Pan-Cancer Analysis of Human Kinome Gene Expression and Promoter DNA Methylation Identifies Dark Kinase Biomarkers in Multiple Cancers
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Retrieval
2.2. Gene Expression Data
2.3. Methylation Data
2.4. t-SNE Plots
2.5. Correlation Analysis
2.6. Survival Analysis
3. Results
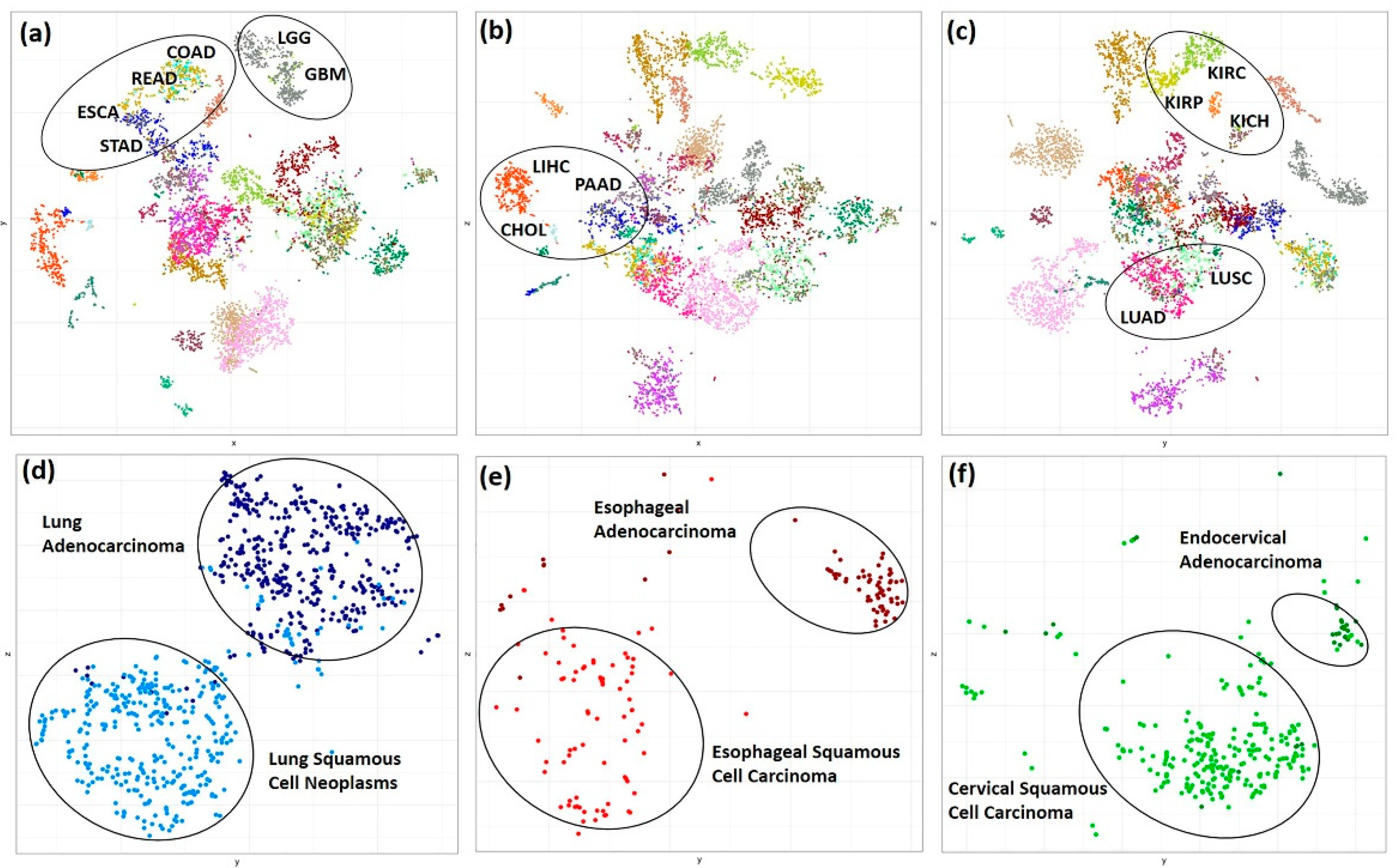
3.1. Unsupervised Clustering of TCGA and GTEx Samples Based on Kinome Profiles Distinguish Cancer Types
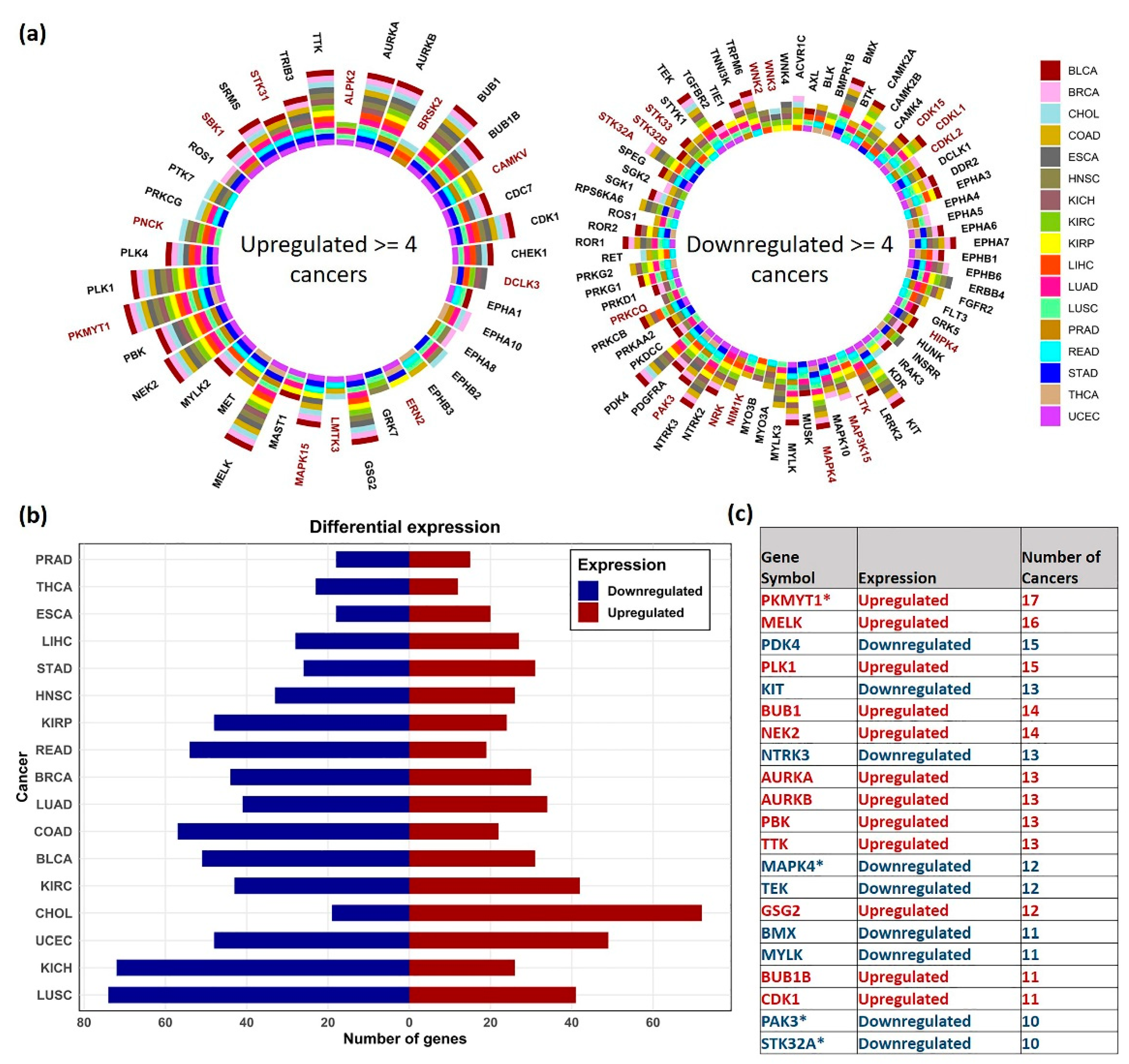
3.2. Differential Gene Expression Analysis
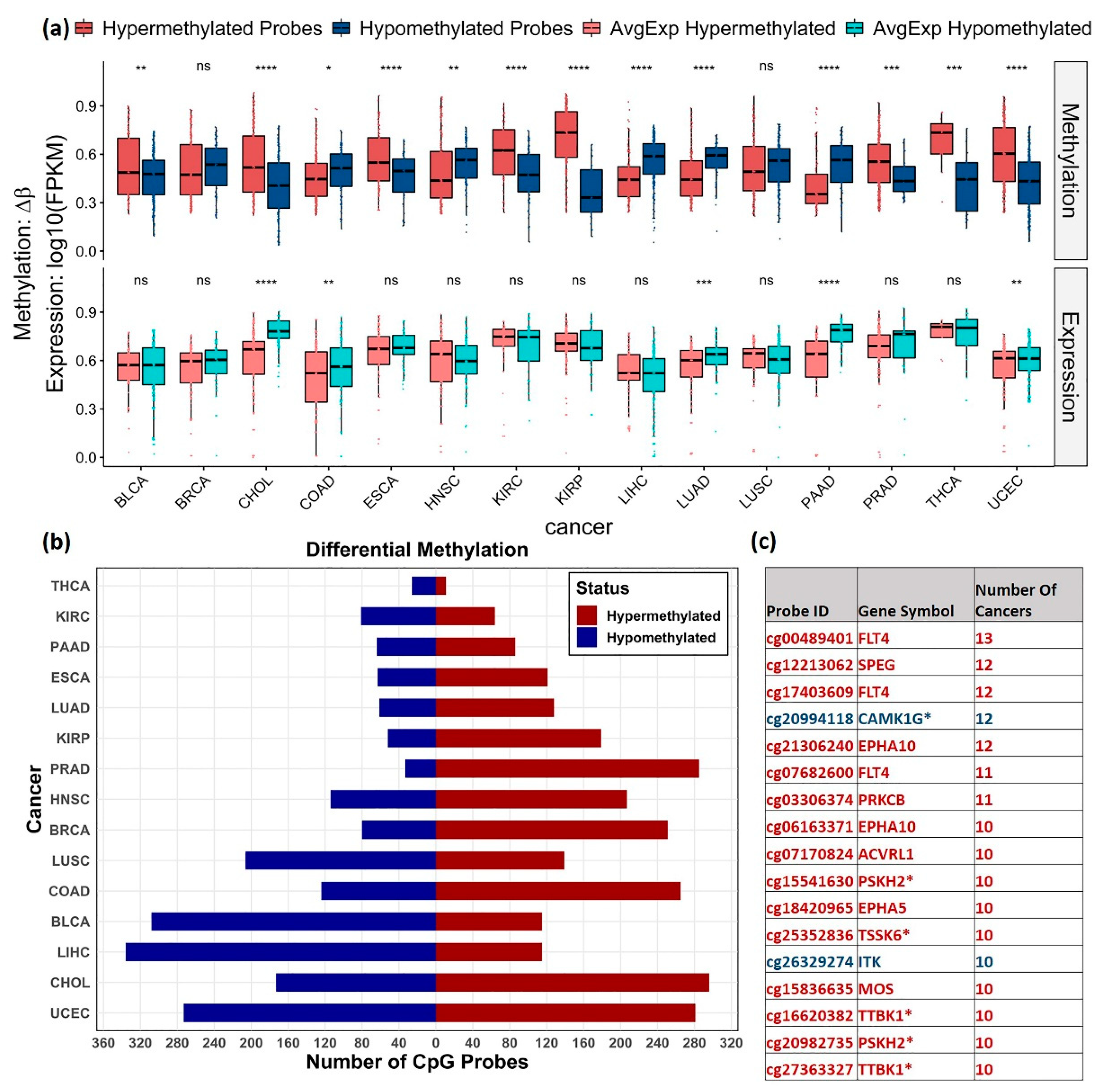
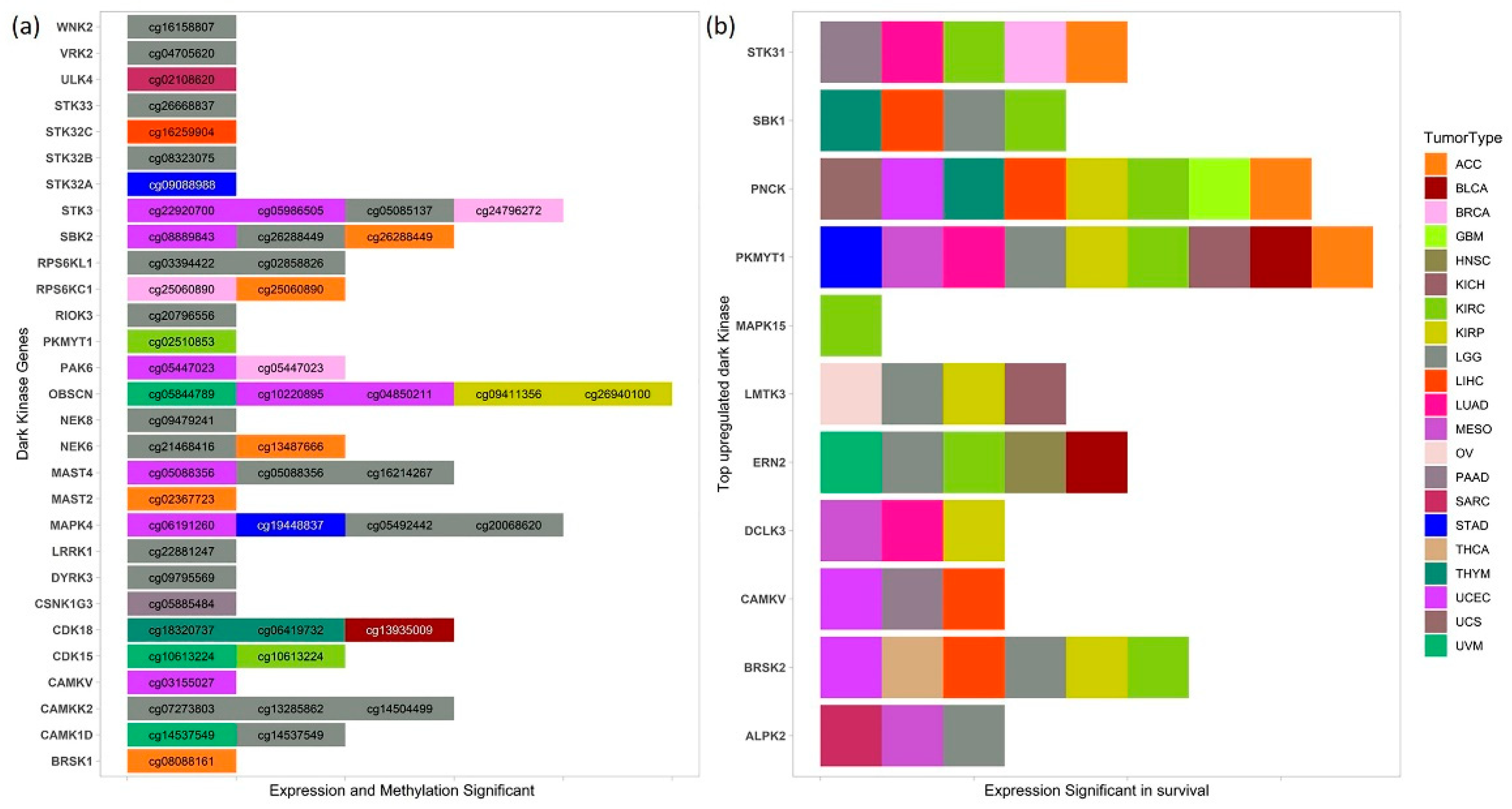
3.3. Differential Methylation Analysis
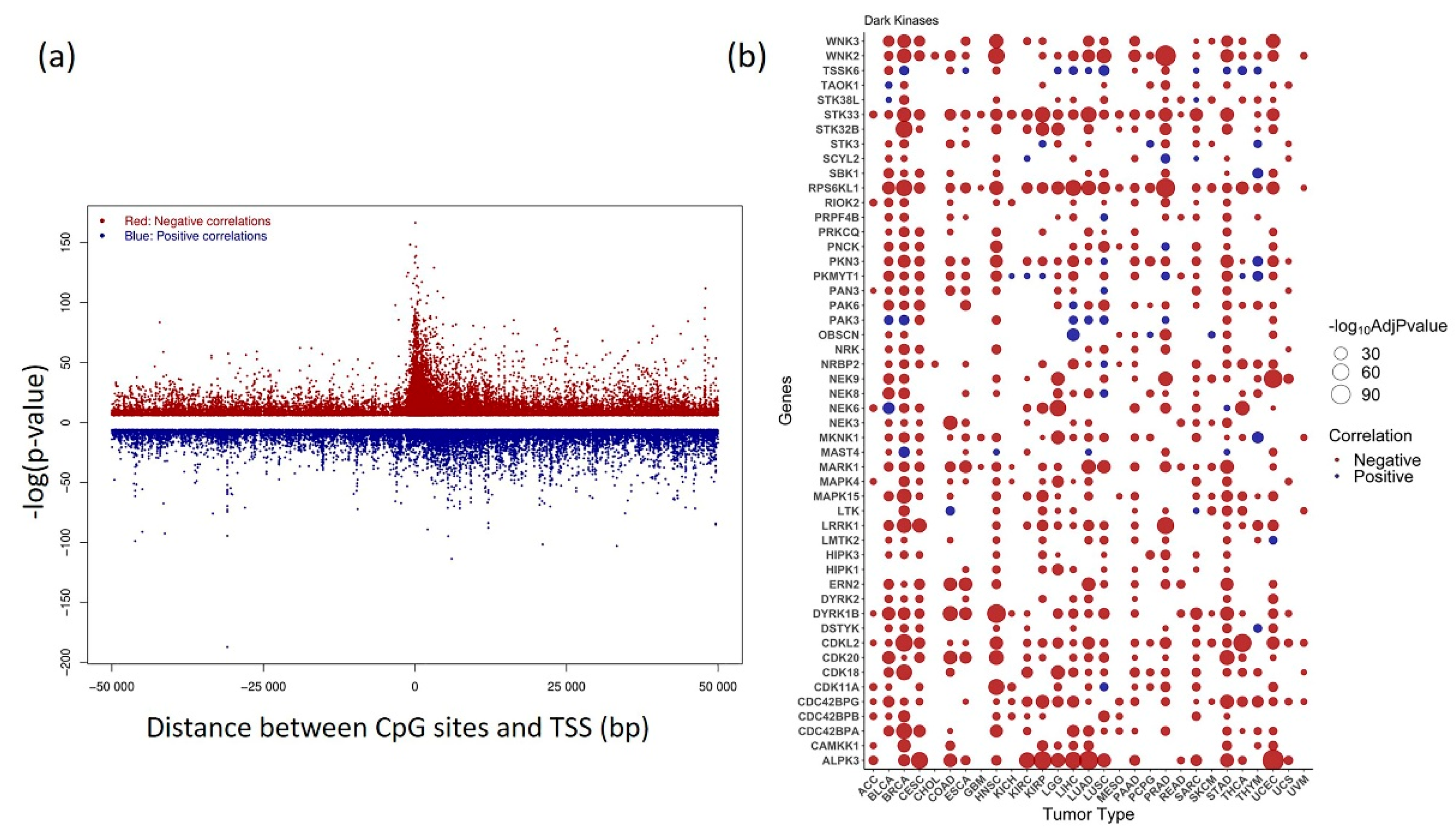
3.4. Correlative Analysis of DNA Methylation and Gene Expression
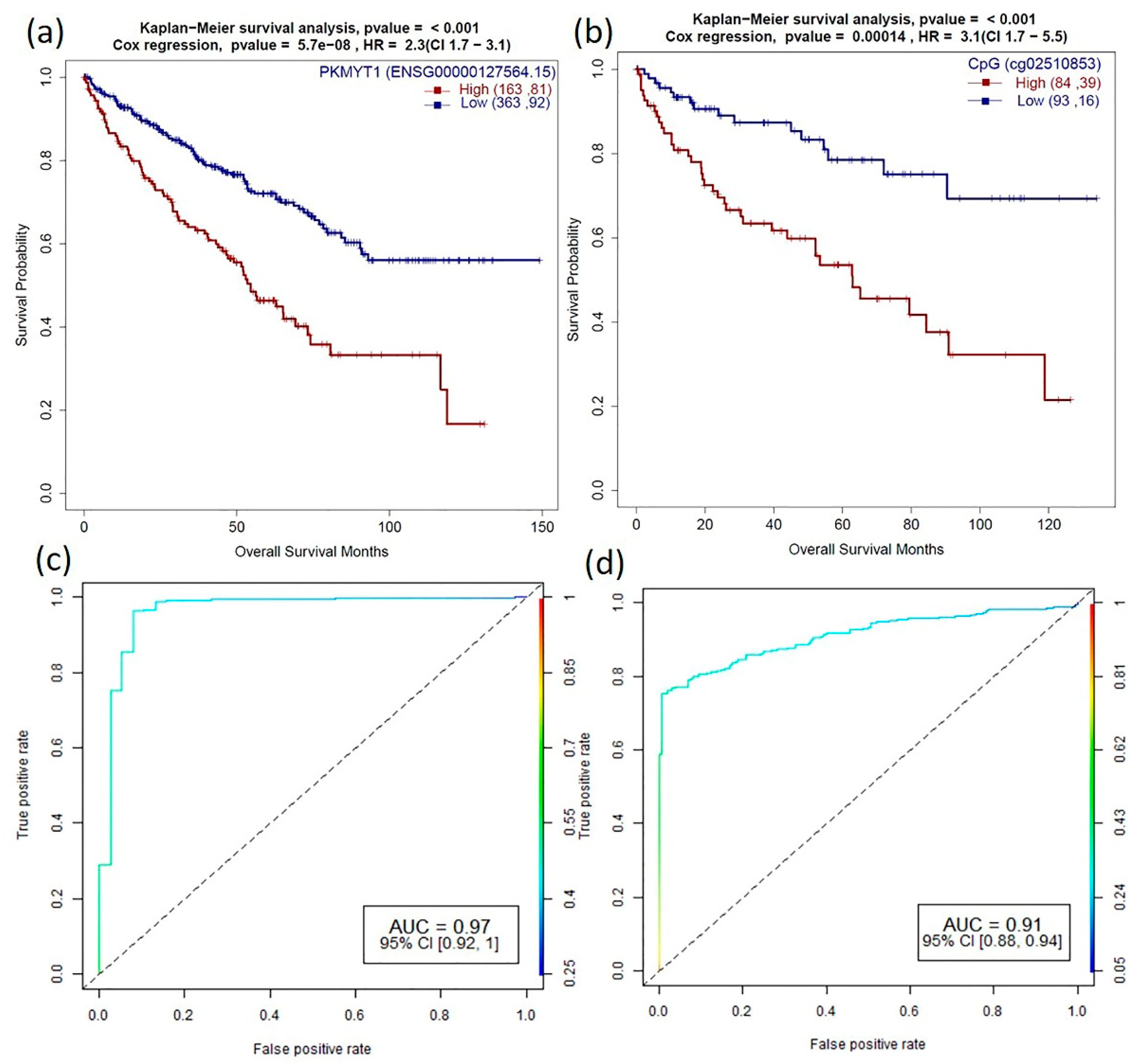
3.5. Survival Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Blume-Jensen, P.; Hunter, T. Oncogenic kinase signalling. Nature 2001, 411, 355–365. [Google Scholar] [CrossRef]
- Kilpinen, S.; Ojala, K.; Kallioniemi, O. Analysis of kinase gene expression patterns across 5681 human tissue samples reveals functional genomic taxonomy of the kinome. PLoS ONE 2010, 5, e15068. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.Y.; Quan, J.; Sodaei, R.; Aguet, F.; Segrè, A.V.; Allen, J.A.; Lanz, T.A.; Reinhart, V.; Crawford, M.; Hasson, S.; et al. A systematic survey of human tissue-specific gene expression and splicing reveals new opportunities for therapeutic target identification and evaluation. bioRxiv 2018. [Google Scholar] [CrossRef]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Wilson, L.J.; Linley, A.; Hammond, D.E.; Hood, F.E.; Coulson, J.M.; MacEwan, D.J.; Ross, S.J.; Slupsky, J.R.; Smith, P.D.; Eyers, P.A.; et al. New Perspectives, Opportunities, and Challenges in Exploring the Human Protein Kinome. Cancer Res. 2018, 78, 15–29. [Google Scholar] [CrossRef]
