Specific Patterns of Blood ILCs in Metastatic Melanoma Patients and Their Modulations in Response to Immunotherapy
Abstract
Simple Summary
Abstract
1. Introduction
2. Patients and Methods
2.1. Patient Characteristics and Study Design
2.2. Definition of Tumor Assessment and Main Clinical Outcomes
2.3. Flow Cytometry and Monoclonal Antibodies
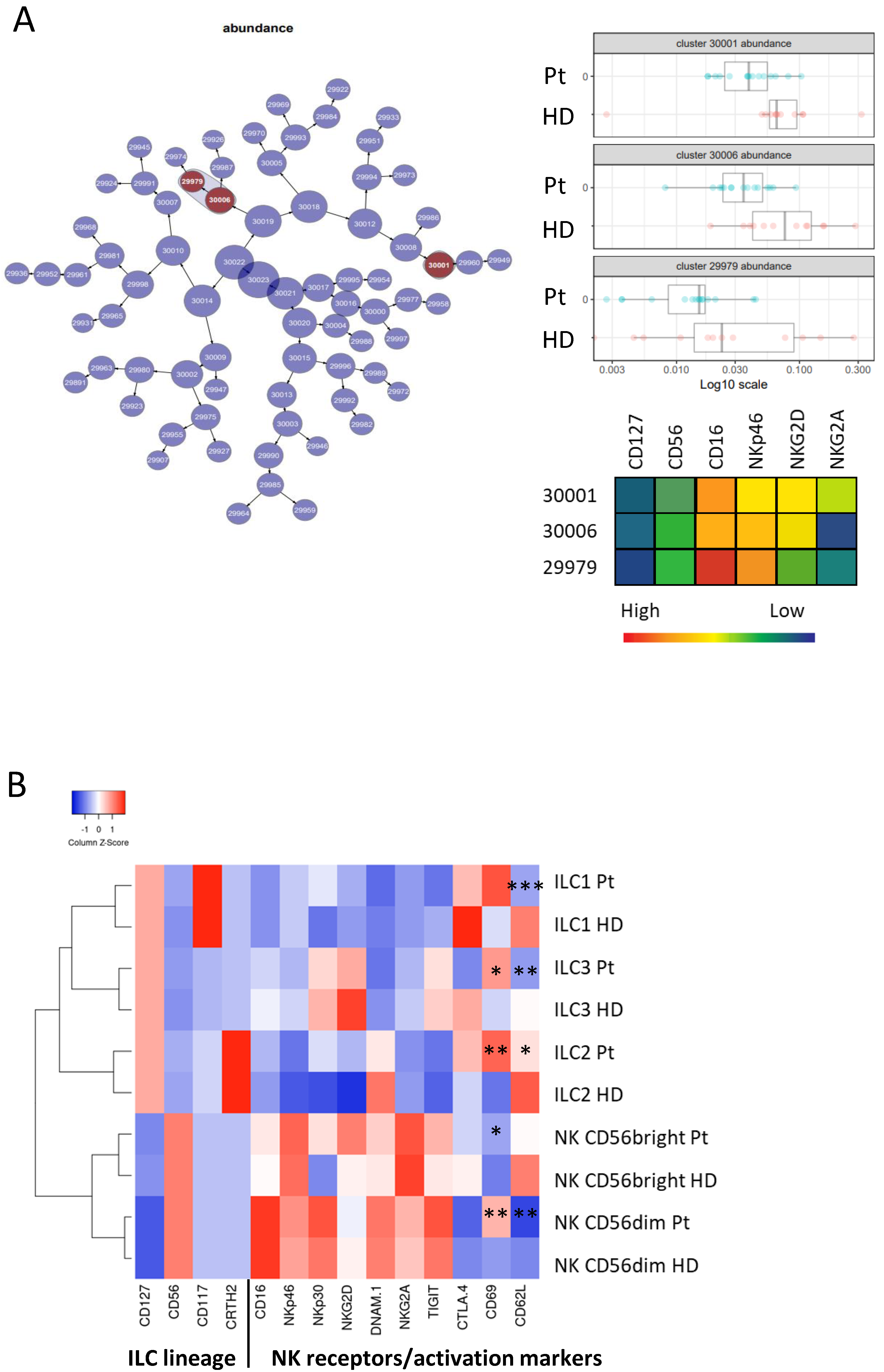
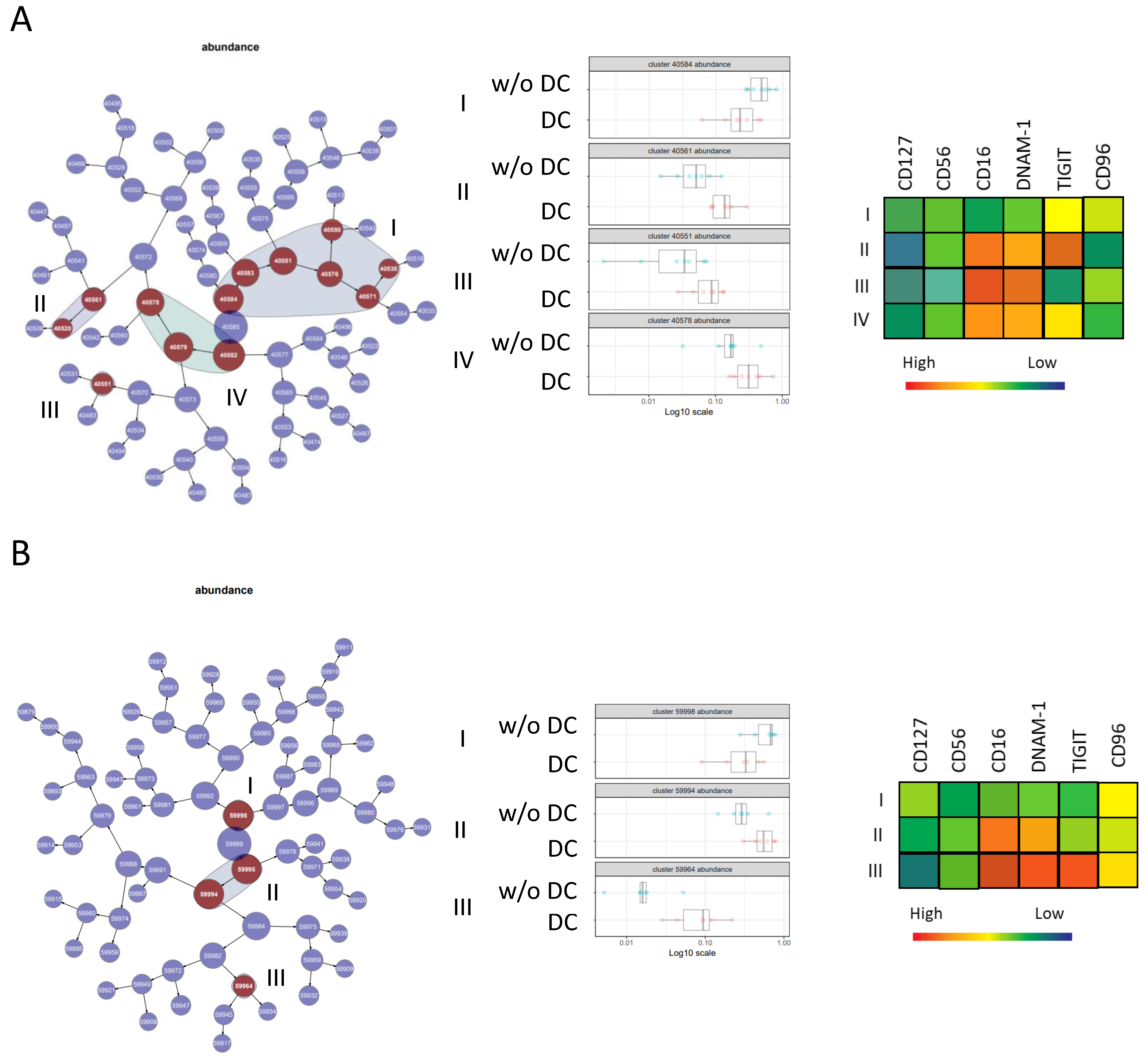
2.4. CITRUS Analysis
2.5. Statistical Analyses
3. Results
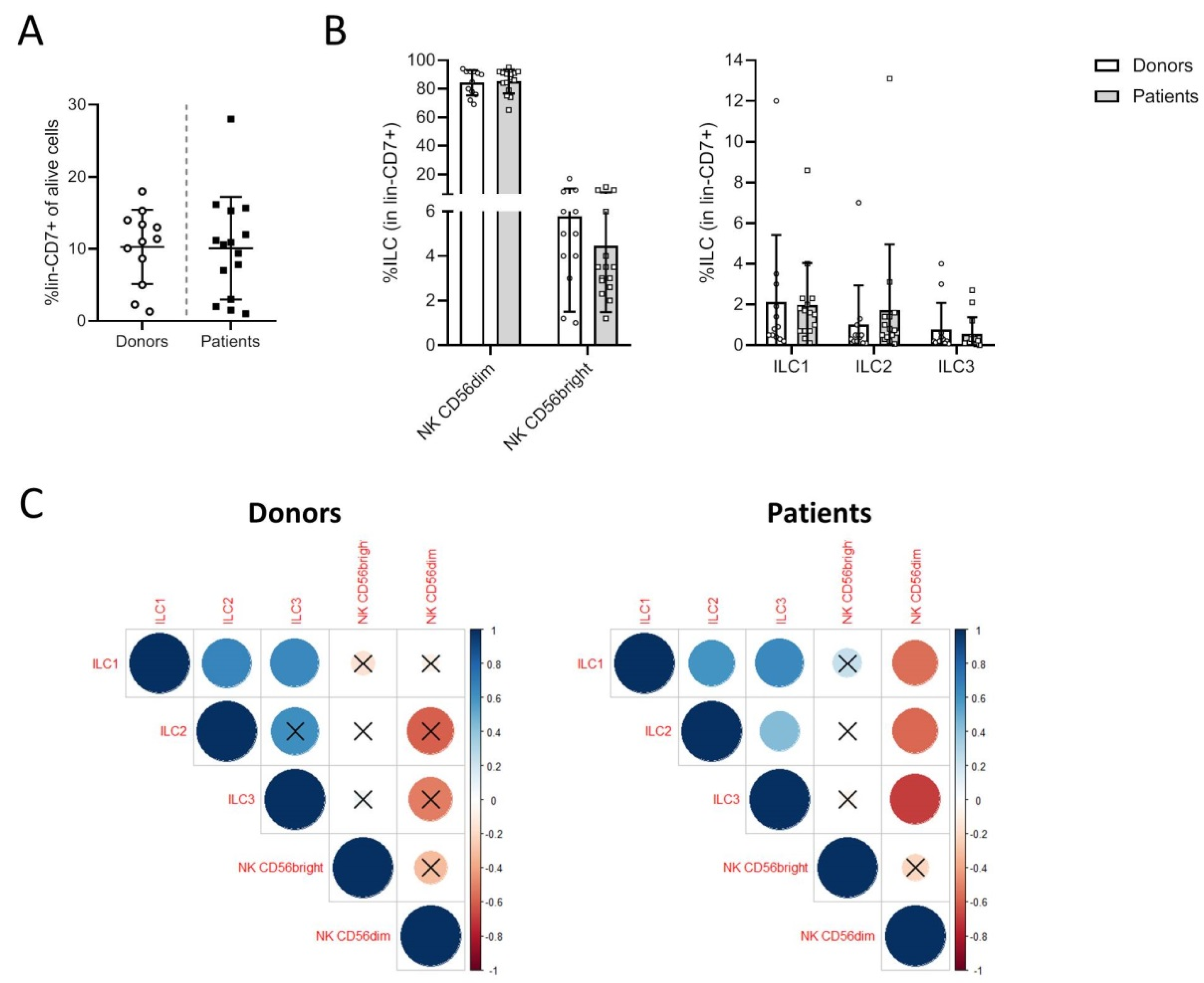
3.1. Melanoma Patients Exhibit a Specific Distribution of Blood ILCs
3.2. Phenotypic Analysis of ILC Subsets in Donors and Patients
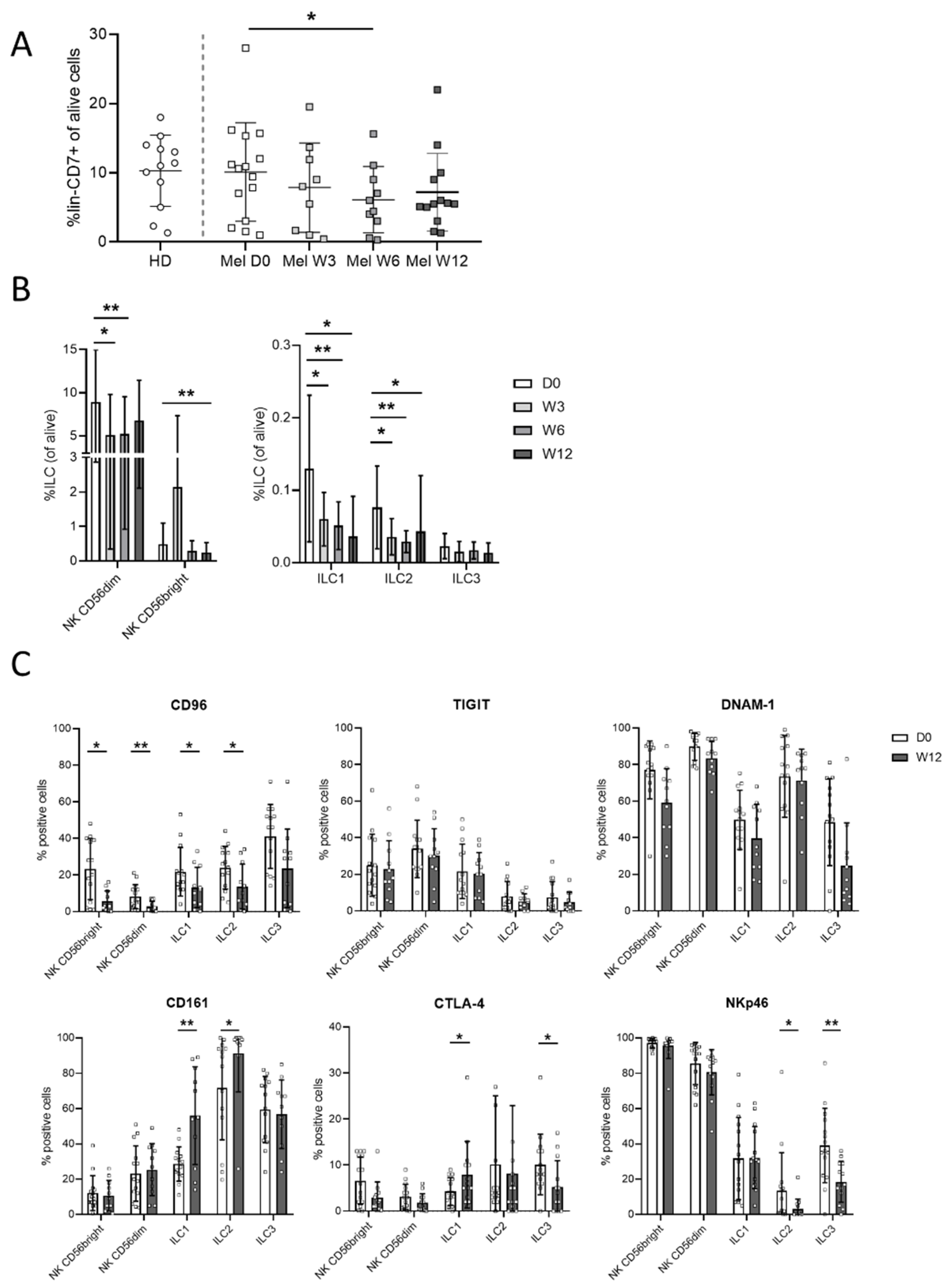
3.3. Ipilimumab Induces Early Changes in ILC Proportions
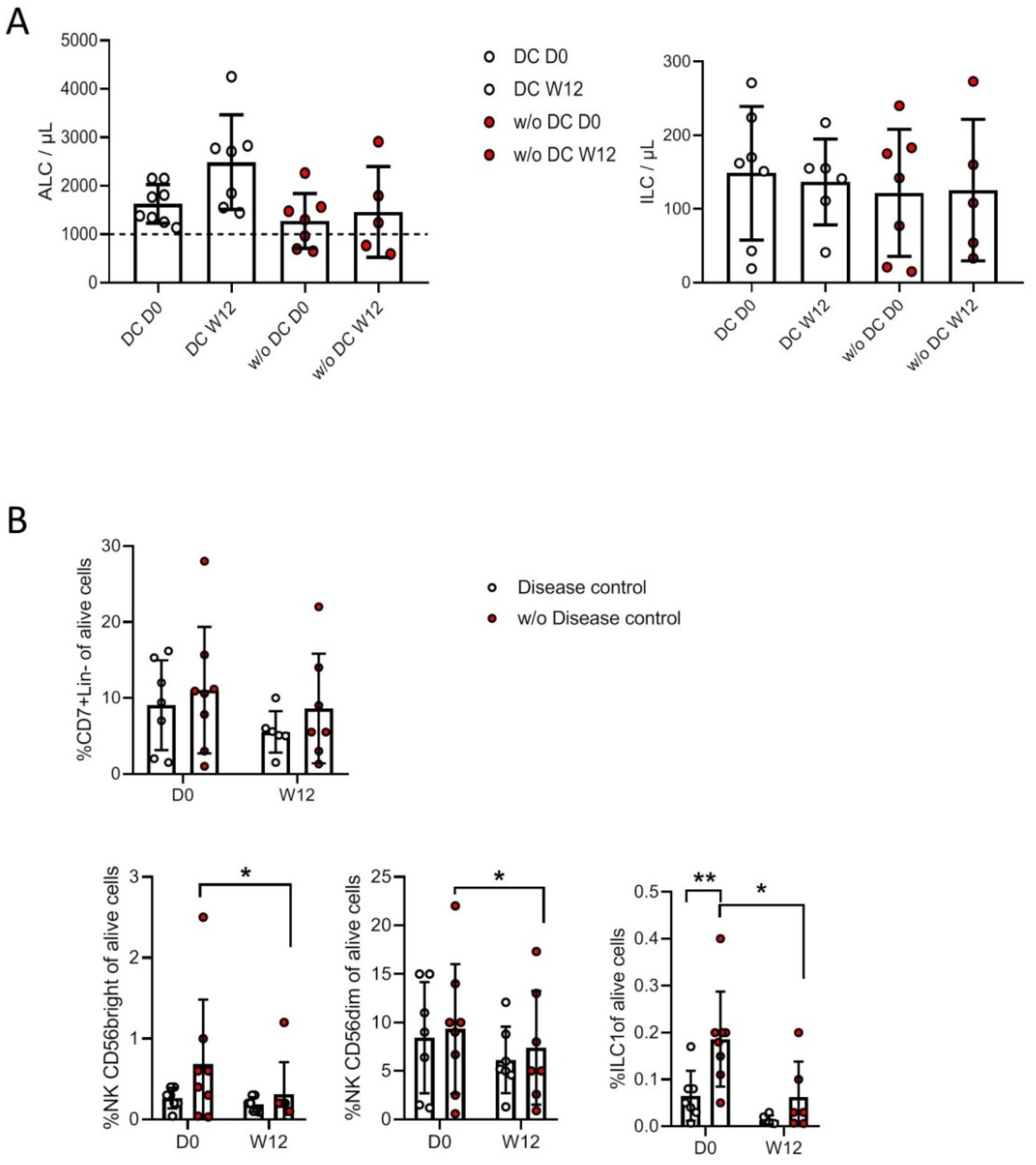
3.4. Correlation with Clinical Status and Response to Treatment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Leach, D.R.; Krummel, M.F.; Allison, J.P. Enhancement of Antitumor Immunity by CTLA-4 Blockade. Science 1996, 271, 1734–1736. [Google Scholar] [CrossRef]
- Postow, M.A.; Chesney, J.; Pavlick, A.C.; Robert, C.; Grossmann, K.; McDermott, D.; Linette, G.P.; Meyer, N.; Giguere, J.K.; Agarwala, S.S.; et al. Nivolumab and Ipilimumab versus Ipilimumab in Untreated Melanoma. N. Engl. J. Med. 2015, 372, 2006–2017. [Google Scholar] [CrossRef]
- Thompson, C.B.; Allison, J.P. The Emerging Role of CTLA-4 as an Immune Attenuator. Immunology 1997, 7, 445–450. [Google Scholar] [CrossRef]
- Gide, T.N.; Wilmott, J.S.; Scolyer, R.A.; Long, G.V. Primary and Acquired Resistance to Immune Checkpoint Inhibitors in Metastatic Melanoma. Clin. Cancer Res. 2018, 24, 1260–1270. [Google Scholar] [CrossRef]
- Caligiuri, M.A. Human natural killer cells. Blood 2008, 112, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Marcus, A.; Gowen, B.G.; Thompson, T.W.; Iannello, A.; Ardolino, M.; Deng, W.; Wang, L.; Shifrin, N.; Raulet, D.H. Recognition of tumors by the innate immune system and natural killer cells. Adv. Immunol. 2014, 122, 91–128. [Google Scholar] [PubMed]
