Rare Germline Pathogenic Variants Identified by Multigene Panel Testing and the Risk of Aggressive Prostate Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Prevalence of Germline Pathogenic Variants in Men with Aggressive Prostate Cancer
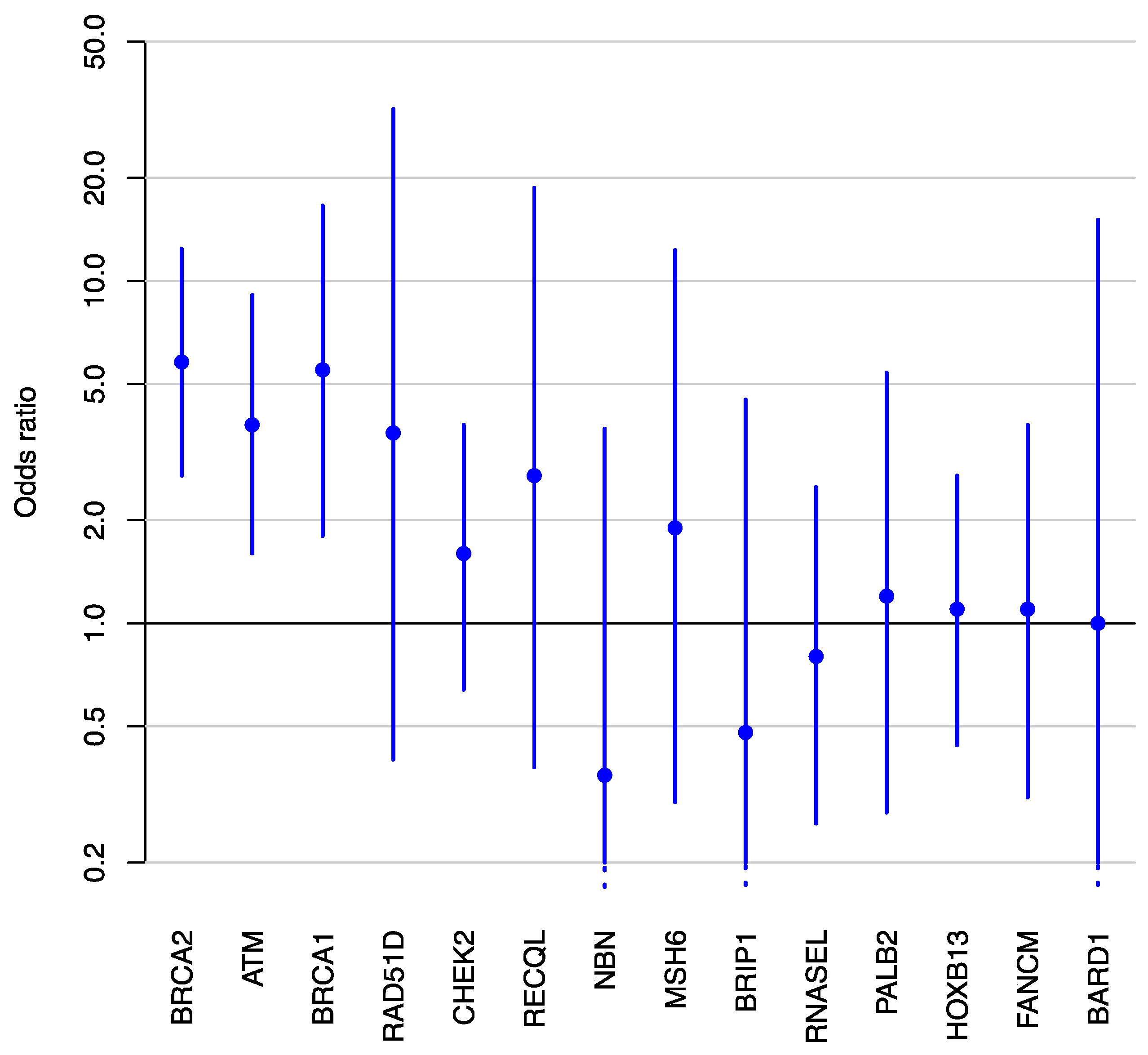
2.2. Associations between Germline Pathogenic Variants and Aggressive Prostate Cancer
3. Discussion
4. Materials and Methods
4.1. Study Subjects
4.2. Gene-Panel Testing
4.3. Variant Calling and Filtering
4.4. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mucci, L.A.; Hjelmborg, J.B.; Harris, J.R.; Czene, K.; Havelick, D.J.; Scheike, T.; Graff, R.E.; Holst, K.; Moller, S.; Unger, R.H.; et al. Familial Risk and Heritability of Cancer Among Twins in Nordic Countries. JAMA 2016, 315, 68–76. [Google Scholar] [CrossRef]
- Conti, D.V.; Darst, B.F.; Moss, L.C.; Saunders, E.J.; Sheng, X.; Chou, A.; Schumacher, F.R.; Olama, A.A.A.; Benlloch, S.; Dadaev, T.; et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat. Genet. 2021, 53, 65–75. [Google Scholar] [CrossRef]
- Ewing, C.M.; Ray, A.M.; Lange, E.M.; Zuhlke, K.A.; Robbins, C.M.; Tembe, W.D.; Wiley, K.E.; Isaacs, S.D.; Johng, D.; Wang, Y.; et al. Germline mutations in HOXB13 and prostate-cancer risk. N. Engl. J. Med. 2012, 366, 141–149. [Google Scholar] [CrossRef]
- MacInnis, R.J.; Severi, G.; Baglietto, L.; Dowty, J.G.; Jenkins, M.A.; Southey, M.C.; Hopper, J.L.; Giles, G.G. Population-based estimate of prostate cancer risk for carriers of the HOXB13 missense mutation G84E. PLoS ONE 2013, 8, e54727. [Google Scholar] [CrossRef]
- Leongamornlert, D.; Mahmud, N.; Tymrakiewicz, M.; Saunders, E.; Dadaev, T.; Castro, E.; Goh, C.; Govindasami, K.; Guy, M.; O’Brien, L.; et al. Germline BRCA1 mutations increase prostate cancer risk. Br. J. Cancer 2012, 106, 1697–1701. [Google Scholar] [CrossRef]
- Agalliu, I.; Karlins, E.; Kwon, E.M.; Iwasaki, L.M.; Diamond, A.; Ostrander, E.A.; Stanford, J.L. Rare germline mutations in the BRCA2 gene are associated with early-onset prostate cancer. Br. J. Cancer 2007, 97, 826–831. [Google Scholar] [CrossRef] [PubMed]
- Kote-Jarai, Z.; Leongamornlert, D.; Saunders, E.; Tymrakiewicz, M.; Castro, E.; Mahmud, N.; Guy, M.; Edwards, S.; O’Brien, L.; Sawyer, E.; et al. BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: Implications for genetic testing in prostate cancer patients. Br. J. Cancer 2011, 105, 1230–1234. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Wang, L.; Taniguchi, K.; Wang, X.; Cunningham, J.M.; McDonnell, S.K.; Qian, C.; Marks, A.F.; Slager, S.L.; Peterson, B.J.; et al. Mutations in CHEK2 associated with prostate cancer risk. Am. J. Hum. Genet. 2003, 72, 270–280. [Google Scholar] [CrossRef]
