Janus Kinases in Leukemia
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Structure and Regulation of Janus Kinases
1.2. JAK Regulation by JH2 Domain
1.3. JAKs in Hematopoiesis
2. JAKs in Leukemia
2.1. Fusion Proteins
2.2. JAK Mutations in Leukemia
2.3. Pathogenic JAK Activation in Leukemia
2.4. STATs in Leukemia
2.5. Mutations in JAK-Related Receptors and CALR
3. Current Status of JAK Inhibitors in MPN/Leukemia
4. Prospects for JAK Targeting in MPN/Leukemia
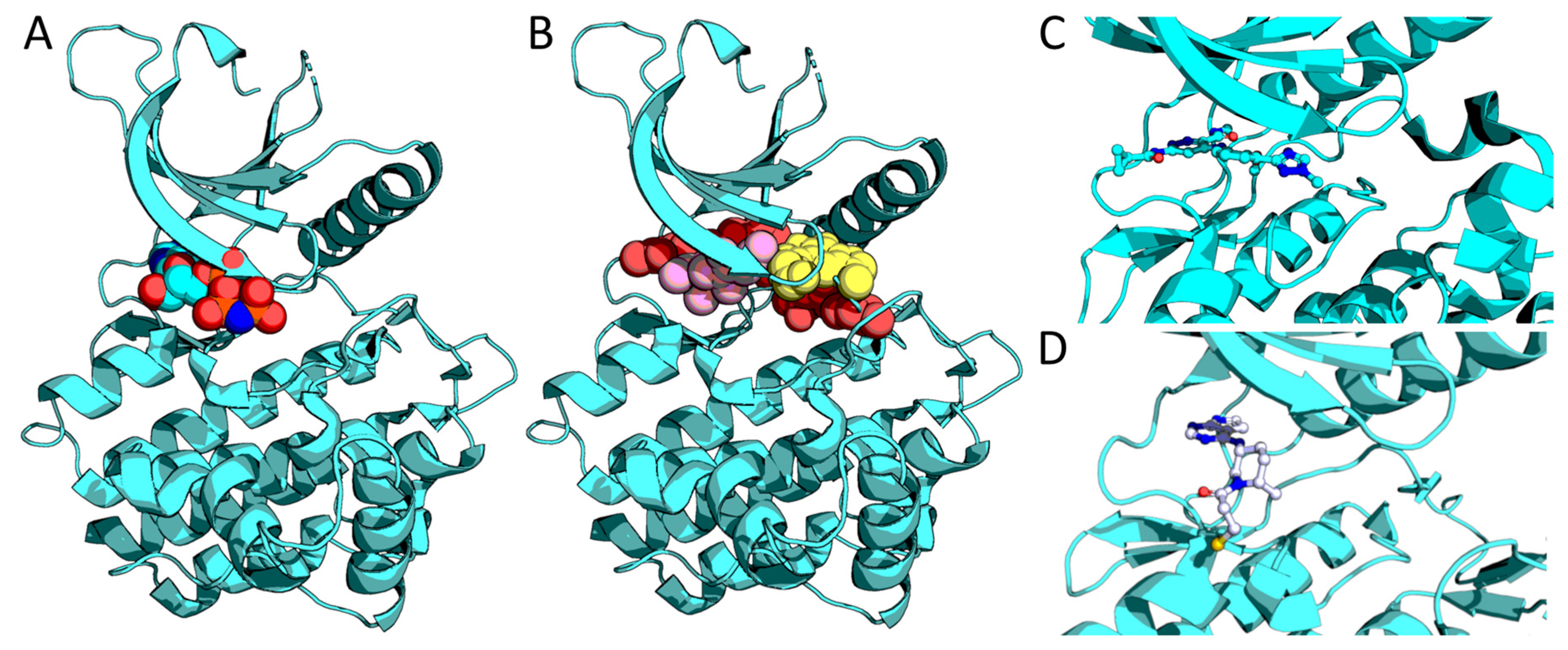
4.1. Pseudokinase Targeting
4.2. Covalent Inhibitors
4.3. Combination Therapy
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ALCL | anaplastic large cell lymphoma |
| ALK | anaplastic lymphoma kinase |
| ALL | acute lymphocytic leukemia |
| AML | acute myeloid leukemia |
| ATL | adult T-cell leukemia/lymphoma |
| CALR | Calreticulin |
| CLL | chronic lymphocytic leukemia |
| CML | chronic myeloid leukemia |
| CMML | chronic myelomonocytic leukemia |
| DS | Down Syndrome |
| G-CSFR | granulocyte colony-stimulating factor receptor |
| GH(R) | growth hormone (receptor) |
| GM-CSFR | granulocyte/macrophage colony-stimulating factor receptor |
| ELAVL1 | ELAVL like RNA binding protein 1 |
| EPO(R) | erythropoietin (receptor) |
| ET | essential thrombocythemia |
| ETV6 | translocation-Ets-leukemia virus |
| FERM | 4.1 protein, ezrin, radixin, moesin domain |
| HSC | hematopoietic stem cell |
| IFN(R) | interferon (receptor) |
| IL(R) | interleukin (receptor) |
| JAK | Janus kinase |
| JH | Janus homology |
| MDS | myelodysplastic syndrome |
| MDM2 | E3 Ubiquitin-Protein Ligase Mdm2 |
| MF | Myelofibrosis |
| NHL | Non-Hodgkin lymphoma |
| NPM1 | nucleophosmin 1 |
| MPN | myeloproliferative neoplasm |
| PCM1 | Pericentriolar material 1 |
| PMF | primary myelofibrosis |
| PRL(R) | prolactin (receptor) |
| PV | polycythemia vera |
| SH2 | Src homology 2 |
| SLL | Small lymphocytic lymphoma |
| SOCS | suppressor of cytokine signaling |
