EBV and the Pathogenesis of NK/T Cell Lymphoma
Abstract
Simple Summary
Abstract
1. Introduction
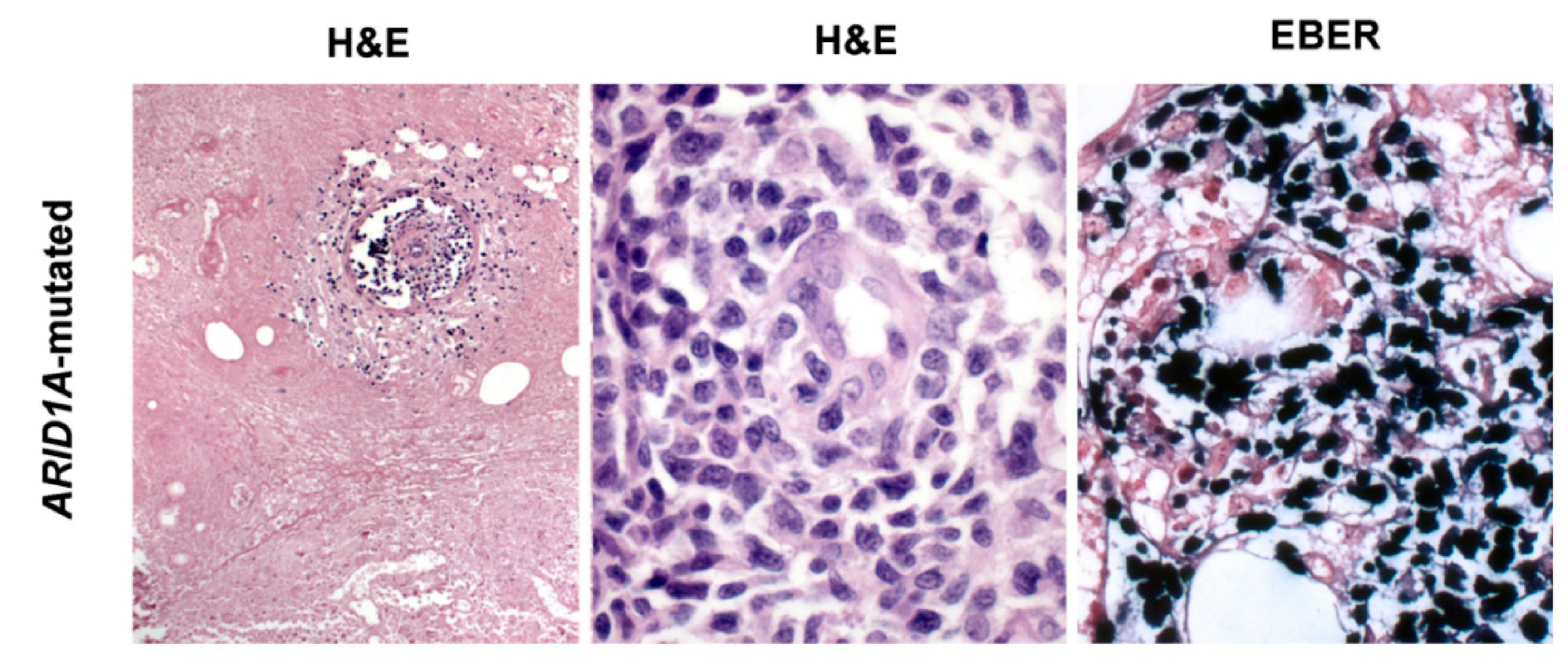
2. Morphological and Immunophenotypical Features of ENKTCL
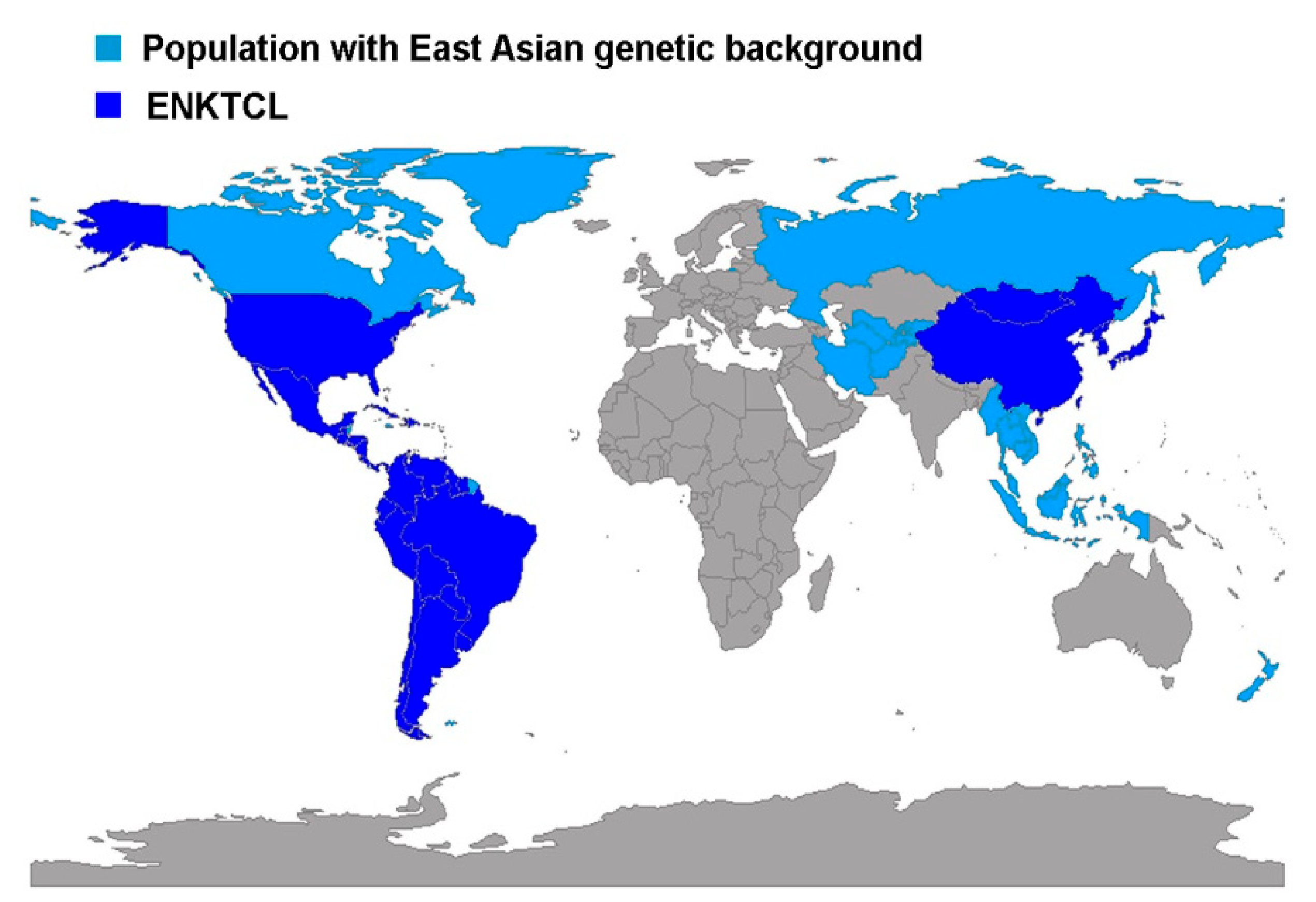
3. ENKTCL Geographic Distribution
4. Epstein-Barr Virus Lymphomagenesis in ENKTCL
4.1. Epstein-Barr Virus Strains and Variations
4.2. LMP1 Variants
| Country | Entity | n | LMP1 Variant | Reference | |
|---|---|---|---|---|---|
| 30 bp del | WT | ||||
| Mexico | ENKTCL | 42 cases | 10 (23.8%) | 32 (76.2%) | [82] |
| Peru | ENKTCL | 27 cases | 0 | 12 (100%) | [78] |
| Argentina | ENKTCL | 12 cases | 5 (41.7%) | 7 (58.3%) | [82] |
| China | ENKTCL | 13 cases | 10 (76.9%) | 3 (23.1%) | [120] |
| China | ENKTCL | 23 cases | 21 (91.3%) | 2 (8.7%) | [84] |
| Mexico | ENKTCL | 23 cases | 6 (26%) | 17 (73.9%) | [6] |
| Malaysia | PTCL | 9 cases | 9 (100%) | 0 | [86] |
| Denmark | PTCL | 18 cases | 11 (61.1%) | 7 (38.9%) | [86] |
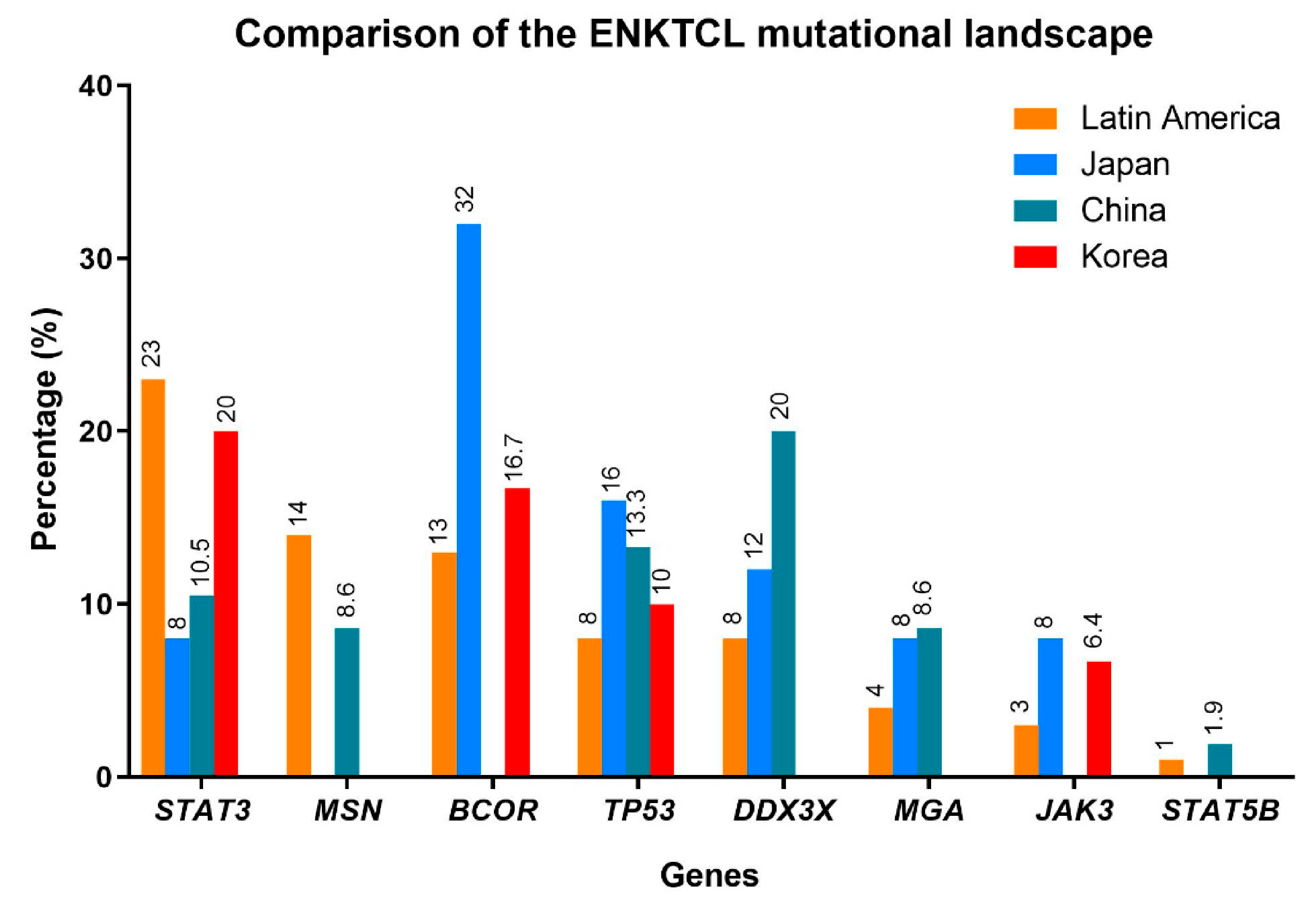
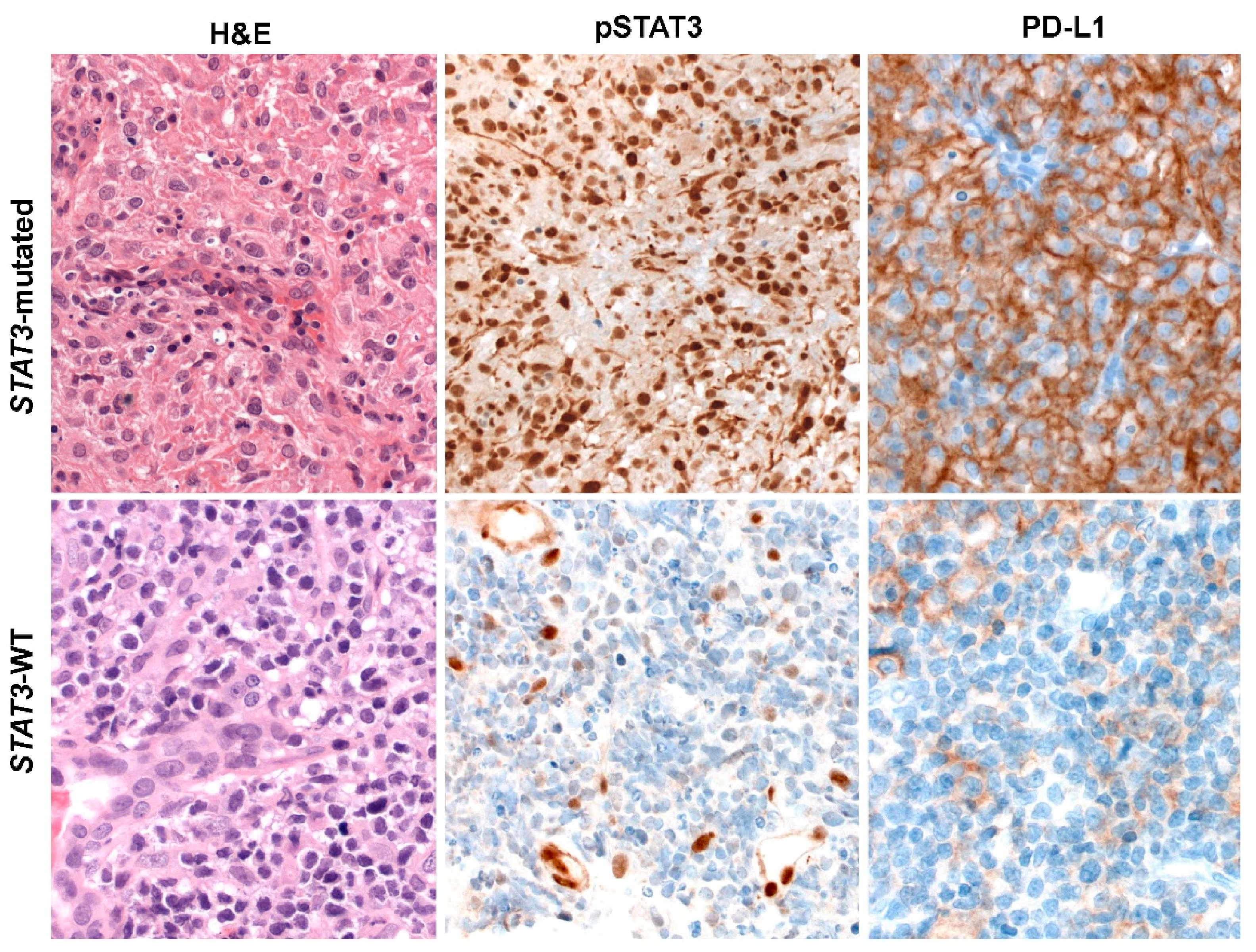
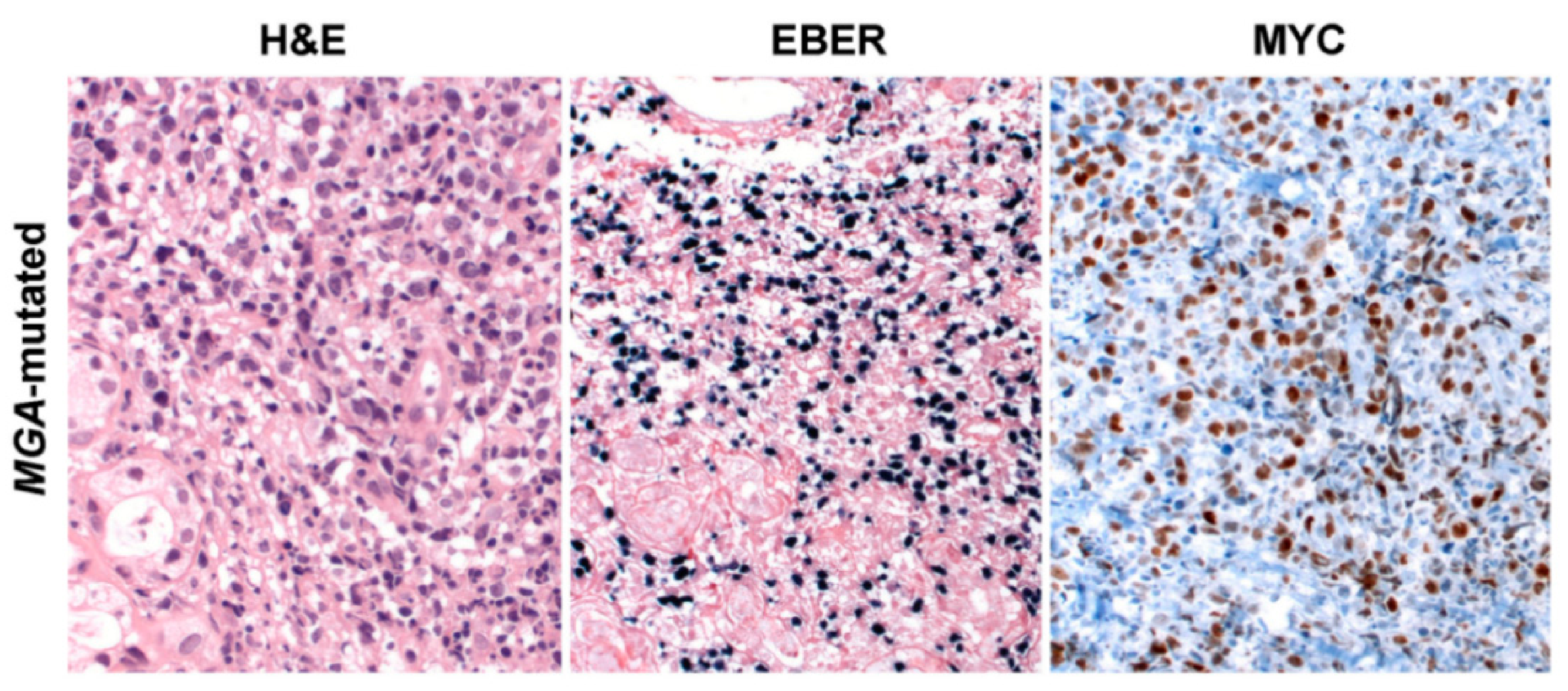
5. ENKTCL Genetic Features
6. ENKTCL Proposed Molecular Classification
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chan, J.K.; Quintanilla-Martinez, L.; Ferry, J.A. Extranodal NK/T-cell Lymphoma, Nasal Type. In WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; Swerdlow, S.H., Ed.; IARC: Lyon, France, 2017; pp. 368–371. [Google Scholar]
- Kanavaros, P.; Briere, J.; Emile, J.F.; Gaulard, P. Epstein-Barr virus in T and natural killer (NK) cell non-Hodgkin’s lymphomas. Leukemia 1996, 10 (Suppl. 2), s84–s87. [Google Scholar]
- Jaffe, E.S.; Harris, N.L.; Stein, H.; Isaacson, P.G. Classification of lymphoid neoplasms: The microscope as a tool for disease discovery. Blood 2008, 112, 4384–4399. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, E.S.; Krenacs, L.; Raffeld, M. Classification of cytotoxic T-cell and natural killer cell lymphomas. Semin. Hematol. 2003, 40, 175–184. [Google Scholar] [CrossRef]
- Lee, J.; Kim, W.S.; Park, Y.H.; Park, S.H.; Park, K.W.; Kang, J.H.; Lee, S.S.; Lee, S.I.; Lee, S.H.; Kim, K.; et al. Nasal-type NK/T cell lymphoma: Clinical features and treatment outcome. Br. J. Cancer 2005, 92, 1226–1230. [Google Scholar] [CrossRef] [PubMed]
- Elenitoba-Johnson, K.S.; Zarate-Osorno, A.; Meneses, A.; Krenacs, L.; Kingma, D.W.; Raffeld, M.; Jaffe, E.S. Cytotoxic granular protein expression, Epstein-Barr virus strain type, and latent membrane protein-1 oncogene deletions in nasal T-lymphocyte/natural killer cell lymphomas from Mexico. Mod. Pathol. 1998, 11, 754–761. [Google Scholar] [PubMed]
- Laurini, J.A.; Perry, A.M.; Boilesen, E.; Diebold, J.; Maclennan, K.A.; Muller-Hermelink, H.K.; Nathwani, B.N.; Armitage, J.O.; Weisenburger, D.D. Classification of non-Hodgkin lymphoma in Central and South America: A review of 1028 cases. Blood 2012, 120, 4795–4801. [Google Scholar] [CrossRef] [PubMed]
- Aviles, A. Nasal NK/T-Cell Lymphoma. A Comparative Analysis of a Mexican Population with the Other Populations of Latin-America. Mediterr. J. Hematol. Infect. Dis. 2015, 7, e2015052. [Google Scholar] [CrossRef] [PubMed]
- Porcu, P.; Caligiuri, M. A sheep in wolf’s clothing. Blood 2011, 117, 1438–1439. [Google Scholar] [CrossRef][Green Version]
- Lamy, T.; Moignet, A.; Loughran, T.P., Jr. LGL leukemia: From pathogenesis to treatment. Blood 2017, 129, 1082–1094. [Google Scholar] [CrossRef]
- Montes-Mojarro, I.A.; Kim, W.Y.; Fend, F.; Quintanilla-Martinez, L. Epstein—Barr virus positive T and NK-cell lymphoproliferations: Morphological features and differential diagnosis. Semin. Diagn. Pathol. 2020, 37, 32–46. [Google Scholar] [CrossRef]
- Chan, J.K.C.; Jaffe, E.S.; Ko, Y.-H. Aggressive NK-cell Leukaemia. In WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; Swerdlow, S.H., Ed.; IARC: Lyon, France, 2017; pp. 353–354. [Google Scholar]
- Jeon, Y.K.; Kim, J.H.; Sung, J.Y.; Han, J.H.; Ko, Y.H. Epstein-Barr virus-positive nodal T/NK-cell lymphoma: An analysis of 15 cases with distinct clinicopathological features. Hum. Pathol. 2015, 46, 981–990. [Google Scholar] [CrossRef]
- Aozasa, K.; Takakuwa, T.; Hongyo, T.; Yang, W.I. Nasal NK/T-cell lymphoma: Epidemiology and pathogenesis. Int. J. Hematol. 2008, 87, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Harabuchi, Y.; Yamanaka, N.; Kataura, A.; Imai, S.; Kinoshita, T.; Mizuno, F.; Osato, T. Epstein-Barr virus in nasal T-cell lymphomas in patients with lethal midline granuloma. Lancet 1990, 335, 128–130. [Google Scholar] [CrossRef]
- Dambaugh, T.; Hennessy, K.; Chamnankit, L.; Kieff, E. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr nuclear antigen 2. Proc. Natl. Acad. Sci. USA 1984, 81, 7632–7636. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Yajima, M.; Ikuta, K. Epstein-Barr virus strain variation and cancer. Cancer Sci. 2019, 110, 1132–1139. [Google Scholar] [CrossRef]
- Lin, G.-W.; Xu, C.; Chen, K.; Huang, H.-Q.; Chen, J.; Song, B.; Chan, J.K.C.; Li, W.; Liu, W.; Shih, L.-Y.; et al. Genetic risk of extranodal natural killer T-cell lymphoma: A genome-wide association study in multiple populations. Lancet Oncol. 2020, 21, 306–316. [Google Scholar] [CrossRef]
- Kanno, H.; Kojya, S.; Li, T.; Ohsawa, M.; Nakatsuka, S.; Miyaguchi, M.; Harabuchi, Y.; Aozasa, K. Low frequency of HLA-A*0201 allele in patients with Epstein-Barr virus-positive nasal lymphomas with polymorphic reticulosis morphology. Int. J. Cancer 2000, 87, 195–199. [Google Scholar] [CrossRef]
- Lee, S.; Park, H.Y.; Kang, S.Y.; Kim, S.J.; Hwang, J.; Lee, S.; Kwak, S.H.; Park, K.S.; Yoo, H.Y.; Kim, W.S.; et al. Genetic alterations of JAK/STAT cascade and histone modification in extranodal NK/T-cell lymphoma nasal type. Oncotarget 2015, 6, 17764–17776. [Google Scholar] [CrossRef] [PubMed]