- Karlsson, Q.; Brook, M.N.; Dadaev, T.; Wakerell, S.; Saunders, E.J.; Muir, K.; Neal, D.E.; Giles, G.G.; MacInnis, R.J.; Thibodeau, S.N.; et al. Rare Germline Variants in ATM Predispose to Prostate Cancer: A PRACTICAL Consortium Study. Eur. Urol. Oncol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, J.; Fraig, M.M.; Metcalf, J.; Turner, W.R.; Bissada, N.K.; Watson, D.K.; Schweinfest, C.W. Defects of DNA mismatch repair in human prostate cancer. Cancer Res. 2001, 61, 4112–4121. [Google Scholar]
- Schaid, D.J.; McDonnell, S.K.; FitzGerald, L.M.; DeRycke, L.; Fogarty, Z.; Giles, G.G.; MacInnis, R.J.; Southey, M.C.; Nguyen-Dumont, T.; Cancel-Tassin, G.; et al. Two-stage Study of Familial Prostate Cancer by Whole-exome Sequencing and Custom Capture Identifies 10 Novel Genes Associated with the Risk of Prostate Cancer. Eur. Urol. 2020. [Google Scholar] [CrossRef]
- Leongamornlert, D.A.; Saunders, E.J.; Wakerell, S.; Whitmore, I.; Dadaev, T.; Cieza-Borrella, C.; Benafif, S.; Brook, M.N.; Donovan, J.L.; Hamdy, F.C.; et al. Germline DNA Repair Gene Mutations in Young-onset Prostate Cancer Cases in the UK: Evidence for a More Extensive Genetic Panel. Eur. Urol. 2019, 76, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Mateo, J.; Walsh, M.F.; De Sarkar, N.; Abida, W.; Beltran, H.; Garofalo, A.; Gulati, R.; Carreira, S.; Eeles, R.; et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N. Engl. J. Med. 2016, 375, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Mijuskovic, M.; Saunders, E.J.; Leongamornlert, D.A.; Wakerell, S.; Whitmore, I.; Dadaev, T.; Cieza-Borrella, C.; Govindasami, K.; Brook, M.N.; Haiman, C.A.; et al. Rare germline variants in DNA repair genes and the angiogenesis pathway predispose prostate cancer patients to develop metastatic disease. Br. J. Cancer 2018, 119, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Nicolosi, P.; Ledet, E.; Yang, S.; Michalski, S.; Freschi, B.; O’Leary, E.; Esplin, E.D.; Nussbaum, R.L.; Sartor, O. Prevalence of Germline Variants in Prostate Cancer and Implications for Current Genetic Testing Guidelines. JAMA Oncol. 2019, 5, 523–528. [Google Scholar] [CrossRef]
- Giri, V.N.; Hegarty, S.E.; Hyatt, C.; O’Leary, E.; Garcia, J.; Knudsen, K.E.; Kelly, W.K.; Gomella, L.G. Germline genetic testing for inherited prostate cancer in practice: Implications for genetic testing, precision therapy, and cascade testing. Prostate 2019, 79, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Dumont, T.; MacInnis, R.J.; Steen, J.A.; Theys, D.; Tsimiklis, H.; Hammet, F.; Mahmoodi, M.; Pope, B.J.; Park, D.J.; Mahmood, K.; et al. Rare germline genetic variants and risk of aggressive prostate cancer. Int. J. Cancer 2020, 147, 2142–2149. [Google Scholar] [CrossRef]
