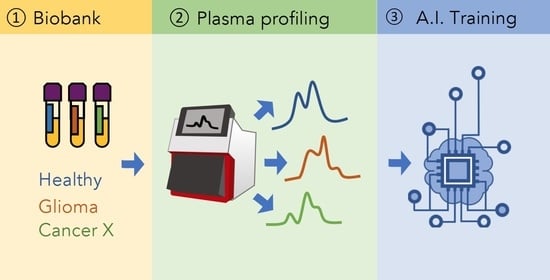
An AI-Powered Blood Test to Detect Cancer Using NanoDSF
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
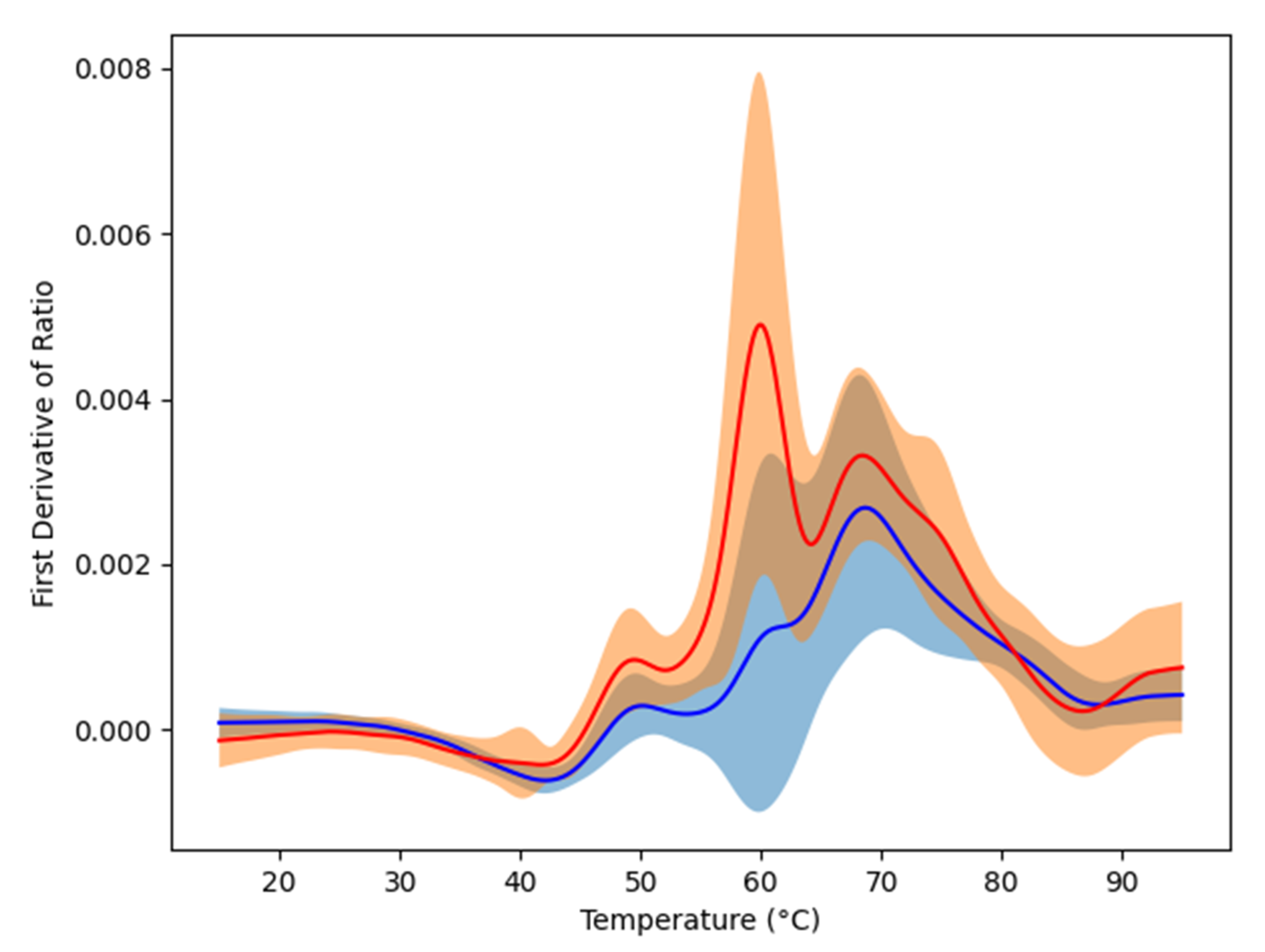
2.2. Plasma Profiling
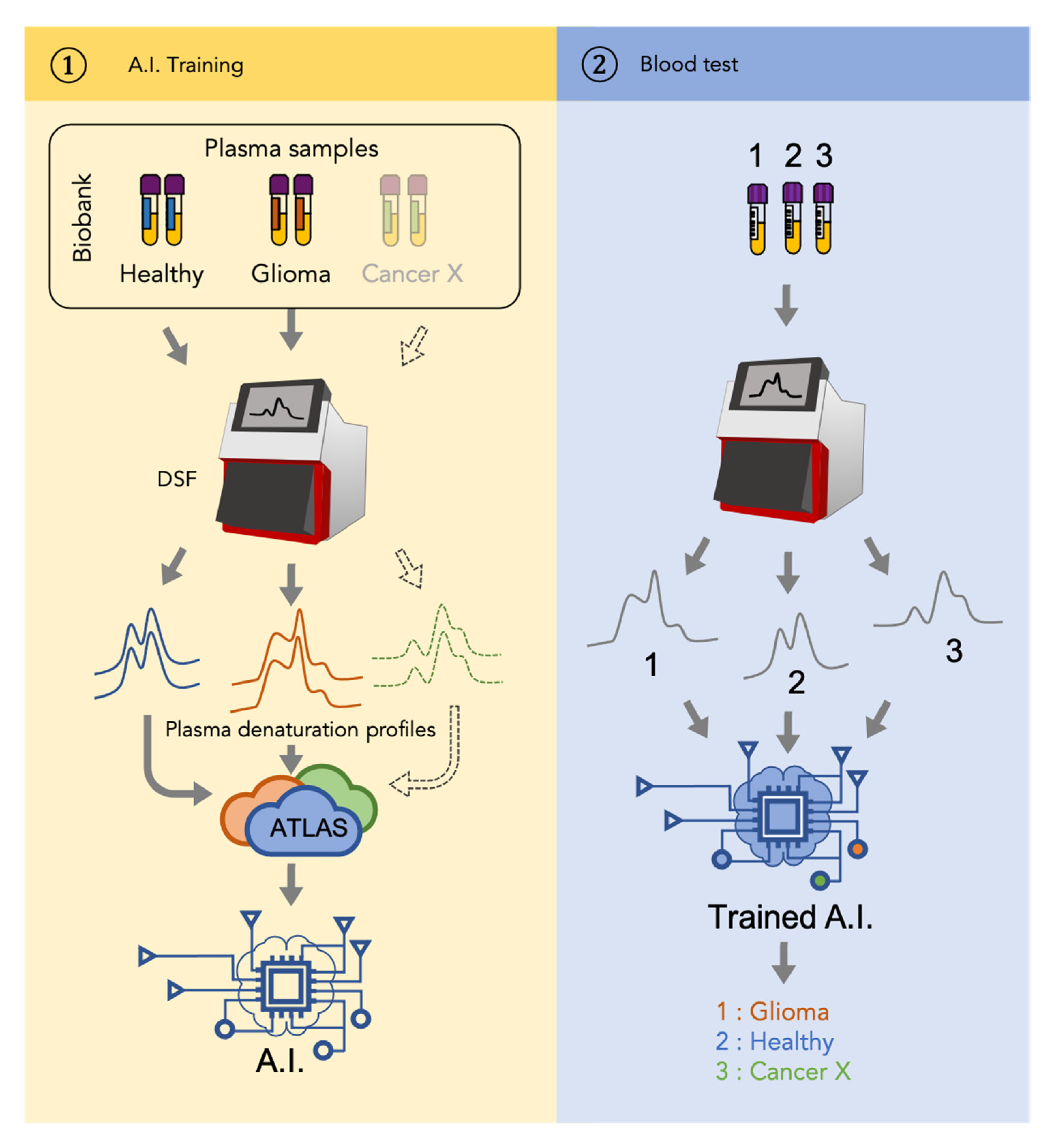
2.3. Plasma Profile Classification Using AI
3. Discussion
4. Methods
4.1. Patients
4.2. Plasma Samples
4.3. Sample Analysis by NanoDSF
4.4. Algorithm Trainings
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Ellingson, B.M.; Chung, C.; Pope, W.B.; Boxerman, J.L.; Kaufmann, T.J. Pseudoprogression, radionecrosis, inflammation or true tumor progression? challenges associated with glioblastoma response assessment in an evolving therapeutic landscape. J. Neurooncol. 2017, 134, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.Y.; Macdonald, D.R.; Reardon, D.A.; Cloughesy, T.F.; Sorensen, A.G.; Galanis, E.; Degroot, J.; Wick, W.; Gilbert, M.R.; Lassman, A.B.; et al. Updated response assessment criteria for high-grade gliomas: Response assessment in neuro-oncology working group. J. Clin. Oncol. 2010, 28, 1963–1972. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, P.O.; Ezraty, B.; Mitchell, J.K.; Devred, F.; Peyrot, V.; Derrick, P.J.; Barras, F.; Makarov, A.A.; Lafitte, D. Calorimetry and mass spectrometry study of oxidized calmodulin interaction with target and differential repair by methionine sulfoxide reductases. Biochimie 2005, 87, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Petrushanko, I.Y.; Lobachev, V.M.; Kononikhin, A.S.; Makarov, A.A.; Devred, F.; Kovacic, H.; Kubatiev, A.A.; Tsvetkov, P.O. Oxidation of Ca2+-Binding Domain of NADPH Oxidase 5 (NOX5): Toward Understanding the Mechanism of Inactivation of NOX5 by ROS. PLoS ONE 2016, 11, e0158726. [Google Scholar] [CrossRef] [PubMed]
- Garbett, N.C.; Mekmaysy, C.S.; DeLeeuw, L.; Chaires, J.B. Clinical application of plasma thermograms. Utility, practical approaches and considerations. Methods 2015, 76, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, P.O.; Devred, F. Plasmatic Signature of Disease by Differential Scanning Calorimetry (DSC). Methods Mol. Biol. 2019, 1964, 45–57. [Google Scholar] [PubMed]