| STAT | signal transducer and activator of transcription |
| TPOR/MPL | Thrombopoietin receptor |
| TYK2 | Tyrosine kinase 2 |
References
- Kiu, H.; Nicholson, S.E. Biology and significance of the JAK/STAT signalling pathways. Growth Factors 2012, 30, 88–106. [Google Scholar] [CrossRef] [PubMed]
- Hammaren, H.M.; Virtanen, A.T.; Raivola, J.; Silvennoinen, O. The regulation of JAKs in cytokine signaling and its breakdown in disease. Cytokine 2019, 118, 48–63. [Google Scholar] [CrossRef] [PubMed]
- Vannucchi, A.M.; Harrison, C.N. Emerging treatments for classical myeloproliferative neoplasms. Blood 2017, 129, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Talpaz, M.; Kiladjian, J.-J. Fedratinib, a newly approved treatment for patients with myeloproliferative neoplasm-associated myelofibrosis. Leukemia 2021, 35, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Eli Lilly and Company FDA Approves OLUMIANT® (baricitinib) 2-mg Tablets for the Treatment of Adults with Moderately-to-Severely Active Rheumatoid Arthritis. Available online: https://investor.lilly.com/news-releases/news-release-details/fda-approves-olumiantr-baricitinib-2-mg-tablets-treatment-adults. (accessed on 25 January 2021).
- AbbVie AbbVie Receives FDA Approval of RINVOQTM (upadacitinib), an Oral JAK Inhibitor for the Treatment of Moderate to Severe Rheumatoid Arthritis. 2019. Available online: https://news.abbvie.com/news/press-releases/abbvie-receives-fda-approval-rinvoq-upadacitinib-an-oral-jak-inhibitor-for-treatment-moderate-to-severe-rheumatoid-arthritis.htm. (accessed on 25 January 2021).
- Pfizer Inc. U.S. FDA Approves Pfizer’s XELJANZ® (tofacitinib) for the Treatment of Active Polyarticular Course Juvenile Idiopathic Arthritis. 2020. Available online: https://investors.pfizer.com/investor-news/press-release-details/2020/U.S.-FDA-Approves-Pfizers-XELJANZ-tofacitinib-for-the-Treatment-of-Active-Polyarticular-Course-Juvenile-Idiopathic-Arthritis/default.aspx. (accessed on 25 January 2021).
