Large Extracellular Vesicle Characterization and Association with Circulating Tumor Cells in Metastatic Castrate Resistant Prostate Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
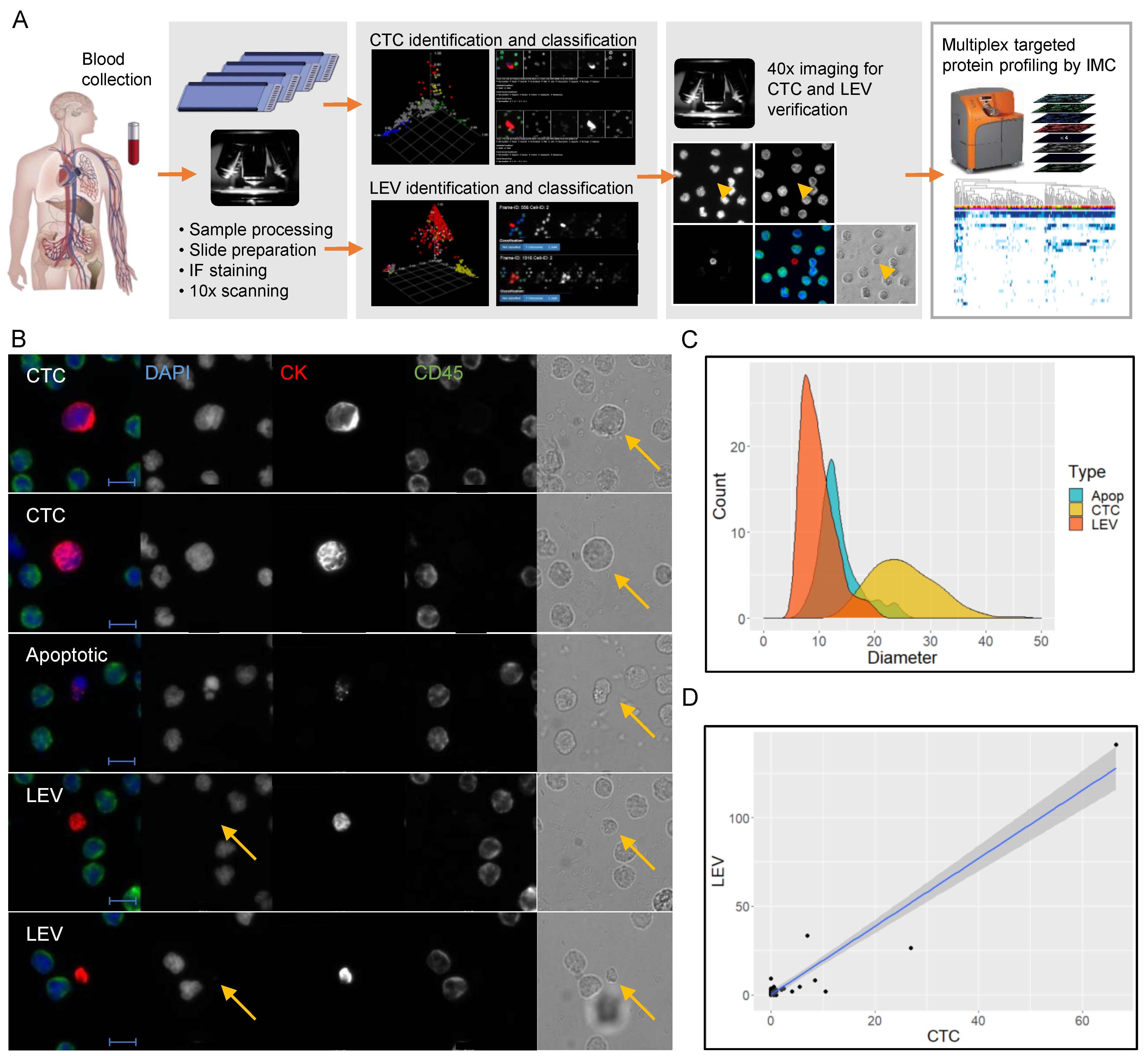
2.2. Identification and Characterization of CTCs and LEVs
2.3. Imaging Mass Cytometry
3. Results
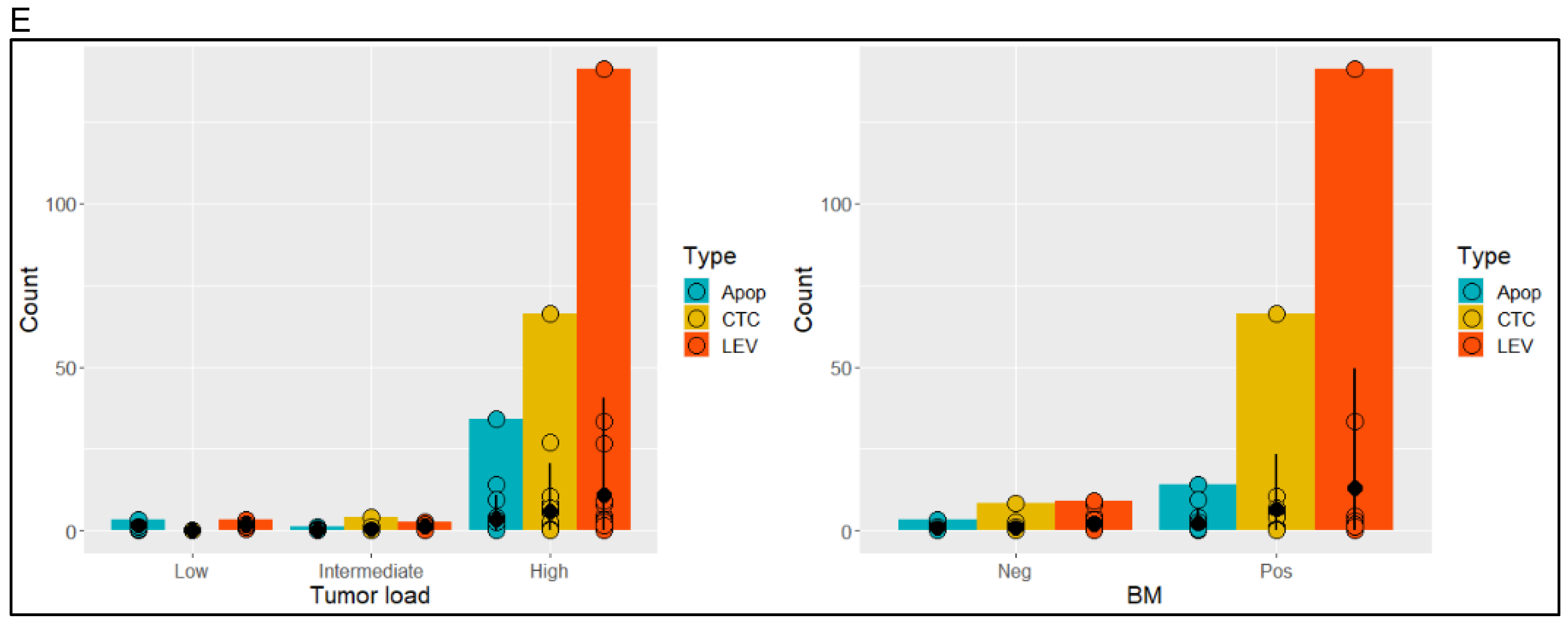
3.1. Identification and Morphometric Characterization of LEVs
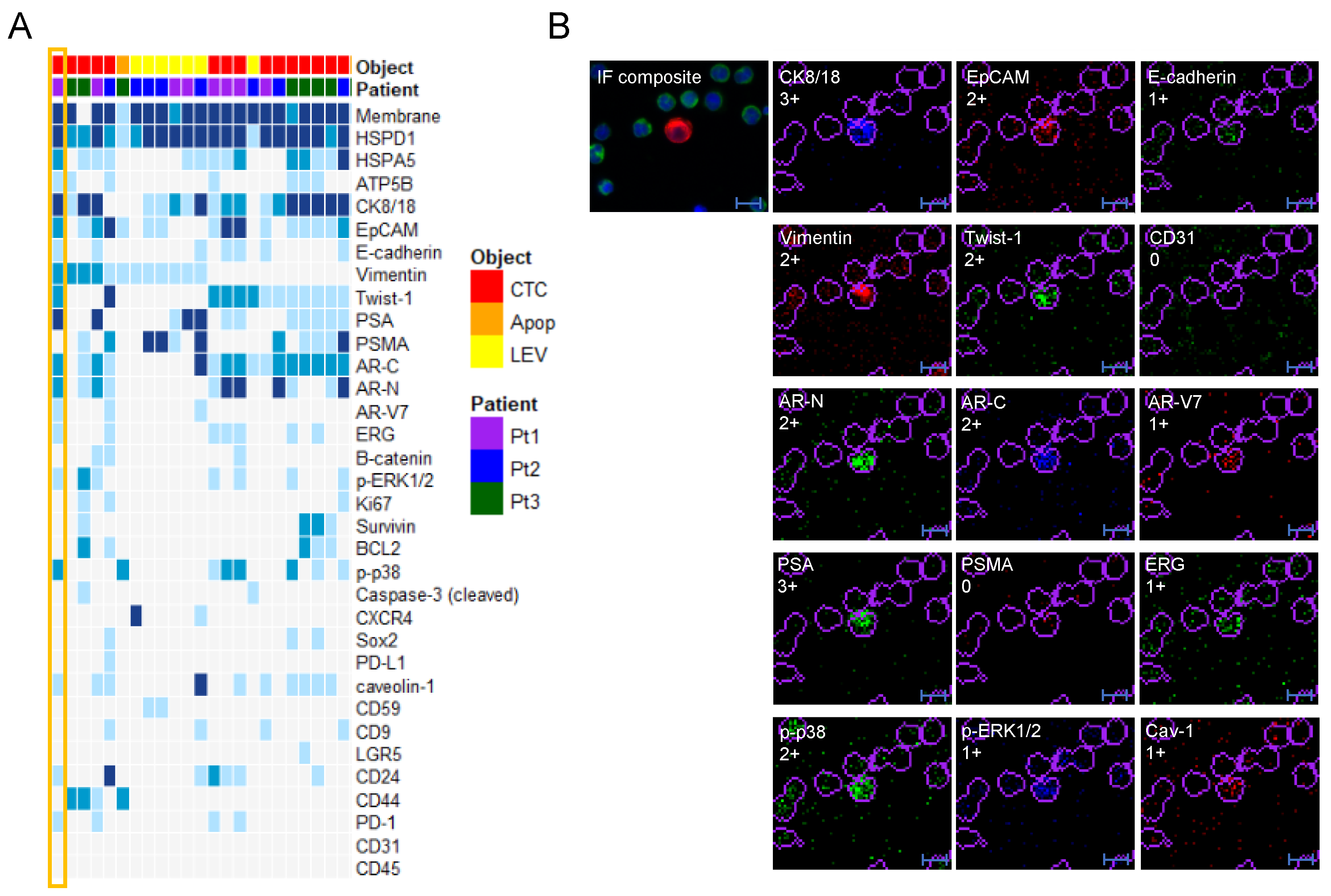
3.2. Protein Profiling of CTCs and LEVs
3.3. Epithelial and Prostate Marker Distribution
3.4. Protein Profile Heterogeneity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhang, Z.; Xia, W.; Cai, J.; Li, Y.; Wu, S. Extracellular vesicles in urologic malignancies-Implementations for future cancer care. Cell Prolif. 2019, 52, e12659. [Google Scholar] [CrossRef]
- Minciacchi, V.R.; Spinelli, C.; Reis-Sobreiro, M.; Cavallini, L.; You, S.; Zandian, M.; Li, X.; Mishra, R.; Chiarugi, P.; Adam, R.M.; et al. MYC Mediates Large Oncosome-Induced Fibroblast Reprogramming in Prostate Cancer. Cancer Res. 2017, 77, 2306–2317. [Google Scholar] [CrossRef] [PubMed]
- Zanetti-Domingues, L.C.; Bonner, S.E.; Iyer, R.S.; Martin-Fernandez, M.L.; Huber, V. Cooperation and Interplay between EGFR Signalling and Extracellular Vesicle Biogenesis in Cancer. Cells 2020, 9, 2639. [Google Scholar] [CrossRef] [PubMed]
- Murali, V.P.; Holmes, C.A. Biomaterial-Based Extracellular Vesicle Delivery for Therapeutic Applications. Acta Biomater. 2021. [Google Scholar] [CrossRef]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef]
- Meehan, B.; Rak, J.; Di Vizio, D. Oncosomes—Large and small: What are they, where they came from? J. Extracell. Vesicles 2016, 5, 33109. [Google Scholar] [CrossRef] [PubMed]
- Vagner, T.; Spinelli, C.; Minciacchi, V.R.; Balaj, L.; Zandian, M.; Conley, A.; Zijlstra, A.; Freeman, M.R.; Demichelis, F.; De, S.; et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J. Extracell. Vesicles 2018, 7, 1505403. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular vesicles in cancer—Implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef]