- Hantschel, O. Unexpected off-targets and paradoxical pathway activation by kinase inhibitors. ACS Chem. Biol. 2015, 10, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; De Carvalho, D.D. Clinical advances in targeting epigenetics for cancer therapy. FEBS J. 2021. [Google Scholar] [CrossRef]
- Klaeger, S.; Heinzlmeir, S.; Wilhelm, M.; Polzer, H.; Vick, B.; Koenig, P.A.; Reinecke, M.; Ruprecht, B.; Petzoldt, S.; Meng, C.; et al. The target landscape of clinical kinase drugs. Science 2017, 358. [Google Scholar] [CrossRef]
- Shrestha, S.; Byrne, D.P.; Harris, J.A.; Kannan, N.; Eyers, P.A. Cataloguing the dead: Breathing new life into pseudokinase research. FEBS J. 2020, 287, 4150–4169. [Google Scholar] [CrossRef]
- Essegian, D.; Khurana, R.; Stathias, V.; Schurer, S.C. The Clinical Kinase Index: A Method to Prioritize Understudied Kinases as Drug Targets for the Treatment of Cancer. Cell Rep. Med. 2020, 1, 100128. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.C.; Ross, K.E.; Baffi, T.R.; Drabkin, H.; Kochut, K.J.; Ruan, Z.; D′Eustachio, P.; McSkimming, D.; Arighi, C.; Chen, C.; et al. Integrative annotation and knowledge discovery of kinase post-translational modifications and cancer-associated mutations through federated protein ontologies and resources. Sci. Rep. 2018, 8, 6518. [Google Scholar] [CrossRef]
- Finan, C.; Gaulton, A.; Kruger, F.A.; Lumbers, R.T.; Shah, T.; Engmann, J.; Galver, L.; Kelley, R.; Karlsson, A.; Santos, R.; et al. The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef]
- Berginski, M.E.; Moret, N.; Liu, C.; Goldfarb, D.; Sorger, P.K.; Gomez, S.M. The Dark Kinase Knowledgebase: An online compendium of knowledge and experimental results of understudied kinases. Nucleic Acids Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Sheetz, J.B.; Mathea, S.; Karvonen, H.; Malhotra, K.; Chatterjee, D.; Niininen, W.; Perttila, R.; Preuss, F.; Suresh, K.; Stayrook, S.E.; et al. Structural Insights into Pseudokinase Domains of Receptor Tyrosine Kinases. Mol. Cell 2020, 79, 390–405.e7. [Google Scholar] [CrossRef]