- Fehniger, T.A.; Cooper, M.A.; Nuovo, G.J.; Cella, M.; Facchetti, F.; Colonna, M.; Caligiuri, M.A. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell-derived IL-2: A potential new link between adaptive and innate immunity. Blood 2003, 101, 3052–3057. [Google Scholar] [CrossRef]
- Frey, M.; Packianathan, N.B.; Fehniger, T.A.; Ross, M.E.; Wang, W.C.; Stewart, C.C. Differential ex-pression and function of L-selectin on CD56bright and CD56dim natural killer cell subsets. J. Immunol. 1998, 161, 400–408. [Google Scholar]
- Lanier, L.L. Turning on Natural Killer Cells. J. Exp. Med. 2000, 191, 1259–1262. [Google Scholar] [CrossRef]
- Moretta, L.; Moretta, A. Unravelling natural killer cell function: Triggering and inhibitory human NK receptors. EMBO J. 2004, 23, 255–259. [Google Scholar] [CrossRef]
- Gilfillan, S.; Chan, C.J.; Cella, M.; Haynes, N.M.; Rapaport, A.S.; Boles, K.S. DNAM-1 promotes ac-tivation of cytotoxic lymphocytes by nonprofessional antigen-presenting cells and tumors. J. Exp. Med. 2008, 205, 2965–2973. [Google Scholar] [CrossRef]
- Bryceson, Y.T.; March, M.E.; Ljunggren, H.-G.; Long, E.O. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood 2006, 107, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Braud, V.M.; Allan, D.S.J.; O’Callaghan, C.A.; Söderström, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H.; et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998, 391, 795–799. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, K.; Dimasi, N.; Wang, J.; Mariuzza, R.A.; Margulies, D.H. Structure and function of natural killer cell receptors: Multiple molecular solutions to self, nonself discrimination. Annu. Rev. Immunol. 2002, 20, 853–885. [Google Scholar] [CrossRef]
- Moretta, L.; Moretta, A. Killer immunoglobulin-like receptors. Curr. Opin. Immunol. 2004, 16, 626–633. [Google Scholar] [CrossRef]
- Albertsson, P.A.; Basse, P.H.; Hokland, M.; Goldfarb, R.H.; Nagelkerke, J.; Nannmark, U.; Kuppen, P.J. NK cells and the tumour microenvironment: Implications for NK-cell function and anti-tumour activity. Trends Immunol. 2003, 24, 603–609. [Google Scholar] [CrossRef]