- Reátegui, E.; van der Vos, K.E.; Lai, C.P.; Zeinali, M.; Atai, N.A.; Aldikacti, B.; Floyd, F.P., Jr.; Khankhel, A.H.; Thapar, V.; Hochberg, F.H.; et al. Engineered nanointerfaces for microfluidic isolation and molecular profiling of tumor-specific extracellular vesicles. Nat. Commun. 2018, 9, 175. [Google Scholar] [CrossRef] [PubMed]
- Shishido, S.N.; Carlsson, A.; Nieva, J.; Bethel, K.; Hicks, J.B.; Bazhenova, L.; Kuhn, P. Circulating tumor cells as a response monitor in stage IV non-small cell lung cancer. J. Transl. Med. 2019, 17, 294. [Google Scholar] [CrossRef]
- Gerdtsson, A.S.; Thiele, J.A.; Shishido, S.N.; Zheng, S.; Schaffer, R.; Bethel, K.; Curley, S.; Lenz, H.J.; Hanna, D.L.; Nieva, J.; et al. Single cell correlation analysis of liquid and solid biopsies in metastatic colorectal cancer. Oncotarget 2019, 10, 7016–7030. [Google Scholar] [CrossRef][Green Version]
- Carlsson, A.; Kuhn, P.; Luttgen, M.S.; Dizon, K.K.; Troncoso, P.; Corn, P.G.; Kolatkar, A.; Hicks, J.B.; Logothetis, C.J.; Zurita, A.J. Paired High-Content Analysis of Prostate Cancer Cells in Bone Marrow and Blood Characterizes Increased Androgen Receptor Expression in Tumor Cell Clusters. Clin. Cancer Res. 2017, 23, 1722–1732. [Google Scholar] [CrossRef]
- Ruiz, C.; Li, J.; Luttgen, M.S.; Kolatkar, A.; Kendall, J.T.; Flores, E.; Topp, Z.; Samlowski, W.E.; McClay, E.; Bethel, K.; et al. Limited genomic heterogeneity of circulating melanoma cells in advanced stage patients. Phys. Biol. 2015, 12, 016008. [Google Scholar] [CrossRef] [PubMed]
- Dago, A.E.; Stepansky, A.; Carlsson, A.; Luttgen, M.; Kendall, J.; Baslan, T.; Kolatkar, A.; Wigler, M.; Bethel, K.; Gross, M.E.; et al. Rapid phenotypic and genomic change in response to therapeutic pressure in prostate cancer inferred by high content analysis of single circulating tumor cells. PLoS ONE 2014, 9, e101777. [Google Scholar] [CrossRef] [PubMed]