- Virtanen, A.T.; Haikarainen, T.; Raivola, J.; Silvennoinen, O. Selective JAKinibs: Prospects in Inflammatory and Autoimmune Diseases. BioDrugs 2019, 33, 15–32. [Google Scholar] [CrossRef] [PubMed]
- Hellgren, G.; Jansson, J.O.; Carlsson, L.M.; Carlsson, B. The growth hormone receptor associates with Jak1, Jak2 and Tyk2 in human liver. Growth Horm. IGF Res. 1999, 9, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Silvennoinen, O.; Witthuhn, B.A.; Quelle, F.W.; Cleveland, J.L.; Yi, T.; Ihle, J.N. Structure of the murine Jak2 protein-tyrosine kinase and its role in interleukin 3 signal transduction. Proc. Natl. Acad. Sci. USA 1993, 90, 8429–8433. [Google Scholar] [CrossRef] [PubMed]
- Wallweber, H.J.A.; Tam, C.; Franke, Y.; Starovasnik, M.A.; Lupardus, P.J. Structural basis of recognition of interferon-α receptor by tyrosine kinase 2. Nat. Struct. Mol. Biol. 2014, 21, 443–448. [Google Scholar] [CrossRef]
- Witthuhn, B.A.; Quelle, F.W.; Silvennoinen, O.; Yi, T.; Tang, B.; Miura, O.; Ihle, J.N. JAK2 associates with the erythropoietin receptor and is tyrosine phosphorylated and activated following stimulation with erythropoietin. Cell 1993, 74, 227–236. [Google Scholar] [CrossRef]
- Babon, J.J.; Lucet, I.S.; Murphy, J.M.; Nicola, N.A.; Varghese, L.N. The molecular regulation of Janus kinase (JAK) activation. Biochem. J. 2014, 462, 1–13. [Google Scholar] [CrossRef]
- Silvennoinen, O.; Hubbard, S.R. Molecular insights into regulation of JAK2 in myeloproliferative neoplasms. Blood 2015, 125, 3388–3392. [Google Scholar] [CrossRef]
- Ivashkiv, L.B.; Hu, X. Signaling by STATs. Arthritis Res. Ther. 2004, 6, 159–168. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moraga, I.; Wernig, G.; Wilmes, S.; Gryshkova, V.; Richter, C.P.; Hong, W.-J.; Sinha, R.; Guo, F.; Fabionar, H.; Wehrman, T.S.; et al. Tuning cytokine receptor signaling by re-orienting dimer geometry with surrogate ligands. Cell 2015, 160, 1196–1208. [Google Scholar] [CrossRef] [PubMed]
- Varghese, L.N.; Ungureanu, D.; Liau, N.P.D.; Young, S.N.; Laktyushin, A.; Hammaren, H.; Lucet, I.S.; Nicola, N.A.; Silvennoinen, O.; Babon, J.J.; et al. Mechanistic insights into activation and SOCS3-mediated inhibition of myeloproliferative neoplasm-associated JAK2 mutants from biochemical and structural analyses. Biochem. J. 2014, 458, 395–405. [Google Scholar] [CrossRef]
- Xu, D.; Qu, C.-K. Protein tyrosine phosphatases in the JAK/STAT pathway. Front. Biosci. 2008, 13, 4925–4932. [Google Scholar] [CrossRef]
- Ali, S.; Nouhi, Z.; Chughtai, N.; Ali, S. SHP-2 regulates SOCS-1-mediated Janus kinase-2 ubiquitination/degradation downstream of the prolactin receptor. J. Biol. Chem. 2003, 278, 52021–52031. [Google Scholar] [CrossRef]
- Feener, E.P.; Rosario, F.; Dunn, S.L.; Stancheva, Z.; Myers, M.G., Jr. Tyrosine phosphorylation of Jak2 in the JH2 domain inhibits cytokine signaling. Mol. Cell. Biol. 2004, 24, 4968–4978. [Google Scholar] [CrossRef]
- Ungureanu, D.; Wu, J.; Pekkala, T.; Niranjan, Y.; Young, C.; Jensen, O.N.; Xu, C.-F.; Neubert, T.A.; Skoda, R.C.; Hubbard, S.R.; et al. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat. Struct. Mol. Biol. 2011, 18, 971–976. [Google Scholar] [CrossRef]