- Rao, C.; Huisman, D.H.; Vieira, H.M.; Frodyma, D.E.; Neilsen, B.K.; Chakraborty, B.; Hight, S.K.; White, M.A.; Fisher, K.W.; Lewis, R.E. A Gene Expression High-Throughput Screen (GE-HTS) for Coordinated Detection of Functionally Similar Effectors in Cancer. Cancers 2020, 12, 3143. [Google Scholar] [CrossRef]
- Spainhour, J.C.; Lim, H.S.; Yi, S.V.; Qiu, P. Correlation Patterns Between DNA Methylation and Gene Expression in The Cancer Genome Atlas. Cancer Inform. 2019, 18. [Google Scholar] [CrossRef] [PubMed]
- Kuang, S.Q.; Bai, H.; Fang, Z.H.; Lopez, G.; Yang, H.; Tong, W.; Wang, Z.Z.; Garcia-Manero, G. Aberrant DNA methylation and epigenetic inactivation of Eph receptor tyrosine kinases and ephrin ligands in acute lymphoblastic leukemia. Blood 2010, 115, 2412–2419. [Google Scholar] [CrossRef] [PubMed]
- Mello, A.A.; Leal, M.F.; Rey, J.A.; Pinto, G.R.; Lamarao, L.M.; Montenegro, R.C.; Alves, A.P.; Assumpcao, P.P.; Borges Bdo, N.; Smith, M.C.; et al. Deregulated Expression of SRC, LYN and CKB Kinases by DNA Methylation and Its Potential Role in Gastric Cancer Invasiveness and Metastasis. PLoS ONE 2015, 10, e0140492. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, J.; Gao, W.; Yuan, B.Z.; Rogers, J.; Reed, E. CHK2 kinase expression is down-regulated due to promoter methylation in non-small cell lung cancer. Mol. Cancer 2004, 3, 14. [Google Scholar] [CrossRef]
- Moret, N.; Liu, C.; Gyori, B.M.; Bachman, J.A.; Steppi, A.; Taujale, R.; Huang, L.-C.; Hug, C.; Berginski, M.; Gomez, S.; et al. Exploring the understudied human kinome for research and therapeutic opportunities. bioRxiv 2020. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Troyanskaya, O.; Cantor, M.; Sherlock, G.; Brown, P.; Hastie, T.; Tibshirani, R.; Botstein, D.; Altman, R.B. Missing value estimation methods for DNA microarrays. Bioinformatics 2001, 17, 520–525. [Google Scholar] [CrossRef]