- Kucuk, C.; Jiang, B.; Hu, X.; Zhang, W.; Chan, J.K.; Xiao, W.; Lack, N.; Alkan, C.; Williams, J.C.; Avery, K.N.; et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from gammadelta-T or NK cells. Nat. Commun. 2015, 6, 6025. [Google Scholar] [CrossRef]
- Jiang, L.; Gu, Z.H.; Yan, Z.X.; Zhao, X.; Xie, Y.Y.; Zhang, Z.G.; Pan, C.M.; Hu, Y.; Cai, C.P.; Dong, Y.; et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat. Genet. 2015, 47, 1061–1066. [Google Scholar] [CrossRef]
- Dobashi, A.; Tsuyama, N.; Asaka, R.; Togashi, Y.; Ueda, K.; Sakata, S.; Baba, S.; Sakamoto, K.; Hatake, K.; Takeuchi, K. Frequent BCOR aberrations in extranodal NK/T-Cell lymphoma, nasal type. Genes Chromosomes Cancer 2016, 55, 460–471. [Google Scholar] [CrossRef]
- Wen, H.; Ma, H.; Cai, Q.; Lin, S.; Lei, X.; He, B.; Wu, S.; Wang, Z.; Gao, Y.; Liu, W.; et al. Recurrent ECSIT mutation encoding V140A triggers hyperinflammation and promotes hemophagocytic syndrome in extranodal NK/T cell lymphoma. Nat. Med. 2018, 24, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.K. Natural killer cell neoplasms. Anat. Pathol. 1998, 3, 77–145. [Google Scholar] [PubMed]
- Hasserjian, R.P.; Harris, N.L. NK-cell lymphomas and leukemias: A spectrum of tumors with variable manifestations and immunophenotype. Am. J. Clin. Pathol. 2007, 127, 860–868. [Google Scholar] [CrossRef]
- Haedicke, W.; Ho, F.C.; Chott, A.; Moretta, L.; Rudiger, T.; Ott, G.; Muller-Hermelink, H.K. Expression of CD94/NKG2A and killer immunoglobulin-like receptors in NK cells and a subset of extranodal cytotoxic T-cell lymphomas. Blood 2000, 95, 3628–3630. [Google Scholar] [CrossRef]
- Lin, C.W.; Chen, Y.H.; Chuang, Y.C.; Liu, T.Y.; Hsu, S.M. CD94 transcripts imply a better prognosis in nasal-type extranodal NK/T-cell lymphoma. Blood 2003, 102, 2623–2631. [Google Scholar] [CrossRef]
- Jhuang, J.Y.; Chang, S.T.; Weng, S.F.; Pan, S.T.; Chu, P.Y.; Hsieh, P.P.; Wei, C.H.; Chou, S.C.; Koo, C.L.; Chen, C.J.; et al. Extranodal natural killer/T-cell lymphoma, nasal type in Taiwan: A relatively higher frequency of T-cell lineage and poor survival for extranasal tumors. Hum. Pathol. 2015, 46, 313–321. [Google Scholar] [CrossRef]
- Kim, W.Y.; Nam, S.J.; Kim, S.; Kim, T.M.; Heo, D.S.; Kim, C.W.; Jeon, Y.K. Prognostic implications of CD30 expression in extranodal natural killer/T-cell lymphoma according to treatment modalities. Leuk. Lymphoma. 2015, 56, 1778–1786. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.S.; Lo, S.T.; Chan, J.K. Peripheral T and putative natural killer cell lymphomas commonly coexpress CD95 and CD95 ligand. Hum. Pathol. 1999, 30, 48–53. [Google Scholar] [CrossRef]
- Chen, Y.; Tan, S.-Y.; Petersson, B.F.; Khor, Y.M.; Gopalakrishnan, S.K.; Tan, D. Occult recurrence of monomorphic epitheliotropic intestinal T-cell lymphoma and the role of MATK gene expression in diagnosis. Hematol. Oncol. 2017, 35, 852–855. [Google Scholar] [CrossRef]
- Roberti, A.; Dobay, M.P.; Bisig, B.; Vallois, D.; Boéchat, C.; Lanitis, E.; Bouchindhomme, B.; Parrens, M.-C.; Bossard, C.; Quintanilla-Martinez, L.; et al. Type II enteropathy-associated T-cell lymphoma features a unique genomic profile with highly recurrent SETD2 alterations. Nat. Commun. 2016, 7, 12602. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Huang, W.; Cao, Z.; Zheng, B.; Liu, X.; Guo, L.; Feng, X. The correlation of clinicopathological features and prognosis in extranodal natural killer/T cell lymphoma: A report of 42 cases in the early stage. Ann. Hematol. 2019, 98, 1467–1476. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wei, Y.; Yan, X.; Li, N.; Song, H.; Yang, L.; Wu, Y.; Xi, Y.-F.; Weng, H.-W.; Li, J.-H.; et al. Survivin is a prognostic marker and therapeutic target for extranodal, nasal-type natural killer/T cell lymphoma. Ann. Transl. Med. 2019, 7, 316. [Google Scholar] [CrossRef]
- Fiore, D.; Cappelli, L.V.; Broccoli, A.; Zinzani, P.L.; Chan, W.C.; Inghirami, G. Peripheral T cell lymphomas: From the bench to the clinic. Nat. Rev. Cancer 2020, 20, 323–342. [Google Scholar] [CrossRef]
- Bellei, M.; Sabattini, E.; Pesce, E.A.; Ko, Y.H.; Kim, W.S.; Cabrera, M.E.; Martinez, V.; Dlouhy, I.; Paes, R.P.; Barrese, T.; et al. Pitfalls and major issues in the histologic diagnosis of peripheral T-cell lymphomas: Results of the central review of 573 cases from the T-Cell Project, an international, cooperative study. Hematol. Oncol. 2017, 35, 630–636. [Google Scholar] [CrossRef]
- Aozasa, K.; Zaki, M.A. Epidemiology and pathogenesis of nasal NK/T-cell lymphoma: A mini-review. Sci. World J. 2011, 11, 422–428. [Google Scholar] [CrossRef]
- Au, W.Y.; Weisenburger, D.D.; Intragumtornchai, T.; Nakamura, S.; Kim, W.S.; Sng, I.; Vose, J.; Armitage, J.O.; Liang, R.; International Peripheral, T.C.L.P. Clinical differences between nasal and extranasal natural killer/T-cell lymphoma: A study of 136 cases from the International Peripheral T-Cell Lymphoma Project. Blood 2009, 113, 3931–3937. [Google Scholar] [CrossRef]
- Peh, S.C. Host ethnicity influences non-Hodgkin’s lymphoma subtype frequency and Epstein-Barr virus association rate: The experience of a multi-ethnic patient population in Malaysia. Histopathology 2001, 38, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Tai, Y.C.; Kim, L.H.; Peh, S.C. High frequency of EBV association and 30-bp deletion in the LMP-1 gene in CD56 lymphomas of the upper aerodigestive tract. Pathol. Int. 2004, 54, 158–166. [Google Scholar] [CrossRef]
- Barrionuevo, C.; Zaharia, M.; Martinez, M.T.; Taxa, L.; Misad, O.; Moscol, A.; Sarria, G.; Guerrero, I.; Casanova, L.; Flores, C.; et al. Extranodal NK/T-cell Lymphoma, Nasal Type: Study of Clinicopathologic and Prognosis Factors in a Series of 78 Cases From Peru. Appl. Immunohistochem. Mol. Morphol. 2007, 15, 38–44. [Google Scholar] [CrossRef]