- Lala, P.K.; Elkashab, M.; Kerbel, R.S.; Parhar, R.S. Cure of human melanoma lung metastases in nude mice with chronic indomethacin therapy combined with multiple rounds of IL-2: Characteristics of killer cells generated in situ. Int. Immunol. 1990, 2, 1149–1158. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Xu, T.; Xiong, M.; Zhou, X.; Wang, Y.; Haydu, L.E.; Ross, M.I.; Gershenwald, J.E.; Prieto, V.G.; Cormier, J.N.; et al. Role of Immune Response, Inflammation, and Tumor Immune Response–Related Cytokines/Chemokines in Melanoma Progression. J. Investig. Dermatol. 2019, 139, 2352–2358.e3. [Google Scholar] [CrossRef]
- Artis, D.; Spits, H. The biology of innate lymphoid cells. Nature 2015, 517, 293–301. [Google Scholar] [CrossRef]
- Constantinides, M.G.; McDonald, B.D.; Verhoef, P.A.; Bendelac, A. A committed precursor to innate lymphoid cells. Nature 2014, 508, 397–401. [Google Scholar] [CrossRef]
- Gronke, K.; Kofoed-Nielsen, M.; Diefenbach, A. Innate lymphoid cells, precursors and plasticity. Immunol. Lett. 2016, 179, 9–18. [Google Scholar] [CrossRef]
- Gasteiger, G.; Fan, X.; Dikiy, S.; Lee, S.Y.; Rudensky, A.Y. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 2015, 350, 981–985. [Google Scholar] [CrossRef]
- Stokic-Trtica, V.; Diefenbach, A.; Klose, C.S.N. NK Cell Development in Times of Innate Lymphoid Cell Diversity. Front. Immunol. 2020, 11, 813. [Google Scholar] [CrossRef]
- Klose, C.S.; Artis, D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat. Immunol. 2016, 17, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Bal, S.M.; Golebski, K.; Spits, H. Plasticity of innate lymphoid cell subsets. Nat. Rev. Immunol. 2020, 20, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Colonna, M. Innate Lymphoid Cells: Diversity, Plasticity, and Unique Functions in Immunity. Immunology 2018, 48, 1104–1117. [Google Scholar] [CrossRef]