- Teschendorff, A.E.; Marabita, F.; Lechner, M.; Bartlett, T.; Tegner, J.; Gomez-Cabrero, D.; Beck, S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 2013, 29, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Morris, T.J.; Butcher, L.M.; Feber, A.; Teschendorff, A.E.; Chakravarthy, A.R.; Wojdacz, T.K.; Beck, S. ChAMP: 450 k Chip Analysis Methylation Pipeline. Bioinformatics 2014, 30, 428–430. [Google Scholar] [CrossRef]
- Zhou, W.; Laird, P.W.; Shen, H. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res. 2017, 45, e22. [Google Scholar] [CrossRef]
- Mishra, N.K.; Niu, M.; Southekal, S.; Bajpai, P.; Elkholy, A.; Manne, U.; Guda, C. Identification of Prognostic Markers in Cholangiocarcinoma Using Altered DNA Methylation and Gene Expression Profiles. Front. Genet. 2020, 11, 522125. [Google Scholar] [CrossRef]
- Sing, T.; Sander, O.; Beerenwinkel, N.; Lengauer, T. ROCR: Visualizing classifier performance in R. Bioinformatics 2005, 21, 3940–3941. [Google Scholar] [CrossRef] [PubMed]
- International Cancer Genome Consortium; Hudson, T.J.; Anderson, W.; Artez, A.; Barker, A.D.; Bell, C.; Bernabe, R.R.; Bhan, M.K.; Calvo, F.; Eerola, I.; et al. International network of cancer genome projects. Nature 2010, 464, 993–998. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Gray, D.; Jubb, A.M.; Hogue, D.; Dowd, P.; Kljavin, N.; Yi, S.; Bai, W.; Frantz, G.; Zhang, Z.; Koeppen, H.; et al. Maternal embryonic leucine zipper kinase/murine protein serine-threonine kinase 38 is a promising therapeutic target for multiple cancers. Cancer Res. 2005, 65, 9751–9761. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhao, X.; Zhang, C.; Wang, W.; Li, F.; Liu, D.; Wu, K.; Zhu, D.; Liu, S.; Shen, C.; et al. Overexpressed PKMYT1 promotes tumor progression and associates with poor survival in esophageal squamous cell carcinoma. Cancer Manag. Res. 2019, 11, 7813–7824. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, R.; Mohyeldin, A.; Thiel, J.; Kornblum, H.I.; Beullens, M.; Nakano, I. MELK-a conserved kinase: Functions, signaling, cancer, and controversy. Clin. Transl. Med. 2015, 4, 11. [Google Scholar] [CrossRef]
- Li, Y.F.; Hsiao, Y.H.; Lai, Y.H.; Chen, Y.C.; Chen, Y.J.; Chou, J.L.; Chan, M.W.; Lin, Y.H.; Tsou, Y.A.; Tsai, M.H.; et al. DNA methylation profiles and biomarkers of oral squamous cell carcinoma. Epigenetics 2015, 10, 229–236. [Google Scholar] [CrossRef]
- Yang, M.; Hlady, R.A.; Zhou, D.; Ho, T.H.; Robertson, K.D. In silico DNA methylation analysis identifies potential prognostic biomarkers in type 2 papillary renal cell carcinoma. Cancer Med. 2019, 8, 5760–5768. [Google Scholar] [CrossRef]
- Ding, W.; Chen, G.; Shi, T. Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis. Epigenetics 2019, 14, 67–80. [Google Scholar] [CrossRef]
- Rajendran, B.K.; Deng, C.X. A comprehensive genomic meta-analysis identifies confirmatory role of OBSCN gene in breast tumorigenesis. Oncotarget 2017, 8, 102263–102276. [Google Scholar] [CrossRef]
- Sanchez-Vega, F.; Gotea, V.; Margolin, G.; Elnitski, L. Pan-cancer stratification of solid human epithelial tumors and cancer cell lines reveals commonalities and tissue-specific features of the CpG island methylator phenotype. Epigenet. Chromatin 2015, 8, 14. [Google Scholar] [CrossRef]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018, 173, 291–304.e296. [Google Scholar] [CrossRef]
- Li, Y.; Kang, K.; Krahn, J.M.; Croutwater, N.; Lee, K.; Umbach, D.M.; Li, L. A comprehensive genomic pan-cancer classification using The Cancer Genome Atlas gene expression data. BMC Genom. 2017, 18, 508. [Google Scholar] [CrossRef]