- Candotti, F.; Oakes, S.A.; Johnston, J.A.; Giliani, S.; Schumacher, R.F.; Mella, P.; Fiorini, M.; Ugazio, A.G.; Badolato, R.; Notarangelo, L.D.; et al. Structural and functional basis for JAK3-deficient severe combined immunodeficiency. Blood 1997, 90, 3996–4003. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Cheng, A.; Candotti, F.; Zhou, Y.-J.; Hymel, A.; Fasth, A.; Notarangelo, L.D.; O’Shea, J.J. Complex Effects of Naturally Occurring Mutations in the JAK3 Pseudokinase Domain: Evidence for Interactions between the Kinase and Pseudokinase Domains. Mol. Cell. Biol. 2000, 20, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Yeh, T.C.; Dondi, E.; Uze, G.; Pellegrini, S. A dual role for the kinase-like domain of the tyrosine kinase Tyk2 in interferon-alpha signaling. Proc. Natl. Acad. Sci. USA 2000, 97, 8991–8996. [Google Scholar] [CrossRef] [PubMed]
- Saharinen, P.; Silvennoinen, O. The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J. Biol. Chem. 2002, 277, 47954–47963. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, S.; Hafer, M.; Vuorio, J.; Tucker, J.A.; Winkelmann, H.; Löchte, S.; Stanly, T.A.; Pulgar Prieto, K.D.; Poojari, C.; Sharma, V.; et al. Mechanism of homodimeric cytokine receptor activation and dysregulation by oncogenic mutations. Science 2020, 367, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Hammaren, H.M.; Ungureanu, D.; Grisouard, J.; Skoda, R.C.; Hubbard, S.R.; Silvennoinen, O. ATP binding to the pseudokinase domain of JAK2 is critical for pathogenic activation. Proc. Natl. Acad. Sci. USA 2015, 112, 4642–4647. [Google Scholar] [CrossRef] [PubMed]
- Hammaren, H.M.; Virtanen, A.T.; Abraham, B.G.; Peussa, H.; Hubbard, S.R.; Silvennoinen, O. Janus kinase 2 activation mechanisms revealed by analysis of suppressing mutations. J. Allergy Clin. Immunol. 2019, 143, 1549–1559.e6. [Google Scholar] [CrossRef] [PubMed]
- Leroy, E.; Balligand, T.; Pecquet, C.; Mouton, C.; Colau, D.; Shiau, A.K.; Dusa, A.; Constantinescu, S.N. Differential effect of inhibitory strategies of the V617 mutant of JAK2 on cytokine receptor signaling. J. Allergy Clin. Immunol. 2019, 144, 224–235. [Google Scholar] [CrossRef]
- Raivola, J.; Hammarén, H.M.; Virtanen, A.T.; Bulleeraz, V.; Ward, A.C.; Silvennoinen, O. Hyperactivation of Oncogenic JAK3 Mutants Depend on ATP Binding to the Pseudokinase Domain. Front. Oncol. 2018, 8, 560. [Google Scholar] [CrossRef] [PubMed]
- Raivola, J.; Haikarainen, T.; Silvennoinen, O. Characterization of JAK1 Pseudokinase Domain in Cytokine Signaling. Cancers 2019, 12, 78. [Google Scholar] [CrossRef]
- Hubbard, S.R. Mechanistic Insights into Regulation of JAK2 Tyrosine Kinase. Front. Endocrinol. 2017, 8, 361. [Google Scholar] [CrossRef]
- Shan, Y.; Gnanasambandan, K.; Ungureanu, D.; Kim, E.T.; Hammaren, H.; Yamashita, K.; Silvennoinen, O.; Shaw, D.E.; Hubbard, S.R. Molecular basis for pseudokinase-dependent autoinhibition of JAK2 tyrosine kinase. Nat. Struct. Mol. Biol. 2014, 21, 579–584. [Google Scholar] [CrossRef]
- Lupardus, P.J.; Ultsch, M.; Wallweber, H.; Bir Kohli, P.; Johnson, A.R.; Eigenbrot, C. Structure of the pseudokinase–kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 8025–8030. [Google Scholar] [CrossRef]
- Akada, H.; Akada, S.; Hutchison, R.E.; Sakamoto, K.; Wagner, K.-U.; Mohi, G. Critical role of Jak2 in the maintenance and function of adult hematopoietic stem cells. Stem Cells 2014, 32, 1878–1889. [Google Scholar] [CrossRef]
- Solar, G.P.; Kerr, W.G.; Zeigler, F.C.; Hess, D.; Donahue, C.; de Sauvage, F.J.; Eaton, D.L. Role of c-mpl in early hematopoiesis. Blood 1998, 92, 4–10. [Google Scholar] [CrossRef]
