabditis elegans Ing-3 gene regulates ionizing radiation-induced germ-cell apoptosis in a p53-associated pathway. Genetics 2009, 181, 473–482. [Google Scholar] [CrossRef]
- Liu, N.; Wang, J.; Wang, J.; Wang, R.; Liu, Z.; Yu, Y.; Lu, H. ING5 is a TIP60 cofactor that acetylates p53 in response to DNA damage. Cancer Res. 2013, 73, 3749–3760. [Google Scholar] [CrossRef]
- Hayflick, L.; Moorhead, P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961, 25, 585–621. [Google Scholar] [CrossRef]
- Garkavtsev, I.; Riabowol, K. Extension of the replicative life span of human diploid fibroblasts by inhibition of the p33ING1 candidate tumor suppressor. Mol. Cell Biol. 1997, 17, 2014–2019. [Google Scholar] [CrossRef]
- Pedeux, R.; Sengupta, S.; Shen, J.C.; Demidov, O.N.; Saito, S.; Onogi, H.; Kumamoto, K.; Wincovitch, S.; Garfield, S.H.; McMenamin, M.; et al. ING2 regulates the onset of replicative senescence by induction of p300-dependent p53 acetylation. Mol. Cell Biol. 2005, 25, 6639–6648. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.B.; Leaver, D.J.; Hermans, S.J.; Kelly, G.L.; Brennan, M.S.; Downer, N.L.; Nguyen, N.; Wichmann, J.; McRae, H.M.; Yang, Y.; et al. Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature 2018, 560, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Luo, J.; Zhang, W.; Gu, W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 2006, 24, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Legube, G.; Linares, L.K.; Tyteca, S.; Caron, C.; Scheffner, M.; Chevillard-Briet, M.; Trouche, D. Role of the histone acetyl transferase TIP60 in the p53 pathway. J. Biol. Chem. 2004, 279, 44825–44833. [Google Scholar] [CrossRef]
- Berns, K.; Hijmans, E.M.; Mullenders, J.; Brummelkamp, T.R.; Velds, A.; Heimerikx, M.; Kerkhoven, R.M.; Madiredjo, M.; Nijkamp, W.; Weigelt, B.; et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 2004, 428, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Sykes, S.M.; Mellert, H.S.; Holbert, M.A.; Li, K.; Marmorstein, R.; Lane, W.S.; McMahon, S.B. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 2006, 24, 841–851. [Google Scholar] [CrossRef]
- Tang, Y.; Zhao, W.; Chen, Y.; Zhao, Y.; Gu, W. Acetylation is indispensable for p53 activation. Cell 2008, 133, 612–626. [Google Scholar] [CrossRef]
- Wang, Y.; Dai, D.L.; Martinka, M.; Li, G. Prognostic significance of nuclear ING3 expression in human cutaneous melanoma. Clin. Cancer Res. 2007, 13, 4111–4116. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.; Soleto, I.; Garcia-Sanz, P.; Moreno-Bueno, G.; Palmero, I. ING4 regulates a secretory phenotype in primary fibroblasts with dual effects on cell proliferation and tumor growth. Oncogene 2014, 33, 1945–1953. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Kawamoto, S.; Ohtani, N.; Hara, E. Impact of senescence-associated secretory phenotype and its potential as a therapeutic target for senescence-associated diseases. Cancer Sci. 2017, 108, 563–569. [Google Scholar] [CrossRef]
- Stedman, W.; Deng, Z.; Lu, F.; Lieberman, P.M. ORC, MCM, and histone hyperacetylation at the Kaposi’s sarcoma-associated herpesvirus latent replication origin. J. Virol. 2004, 78, 12566–12575. [Google Scholar] [CrossRef]
- Feng, Y.; Vlassis, A.; Roques, C.; Lalonde, M.E.; Gonzalez-Aguilera, C.; Lambert, J.P.; Lee, S.B.; Zhao, X.; Alabert, C.; Johansen, J.V.; et al. BRPF3-HBO1 regulates replication origin activation and histone H3K14 acetylation. EMBO J. 2016, 35, 176–192. [Google Scholar] [CrossRef]