- Vivier, E.; Artis, D.; Colonna, M.; Diefenbach, A.; Di Santo, J.P.; Eberl, G.; Koyasu, S.; Locksley, R.M.; McKenzie, A.N.; Mebius, R.E.; et al. Innate Lymphoid Cells: 10 Years On. Cell 2018, 174, 1054–1066. [Google Scholar] [CrossRef] [PubMed]
- Tikoo, S.; Jain, R.; Kurz, A.R.; Weninger, W. The lymphoid cell network in the skin. Immunol. Cell Biol. 2018, 96, 485–496. [Google Scholar] [CrossRef]
- Stamatiades, E.G.; Li, M.O. Tissue-resident cytotoxic innate lymphoid cells in tumor immunosurveillance. Semin. Immunol. 2019, 41, 101269. [Google Scholar] [CrossRef]
- Robert, C.; Thomas, L.; Bondarenko, I.; O’Day, S.; Weber, J.; Garbe, C.; Lebbe, C.; Baurain, J.-F.; Testori, A.; Grob, J.-J.; et al. Ipilimumab plus Dacarbazine for Previously Untreated Metastatic Melanoma. N. Engl. J. Med. 2011, 364, 2517–2526. [Google Scholar] [CrossRef] [PubMed]
- Felix, J.; Lambert, J.; Roelens, M.; Maubec, E.; Guermouche, H.; Pagès, C.; Sidina, I.; Cordeiro, D.J.; Maki, G.; Chasset, F.; et al. Ipilimumab reshapes T cell memory subsets in melanoma patients with clinical response. OncoImmunology 2016, 5, 1136045. [Google Scholar] [CrossRef] [PubMed]
- Apraiz, A.; Benedicto, A.; Marquez, J.; Agüera-Lorente, A.; Asumendi, A.; Olaso, E.; Arteta, B. Innate Lymphoid Cells in the Malignant Melanoma Microenvironment. Cancers 2020, 12, 3177. [Google Scholar] [CrossRef] [PubMed]
- Mirjacic Martinovic, K.M.; Babovic, N.; Dzodic, R.R.; Jurisic, V.B.; Tanic, N.T.; Konjevic, G.M. Decreased expression of NKG2D, NKp46, DNAM-1 receptors, and intracellular perforin and STAT-1 effector molecules in NK cells and their dim and bright subsets in metastatic melanoma patients. Melanoma Res. 2014, 24, 295–304. [Google Scholar] [CrossRef]
- Fregni, G.; Messaoudene, M.; Fourmentraux-Neves, E.; Mazouz-Dorval, S.; Chanal, J.; Maubec, E.; Marinho, E.; Scheer-Senyarich, I.; Cremer, I.; Avril, M.-F.; et al. Phenotypic and Functional Characteristics of Blood Natural Killer Cells from Melanoma Patients at Different Clinical Stages. PLoS ONE 2013, 8, e76928. [Google Scholar] [CrossRef]