- Quintanilla-Martinez, L.; Franklin, J.L.; Guerrero, I.; Krenacs, L.; Naresh, K.N.; Rama-Rao, C.; Bhatia, K.; Raffeld, M.; Magrath, I.T. Histological and immunophenotypic profile of nasal NK/T cell lymphomas from Peru: High prevalence of p53 overexpression. Hum. Pathol. 1999, 30, 849–855. [Google Scholar] [CrossRef]
- Arber, D.A.; Weiss, L.M.; Albujar, P.F.; Chen, Y.Y.; Jaffe, E.S. Nasal lymphomas in Peru. High incidence of T-cell immunophenotype and Epstein-Barr virus infection. Am. J. Surg. Pathol. 1993, 17, 392–399. [Google Scholar] [CrossRef]
- Barros, M.H.; Vera-Lozada, G.; Soares, F.A.; Niedobitek, G.; Hassan, R. Tumor microenvironment composition in pediatric classical Hodgkin lymphoma is modulated by age and Epstein-Barr virus infection. Int. J. Cancer 2012, 131, 1142–1152. [Google Scholar] [CrossRef] [PubMed]
- Altemani, A.; Barbosa, A.C.; Kulka, M.; Takahashi, T.; Endo, L.; Vassallo, J.; Lorand-Metze, I. Characteristics of nasal T/NK-cell lymphoma among Brazilians. Neoplasma 2002, 49, 55–60. [Google Scholar] [PubMed]
- Bellei, M.; Chiattone, C.S.; Luminari, S.; Pesce, E.A.; Cabrera, M.E.; de Souza, C.A.; Gabús, R.; Zoppegno, L.; Zoppegno, L.; Milone, J.; et al. T-cell lymphomas in South america and europe. Rev. Bras. Hematol. E Hemoter. 2012, 34, 42–47. [Google Scholar] [CrossRef]
- Schwartz, E.J.; Molina-Kirsch, H.; Zhao, S.; Marinelli, R.J.; Warnke, R.A.; Natkunam, Y. Immunohistochemical Characterization of Nasal-Type Extranodal NK/T-Cell Lymphoma Using a Tissue Microarray: An Analysis of 84 Cases. Am. J. Clin. Pathol. 2008, 130, 343–351. [Google Scholar] [CrossRef]
- Cabrera, M.E.; Eizuru, Y.; Itoh, T.; Koriyama, C.; Tashiro, Y.; Ding, S.; Rey, S.; Akiba, S.; Corvalan, A. Nasal natural killer/T-cell lymphoma and its association with type “i”/XhoI loss strain Epstein-Barr virus in Chile. J. Clin. Pathol. 2007, 60, 656–660. [Google Scholar] [CrossRef]
- Quintanilla-Martinez, L.; Kremer, M.; Keller, G.; Nathrath, M.; Gamboa-Dominguez, A.; Meneses, A.; Luna-Contreras, L.; Cabras, A.; Hoefler, H.; Mohar, A.; et al. p53 Mutations in nasal natural killer/T-cell lymphoma from Mexico: Association with large cell morphology and advanced disease. Am. J. Pathol. 2001, 159, 2095–2105. [Google Scholar] [CrossRef]
- Sanchez-Romero, C.; Paes de Almeida, O.; Rendon Henao, J.; Carlos, R. Extranodal NK/T-Cell Lymphoma, Nasal Type in Guatemala: An 86-Case Series Emphasizing Clinical Presentation and Microscopic Characteristics. Head Neck. Pathol. 2019, 13, 624–634. [Google Scholar] [CrossRef]
- Haverkos, B.M.; Pan, Z.; Gru, A.A.; Freud, A.G.; Rabinovitch, R.; Xu-Welliver, M.; Otto, B.; Barrionuevo, C.; Baiocchi, R.A.; Rochford, R.; et al. Extranodal NK/T Cell Lymphoma, Nasal Type (ENKTL-NT): An Update on Epidemiology, Clinical Presentation, and Natural History in North American and European Cases. Curr. Hematol. Malig. Rep. 2016, 11, 514–527. [Google Scholar] [CrossRef]
- Harabuchi, Y.; Takahara, M.; Kishibe, K.; Nagato, T.; Kumai, T. Extranodal Natural Killer/T-Cell Lymphoma, Nasal Type: Basic Science and Clinical Progress. Front. Pediatr. 2019, 7, 141. [Google Scholar] [CrossRef]
- Winter, S.; Martin, E.; Boutboul, D.; Lenoir, C.; Boudjemaa, S.; Petit, A.; Picard, C.; Fischer, A.; Leverger, G.; Latour, S. Loss of RASGRP1 in humans impairs T-cell expansion leading to Epstein-Barr virus susceptibility. EMBO Mol. Med. 2018, 10, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.Y.; Ng, A.Y.J.; Cheng, C.L.; Nairismagi, M.L.; Venkatesh, B.; Cheah, D.M.Z.; Li, S.T.; Chan, S.H.; Ngeow, J.; Laurensia, Y.; et al. Whole exome sequencing identifies recessive germline mutations in FAM160A1 in familial NK/T cell lymphoma. Blood Cancer J. 2018, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- South, A. rworldmap: A New R package for Mapping Global Data. R J. 2011, 3, 35–43. [Google Scholar] [CrossRef]
- Tzellos, S.; Farrell, P.J. Epstein-barr virus sequence variation-biology and disease. Pathogens 2012, 1, 156–174. [Google Scholar] [CrossRef] [PubMed]
- Haahr, S.; Plesner, A.M.; Vestergaard, B.F.; Höllsberg, P. A role of late Epstein-Barr virus infection in multiple sclerosis. Acta Neurol. Scand. 2004, 109, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Henle, G.; Henle, W.; Clifford, P.; Diehl, V.; Kafuko, G.W.; Kirya, B.G.; Klein, G.; Morrow, R.H.; Munube, G.M.; Pike, P.; et al. Antibodies to Epstein-Barr virus in Burkitt’s lymphoma and control groups. J. Natl. Cancer Inst. 1969, 43, 1147–1157. [Google Scholar] [PubMed]
- Kassel, S.H.; Echevarria, R.A.; Guzzo, F.P. Midline malignant reticulosis (so-called lethal midline granuloma). Cancer 1969, 23, 920–935. [Google Scholar] [CrossRef]
- Cohen, J.I. Epstein-Barr virus infection. N. Engl. J. Med. 2000, 343, 481–492. [Google Scholar] [CrossRef]
- Taylor, G.S.; Long, H.M.; Brooks, J.M.; Rickinson, A.B.; Hislop, A.D. The immunology of Epstein-Barr virus-induced disease. Annu. Rev. Immunol. 2015, 33, 787–821. [Google Scholar] [CrossRef]
- Luzuriaga, K.; Sullivan, J.L. Infectious Mononucleosis. N. Engl. J. Med. 2010, 362, 1993–2000. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Chen, K.; Young, K.H. Epstein-Barr virus-positive T/NK-cell lymphoproliferative disorders. Exp. Mol. Med. 2015, 47, e133. [Google Scholar] [CrossRef]
- Anagnostopoulos, I.; Hummel, M.; Kreschel, C.; Stein, H. Morphology, immunophenotype, and distribution of latently and/or productively Epstein-Barr virus-infected cells in acute infectious mononucleosis: Implications for the interindividual infection route of Epstein-Barr virus. Blood 1995, 85, 744–750. [Google Scholar] [CrossRef] [PubMed]