- Tsvetkov, P.O.; Tabouret, E.; Roman, A.Y.; Romain, S.; Bequet, C.; Ishimbaeva, O.; Honoré, S.; Figarella-Branger, D.; Chinot, O.; Devred, F. Differential scanning calorimetry of plasma in glioblastoma: Toward a new prognostic/monitoring tool. Oncotarget 2018, 9, 9391–9399. [Google Scholar] [CrossRef] [PubMed]
- Garnier, C.; Devred, F.; Byrne, D.; Puppo, R.; Roman, A.Y.; Malesinski, S.; Golovin, A.V.; Lebrun, R.; Ninkina, N.N.; Tsvetkov, P.O. Zinc binding to RNA recognition motif of TDP-43 induces the formation of amyloid-like aggregates. Sci. Rep. 2017, 7, 6812. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, P.O.; Roman, A.Y.; Baksheeva, V.E.; Nazipova, A.A.; Shevelyova, M.P.; Vladimirov, V.I.; Buyanova, M.F.; Zinchenko, D.V.; Zamyatnin, A.A., Jr.; Devred, F.; et al. Functional Status of Neuronal Calcium Sensor-1 Is Modulated by Zinc Binding. Front. Mol. Neurosci. 2018, 11, 459. [Google Scholar] [CrossRef] [PubMed]
- Garbett, N.C.; Mekmaysy, C.S.; Helm, C.W.; Jenson, A.B.; Chaires, J.B. Differential scanning calorimetry of blood plasma for clinical diagnosis and monitoring. Exp. Mol. Pathol. 2009, 86, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: Berlin/Heidelberg, Germany, 2016; ISBN 9781493938438. [Google Scholar]
- Sala, A.; Anderson, D.J.; Brennan, P.M.; Butler, H.J.; Cameron, J.M.; Jenkinson, M.D.; Rinaldi, C.; Theakstone, A.G.; Baker, M.J. Biofluid diagnostics by FTIR spectroscopy: A platform technology for cancer detection. Cancer Lett. 2020, 477, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Saenz-Antoñanzas, A.; Auzmendi-Iriarte, J.; Carrasco-Garcia, E.; Moreno-Cugnon, L.; Ruiz, I.; Villanua, J.; Egaña, L.; Otaegui, D.; Samprón, N.; Matheu, A. Liquid Biopsy in Glioblastoma: Opportunities, Applications and Challenges. Cancers 2019, 11, 950. [Google Scholar] [CrossRef] [PubMed]