- Hara, T.; Miyajima, A. Function and Signal Transduction Mediated by the Interleukin 3 Receptor System in Hematopoiesis. Stem Cells 1996, 14, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, S.E.; Novak, U.; Ziegler, S.F.; Layton, J.E. Distinct regions of the granulocyte colony-stimulating factor receptor are required for tyrosine phosphorylation of the signaling molecules JAK2, Stat3, and p42, p44MAPK. Blood 1995, 86, 3698–3704. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Itoh, T.; Arai, K. Roles of JAK kinases in human GM-CSF receptor signal transduction. J. Allergy Clin. Immunol. 1996, 98, S183–S191. [Google Scholar] [CrossRef]
- Wang, X.; Lupardus, P.; Laporte, S.L.; Garcia, K.C. Structural biology of shared cytokine receptors. Annu. Rev. Immunol. 2009, 27, 29–60. [Google Scholar] [CrossRef] [PubMed]
- Haan, C.; Rolvering, C.; Raulf, F.; Kapp, M.; Druckes, P.; Thoma, G.; Behrmann, I.; Zerwes, H.-G. Jak1 has a dominant role over Jak3 in signal transduction through gammac-containing cytokine receptors. Chem. Biol. 2011, 18, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Baldridge, M.T.; King, K.Y.; Boles, N.C.; Weksberg, D.C.; Goodell, M.A. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature 2010, 465, 793–797. [Google Scholar] [CrossRef]
- de Bruin, A.M.; Demirel, Ö.; Hooibrink, B.; Brandts, C.H.; Nolte, M.A. Interferon-γ impairs proliferation of hematopoietic stem cells in mice. Blood 2013, 121, 3578–3585. [Google Scholar] [CrossRef] [PubMed]
- Trumpp, A.; Essers, M.; Wilson, A. Awakening dormant haematopoietic stem cells. Nat. Rev. Immunol. 2010, 10, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Tomasson, M.H.; Xiang, Z.; Walgren, R.; Zhao, Y.; Kasai, Y.; Miner, T.; Ries, R.E.; Lubman, O.; Fremont, D.H.; McLellan, M.D.; et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood 2008, 111, 4797–4808. [Google Scholar] [CrossRef] [PubMed]
- Tai, E.W.; Ward, K.C.; Bonaventure, A.; Siegel, D.A.; Coleman, M.P. Survival among children diagnosed with acute lymphoblastic leukemia in the United States, by race and age, 2001 to 2009: Findings from the CONCORD-2 study. Cancer 2017, 123, 5178–5189. [Google Scholar] [CrossRef] [PubMed]
- Carroll, W.L.; Raetz, E.A. Clinical and laboratory biology of childhood acute lymphoblastic leukemia. J. Pediatr. 2012, 160, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Kreipe, H.; Hussein, K.; Göhring, G.; Schlegelberger, B. Progression of myeloproliferative neoplasms to myelofibrosis and acute leukaemia. J. Hematopathol. 2011, 4, 61–68. [Google Scholar] [CrossRef][Green Version]
- Lacronique, V.; Boureux, A.; Valle, V.D.; Poirel, H.; Quang, C.T.; Mauchauffe, M.; Berthou, C.; Lessard, M.; Berger, R.; Ghysdael, J.; et al. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science 1997, 278, 1309–1312. [Google Scholar] [CrossRef] [PubMed]
- Peeters, P.; Raynaud, S.D.; Cools, J.; Wlodarska, I.; Grosgeorge, J.; Philip, P.; Monpoux, F.; Van Rompaey, L.; Baens, M.; Van den Berghe, H.; et al. Fusion of TEL, the ETS-variant gene 6 (ETV6), to the receptor-associated kinase JAK2 as a result of t(9;12) in a lymphoid and t(9;15;12) in a myeloid leukemia. Blood 1997, 90, 2535–2540. [Google Scholar] [CrossRef] [PubMed]
- Boer, J.M.; den Boer, M.L. BCR-ABL1-like acute lymphoblastic leukaemia: From bench to bedside. Eur. J. Cancer 2017, 82, 203–218. [Google Scholar] [CrossRef]
- Mullighan, C.G.; Zhang, J.; Harvey, R.C.; Collins-Underwood, J.R.; Schulman, B.A.; Phillips, L.A.; Tasian, S.K.; Loh, M.L.; Su, X.; Liu, W.; et al. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 2009, 106, 9414–9418. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L.; Loriaux, M.; Huntly, B.J.P.; Loh, M.L.; Beran, M.; Stoffregen, E.; Berger, R.; Clark, J.J.; Willis, S.G.; Nguyen, K.T.; et al. The JAK2V617F activating mutation occurs in chronic myelomonocytic leukemia and acute myeloid leukemia, but not in acute lymphoblastic leukemia or chronic lymphocytic leukemia. Blood 2005, 106, 3377–3379. [Google Scholar] [CrossRef]