- Fregni, G.; Perier, A.; Pittari, G.; Jacobelli, S.; Sastre, X.; Gervois, N.; Allard, M.; Bercovici, N.; Avril, M.F.; Caignard, A. Unique Functional Status of Natural Killer Cells in Metastatic Stage IV Melanoma Patients and Its Modulation by Chemotherapy. Clin. Cancer Res. 2011, 17, 2628–2637. [Google Scholar] [CrossRef]
- Federici, C.; Shahaj, E.; Cecchetti, S.; Camerini, S.; Casella, M.; Iessi, E.; Camisaschi, C.; Paolino, G.; Calvieri, S.; Ferro, S.; et al. Natural-killer-derived extracellular vesicles: Immune sensors and interactors. Front. Immunol. 2020, 11, 262. [Google Scholar] [CrossRef]
- Sabry, M.; Zubiak, A.; Hood, S.P.; Simmonds, P.; Arellano-Ballestero, H.; Cournoyer, E. Tumor- and cytokine-primed human natural killer cells exhibit distinct phenotypic and transcriptional signa-tures. PLoS ONE 2019, 14, e0218674. [Google Scholar] [CrossRef]
- Luetke-Eversloh, M.; Killig, M.; Romagnani, C. Signatures of human NK cell development and ter-minal differentiation. Front. Immunol. 2013, 4, 499. [Google Scholar] [CrossRef]
- Juelke, K.; Killig, M.; Luetke-Eversloh, M.; Parente, E.; Gruen, J.; Morandi, B.; Ferlazzo, G.; Thiel, A.; Schmitt-Knosalla, I.; Romagnani, C. CD62L expression identifies a unique subset of polyfunctional CD56dim NK cells. Blood 2010, 116, 1299–1307. [Google Scholar] [CrossRef]
- Tietze, J.K.; Angelova, D.; Heppt, M.V.; Ruzicka, T.; Berking, C. Low baseline levels of NK cells may predict a positive response to ipilimumab in melanoma therapy. Exp. Dermatol. 2017, 26, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Romano, E.; Kusio-Kobialka, M.; Foukas, P.G.; Baumgaertner, P.; Meyer, C.; Ballabeni, P. Ipili-mumab-dependent cell-mediated cytotoxicity of regulatory T cells ex vivo by nonclassical mono-cytes in melanoma patients. Proc. Natl. Acad. Sci. USA 2015, 112, 6140–6145. [Google Scholar] [CrossRef]
- Pistillo, M.P.; Carosio, R.; Grillo, F.; Fontana, V.; Mastracci, L.; Morabito, A. Phenotypic character-ization of tumor CTLA-4 expression in melanoma tissues and its possible role in clinical response to Ipilimumab. Clin. Immunol. 2020, 215, 108428. [Google Scholar] [CrossRef] [PubMed]