- Hudnall, S.D.; Ge, Y.; Wei, L.; Yang, N.P.; Wang, H.Q.; Chen, T. Distribution and phenotype of Epstein-Barr virus-infected cells in human pharyngeal tonsils. Mod. Pathol. 2005, 18, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Hatton, O.L.; Harris-Arnold, A.; Schaffert, S.; Krams, S.M.; Martinez, O.M. The interplay between Epstein-Barr virus and B lymphocytes: Implications for infection, immunity, and disease. Immunol. Res. 2014, 58, 268–276. [Google Scholar] [CrossRef]
- Smith, N.A.; Coleman, C.B.; Gewurz, B.E.; Rochford, R. CD21 (Complement Receptor 2) Is the Receptor for Epstein-Barr Virus Entry into T Cells. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Tabiasco, J.; Vercellone, A.; Meggetto, F.; Hudrisier, D.; Brousset, P.; Fournié, J.J. Acquisition of viral receptor by NK cells through immunological synapse. J. Immunol. 2003, 170, 5993–5998. [Google Scholar] [CrossRef]
- Isobe, Y.; Sugimoto, K.; Yang, L.; Tamayose, K.; Egashira, M.; Kaneko, T.; Takada, K.; Oshimi, K. Epstein-Barr virus infection of human natural killer cell lines and peripheral blood natural killer cells. Cancer Res. 2004, 64, 2167–2174. [Google Scholar] [CrossRef]
- Kang, M.-S.; Kieff, E. Epstein-Barr virus latent genes. Exp. Mol. Med. 2015, 47, e131. [Google Scholar] [CrossRef] [PubMed]
- Price, A.M.; Luftig, M.A. To be or not IIb: A multi-step process for Epstein-Barr virus latency establishment and consequences for B cell tumorigenesis. PLoS Pathog. 2015, 11, e1004656. [Google Scholar] [CrossRef]
- Babcock, G.J.; Hochberg, D.; Thorley-Lawson, A.D. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 2000, 13, 497–506. [Google Scholar] [CrossRef]
- Klein, E.; Kis, L.L.; Klein, G. Epstein-Barr virus infection in humans: From harmless to life endangering virus-lymphocyte interactions. Oncogene 2007, 26, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Smatti, M.K.; Al-Sadeq, D.W.; Ali, N.H.; Pintus, G.; Abou-Saleh, H.; Nasrallah, G.K. Epstein-Barr Virus Epidemiology, Serology, and Genetic Variability of LMP-1 Oncogene Among Healthy Population: An Update. Front. Oncol. 2018, 8, 211. [Google Scholar] [CrossRef]
- Young, L.S.; Arrand, J.R.; Murray, P.G. EBV Gene Expression and Regulation. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Nemerow, G.R.; Mold, C.; Schwend, V.K.; Tollefson, V.; Cooper, N.R. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: Sequence homology of gp350 and C3 complement fragment C3d. J. Virol. 1987, 61, 1416–1420. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A. EBV Persistence--Introducing the Virus. Curr. Top. Microbiol. Immunol. 2015, 390, 151–209. [Google Scholar] [CrossRef] [PubMed]
- Young, L.S.; Yap, L.F.; Murray, P.G. Epstein-Barr virus: More than 50 years old and still providing surprises. Nat. Rev. Cancer 2016, 16, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Young, L.; Alfieri, C.; Hennessy, K.; Evans, H.; O’Hara, C.; Anderson, K.C.; Ritz, J.; Shapiro, R.S.; Rickinson, A.; Kieff, E.; et al. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N. Engl. J. Med. 1989, 321, 1080–1085. [Google Scholar] [CrossRef]
- Xiong, J.; Cui, B.W.; Wang, N.; Dai, Y.T.; Zhang, H.; Wang, C.F.; Zhong, H.J.; Cheng, S.; Ou-Yang, B.S.; Hu, Y.; et al. Genomic and Transcriptomic Characterization of Natural Killer T Cell Lymphoma. Cancer Cell 2020, 37, 403–419.e406. [Google Scholar] [CrossRef]
- Montes-Mojarro, I.A.; Chen, B.J.; Ramirez-Ibarguen, A.F.; Quezada-Fiallos, C.M.; Perez-Baez, W.B.; Duenas, D.; Casavilca-Zambrano, S.; Ortiz-Mayor, M.; Rojas-Bilbao, E.; Garcia-Rivello, H.; et al. Mutational profile and EBV strains of extranodal NK/T-cell lymphoma, nasal type in Latin America. Mod. Pathol. 2020, 33, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Park, E.-R.; Park, S.-H.; Lin, Z.; Kim, Y.-S. Characteristics of Epstein-Barr virus isolated from the malignant lymphomas in Korea. J. Med. Virol. 2002, 67, 59–66. [Google Scholar] [CrossRef]
- Chiang, A.K.; Wong, K.Y.; Liang, A.C.; Srivastava, G. Comparative analysis of Epstein-Barr virus gene polymorphisms in nasal T/NK-cell lymphomas and normal nasal tissues: Implications on virus strain selection in malignancy. Int. J. Cancer 1999, 80, 356–364. [Google Scholar] [CrossRef]
- Wu, S.-J.; Lay, J.-D.; Chen, C.-L.; Chen, J.-Y.; Liu, M.-Y.; Su, I.-J. Genomic analysis of Epstein-Barr virus in nasal and peripheral T-cell lymphoma: A comparison with nasopharyngeal carcinoma in an endemic area. J. Med. Virol. 1996, 50, 314–321. [Google Scholar] [CrossRef]
- Sandvej, K.; Peh, S.; Andresen, B.; Pallesen, G. Identification of potential hot spots in the carboxy-terminal part of the Epstein-Barr virus (EBV) BNLF-1 gene in both malignant and benign EBV-associated diseases: High frequency of a 30-bp deletion in Malaysian and Danish peripheral T-cell lymphomas. Blood 1994, 84, 4053–4060. [Google Scholar] [CrossRef]
- Rickinson, A.B.; Young, L.S.; Rowe, M. Influence of the Epstein-Barr virus nuclear antigen EBNA 2 on the growth phenotype of virus-transformed B cells. J. Virol. 1987, 61, 1310–1317. [Google Scholar] [CrossRef]
- Fassone, L.; Cingolani, A.; Martini, M.; Migliaretti, G.; Oreste, P.L.; Capello, D.; Gloghini, A.; Vivenza, D.; Dolcetti, R.; Carbone, A.; et al. Characterization of Epstein-Barr virus genotype in AIDS-related non-Hodgkin’s lymphoma. AIDS Res. Hum. Retrovir. 2002, 18, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Panea, R.I.; Love, C.L.; Shingleton, J.R.; Reddy, A.; Bailey, J.A.; Moormann, A.M.; Otieno, J.A.; Ong’echa, J.M.; Oduor, C.I.; Schroeder, K.M.S.; et al. The whole-genome landscape of Burkitt lymphoma subtypes. Blood 2019, 134, 1598–1607. [Google Scholar] [CrossRef] [PubMed]