- Darst, B.F.; Dadaev, T.; Saunders, E.; Sheng, X.; Wan, P.; Pooler, L.; Xia, L.Y.; Chanock, S.; Berndt, S.I.; Gapstur, S.M.; et al. Germline sequencing DNA repair genes in 5,545 men with aggressive and non-aggressive prostate cancer. J. Natl. Cancer Inst. 2020. [Google Scholar] [CrossRef]
- Wu, Y.S.; Yu, H.J.; Zheng, S.L.; Na, R.; Mamawala, M.; Landis, T.; Wiley, K.; Petkewicz, J.; Shah, S.; Shi, Z.Q.; et al. A comprehensive evaluation of CHEK2 germline mutations in men with prostate cancer. Prostate 2018, 78, 607–615. [Google Scholar] [CrossRef]
- Hurwitz, L.M.; Agalliu, I.; Albanes, D.; Barry, K.H.; Berndt, S.I.; Cai, Q.; Chen, C.; Cheng, I.; Genkinger, J.M.; Giles, G.G.; et al. Recommended definitions of aggressive prostate cancer for etiologic epidemiologic research. J. Natl. Cancer Inst. 2020. [Google Scholar] [CrossRef]
- Mateo, J.; Carreira, S.; Sandhu, S.; Miranda, S.; Mossop, H.; Perez-Lopez, R.; Nava Rodrigues, D.; Robinson, D.; Omlin, A.; Tunariu, N.; et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N. Engl. J. Med. 2015, 373, 1697–1708. [Google Scholar] [CrossRef]
- Cheng, H.H.; Pritchard, C.C.; Boyd, T.; Nelson, P.S.; Montgomery, B. Biallelic Inactivation of BRCA2 in Platinum-sensitive Metastatic Castration-resistant Prostate Cancer. Eur. Urol. 2016, 69, 992–995. [Google Scholar] [CrossRef] [PubMed]
- Vallee, M.P.; Francy, T.C.; Judkins, M.K.; Babikyan, D.; Lesueur, F.; Gammon, A.; Goldgar, D.E.; Couch, F.J.; Tavtigian, S.V. Classification of missense substitutions in the BRCA genes: A database dedicated to Ex-UVs. Hum. Mutat. 2012, 33, 22–28. [Google Scholar] [CrossRef]
- Guo, M.H.; Plummer, L.; Chan, Y.-M.; Hirschhorn, J.N.; Lippincott, M.F. Burden Testing of Rare Variants Identified through Exome Sequencing via Publicly Available Control Data. Am. J. Hum. Genet. 2018, 103, 522–534. [Google Scholar] [CrossRef] [PubMed]
- Povysil, G.; Petrovski, S.; Hostyk, J.; Aggarwal, V.; Allen, A.S.; Goldstein, D.B. Rare-variant collapsing analyses for complex traits: Guidelines and applications. Nat. Rev. Genet. 2019, 20, 747–759. [Google Scholar] [CrossRef] [PubMed]
- Le Calvez-Kelm, F.; Lesueur, F.; Damiola, F.; Vallée, M.; Voegele, C.; Babikyan, D.; Durand, G.; Forey, N.; McKay-Chopin, S.; Robinot, N. Rare, evolutionarily unlikely missense substitutions in CHEK2 contribute to breast cancer susceptibility: Results from a breast cancer family registry case-control mutation-screening study. Breast Cancer Res. 2011, 13, R6. [Google Scholar] [CrossRef]