- Laurent, S.; Queirolo, P.; Boero, S.; Salvi, S.; Piccioli, P.; Boccardo, S.; Minghelli, S.; Morabito, A.; Fontana, V.; Pietra, G.; et al. The engagement of CTLA-4 on primary melanoma cell lines induces antibody-dependent cellular cytotoxicity and TNF-α production. J. Transl. Med. 2013, 11, 108. [Google Scholar] [CrossRef]
- Passariello, M.; Camorani, S.; Vetrei, C.; Ricci, S.; Cerchia, L.; De Lorenzo, C. Ipilimumab and Its Derived EGFR Aptamer-Based Conjugate Induce Efficient NK Cell Activation against Cancer Cells. Cancers 2020, 12, 331. [Google Scholar] [CrossRef]
- Hawke, L.G.; Whitford, M.K.M.; Ormiston, M.L. The Production of Pro-angiogenic VEGF-A Isoforms by Hypoxic Human NK Cells Is Independent of Their TGF-beta-Mediated Conversion to an ILC1-Like Phenotype. Front. Immunol. 2020, 11, 1903. [Google Scholar] [CrossRef]
- Gao, Y.; Souza-Fonseca-Guimaraes, F.; Bald, T.; Ng, S.S.; Young, A.; Ngiow, S.F. Tumor immuno-evasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 2017, 18, 1004–1015. [Google Scholar] [CrossRef] [PubMed]
- Bie, Q.; Zhang, P.; Su, Z.; Zheng, D.; Ying, X.; Wu, Y.; Yang, H.; Chen, D.; Wang, S.; Xu, H. Polarization of ILC2s in Peripheral Blood Might Contribute to Immunosuppressive Microenvironment in Patients with Gastric Cancer. J. Immunol. Res. 2014, 2014, 1–10. [Google Scholar] [CrossRef]
- de Weerdt, I.; van Hoeven, V.; Munneke, J.M.; Endstra, S.; Hofland, T.; Hazenberg, M.D. Innate lymphoid cells are expanded and functionally altered in chronic lymphocytic leukemia. Haematologica 2016, 101, e461–e464. [Google Scholar] [CrossRef]
- Schuijs, M.J.; Png, S.; Richard, A.C.; Tsyben, A.; Hamm, G.; Stockis, J.; Garcia, C.; Pinaud, S.; Nicholls, A.; Ros, X.R.; et al. ILC2-driven innate immune checkpoint mechanism antagonizes NK cell antimetastatic function in the lung. Nat. Immunol. 2020, 21, 998–1009. [Google Scholar] [CrossRef]
- Long, A.; Dominguez, D.; Qin, L.; Chen, S.; Fan, J.; Zhang, M.; Fang, D.; Zhang, Y.; Kuzel, T.M.; Zhang, B. Type 2 Innate Lymphoid Cells Impede IL-33–Mediated Tumor Suppression. J. Immunol. 2018, 201, 3456–3464. [Google Scholar] [CrossRef]