- Gerdtsson, E.; Pore, M.; Thiele, J.A.; Gerdtsson, A.S.; Malihi, P.D.; Nevarez, R.; Kolatkar, A.; Velasco, C.R.; Wix, S.; Singh, M.; et al. Multiplex protein detection on circulating tumor cells from liquid biopsies using imaging mass cytometry. Converg. Sci. Phys. Oncol. 2018, 4, 015002. [Google Scholar] [CrossRef]
- Coumans, F.A.; Doggen, C.J.; Attard, G.; de Bono, J.S.; Terstappen, L.W. All circulating EpCAM+CK+CD45- objects predict overall survival in castration-resistant prostate cancer. Ann. Oncol. 2010, 21, 1851–1857. [Google Scholar] [CrossRef]
- Minciacchi, V.R.; Zijlstra, A.; Rubin, M.A.; Di Vizio, D. Extracellular vesicles for liquid biopsy in prostate cancer: Where are we and where are we headed? Prostate Cancer Prostatic Dis. 2017, 20, 251–258. [Google Scholar] [CrossRef]
- Di Vizio, D.; Kim, J.; Hager, M.H.; Morello, M.; Yang, W.; Lafargue, C.J.; True, L.D.; Rubin, M.A.; Adam, R.M.; Beroukhim, R.; et al. Oncosome formation in prostate cancer: Association with a region of frequent chromosomal deletion in metastatic disease. Cancer Res. 2009, 69, 5601–5609. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, A.M.; Harzstark, A.L.; Corn, P.G.; Wen, S.; Araujo, S.C.; Tu, S.; Pagliaro, L.C.; Kim, J.; Millikan, R.E.; Ryan, C.; et al. Platinum-based chemotherapy for variant castrate-resistant prostate cancer. Clin. Cancer Res. 2013, 19, 3621–3630. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, A.M.; Shen, L.; Tapia, E.L.N.; Lu, J.; Chen, H.; Zhang, J.; Wu, G.; Wang, X.; Troncoso, P.; Corn, P.; et al. Combined Tumor Suppressor Defects Characterize Clinically Defined Aggressive Variant Prostate Cancers. Clin. Cancers Res. 2016, 22, 1520–1530. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Lee, M.; Kolatkar, A.; McCormick, M.; Dago, A.D.; Kendall, J.; Carlsson, N.A.; Bethel, K.; Greenspan, E.J.; Hwang, S.E.; Waitman, K.R.; et al. Effect of Blood Collection Tube Type and Time to Processing on the Enumeration and High-Content Characterization of Circulating Tumor