- Tavtigian, S.V.; Oefner, P.J.; Babikyan, D.; Hartmann, A.; Healey, S.; Le Calvez-Kelm, F.; Lesueur, F.; Byrnes, G.B.; Chuang, S.-C.; Forey, N.; et al. Rare, evolutionarily unlikely missense substitutions in ATM confer increased risk of breast cancer. Am. J. Hum. Genet. 2009, 85, 427–446. [Google Scholar] [CrossRef] [PubMed]
- Wiltshire, T.; Ducy, M.; Foo, T.K.; Hu, C.; Lee, K.Y.; Belur Nagaraj, A.; Rodrigue, A.; Gomes, T.T.; Simard, J.; Monteiro, A.N.A.; et al. Functional characterization of 84 PALB2 variants of uncertain significance. Genet. Med. 2020, 22, 622–632. [Google Scholar] [CrossRef] [PubMed]
- Boonen, R.; Rodrigue, A.; Stoepker, C.; Wiegant, W.W.; Vroling, B.; Sharma, M.; Rother, M.B.; Celosse, N.; Vreeswijk, M.P.G.; Couch, F.; et al. Functional analysis of genetic variants in the high-risk breast cancer susceptibility gene PALB2. Nat Commun. 2019, 10, 5296. [Google Scholar] [CrossRef] [PubMed]
- Rodrigue, A.; Margaillan, G.; Torres Gomes, T.; Coulombe, Y.; Montalban, G.; da Costa, E.S.C.S.; Milano, L.; Ducy, M.; De-Gregoriis, G.; Dellaire, G.; et al. A global functional analysis of missense mutations reveals two major hotspots in the PALB2 tumor suppressor. Nucleic. Acids Res. 2019, 47, 10662–10677. [Google Scholar] [CrossRef]
- Southey, M.C.; Rewse, A.; Nguyen-Dumont, T. PALB2 Genetic Variants: Can Functional Assays Assist Translation? Trends Cancer 2020, 6, 263–265. [Google Scholar] [CrossRef]
- Milne, R.L.; Fletcher, A.S.; MacInnis, R.J.; Hodge, A.M.; Hopkins, A.H.; Bassett, J.K.; Bruinsma, F.J.; Lynch, B.M.; Dugue, P.A.; Jayasekara, H.; et al. Cohort Profile: The Melbourne Collaborative Cohort Study (Health 2020). Int. J. Epidemiol. 2017, 46, 1757–1757i. [Google Scholar] [CrossRef] [PubMed]
- Papa, N.P.; MacInnis, R.J.; English, D.R.; Bolton, D.; Davis, I.D.; Lawrentschuk, N.; Millar, J.L.; Pedersen, J.; Severi, G.; Southey, M.C.; et al. Ejaculatory frequency and the risk of aggressive prostate cancer: Findings from a case-control study. Urol. Oncol. 2017, 35, 530.e7–530.e13. [Google Scholar] [CrossRef] [PubMed]
- Giles, G.G.; Severi, G.; Sinclair, R.; English, D.R.; McCredie, M.R.; Johnson, W.; Boyle, P.; Hopper, J.L. Androgenetic alopecia and prostate cancer: Findings from an Australian case-control study. Cancer Epidemiol. Biomarkers Prev. 2002, 11, 549–553. [Google Scholar] [PubMed]
- Aspree Investigator Group. Study design of ASPirin in Reducing Events in the Elderly (ASPREE): A randomized, controlled trial. Contemp. Clin. Trials 2013, 36, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Lockery, J.E.; Collyer, T.A.; Abhayaratna, W.P.; Fitzgerald, S.M.; McNeil, J.J.; Nelson, M.R.; Orchard, S.G.; Reid, C.; Stocks, N.P.; Trevaks, R.E.; et al. Recruiting general practice patients for large clinical trials: Lessons from the Aspirin in Reducing Events in the Elderly (ASPREE) study. Med. J. Aust. 2019, 210, 168–173. [Google Scholar] [CrossRef]