- Moral, J.A.; Leung, J.; Rojas, L.A.; Ruan, J.; Zhao, J.; Sethna, Z.; Ramnarain, A.; Gasmi, B.; Gururajan, M.; Redmond, D.; et al. ILC2s amplify PD-1 blockade by activating tissue-specific cancer immunity. Nature 2020, 579, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Lakshmikanth, T.; Burke, S.; Ali, T.H.; Kimpfler, S.; Ursini, F.; Ruggeri, L. NCRs and DNAM-1 me-diate NK cell recognition and lysis of human and mouse melanoma cell lines in vitro and in vivo. J. Clin. Investig. 2009, 119, 1251–1263. [Google Scholar] [CrossRef]
- Chan, C.J.; Andrews, D.M.; McLaughlin, N.M.; Yagita, H.; Gilfillan, S.; Colonna, M. DNAM-1/CD155 interactions promote cytokine and NK cell-mediated suppression of poorly immunogenic mela-noma metastases. J. Immunol. 2010, 184, 902–911. [Google Scholar] [CrossRef]
- Roman Aguilera, A.; Lutzky, V.P.; Mittal, D.; Li, X.Y.; Stannard, K.; Takeda, K. CD96 targeted an-tibodies need not block CD96-CD155 interactions to promote NK cell anti-metastatic activity. Oncoimmunology 2018, 7, e1424677. [Google Scholar] [CrossRef]
- Chan, C.J.; Martinet, L.; Gilfillan, S.; Souza-Fonseca-Guimaraes, F.; Chow, M.T.; Town, L. The re-ceptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat. Immunol. 2014, 15, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Okumura, G.; Iguchi-Manaka, A.; Murata, R.; Yamashita-Kanemaru, Y.; Shibuya, A.; Shibuya, K. Tu-mor-derived soluble CD155 inhibits DNAM-1-mediated antitumor activity of natural killer cells. J. Exp. Med. 2020, 217, e20191290. [Google Scholar] [CrossRef]
- Chauvin, J.-M.; Ka, M.; Pagliano, O.; Menna, C.; Ding, Q.; DeBlasio, R.; Sanders, C.; Hou, J.; Li, X.-Y.; Ferrone, S.; et al. IL15 Stimulation with TIGIT Blockade Reverses CD155-mediated NK-Cell Dysfunction in Melanoma. Clin. Cancer Res. 2020, 26, 5520–5533. [Google Scholar] [CrossRef] [PubMed]