- Correia, S.; Bridges, R.; Wegner, F.; Venturini, C.; Palser, A.; Middeldorp, J.M.; Cohen, J.I.; Lorenzetti, M.A.; Bassano, I.; White, R.E.; et al. Sequence Variation of Epstein-Barr Virus: Viral Types, Geography, Codon Usage, and Diseases. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Kaymaz, Y.; Oduor, C.I.; Aydemir, O.; Luftig, M.A.; Otieno, J.A.; Ong’echa, J.M.; Bailey, J.A.; Moormann, A.M. Epstein-Barr Virus Genomes Reveal Population Structure and Type 1 Association with Endemic Burkitt Lymphoma. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Chang, C.M.; Yu, K.J.; Mbulaiteye, S.M.; Hildesheim, A.; Bhatia, K. The extent of genetic diversity of Epstein-Barr virus and its geographic and disease patterns: A need for reappraisal. Virus Res. 2009, 143, 209–221. [Google Scholar] [CrossRef]
- Kingma, D.W.; Weiss, W.B.; Jaffe, E.S.; Kumar, S.; Frekko, K.; Raffeld, M. Epstein-Barr virus latent membrane protein-1 oncogene deletions: Correlations with malignancy in Epstein-Barr virus--associated lymphoproliferative disorders and malignant lymphomas. Blood 1996, 88, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Hofscheier, A.; Ponciano, A.; Bonzheim, I.; Adam, P.; Lome-Maldonado, C.; Vela, T.; Cortes, E.; Ortiz-Hidalgo, C.; Fend, F.; Quintanilla-Martinez, L. Geographic variation in the prevalence of Epstein-Barr virus-positive diffuse large B-cell lymphoma of the elderly: A comparative analysis of a Mexican and a German population. Mod. Pathol. 2011, 24, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Dirnhofer, S.; Angeles-Angeles, A.; Ortiz-Hidalgo, C.; Reyes, E.; Gredler, E.; Krugmann, J.; Fend, F.; Quintanilla-Martinez, L. High prevalence of a 30-base pair deletion in the Epstein-Barr virus (EBV) latent membrane protein 1 gene and of strain type B EBV in Mexican classical Hodgkin’s disease and reactive lymphoid tissue. Hum. Pathol. 1999, 30, 781–787. [Google Scholar] [CrossRef]
- Hu, L.F.; Zabarovsky, E.R.; Chen, F.; Cao, S.L.; Ernberg, I.; Klein, G.; Winberg, G. Isolation and sequencing of the Epstein-Barr virus BNLF-1 gene (LMP1) from a Chinese nasopharyngeal carcinoma. J. Gen. Virol. 1991, 72 Pt 10, 2399–2409. [Google Scholar] [CrossRef]
- See, H.S.; Yap, Y.Y.; Yip, W.K.; Seow, H.F. Epstein-Barr virus latent membrane protein-1 (LMP-1) 30-bp deletion and Xho I-loss is associated with type III nasopharyngeal carcinoma in Malaysia. World J. Surg. Oncol. 2008, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.; Xie, Z.; Liu, C.; Huang, Z.; Xu, J. Analysis of EBNA-1 and LMP-1 variants in diseases associated with EBV infection in Chinese children. Virol. J. 2012, 9, 13. [Google Scholar] [CrossRef]
- Do, N.V.; Ingemar, E.; Phi, P.T.; Jenny, A.; Chinh, T.T.; Zeng, Y.; Hu, L. A major EBNA1 variant from Asian EBV isolates shows enhanced transcriptional activity compared to prototype B95.8. Virus Res. 2008, 132, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Lung, M.L.; Lam, W.P.; Sham, J.; Choy, D.; Yong-Sheng, Z.; Guo, H.Y.; Ng, M.H. Detection and prevalence of the “f” variant of Epstein-Barr virus in southern China. Virology 1991, 185, 67–71. [Google Scholar] [CrossRef]
- Abdirad, A.; Ghaderi-Sohi, S.; Shuyama, K.; Koriyama, C.; Nadimi-Barforoosh, H.; Emami, S.; Mosavi-Jarrahi, A.; Nahvijou, A.; Akiba, S. Epstein-Barr virus associated gastric carcinoma: A report from Iran in the last four decades. Diagn. Pathol. 2007, 2, 25. [Google Scholar] [CrossRef]
- Kaye, K.M.; Izumi, K.M.; Kieff, E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 1993, 90, 9150–9154. [Google Scholar] [CrossRef]
- Young, L.S.; Rickinson, A.B. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 2004, 4, 757–768. [Google Scholar] [CrossRef]
- Gires, O.; Zimber-Strobl, U.; Gonnella, R.; Ueffing, M.; Marschall, G.; Zeidler, R.; Pich, D.; Hammerschmidt, W. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 1997, 16, 6131–6140. [Google Scholar] [CrossRef]
- Martin, J.; Sugden, B. The latent membrane protein oncoprotein resembles growth factor receptors in the properties of its turnover. Cell Growth Differ 1991, 2, 653–660. [Google Scholar]
- Eliopoulos, A.G.; Young, L.S. LMP1 structure and signal transduction. Semin. Cancer Biol. 2001, 11, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Fennewald, S.; van Santen, V.; Kieff, E. Nucleotide sequence of an mRNA transcribed in latent growth-transforming virus infection indicates that it may encode a membrane protein. J. Virol. 1984, 51, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Coffin, W.F., 3rd; Erickson, K.D.; Hoedt-Miller, M.; Martin, J.M. The cytoplasmic amino-terminus of the Latent Membrane Protein-1 of Epstein-Barr Virus: Relationship between transmembrane orientation and effector functions of the carboxy-terminus and transmembrane domain. Oncogene 2001, 20, 5313–5330. [Google Scholar] [CrossRef]
- Zhang, X.S.; Song, K.H.; Mai, H.Q.; Jia, W.H.; Feng, B.J.; Xia, J.C.; Zhang, R.H.; Huang, L.X.; Yu, X.J.; Feng, Q.S.; et al. The 30-bp deletion variant: A polymorphism of latent membrane protein 1 prevalent in endemic and non-endemic areas of nasopharyngeal carcinomas in China. Cancer Lett. 2002, 176, 65–73. [Google Scholar] [CrossRef]
- Hadhri-Guiga, B.; Khabir, A.-M.; Mokdad-Gargouri, R.; Ghorbel, A.-M.; Drira, M.; Daoud, J.; Frikha, M.; Jlidi, R.; Gargouri, A. Various 30 and 69 bp deletion variants of the Epstein-Barr virus LMP1 may arise by homologous recombination in nasopharyngeal carcinoma of Tunisian patients. Virus Res. 2006, 115, 24–30. [Google Scholar] [CrossRef]
- Edwards, R.H.; Seillier-Moiseiwitsch, F.; Raab-Traub, N. Signature amino acid changes in latent membrane protein 1 distinguish Epstein-Barr virus strains. Virology 1999, 261, 79–95. [Google Scholar] [CrossRef] [PubMed]
- Neves, M.; Marinho-Dias, J.; Ribeiro, J.; Sousa, H. Epstein-Barr virus strains and variations: Geographic or disease-specific variants? J. Med. Virol. 2017, 89, 373–387. [Google Scholar] [CrossRef]