- Abrams, Z.B.; Zucker, M.; Wang, M.; Asiaee Taheri, A.; Abruzzo, L.V.; Coombes, K.R. Thirty biologically interpretable clusters of transcription factors distinguish cancer type. BMC Genom. 2018, 19, 738. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.W.; Karakasheva, T.A.; Lee, D.J.; Lee, J.S.; Long, Q.; Bass, A.J.; Wong, K.K.; Rustgi, A.K. Comparative transcriptomes of adenocarcinomas and squamous cell carcinomas reveal molecular similarities that span classical anatomic boundaries. PLoS Genet. 2017, 13, e1006938. [Google Scholar] [CrossRef]
- Badouel, C.; Chartrain, I.; Blot, J.; Tassan, J.P. Maternal embryonic leucine zipper kinase is stabilized in mitosis by phosphorylation and is partially degraded upon mitotic exit. Exp. Cell Res. 2010, 316, 2166–2173. [Google Scholar] [CrossRef]
- Giuliano, C.J.; Lin, A.; Smith, J.C.; Palladino, A.C.; Sheltzer, J.M. MELK expression correlates with tumor mitotic activity but is not required for cancer growth. Elife 2018, 7. [Google Scholar] [CrossRef]
- Beke, L.; Kig, C.; Linders, J.T.; Boens, S.; Boeckx, A.; van Heerde, E.; Parade, M.; De Bondt, A.; Van den Wyngaert, I.; Bashir, T.; et al. MELK-T1, a small-molecule inhibitor of protein kinase MELK, decreases DNA-damage tolerance in proliferating cancer cells. Biosci. Rep. 2015, 35. [Google Scholar] [CrossRef]
- Chung, S.; Suzuki, H.; Miyamoto, T.; Takamatsu, N.; Tatsuguchi, A.; Ueda, K.; Kijima, K.; Nakamura, Y.; Matsuo, Y. Development of an orally-administrative MELK-targeting inhibitor that suppresses the growth of various types of human cancer. Oncotarget 2012, 3, 1629–1640. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Cuellar, R.A.; Berndt, N.; Lee, H.E.; Olesen, S.H.; Martin, M.P.; Jensen, J.T.; Georg, G.I.; Schonbrunn, E. Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors. J. Med. Chem. 2017, 60, 7863–7875. [Google Scholar] [CrossRef]