- Lepletier, A.; Madore, J.; O’Donnell, J.S.; Johnston, R.L.; Li, X.Y.; McDonald, E. Tumor CD155 Ex-pression Is Associated with Resistance to Anti-PD1 Immunotherapy in Metastatic Melanoma. Clin. Cancer Res. 2020, 26, 3671–3681. [Google Scholar] [PubMed]
- O’Donnell, J.S.; Madore, J.; Li, X.-Y.; Smyth, M.J. Tumor intrinsic and extrinsic immune functions of CD155. Semin. Cancer Biol. 2020, 65, 189–196. [Google Scholar] [CrossRef]
- Blake, S.J.; Stannard, K.; Liu, J.; Allen, S.; Yong, M.C.; Mittal, D.; Aguilera, A.R.; Miles, J.J.; Lutzky, V.P.; De Andrade, L.F.; et al. Suppression of Metastases Using a New Lymphocyte Checkpoint Target for Cancer Immunotherapy. Cancer Discov. 2016, 6, 446–459. [Google Scholar] [CrossRef] [PubMed]
- André, P.; Denis, C.; Soulas, C.; Bourbon-Caillet, C.; Lopez, J.; Arnoux, T.; Bléry, M.; Bonnafous, C.; Gauthier, L.; Morel, A.; et al. Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells. Cell 2018, 175, 1731–1743.e13. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rethacker, L.; Roelens, M.; Bejar, C.; Maubec, E.; Moins-Teisserenc, H.; Caignard, A. Specific Patterns of Blood ILCs in Metastatic Melanoma Patients and Their Modulations in Response to Immunotherapy. Cancers 2021, 13, 1446. https://doi.org/10.3390/cancers13061446
Rethacker L, Roelens M, Bejar C, Maubec E, Moins-Teisserenc H, Caignard A. Specific Patterns of Blood ILCs in Metastatic Melanoma Patients and Their Modulations in Response to Immunotherapy. Cancers. 2021; 13(6):1446. https://doi.org/10.3390/cancers13061446
Chicago/Turabian StyleRethacker, Louise, Marie Roelens, Claudia Bejar, Eve Maubec, Hélène Moins-Teisserenc, and Anne Caignard. 2021. "Specific Patterns of Blood ILCs in Metastatic Melanoma Patients and Their Modulations in Response to Immunotherapy" Cancers 13, no. 6: 1446. https://doi.org/10.3390/cancers13061446
APA StyleRethacker, L., Roelens, M., Bejar, C., Maubec, E., Moins-Teisserenc, H., & Caignard, A. (2021). Specific Patterns of Blood ILCs in Metastatic Melanoma Patients and Their Modulations in Response to Immunotherapy. Cancers, 13(6), 1446. https://doi.org/10.3390/cancers13061446