- Garkavtsev, I.; Kozin, S.V.; Chernova, O.; Xu, L.; Winkler, F.; Brown, E.; Barnett, G.H.; Jain, R.K. The candidate tumour suppressor protein ING4 regulates brain tumour growth and angiogenesis. Nature 2004, 428, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Huang, Y.; Hou, P.; Zhang, Z.; Zhang, Y.; Wang, W.; Sun, G.; Xu, L.; Zhou, J.; Bai, J.; et al. ING4 suppresses tumor angiogenesis and functions as a prognostic marker in human colorectal cancer. Oncotarget 2016, 7, 79017–79031. [Google Scholar] [CrossRef]
- Yan, R.; He, L.; Li, Z.; Han, X.; Liang, J.; Si, W.; Chen, Z.; Li, L.; Xie, G.; Li, W.; et al. SCF(JFK) is a bona fide E3 ligase for ING4 and a potent promoter of the angiogenesis and metastasis of breast cancer. Genes Dev. 2015, 29, 672–685. [Google Scholar] [CrossRef] [PubMed]
- Ozer, A.; Wu, L.C.; Bruick, R.K. The candidate tumor suppressor ING4 represses activation of the hypoxia inducible factor (HIF). Proc. Natl. Acad. Sci. USA 2005, 102, 7481–7486. [Google Scholar] [CrossRef]
- Tallen, G.; Farhangi, S.; Tamannai, M.; Holtkamp, N.; Mangoldt, D.; Shah, S.; Suzuki, K.; Truss, M.; Henze, G.; Riabowol, K.; et al. The inhibitor of growth 1 (ING1) proteins suppress angiogenesis and differentially regulate angiopoietin expression in glioblastoma cells. Oncol. Res. 2009, 18, 95–105. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Blondel, A.; Benberghout, A.; Pedeux, R.; Ricordel, C. Exploiting ING2 epigenetic modulation as a therapeutic opportunity for non-small cell lung cancer. Cancers 2019, 11, 1601. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Smith, J.S.; Meijer, M.; Williams, T.J.; Bachman, N.; Boeke, J.D.; Young, D. Pho23 is associated with the Rpd3 histone deacetylase and is required for its normal function in regulation of gene expression and silencing in saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 24068–24074. [Google Scholar] [CrossRef] [PubMed]
- Howe, L.; Kusch, T.; Muster, N.; Chaterji, R.; Yates, J.R., 3rd; Workman, J.L. Yng1p modulates the activity of Sas3p as a component of the yeast NuA3 histone acetyltransferase complex. Mol. Cell Biol. 2002, 22, 5047–5053. [Google Scholar] [CrossRef][Green Version]
- Nourani, A.; Doyon, Y.; Utley, R.T.; Allard, S.; Lane, W.S.; Côté, J. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell Biol. 2001, 21, 7629–7640. [Google Scholar] [CrossRef]
- Nourani, A.; Howe, L.; Pray-Grant, M.G.; Workman, J.L.; Grant, P.A.; Côté, J. Opposite role of yeast ing family members in p53-dependent transcriptional activation. J. Biol. Chem. 2003, 278, 19171–19175. [Google Scholar] [CrossRef]
- Choy, J.S.; Tobe, B.T.; Huh, J.H.; Kron, S.J. Yng2p-dependent NuA4 histone H4 acetylation activity is required for mitotic and meiotic progression. J. Biol. Chem. 2001, 276, 43653–43662. [Google Scholar] [CrossRef] [PubMed]
- Choy, J.S.; Kron, S.J. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol. Cell Biol. 2002, 22, 8215–8225. [Google Scholar] [CrossRef]
- Le Masson, I.; Yu, D.Y.; Jensen, K.; Chevalier, A.; Courbeyrette, R.; Boulard, Y.; Smith, M.M.; Mann, C. Yaf9, a novel NuA4 histone acetyltransferase subunit, is required for the cellular response to spindle stress in yeast. Mol. Cell Biol. 2003, 23, 6086–6102. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; He, D.; Backues, S.K.; Freeberg, M.A.; Liu, X.; Kim, J.K.; Klionsky, D.J. Transcriptional regulation by pho23 modulates the frequency of autophagosome formation. Curr. Biol. 2014, 24, 1314–1322. [Google Scholar] [CrossRef]
- Chen, J.Q.; Li, Y.; Pan, X.; Lei, B.K.; Chang, C.; Liu, Z.X.; Lu, H. The fission yeast inhibitor of growth (ING) protein Png1p functions in response to DNA damage. J. Biol. Chem. 2010, 285, 15786–15793. [Google Scholar] [CrossRef]
- Abad, M.; Menendez, C.; Fuchtbauer, A.; Serrano, M.; Fuchtbauer, E.M.; Palmero, I. ING1 mediates p53 accumulation and chromatin modification in response to oncogenic stress. J. Biol. Chem. 2007, 282, 31060–31067. [Google Scholar] [CrossRef] [PubMed]