- Chow, J.P.; Poon, R.Y. The CDK1 inhibitory kinase MYT1 in DNA damage checkpoint recovery. Oncogene 2013, 32, 4778–4788. [Google Scholar] [CrossRef]
- Toledo, C.M.; Ding, Y.; Hoellerbauer, P.; Davis, R.J.; Basom, R.; Girard, E.J.; Lee, E.; Corrin, P.; Hart, T.; Bolouri, H.; et al. Genome-wide CRISPR-Cas9 Screens Reveal Loss of Redundancy between PKMYT1 and WEE1 in Glioblastoma Stem-like Cells. Cell Rep. 2015, 13, 2425–2439. [Google Scholar] [CrossRef] [PubMed]
- Oprea, T.I.; Bologa, C.G.; Brunak, S.; Campbell, A.; Gan, G.N.; Gaulton, A.; Gomez, S.M.; Guha, R.; Hersey, A.; Holmes, J.; et al. Unexplored therapeutic opportunities in the human genome. Nat. Rev. Drug Discov. 2018, 17, 317–332. [Google Scholar] [CrossRef]
- Tamir, T.Y.; Bowman, B.M.; Agajanian, M.J.; Goldfarb, D.; Schrank, T.P.; Stohrer, T.; Hale, A.E.; Siesser, P.F.; Weir, S.J.; Murphy, R.M.; et al. Gain-of-function genetic screen of the kinome reveals BRSK2 as an inhibitor of the NRF2 transcription factor. J. Cell Sci. 2020, 133. [Google Scholar] [CrossRef]
- Saiyin, H.; Na, N.; Han, X.; Fang, Y.; Wu, Y.; Lou, W.; Yang, X. BRSK2 induced by nutrient deprivation promotes Akt activity in pancreatic cancer via downregulation of mTOR activity. Oncotarget 2017, 8, 44669–44681. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, J.; Cai, S.; Chen, G.; Xiao, N.; Fu, Y.; Chen, Q.; Qiu, S. PNCK depletion inhibits proliferation and induces apoptosis of human nasopharyngeal carcinoma cells in vitro and in vivo. J. Cancer 2019, 10, 6925–6932. [Google Scholar] [CrossRef]
- Deb, T.B.; Zuo, A.H.; Barndt, R.J.; Sengupta, S.; Jankovic, R.; Johnson, M.D. Pnck overexpression in HER-2 gene-amplified breast cancer causes Trastuzumab resistance through a paradoxical PTEN-mediated process. Breast Cancer Res. Treat. 2015, 150, 347–361. [Google Scholar] [CrossRef]