Cells Using the High-Definition Single-Cell Assay. Arch. Pathol. Lab. Med. 2018, 142, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Malihi, P.D.; Graf, R.P.; Rodriguez, A.; Ramesh, N.; Lee, J.; Sutton, R.; Jiles, R.; Ruiz Velasco, C.; Sei, E.; Kolatkar, A.; et al. Single-cell circulating tumor cell analysis reveals genomic instability as a distinctive feature of aggressive prostate cancer. Clin. Cancer Res. 2020, 26, 4143–4153. [Google Scholar] [CrossRef] [PubMed]
- Corn, P.G.; Heath, E.I.; Zurita, A.; Ramesh, N.; Xiao, L.; Sei, E.; Li-Ning-Tapia, E.; Tu, S.M.; Subudhi, S.K.; Wang, J.; et al. Cabazitaxel plus carboplatin for the treatment of men with metastatic castration-resistant prostate cancers: A randomised, open-label, phase 1-2 trial. Lancet Oncol. 2019, 20, 1432–1443. [Google Scholar] [CrossRef]
- Malihi, P.D.; Morikado, M.; Welter, L.; Liu, S.T.; Miller, E.T.; Cadaneanu, R.M.; Knudsen, B.S.; Lewis, M.S.; Carlsson, A.; Velasco, C.R.; et al. Clonal diversity revealed by morphoproteomic and copy number profiles of single prostate cancer cells at diagnosis. Converg. Sci. Phys. Oncol. 2018, 4, 015003. [Google Scholar] [CrossRef] [PubMed]
- Minciacchi, V.R.; You, S.; Spinelli, C.; Morley, S.; Zandian, M.; Aspuria, P.J.; Cavallini, L.; Ciardiello, C.; Reis Sobreiro, M.; Morello, M.; et al. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget 2015, 6, 11327–11341. [Google Scholar] [CrossRef]
- Onstenk, W.; Sieuwerts, A.M.; Kraan, J.; Van, M.; Nieuweboer, A.J.; Mathijssen, R.H.; Hamberg, P.; Meulenbeld, H.J.; De Laere, B.; Dirix, L.Y.; et al. Efficacy of Cabazitaxel in Castration-resistant Prostate Cancer Is Independent of the Presence of AR-V7 in Circulating Tumor Cells. Eur. Urol. 2015, 68, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Scher, H.I.; Graf, R.P.; Schreiber, N.A.; McLaughlin, B.; Lu, D.; Louw, J.; Danila, D.C.; Dugan, L.; Johnson, A.; Heller, G.; et al. Nuclear-specific AR-V7 Protein Localization is Necessary to Guide Treatment Selection in Metastatic Castration-resistant Prostate cancer. Eur. Urol. 2017, 71, 874–882. [Google Scholar] [CrossRef]