- Roberts, K.G.; Li, Y.; Payne-Turner, D.; Harvey, R.C.; Yang, Y.-L.; Pei, D.; McCastlain, K.; Ding, L.; Lu, C.; Song, G.; et al. Targetable Kinase-Activating Lesions in Ph-like Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2014, 371, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Crescenzo, R.; Abate, F.; Lasorsa, E.; Tabbo’, F.; Gaudiano, M.; Chiesa, N.; Di Giacomo, F.; Spaccarotella, E.; Barbarossa, L.; Ercole, E.; et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell 2015, 27, 516–532. [Google Scholar] [CrossRef]
- Tron, A.E.; Keeton, E.K.; Ye, M.; Casas-Selves, M.; Chen, H.; Dillman, K.S.; Gale, R.E.; Stengel, C.; Zinda, M.; Linch, D.C.; et al. Next-generation sequencing identifies a novel ELAVL1-TYK2 fusion gene in MOLM-16, an AML cell line highly sensitive to the PIM kinase inhibitor AZD1208. Leuk. Lymphoma 2016, 57, 2927–2929. [Google Scholar] [CrossRef] [PubMed]
- Velusamy, T.; Kiel, M.J.; Sahasrabuddhe, A.A.; Rolland, D.; Dixon, C.A.; Bailey, N.G.; Betz, B.L.; Brown, N.A.; Hristov, A.C.; Wilcox, R.A.; et al. A novel recurrent NPM1-TYK2 gene fusion in cutaneous CD30-positive lymphoproliferative disorders. Blood 2014, 124, 3768–3771. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Z.; Zhao, Y.; Mitaksov, V.; Fremont, D.H.; Kasai, Y.; Molitoris, A.; Ries, R.E.; Miner, T.L.; McLellan, M.D.; DiPersio, J.F.; et al. Identification of somatic JAK1 mutations in patients with acute myeloid leukemia. Blood 2008, 111, 4809–4812. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, B.; Hu, L.; Ning, H.; Jiang, M.; Wang, D.; Liu, T.; Zhang, B.; Chen, H. Identification of a novel functional JAK1 S646P mutation in acute lymphoblastic leukemia. Oncotarget 2017, 8, 34687–34697. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jeong, E.G.; Kim, M.S.; Nam, H.K.; Min, C.K.; Lee, S.; Chung, Y.J.; Yoo, N.J.; Lee, S.H. Somatic mutations of JAK1 and JAK3 in acute leukemias and solid cancers. Clin. Cancer Res. 2008, 14, 3716–3721. [Google Scholar] [CrossRef] [PubMed]
- Staerk, J.; Kallin, A.; Demoulin, J.-B.; Vainchenker, W.; Constantinescu, S.N. JAK1 and Tyk2 activation by the homologous polycythemia vera JAK2 V617F mutation: Cross-talk with IGF1 receptor. J. Biol. Chem. 2005, 280, 41893–41899. [Google Scholar] [CrossRef]
- Flex, E.; Petrangeli, V.; Stella, L.; Chiaretti, S.; Hornakova, T.; Knoops, L.; Ariola, C.; Fodale, V.; Clappier, E.; Paoloni, F.; et al. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. J. Exp. Med. 2008, 205, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Hanratty, W.P.; Dearolf, C.R. The Drosophila Tumorous-lethal hematopoietic oncogene is a dominant mutation in the hopscotch locus. Mol. Gen. Genet. 1993, 238, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.A.; Binari, R.; Nahreini, T.S.; Gilman, M.; Perrimon, N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995, 14, 2857–2865. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Rose, P.; Barber, D.; Hanratty, W.P.; Lee, S.; Roberts, T.M.; D’Andrea, A.D.; Dearolf, C.R. Mutation in the Jak kinase JH2 domain hyperactivates Drosophila and mammalian Jak-Stat pathways. Mol. Cell. Biol. 1997, 17, 1562–1571. [Google Scholar] [CrossRef] [PubMed]
- Ha, J.-S.; Jeon, D.-S. Possible new LNK mutations in myeloproliferative neoplasms. Am. J. Hematol. 2011, 86, 866–868. [Google Scholar] [CrossRef] [PubMed]