- Fink, D.; Yau, T.; Nabbi, A.; Wagner, B.; Wagner, C.; Hu, S.M.; Lang, V.; Handschuh, S.; Riabowol, K.; Rulicke, T. Loss of ING3 expression results in growth retardation and embryonic death. Cancers 2019, 12, 80. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Fisher, J.B.; Koprowski, S.; McAllister, D.; Kim, M.S.; Lough, J. Homozygous disruption of the TIP60 gene causes early embryonic lethality. Dev. Dyn. 2009, 238, 2912–2921. [Google Scholar] [CrossRef]
- Gorrini, C.; Squatrito, M.; Luise, C.; Syed, N.; Perna, D.; Wark, L.; Martinato, F.; Sardella, D.; Verrecchia, A.; Bennett, S.; et al. TIP60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature 2007, 448, 1063–1067. [Google Scholar] [CrossRef] [PubMed]
- Perkins, N.D. The diverse and complex roles of NF-kappaB subunits in cancer. Nat. Rev. Cancer 2012, 12, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Coles, A.H.; Gannon, H.; Cerny, A.; Kurt-Jones, E.; Jones, S.N. Inhibitor of growth-4 promotes IkappaB promoter activation to suppress NF-kappaB signaling and innate immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 11423–11428. [Google Scholar] [CrossRef] [PubMed]
- Trinh, D.A.; Shirakawa, R.; Kimura, T.; Sakata, N.; Goto, K.; Horiuchi, H. Inhibitor of growth 4 (ING4) is a positive regulator of rRNA synthesis. Sci. Rep. 2019, 9, 17235. [Google Scholar] [CrossRef] [PubMed]
- Linzen, U.; Lilischkis, R.; Pandithage, R.; Schilling, B.; Ullius, A.; Luscher-Firzlaff, J.; Kremmer, E.; Luscher, B.; Vervoorts, J. ING5 is phosphorylated by CDK2 and controls cell proliferation independently of p53. PLoS ONE 2015, 10, e0123736. [Google Scholar] [CrossRef]
- Mulder, K.W.; Wang, X.; Escriu, C.; Ito, Y.; Schwarz, R.F.; Gillis, J.; Sirokmany, G.; Donati, G.; Uribe-Lewis, S.; Pavlidis, P.; et al. Diverse epigenetic strategies interact to control epidermal differentiation. Nat. Cell Biol. 2012, 14, 753–763. [Google Scholar] [CrossRef]
- Wagner, M.J.; Helbing, C.C. Multiple variants of the ING1 and ING2 tumor suppressors are differentially expressed and thyroid hormone-responsive in xenopus laevis. Gen. Comp. Endocrinol. 2005, 144, 38–50. [Google Scholar] [CrossRef]
- Gunduz, M.; Ouchida, M.; Fukushima, K.; Hanafusa, H.; Etani, T.; Nishioka, S.; Nishizaki, K.; Shimizu, K. Genomic structure of the human ING1 gene and tumor-specific mutations detected in head and neck squamous cell carcinomas. Cancer Res. 2000, 60, 3143–3146. [Google Scholar]
- Zeremski, M.; Hill, J.E.; Kwek, S.S.; Grigorian, I.A.; Gurova, K.V.; Garkavtsev, I.V.; Diatchenko, L.; Koonin, E.V.; Gudkov, A.V. Structure and regulation of the mouse Ing1 gene. Three alternative transcripts encode two PHD finger proteins that have opposite effects on p53 function. J. Biol. Chem. 1999, 274, 32172–32181. [Google Scholar] [CrossRef]
- Yuan, H.; Rossetto, D.; Mellert, H.; Dang, W.; Srinivasan, M.; Johnson, J.; Hodawadekar, S.; Ding, E.C.; Speicher, K.; Abshiru, N.; et al. MYST protein acetyltransferase activity requires active site lysine autoacetylation. EMBO J. 2012, 31, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef] [PubMed]
- Satpathy, S.; Nabbi, A.; Riabowol, K. Regulating chromatin regulators: Post-translational modification of the ING family of epigenetic regulators. Biochem. J. 2013, 450, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. Phosphositeplus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef] [PubMed]
- Irving-Hooper, B.K.; Binda, O. A phosphotyrosine switch controls the association of histone mark readers with methylated proteins. Biochemistry 2016, 55, 1631–1634. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).