- Graf, R.P.; Hullings, M.; Barnett, E.S.; Carbone, E.; Dittamore, R.; Scher, H.I. Clinical Utility of the Nuclear-localized AR-V7 Biomarker in Circulating Tumor Cells in Improving Physician Treatment Choice in Castration-resistant Prostate Cancer. Eur. Urol. 2020, 77, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Rycaj, K.; Chen, X.; Tang, D.G. Cancer stem cells and cell size: A causal link? Semin. Cancer Biol. 2015, 35, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Di Vizio, D.; Morello, M.; Dudley, A.C.; Schow, P.W.; Adam, R.M.; Morley, S.; Mulholland, D.; Rotinen, M.; Hager, M.H.; Insabato, L.; et al. Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease. Am. J. Pathol. 2012, 181, 1573–1584. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Lee, T.H.; Spinelli, C.; Chennakrishnaiah, S.; D’Asti, E.; Rak, J. Extracellular vesicle communication pathways as regulatory targets of oncogenic transformation. Semin. Cell Dev. Biol. 2017, 67, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed]
- Nanou, A.; Zeune, L.L.; Terstappen, L.W.M.M. Leukocyte-Derived Extracellular Vesicles in Blood with and without EpCAM Enrichment. Cells 2019, 8, 937. [Google Scholar] [CrossRef]
- Lee, K.; Fraser, K.; Ghaddar, B.; Yang, K.; Kim, E.; Balaj, L.; Chiocca, E.A.; Breakefield, X.O.; Lee, H.; Weissleder, R. Multiplexed Profiling of Single Extracellular Vesicles. ACS Nano 2018, 12, 494–503. [Google Scholar] [CrossRef] [PubMed]
- Beekman, P.; Enciso-Martinez, A.; Rho, H.S.; Pujari, S.P.; Lenferink, A.; Zuilhof, H.; Terstappen, L.W.M.M.; Otto, C.; Le Gac, S. Immuno-capture of extracellular vesicles for individual multi-modal characterization using AFM, SEM and Raman spectroscopy. Lab Chip 2019, 19, 2526–2536. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Spinelli, C.; Montermini, L.; Rak, J. Oncogenic regulation of extracellular vesicle proteome and heterogeneity. Proteomics 2019, 19, e1800169. [Google Scholar] [CrossRef]