- Müller Bark, J.; Kulasinghe, A.; Chua, B.; Day, B.W.; Punyadeera, C. Circulating biomarkers in patients with glioblastoma. Br. J. Cancer 2020, 122, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gole, J.; Gore, A.; He, Q.; Lu, M.; Min, J.; Yuan, Z.; Yang, X.; Jiang, Y.; Zhang, T.; et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat. Commun. 2020, 11, 3475. [Google Scholar] [CrossRef] [PubMed]
- Palmirotta, R.; Lovero, D.; Cafforio, P.; Felici, C.; Mannavola, F.; Pellè, E.; Quaresmini, D.; Tucci, M.; Silvestris, F. Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Ther. Adv. Med. Oncol. 2018, 10. [Google Scholar] [CrossRef]
- Miller, A.M.; Shah, R.H.; Pentsova, E.I.; Pourmaleki, M.; Briggs, S.; Distefano, N.; Zheng, Y.; Skakodub, A.; Mehta, S.A.; Campos, C.; et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature 2019, 565, 654–658. [Google Scholar] [CrossRef] [PubMed]
- Chagovetz, A.A.; Jensen, R.L.; Recht, L.; Glantz, M.; Chagovetz, A.M. Preliminary use of differential scanning calorimetry of cerebrospinal fluid for the diagnosis of glioblastoma multiforme. J. Neuro-Oncol. 2011, 105, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Chagovetz, A.A.; Quinn, C.; Damarse, N.; Hansen, L.D.; Chagovetz, A.M.; Jensen, R.L. Differential scanning calorimetry of gliomas: A new tool in brain cancer diagnostics? Neurosurgery 2013, 73, 289–295. [Google Scholar] [CrossRef] [PubMed]
| Factors | N | % |
|---|---|---|
| Age (median, min–max) | 49.3 (19.6–77.5) | |
| Gender (Men/women) | 45/39 | 54/46 |
| KPS * (median, min–max) | 80 (50–100) | |
| 50 | 1 | 1 |
| 60 | 10 | 13 |
| 70 | 17 | 21 |
| 80 | 23 | 29 |
| 90 | 20 | 25 |
| 100 | 9 | 11 |
| Steroids | 51 | 64 |
| Histology | ||
| Oligodendroglioma | ||
| Grade II | 2 | 2 |
| Grade III | 20 | 24 |
| Astrocytoma IDHmut * | ||
| Grade II | 3 | 4 |
| Grade III | 19 | 23 |
| Grade IV | 3 | 4 |
| Astrocytoma IDHwt * | ||
| Grade II | 7 | 8 |