- Li, S.N.; Chang, Y.S.; Liu, S.T. Effect of a 10-amino acid deletion on the oncogenic activity of latent membrane protein 1 of Epstein-Barr virus. Oncogene 1996, 12, 2129–2135. [Google Scholar]
- Knecht, H.; Bachmann, E.; Brousset, P.; Sandvej, K.; Nadal, D.; Bachmann, F.; Odermatt, B.F.; Delsol, G.; Pallesen, G. Deletions within the LMP1 oncogene of Epstein-Barr virus are clustered in Hodgkin’s disease and identical to those observed in nasopharyngeal carcinoma. Blood 1993, 82, 2937–2942. [Google Scholar] [CrossRef] [PubMed]
- Itakura, O.; Yamada, S.; Narita, M.; Kikuta, H. High prevalence of a 30-base pair deletion and single-base mutations within the carboxy terminal end of the LMP-1 oncogene of Epstein-Barr virus in the Japanese population. Oncogene 1996, 13, 1549–1553. [Google Scholar]
- Mori, S.; Itoh, T.; Tokunaga, M.; Eizuru, Y. Deletions and single-base mutations within the carboxy-terminal region of the latent membrane protein 1 oncogene in Epstein-Barr virus-related gastric cancers of southern Japan. J. Med. Virol. 1999, 57, 152–158. [Google Scholar] [CrossRef]
- da Costa, V.G.; Marques-Silva, A.C.; Moreli, M.L. The Epstein-Barr virus latent membrane protein-1 (LMP1) 30-bp deletion and XhoI-polymorphism in nasopharyngeal carcinoma: A meta-analysis of observational studies. Syst. Rev. 2015, 4, 46. [Google Scholar] [CrossRef] [PubMed]
- Okuno, Y.; Murata, T.; Sato, Y.; Muramatsu, H.; Ito, Y.; Watanabe, T.; Okuno, T.; Murakami, N.; Yoshida, K.; Sawada, A.; et al. Defective Epstein-Barr virus in chronic active infection and haematological malignancy. Nat. Microbiol. 2019, 4, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.J.; Han, B.W.; Cai, Q.Q.; Zuo, X.Y.; Xia, T.; Chen, J.R.; Feng, L.N.; Lim, J.Q.; Chen, S.W.; Zeng, M.S.; et al. Genomic and transcriptomic landscapes of Epstein-Barr virus in extranodal natural killer T-cell lymphoma. Leukemia 2019, 33, 1451–1462. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-H.; Wu, Q.-L.; Zong, Y.-S.; Feng, Y.-F.; Hou, J.-H. Nasopharyngeal Extranodal NK/T-Cell Lymphoma, Nasal Type: Retrospective Study of 18 Consecutive Cases in Guangzhou, China. Int. J. Surg. Pathol. 2011, 19, 51–61. [Google Scholar] [CrossRef]
- Han, A.J.; Zong, Y.S.; Zhang, M.; Cao, S.M.; Lin, S.X.; Liang, Y.J. Analysis of Epstein-Barr virus with BamHI “f” variant and XhoI-loss of LMP1 gene in nasopharyngeal carcinoma. Zhonghua Bing Li Xue Za Zhi 2003, 32, 534–538. [Google Scholar]
- Khanim, F.; Yao, Q.Y.; Niedobitek, G.; Sihota, S.; Rickinson, A.B.; Young, L. Analysis of Epstein-Barr Virus gene polymorphisms in normal donors and in virus-associated tumors from different geographic locations. Blood 1996, 88, 3491–3501. [Google Scholar] [CrossRef]
- Feederle, R.; Klinke, O.; Kutikhin, A.; Poirey, R.; Tsai, M.H.; Delecluse, H.J. Epstein-Barr Virus: From the Detection of Sequence Polymorphisms to the Recognition of Viral Types. Curr. Top. Microbiol. Immunol. 2015, 390, 119–148. [Google Scholar] [CrossRef]
- Xiong, J.; Zhao, W.-L. Advances in multiple omics of natural-killer/T cell lymphoma. J. Hematol. Oncol. 2018, 11, 134. [Google Scholar] [CrossRef] [PubMed]
- de Mel, S.; Hue, S.S.-S.; Jeyasekharan, A.D.; Chng, W.-J.; Ng, S.-B. Molecular pathogenic pathways in extranodal NK/T cell lymphoma. J. Hematol. Oncol. 2019, 12, 33. [Google Scholar] [CrossRef]
- Karube, K.; Nakagawa, M.; Tsuzuki, S.; Takeuchi, I.; Honma, K.; Nakashima, Y.; Shimizu, N.; Ko, Y.H.; Morishima, Y.; Ohshima, K.; et al. Identification of FOXO3 and PRDM1 as tumor-suppressor gene candidates in NK-cell neoplasms by genomic and functional analyses. Blood 2011, 118, 3195–3204. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, J.; Kucuk, C.; deLeeuw, R.J.; Srivastava, G.; Tam, W.; Geng, H.; Klinkebiel, D.; Christman, J.K.; Patel, K.; Cao, K.; et al. Genomic analyses reveal global functional alterations that promote tumor growth and novel tumor suppressor genes in natural killer-cell malignancies. Leukemia 2009, 23, 1139–1151. [Google Scholar] [CrossRef]
- Huang, Y.; de Reynies, A.; de Leval, L.; Ghazi, B.; Martin-Garcia, N.; Travert, M.; Bosq, J.; Briere, J.; Petit, B.; Thomas, E.; et al. Gene expression profiling identifies emerging oncogenic pathways operating in extranodal NK/T-cell lymphoma, nasal type. Blood 2010, 115, 1226–1237. [Google Scholar] [CrossRef]
- Ko, Y.H.; Choi, K.E.; Han, J.H.; Kim, J.M.; Ree, H.J. Comparative genomic hybridization study of nasal-type NK/T-cell lymphoma. Cytometry 2001, 46, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Li, M.; Huang, X.; Xu, J.; Gao, Z.; Liu, C. High-resolution genome-wide analysis identified recurrent genetic alterations in NK/T-cell lymphoma, nasal type, which are associated with disease progression. Med. Oncol. 2014, 31, 71. [Google Scholar] [CrossRef]
- Nakashima, Y.; Tagawa, H.; Suzuki, R.; Karnan, S.; Karube, K.; Ohshima, K.; Muta, K.; Nawata, H.; Morishima, Y.; Nakamura, S.; et al. Genome-wide array-based comparative genomic hybridization of natural killer cell lymphoma/leukemia: Different genomic alteration patterns of aggressive NK-cell leukemia and extranodal Nk/T-cell lymphoma, nasal type. Genes Chromosomes Cancer 2005, 44, 247–255. [Google Scholar] [CrossRef]
- Yang, C.F.; Hsu, C.Y.; Ho, D.M. Aggressive natural killer (NK)-cell leukaemia and extranodal NK/T-cell lymphoma are two distinct diseases that differ in their clinical presentation and cytogenetic findings. Histopathology 2018, 72, 955–964. [Google Scholar] [CrossRef]
- Ng, S.B.; Chung, T.H.; Kato, S.; Nakamura, S.; Takahashi, E.; Ko, Y.H.; Khoury, J.D.; Yin, C.C.; Soong, R.; Jeyasekharan, A.D.; et al. Epstein-Barr virus-associated primary nodal T/NK-cell lymphoma shows a distinct molecular signature and copy number changes. Haematologica 2018, 103, 278–287. [Google Scholar] [CrossRef]