- McNeil, J.J.; Woods, R.L.; Nelson, M.R.; Murray, A.M.; Reid, C.M.; Kirpach, B.; Storey, E.; Shah, R.C.; Wolfe, R.S.; Tonkin, A.M.; et al. Baseline Characteristics of Participants in the ASPREE (ASPirin in Reducing Events in the Elderly) Study. J. Gerontol. A Biol. Sci. Med. Sci. 2017, 72, 1586–1593. [Google Scholar] [CrossRef]
- McNeil, J.J.; Woods, R.L.; Nelson, M.R.; Reid, C.M.; Kirpach, B.; Wolfe, R.; Storey, E.; Shah, R.C.; Lockery, J.E.; Tonkin, A.M.; et al. Effect of Aspirin on Disability-free Survival in the Healthy Elderly. N. Engl. J. Med. 2018, 379, 1499–1508. [Google Scholar] [CrossRef]
- Lacaze, P.; Sebra, R.; Riaz, M.; Tiller, J.; Revote, J.; Phung, J.; Parker, E.J.; Orchard, S.G.; Lockery, J.E.; Wolfe, R.; et al. Medically actionable pathogenic variants in a population of 13,131 healthy elderly individuals. Genet. Med. 2020, 22, 1883–1886. [Google Scholar] [CrossRef]
- Hammet, F.; Mahmood, K.; Green, T.R.; Nguyen-Dumont, T.; Southey, M.C.; Buchanan, D.D.; Lonie, A.; Nathanson, K.L.; Couch, F.J.; Pope, B.J.; et al. Hi-Plex2: A simple and robust approach to targeted sequencing-based genetic screening. Biotechniques 2019. [Google Scholar] [CrossRef]
- Lai, Z.; Markovets, A.; Ahdesmaki, M.; Chapman, B.; Hofmann, O.; McEwen, R.; Johnson, J.; Dougherty, B.; Barrett, J.C.; Dry, J.R. VarDict: A novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic. Acids Res. 2016, 44, e108. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 23 March 2021).
| Variables and Values | Aggressive PrCa Cases, Number (%) |
|---|---|
| Study | non-missing = 833 |
| ASPREE a | 0 (0%) |
| APC a | 322 (39%) |
| EOPCFS a | 185 (22%) |
| MCCS a | 140 (17%) |
| RFPCS a | 186 (22%) |
| Age at diagnosis in years | non-missing = 833 |
| <60 | 258 (31%) |
| 60–64 | 147 (18%) |
| 65–69 | 262 (31%) |
| ≥70 | 166 (20%) |
| Gleason score | non-missing = 628 |
| 2 | 1 (0%) |
| 3 | 0 (0%) |
| 4 | 5 (1%) |
| 5 | 16 (3%) |
| 6 | 41 (7%) |
| 7 | 105 (17%) |
| 8 | 189 (30%) |
| 9 | 251 (40%) |
| 10 | 20 (3%) |
| Died from prostate cancer | non-missing = 799 |
| No | 324 (41%) |
| Yes | 475 (59%) |
| Gene | Cases (n = 833) | Controls b (n = 7255) | Adjusted OR (95% CI) | p-Value | ||
|---|---|---|---|---|---|---|
| Number of Carriers | % | Number of Carriers | % | |||
| ATM | 14 | 1.7% | 25 | 0.3% | 3.8 (1.6–9.1) | 0.0021 |
| BARD1 | 1 | 0.1% | 8 | 0.1% | 1.0 (0.07–15.1) | 0.97 |
| BRCA1 | 5 | 0.6% | 12 | 0.2% | 5.5 (1.8–16.6) | 0.0023 |