- Taha, E.A.; Ono, K.; Eguchi, T. Roles of Extracellular HSPs as Biomarkers in Immune Surveillance and Immune Evasion. Int. J. Mol. Sci. 2019, 20, 4588. [Google Scholar] [CrossRef]
- Antonio, D.M. Extracellular heat shock proteins, cellular export vesicles, and the Stress Observation System: A form of communication during injury, infection, and cell damage. Cell Stress Chaperones 2011, 16, 235–249. [Google Scholar]
- Ghosh, A.; Davey, M.; Chute, I.C.; Griffiths, S.G.; Lewis, S.; Chacko, S.; Barnett, D.; Crapoulet, N.; Fournier, S.; Joy, A.; et al. Rapid isolation of extracellular vesicles from cell culture and biological fluids using a synthetic peptide with specific affinity for heat shock proteins. PLoS ONE 2014, 9, e110443. [Google Scholar] [CrossRef]
- Larson, C.J.; Moreno, J.G.; Pienta, K.J.; Gross, S.; Repollet, M.; O’hara, S.M.; Russell, T.; Terstappen, L.W. Apoptosis of circulating tumor cells in prostate cancer patients. Cytom. Part A 2004, 62, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Lepage, C.C.; Thompson, L.L.; Larson, B.; McManus, K.J. An automated single cell quantitative imaging microscopy approach to assess micronucleus formation, genotoxicity and chromosome instability. Cells 2020, 9, 344. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Chung, J.; Balaj, L.; Charest, A.; Bigner, D.D.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Weissleder, R.; Lee, H. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med. 2012, 18, 1835–1840. [Google Scholar] [CrossRef] [PubMed]


| Feature | Distribution | |
|---|---|---|
| Age | Median 67.5, Range 56–81 | |
| Race | Caucasian | n = 36 (82%) |
| African American | n = 3 (7%) | |
| Hispanic | n = 2 (5%) | |
| NA | n = 3 (7%) | |
| ECOG | 0 | n = 12 (27%) |
| 1–2 | n = 29 (66%) | |
| NA | n = 3 (7%) | |
| Clinicopathological AVPC (AVPC-C) | Yes | n = 16 (36%) |
| No | n = 25 (57%) | |
| NA | n = 3 (7%) | |
| Prior therapy | Docetaxel | n = 11 (25%) |