- Huang, Y.; de Leval, L.; Gaulard, P. Molecular underpinning of extranodal NK/T-cell lymphoma. Best Pract. Res. Clin. Haematol. 2013, 26, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Song, T.L.; Nairismagi, M.L.; Laurensia, Y.; Lim, J.Q.; Tan, J.; Li, Z.M.; Pang, W.L.; Kizhakeyil, A.; Wijaya, G.C.; Huang, D.C.; et al. Oncogenic activation of the STAT3 pathway drives PD-L1 expression in natural killer/T-cell lymphoma. Blood 2018, 132, 1146–1158. [Google Scholar] [CrossRef]
- Sim, S.H.; Kim, S.; Kim, T.M.; Jeon, Y.K.; Nam, S.J.; Ahn, Y.O.; Keam, B.; Park, H.H.; Kim, D.W.; Kim, C.W.; et al. Novel JAK3-Activating Mutations in Extranodal NK/T-Cell Lymphoma, Nasal Type. Am. J. Pathol. 2017, 187, 980–986. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, X.; Xue, W.; Zhang, Y.; Li, C.; Song, Y.; Mei, M.; Lu, L.; Wang, Y.; Zhou, Z.; et al. Recurrent GNAQ mutation encoding T96S in natural killer/T cell lymphoma. Nat. Commun. 2019, 10, 4209. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Liang, A.C.; Lu, L.; Au, W.Y.; Kwong, Y.L.; Liang, R.H.; Srivastava, G. Frequent deletion of Fas gene sequences encoding death and transmembrane domains in nasal natural killer/T-cell lymphoma. Am. J. Pathol. 2002, 161, 2123–2131. [Google Scholar] [CrossRef]
- Takakuwa, T.; Dong, Z.; Nakatsuka, S.; Kojya, S.; Harabuchi, Y.; Yang, W.I.; Nagata, S.; Aozasa, K. Frequent mutations of Fas gene in nasal NK/T cell lymphoma. Oncogene 2002, 21, 4702–4705. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, N.; Inagaki, N.; Mizumura, S.; Sugimoto, K.J.; Sakajiri, S.; Ohyanagi-Hara, M.; Oshimi, K. Methylation status analysis of cell cycle regulatory genes (p16INK4A, p15INK4B, p21Waf1/Cip1, p27Kip1 and p73) in natural killer cell disorders. Eur. J. Haematol. 2005, 74, 424–429. [Google Scholar] [CrossRef]
- Küçük, C.; Hu, X.; Jiang, B.; Klinkebiel, D.; Geng, H.; Gong, Q.; Bouska, A.; Iqbal, J.; Gaulard, P.; McKeithan, T.W.; et al. Global Promoter Methylation Analysis Reveals Novel Candidate Tumor Suppressor Genes in Natural Killer Cell Lymphoma. Clin. Cancer Res. 2015, 21, 1699. [Google Scholar] [CrossRef]
- Loong, K.T. EZH2 Mediates Resistance to Apoptosis in Nktl By Activating Nfkb Signaling Through Repression Of TNFAIP3/A20 By H3K27 Trimethylation. Blood 2013, 122, 1278. [Google Scholar] [CrossRef]
- Yan, J.; Ng, S.B.; Tay, J.L.; Lin, B.; Koh, T.L.; Tan, J.; Selvarajan, V.; Liu, S.C.; Bi, C.; Wang, S.; et al. EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood 2013, 121, 4512–4520. [Google Scholar] [CrossRef]
- Ng, S.B.; Selvarajan, V.; Huang, G.; Zhou, J.; Feldman, A.L.; Law, M.; Kwong, Y.L.; Shimizu, N.; Kagami, Y.; Aozasa, K.; et al. Activated oncogenic pathways and therapeutic targets in extranodal nasal-type NK/T cell lymphoma revealed by gene expression profiling. J. Pathol. 2011, 223, 496–510. [Google Scholar] [CrossRef] [PubMed]
- Paik, J.H.; Jang, J.Y.; Jeon, Y.K.; Kim, W.Y.; Kim, T.M.; Heo, D.S.; Kim, C.W. MicroRNA-146a downregulates NFkappaB activity via targeting TRAF6 and functions as a tumor suppressor having strong prognostic implications in NK/T cell lymphoma. Clin. Cancer Res. 2011, 17, 4761–4771. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, Y.; Tagawa, H.; Takahashi, N.; Watanabe, A.; Guo, Y.M.; Iwamoto, K.; Yamashita, J.; Saitoh, H.; Kameoka, Y.; Shimizu, N.; et al. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood 2009, 114, 3265–3275. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhao, Y.; Shi, H.; Ma, C.; Wei, L. LMP-1 induces survivin expression to inhibit cell apoptosis through the NF-κB and PI3K/Akt signaling pathways in nasal NK/T-cell lymphoma. Oncol. Rep. 2015, 33, 2253–2260. [Google Scholar] [CrossRef] [PubMed]
- Selvarajan, V.; Osato, M.; Nah, G.S.S.; Yan, J.; Chung, T.H.; Voon, D.C.; Ito, Y.; Ham, M.F.; Salto-Tellez, M.; Shimizu, N.; et al. RUNX3 is oncogenic in natural killer/T-cell lymphoma and is transcriptionally regulated by MYC. Leukemia 2017, 31, 2219–2227. [Google Scholar] [CrossRef]
- Dirmeier, U.; Hoffmann, R.; Kilger, E.; Schultheiss, U.; Briseño, C.; Gires, O.; Kieser, A.; Eick, D.; Sugden, B.; Hammerschmidt, W. Latent membrane protein 1 of Epstein-Barr virus coordinately regulates proliferation with control of apoptosis. Oncogene 2005, 24, 1711–1717. [Google Scholar] [CrossRef]
- Nagato, T.; Ohkuri, T.; Ohara, K.; Hirata, Y.; Kishibe, K.; Komabayashi, Y.; Ueda, S.; Takahara, M.; Kumai, T.; Ishibashi, K.; et al. Programmed death-ligand 1 and its soluble form are highly expressed in nasal natural killer/T-cell lymphoma: A potential rationale for immunotherapy. Cancer Immunol. Immunother. 2017, 66, 877–890. [Google Scholar] [CrossRef]
- Bi, X.-W.; Wang, H.; Zhang, W.-W.; Wang, J.-H.; Liu, W.-J.; Xia, Z.-J.; Huang, H.-Q.; Jiang, W.-Q.; Zhang, Y.-J.; Wang, L. PD-L1 is upregulated by EBV-driven LMP1 through NF-κB pathway and correlates with poor prognosis in natural killer/T-cell lymphoma. J. Hematol. Oncol. 2016, 9, 109. [Google Scholar] [CrossRef]
- Jo, J.-C.; Kim, M.; Choi, Y.; Kim, H.-J.; Kim, J.E.; Chae, S.W.; Kim, H.; Cha, H.J. Expression of programmed cell death 1 and programmed cell death ligand 1 in extranodal NK/T-cell lymphoma, nasal type. Ann. Hematol. 2017, 96, 25–31. [Google Scholar] [CrossRef]
- Iqbal, J.; Weisenburger, D.D.; Chowdhury, A.; Tsai, M.Y.; Srivastava, G.; Greiner, T.C.; Kucuk, C.; Deffenbacher, K.; Vose, J.; Smith, L.; et al. Natural killer cell lymphoma shares strikingly similar molecular features with a group of non-hepatosplenic gammadelta T-cell lymphoma and is highly sensitive to a novel aurora kinase A inhibitor in vitro. Leukemia 2011, 25, 348–358. [Google Scholar] [CrossRef]