| BRCA2 | 19 | 2.3% | 24 | 0.3% | 5.8 (2.7–12.4) | <0.0001 |
| BRIP1 | 1 | 0.1% | 14 | 0.2% | 0.48 (0.05–4.5) | 0.53 |
| CDH1 | 0 | 0% | 0 | 0% | - | - |
| CHEK2 | 8 | 1% | 41 | 0.6% | 1.6 (0.64–3.8) | 0.32 |
| FANCM | 4 | 0.5% | 23 | 0.3% | 1.1 (0.31–3.8) | 0.89 |
| HOXB13 | 11 | 1.3% | 18 | 0.2% | 1.1 (0.44–2.7) | 0.84 |
| MLH1 | 0 | 0% | 1 | 0% | - | - |
| MRE11A | 0 | 0% | 6 | 0.1% | - | - |
| MSH2 | 1 | 0.1% | 0 | 0% | - | - |
| MSH6 | 3 | 0.4% | 6 | 0.1% | 1.9 (0.3–12.3) | 0.49 |
| MUTYH | 0 | 0% | 0 | 0% | - | - |
| NBN | 1 | 0.1% | 10 | 0.1% | 0.36 (0.03–3.7) | 0.39 |
| NF1 | 0 | 0% | 2 | 0% | - | - |
| PALB2 | 4 | 0.5% | 13 | 0.2% | 1.2 (0.28–5.4) | 0.79 |
| PMS2 | 2 | 0.2% | 0 | 0% | - | - |
| PTEN | 0 | 0% | 0 | 0% | - | - |
| RAD50 | 0 | 0% | 14 | 0.2% | - | - |
| RAD51C | 0 | 0% | 6 | 0.1% | - | - |
| RAD51D | 1 | 0.1% | 6 | 0.1% | 3.6 (0.4–31.8) | 0.25 |
| RECQL | 2 | 0.2% | 5 | 0.1% | 2.7 (0.38–18.7) | 0.33 |
| RNASEL | 5 | 0.6% | 23 | 0.3% | 0.8 (0.26–2.5) | 0.7 |
| STK11 | 0 | 0% | 0 | 0% | - | - |
| TP53 | 0 | 0% | 2 | 0% | - | - |
| Total | 82 | 9.8% | 259 | 3.6% | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen-Dumont, T.; Dowty, J.G.; MacInnis, R.J.; Steen, J.A.; Riaz, M.; Dugué, P.-A.; Renault, A.-L.; Hammet, F.; Mahmoodi, M.; Theys, D.; et al. Rare Germline Pathogenic Variants Identified by Multigene Panel Testing and the Risk of Aggressive Prostate Cancer. Cancers 2021, 13, 1495. https://doi.org/10.3390/cancers13071495
Nguyen-Dumont T, Dowty JG, MacInnis RJ, Steen JA, Riaz M, Dugué P-A, Renault A-L, Hammet F, Mahmoodi M, Theys D, et al. Rare Germline Pathogenic Variants Identified by Multigene Panel Testing and the Risk of Aggressive Prostate Cancer. Cancers. 2021; 13(7):1495. https://doi.org/10.3390/cancers13071495
Chicago/Turabian StyleNguyen-Dumont, Tú, James G. Dowty, Robert J. MacInnis, Jason A. Steen, Moeen Riaz, Pierre-Antoine Dugué, Anne-Laure Renault, Fleur Hammet, Maryam Mahmoodi, Derrick Theys, and et al. 2021. "Rare Germline Pathogenic Variants Identified by Multigene Panel Testing and the Risk of Aggressive Prostate Cancer" Cancers 13, no. 7: 1495. https://doi.org/10.3390/cancers13071495
APA StyleNguyen-Dumont, T., Dowty, J. G., MacInnis, R. J., Steen, J. A., Riaz, M., Dugué, P.-A., Renault, A.-L., Hammet, F., Mahmoodi, M., Theys, D., Tsimiklis, H., Severi, G., Bolton, D., Lacaze, P., Sebra, R., Schadt, E., McNeil, J., Giles, G. G., Milne, R. L., & Southey, M. C. (2021). Rare Germline Pathogenic Variants Identified by Multigene Panel Testing and the Risk of Aggressive Prostate Cancer. Cancers, 13(7), 1495. https://doi.org/10.3390/cancers13071495