| Grade III | 11 | 13 |
| Grade IV | 19 | 22 |
| Surgery | ||
| Gross total resection | 33 | 41 |
| Partial resection | 48 | 59 |
| Adjuvant treatment | ||
| Radiotherapy alone | 10 | 12 |
| Chemotherapy alone | 12 | 14 |
| Radiotherapy + chemotherapy | 57 | 68 |
| None | 5 | 6 |
| Algorithm | Original Algorithms | FN Minimized | ||||
|---|---|---|---|---|---|---|
| LR | SVM | NN | RF | AdaBoost | fnf-SVM | |
| Accuracy (%) | 89.80 | 87.07 | 92.52 | 89.12 | 92.52 | 87.07 |
| False positives (n) | 4 | 13 | 5 | 8 | 6 | 18 |
| False negatives (n) | 11 | 6 | 6 | 8 | 5 | 1 |
| Sensitivity | 0.87 | 0.93 | 0.93 | 0.90 | 0.94 | 0.99 |
| Specificity | 0.94 | 0.79 | 0.92 | 0.87 | 0.90 | 0.71 |
| Precision | 0.95 | 0.86 | 0.94 | 0.90 | 0.93 | 0.82 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsvetkov, P.O.; Eyraud, R.; Ayache, S.; Bougaev, A.A.; Malesinski, S.; Benazha, H.; Gorokhova, S.; Buffat, C.; Dehais, C.; Sanson, M.; et al. An AI-Powered Blood Test to Detect Cancer Using NanoDSF. Cancers 2021, 13, 1294. https://doi.org/10.3390/cancers13061294
Tsvetkov PO, Eyraud R, Ayache S, Bougaev AA, Malesinski S, Benazha H, Gorokhova S, Buffat C, Dehais C, Sanson M, et al. An AI-Powered Blood Test to Detect Cancer Using NanoDSF. Cancers. 2021; 13(6):1294. https://doi.org/10.3390/cancers13061294
Chicago/Turabian StyleTsvetkov, Philipp O., Rémi Eyraud, Stéphane Ayache, Anton A. Bougaev, Soazig Malesinski, Hamed Benazha, Svetlana Gorokhova, Christophe Buffat, Caroline Dehais, Marc Sanson, and et al. 2021. "An AI-Powered Blood Test to Detect Cancer Using NanoDSF" Cancers 13, no. 6: 1294. https://doi.org/10.3390/cancers13061294
APA StyleTsvetkov, P. O., Eyraud, R., Ayache, S., Bougaev, A. A., Malesinski, S., Benazha, H., Gorokhova, S., Buffat, C., Dehais, C., Sanson, M., Bielle, F., Figarella Branger, D., Chinot, O., Tabouret, E., & Devred, F. (2021). An AI-Powered Blood Test to Detect Cancer Using NanoDSF. Cancers, 13(6), 1294. https://doi.org/10.3390/cancers13061294