| Abi. and/or Enza. * | n = 28 (64%) | |
| Other | n = 10 (23%) | |
| NA | n = 3 (7%) | |
| Bone marrow positive | Yes | n = 15 (34%) |
| No | n = 22 (50%) | |
| NA | n = 7 (16%) | |
| Tumor load | High | n = 23 (52%) |
| Intermediate | n = 12 (27%) | |
| Low | n = 6 (14%) | |
| NA | n = 3 (7%) | |
| PSA (ng/mL) | median 40.3, range 0.1–681.6 | |
| PFS (months) | median 6.1, range 1.1–11.7 | |
| OS (months) | median 22.6, range 4.7–39.7 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gerdtsson, A.S.; Setayesh, S.M.; Malihi, P.D.; Ruiz, C.; Carlsson, A.; Nevarez, R.; Matsumoto, N.; Gerdtsson, E.; Zurita, A.; Logothetis, C.; et al. Large Extracellular Vesicle Characterization and Association with Circulating Tumor Cells in Metastatic Castrate Resistant Prostate Cancer. Cancers 2021, 13, 1056. https://doi.org/10.3390/cancers13051056
Gerdtsson AS, Setayesh SM, Malihi PD, Ruiz C, Carlsson A, Nevarez R, Matsumoto N, Gerdtsson E, Zurita A, Logothetis C, et al. Large Extracellular Vesicle Characterization and Association with Circulating Tumor Cells in Metastatic Castrate Resistant Prostate Cancer. Cancers. 2021; 13(5):1056. https://doi.org/10.3390/cancers13051056
Chicago/Turabian StyleGerdtsson, Anna S., Sonia M. Setayesh, Paymaneh D. Malihi, Carmen Ruiz, Anders Carlsson, Rafael Nevarez, Nicholas Matsumoto, Erik Gerdtsson, Amado Zurita, Christopher Logothetis, and et al. 2021. "Large Extracellular Vesicle Characterization and Association with Circulating Tumor Cells in Metastatic Castrate Resistant Prostate Cancer" Cancers 13, no. 5: 1056. https://doi.org/10.3390/cancers13051056
APA StyleGerdtsson, A. S., Setayesh, S. M., Malihi, P. D., Ruiz, C., Carlsson, A., Nevarez, R., Matsumoto, N., Gerdtsson, E., Zurita, A., Logothetis, C., Corn, P. G., Aparicio, A. M., Hicks, J., & Kuhn, P. (2021). Large Extracellular Vesicle Characterization and Association with Circulating Tumor Cells in Metastatic Castrate Resistant Prostate Cancer. Cancers, 13(5), 1056. https://doi.org/10.3390/cancers13051056