- Chen, Y.P.; Chang, K.C.; Su, W.C.; Chen, T.Y. The expression and prognostic significance of platelet-derived growth factor receptor alpha in mature T- and natural killer-cell lymphomas. Ann. Hematol. 2008, 87, 985–990. [Google Scholar] [CrossRef]
- Wang, L.; Wang, H.; Li, P.-f.; Lu, Y.; Xia, Z.-j.; Huang, H.-q.; Zhang, Y.-j. CD38 expression predicts poor prognosis and might be a potential therapy target in extranodal NK/T cell lymphoma, nasal type. Ann. Hematol. 2015, 94, 1381–1388. [Google Scholar] [CrossRef] [PubMed]
- Hoshida, Y.; Hongyo, T.; Jia, X.; He, Y.; Hasui, K.; Dong, Z.; Luo, W.J.; Ham, M.F.; Nomura, T.; Aozasa, K. Analysis of p53, K-ras, c-kit, and beta-catenin gene mutations in sinonasal NK/T cell lymphoma in northeast district of China. Cancer Sci. 2003, 94, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Takahara, M.; Kishibe, K.; Bandoh, N.; Nonaka, S.; Harabuchi, Y. P53, N- and K-Ras, and beta-catenin gene mutations and prognostic factors in nasal NK/T-cell lymphoma from Hokkaido, Japan. Hum. Pathol. 2004, 35, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Farazi, T.A.; Spitzer, J.I.; Morozov, P.; Tuschl, T. miRNAs in human cancer. J. Pathol. 2011, 223, 102–115. [Google Scholar] [CrossRef]
- Lai, E.C. microRNAs: Runts of the genome assert themselves. Curr. Biol. 2003, 13, R925–R936. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Broderick, J.A.; Zamore, P.D. MicroRNA therapeutics. Gene Ther. 2011, 18, 1104–1110. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Doss, C.G.P.; Lee, S.-S. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol. Ther. 2017, 8, 132–143. [Google Scholar] [CrossRef]
- Ott, G.; Rosenwald, A.; Campo, E. Understanding MYC-driven aggressive B-cell lymphomas: Pathogenesis and classification. Blood 2013, 122, 3884–3891. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Martin, M.; Ferrando, A. The NOTCH1-MYC highway toward T-cell acute lymphoblastic leukemia. Blood 2017, 129, 1124–1133. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-H.; Bi, X.-W.; Li, P.-F.; Xia, Z.-J.; Huang, H.-Q.; Jiang, W.-Q.; Zhang, Y.-J.; Wang, L. Overexpression of MYC and BCL2 Predicts Poor Prognosis in Patients with Extranodal NK/T-cell Lymphoma, Nasal Type. J. Cancer 2017, 8, 793–800. [Google Scholar] [CrossRef] [PubMed]
| Country | Entity | n | EBV Strain | Reference | |
|---|---|---|---|---|---|
| Type A | Type B | ||||
| China | ENKTCL | 31 cases | 29 (93.5%) | 2 (6.5%) | [81] |
| Mexico | ENKTCL | 42 cases | 39 (93%) | 3 (7%) | [82] |
| Peru | ENKTCL | 27 cases | 15 (88%) | 3 (12%) | [82] |
| Argentina | ENKTCL | 12 cases | 11 (92%) | 1 (8%) | [82] |
| Korea | T cell NHL | 15 cases | 14 (93.3%) | 1 (6.7%) | [83] |
| China | ENKTCL | 16 cases | 16 (100%) | 0 | [84] |
| Mexico | ENKTCL | 23 cases | 21 (91%) | 2 (9%) | [6] |
| China/Taiwan | Nasal and extranasal PTCL | 19 cases | 19 (100%) | 1 (5.3%) | [85] |
| Malaysia | PTCL | 9 cases | 9 (100%) | 0 | [86] |
| Denmark | PTCL | 18 cases | 15 (83.3%) | 3 (16.7%) | [86] |
| Genetic Alteration | Reference | |||
|---|---|---|---|---|
| Chromosomal abnormalities | Losses of 6q21–6q25 (40–50%) | POPDC3, PREP, PRDM1, ATG5, AIM1 and HACE1 | [126,127] | |
| Other chromosomal alterations | Losses in 5p13, 11q22-q23,11q24-25, 12q3, 13q14, 14q21, 15q24, 17p13, 17p4 and 19q13 Gains in 1q21-q44, 2q13-14, 2q31-q32, 2q5, 3q26, 6p25-p11, 7q34, 7q35-q36, 8q24, 10q3, 13q14, 13q4 and 20q11. | [129,130,134] | ||
| Recurrent mutations | JAK-STAT signaling pathway | STAT3, STAT5b, JAK3, | [21,135,136] | |
| RNA helicase family | DDX3X | [22] | ||
| Tumor suppressors | TP53, MGA | [22,23,50] | ||
| RAS-MAPK signaling pathway | NOTCH3, EPHA1, PTPRQ, PTPRK, GNAQ | [81,137] | ||
| Apoptosis | FAS | [138,139] | ||
| Epigenetic modifiers | ARID1A, ASXL1, BCOR, KMT2D, MLL2, EP300 | [23,124,125] | ||
| Epigenetic alterations | Hyper methylation | Cell cycle regulators: CDKN2A, CDKN2B, CDKN1A Tumor suppressors: BCL2L11 (BIM), DAPK1, PTPN6 (SHP1), TET2, SOCS6, and ASNS. | [140] [141] | |
| Histone modifications | EZH2: histone methyltransferase, aberrant overexpressed in ENKTCL, leading to activation of NF-kB signaling pathway. | [142,143,144] | ||
| mi-RNAs | Downregulated | miR-26a, miR-26b, miR-28-5, miR-101 and miR-363. De-regulated miR-146a: leading to inhibition of TRAF6, downregulating NF-kB signaling. | [144,145] | |
| Upregulated | miR-155 and miR21 | [137,146] | ||
| Gene Overexpression | Survivin: | Induced by LMP1, EBV latent viral proteins | [147] | |
| MYC: | Upregulation possibly through LMP1 latent viral protein. | [81,148,149] | ||
| PD-L1: | Overexpression of the cell death ligand favoring immune evasion | [150,151,152] | ||
| RUNX3: | Mediated by MYC, resulting in decreased apoptosis and increase cell proliferation | [148] | ||
| AURKA: | Increased cell proliferation | [153] | ||
| PDGFRA: | Overexpression of PDGFRα but absence of genomic alteration | [154] | ||
| Other | CD38 | Transmembrane protein associated with poor outcome | [155] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montes-Mojarro, I.A.; Fend, F.; Quintanilla-Martinez, L. EBV and the Pathogenesis of NK/T Cell Lymphoma. Cancers 2021, 13, 1414. https://doi.org/10.3390/cancers13061414
Montes-Mojarro IA, Fend F, Quintanilla-Martinez L. EBV and the Pathogenesis of NK/T Cell Lymphoma. Cancers. 2021; 13(6):1414. https://doi.org/10.3390/cancers13061414
Chicago/Turabian StyleMontes-Mojarro, Ivonne A., Falko Fend, and Leticia Quintanilla-Martinez. 2021. "EBV and the Pathogenesis of NK/T Cell Lymphoma" Cancers 13, no. 6: 1414. https://doi.org/10.3390/cancers13061414
APA StyleMontes-Mojarro, I. A., Fend, F., & Quintanilla-Martinez, L. (2021). EBV and the Pathogenesis of NK/T Cell Lymphoma. Cancers, 13(6), 1414. https://doi.org/10.3390/cancers13061414