- Mishra, N.K.; Southekal, S.; Guda, C. Survival Analysis of Multi-Omics Data Identifies Potential Prognostic Markers of Pancreatic Ductal Adenocarcinoma. Front. Genet. 2019, 10, 624. [Google Scholar] [CrossRef] [PubMed]
- Capra, M.; Nuciforo, P.G.; Confalonieri, S.; Quarto, M.; Bianchi, M.; Nebuloni, M.; Boldorini, R.; Pallotti, F.; Viale, G.; Gishizky, M.L.; et al. Frequent alterations in the expression of serine/threonine kinases in human cancers. Cancer Res. 2006, 66, 8147–8154. [Google Scholar] [CrossRef]
- Xia, L.; Wang, H.; Xiao, H.; Lan, B.; Liu, J.; Yang, Z. EEF1A2 and ERN2 could potentially discriminate metastatic status of mediastinal lymph node in lung adenocarcinomas harboring EGFR 19Del/L858R mutations. Thorac. Cancer 2020, 11, 2755–2766. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Southekal, S.; Mishra, N.K.; Guda, C. Pan-Cancer Analysis of Human Kinome Gene Expression and Promoter DNA Methylation Identifies Dark Kinase Biomarkers in Multiple Cancers. Cancers 2021, 13, 1189. https://doi.org/10.3390/cancers13061189
Southekal S, Mishra NK, Guda C. Pan-Cancer Analysis of Human Kinome Gene Expression and Promoter DNA Methylation Identifies Dark Kinase Biomarkers in Multiple Cancers. Cancers. 2021; 13(6):1189. https://doi.org/10.3390/cancers13061189
Chicago/Turabian StyleSouthekal, Siddesh, Nitish Kumar Mishra, and Chittibabu Guda. 2021. "Pan-Cancer Analysis of Human Kinome Gene Expression and Promoter DNA Methylation Identifies Dark Kinase Biomarkers in Multiple Cancers" Cancers 13, no. 6: 1189. https://doi.org/10.3390/cancers13061189
APA StyleSouthekal, S., Mishra, N. K., & Guda, C. (2021). Pan-Cancer Analysis of Human Kinome Gene Expression and Promoter DNA Methylation Identifies Dark Kinase Biomarkers in Multiple Cancers. Cancers, 13(6), 1189. https://doi.org/10.3390/cancers13061189





_Guda.png)