- Lasho, T.L.; Tefferi, A.; Finke, C.; Pardanani, A. Clonal hierarchy and allelic mutation segregation in a myelofibrosis patient with two distinct LNK mutations. Leukemia 2011, 25, 1056–1058. [Google Scholar] [CrossRef][Green Version]
- Pardanani, A.D.; Levine, R.L.; Lasho, T.; Pikman, Y.; Mesa, R.A.; Wadleigh, M.; Steensma, D.P.; Elliott, M.A.; Wolanskyj, A.P.; Hogan, W.J.; et al. MPL515 mutations in myeloproliferative and other myeloid disorders: A study of 1182 patients. Blood 2006, 108, 3472–3476. [Google Scholar] [CrossRef]
- Baxter, E.J.; Scott, L.M.; Campbell, P.J.; East, C.; Fourouclas, N.; Swanton, S.; Vassiliou, G.S.; Bench, A.J.; Boyd, E.M.; Curtin, N.; et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 2005, 365, 1054–1061. [Google Scholar] [CrossRef]
- Kralovics, R.; Passamonti, F.; Buser, A.S.; Teo, S.-S.; Tiedt, R.; Passweg, J.R.; Tichelli, A.; Cazzola, M.; Skoda, R.C. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 2005, 352, 1779–1790. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L.; Wadleigh, M.; Cools, J.; Ebert, B.L.; Wernig, G.; Huntly, B.J.P.; Boggon, T.J.; Wlodarska, I.; Clark, J.J.; Moore, S.; et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 2005, 7, 387–397. [Google Scholar] [CrossRef]
- Goyal, H.; Chachoua, I.; Pecquet, C.; Vainchenker, W.; Constantinescu, S.N. A p53-JAK-STAT connection involved in myeloproliferative neoplasm pathogenesis and progression to secondary acute myeloid leukemia. Blood Rev. 2020, 42, 100712. [Google Scholar] [CrossRef] [PubMed]
- Pastore, F.; Krishnan, A.; Hammarén, H.M.; Silvennoinen, O.; Yan, B.; Levine, R.L. JAK2S523L, a novel gain-of-function mutation in a critical autoregulatory residue in JAK2V617F−MPNs. Blood Adv. 2020, 4, 4554–4559. [Google Scholar] [CrossRef]
- Chen, C.; Li, F.; Ma, M.-M.; Zhang, S.; Liu, Y.; Yan, Z.-L.; Chen, W.; Cao, J.; Zeng, L.-Y.; Wang, X.-Y.; et al. Roles of T875N somatic mutation in the activity, structural stability of JAK2 and the transformation of OCI-AML3 cells. Int. J. Biol. Macromol. 2019, 137, 1030–1040. [Google Scholar] [CrossRef]
- Wang, S.; Yan, J.; Zhou, G.; Heintzelman, R.; Hou, J.S. Myeloproliferative Neoplasm or Reactive Process? A Rare Case of Acute Myeloid Leukemia and Transient Posttreatment Megakaryocytic Hyperplasia with JAK-2 Mutation. Case Rep. Hematol. 2016, 2016, 6054017. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.M. Lymphoid malignancies: Another face to the Janus kinases. Blood Rev. 2013, 27, 63–70. [Google Scholar] [CrossRef]
- Alhuraiji, A.; Naqvi, K.; Huh, Y.O.; Ho, C.; Verstovsek, S.; Bose, P. Acute lymphoblastic leukemia secondary to myeloproliferative neoplasms or after lenalidomide exposure. Clin. Case Rep. 2017, 6, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Mullighan, C.G.; Harvey, R.C.; Wu, G.; Chen, X.; Edmonson, M.; Buetow, K.H.; Carroll, W.L.; Chen, I.-M.; Devidas, M.; et al. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: A report from the Children’s Oncology Group. Blood 2011, 118, 3080–3087. [Google Scholar] [CrossRef]
- Hitzler, J.K.; Zipursky, A. Origins of leukaemia in children with Down syndrome. Nat. Rev. Cancer 2005, 5, 11–20. [Google Scholar] [CrossRef]
- Bercovich, D.; Ganmore, I.; Scott, L.M.; Wainreb, G.; Birger, Y.; Elimelech, A.; Shochat, C.; Cazzaniga, G.; Biondi, A.; Basso, G.; et al. Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down’s syndrome. Lancet 2008, 372, 1484–1492. [Google Scholar] [CrossRef]
- Degryse, S.; Bornschein, S.; de Bock, C.E.; Leroy, E.; Vanden Bempt, M.; Demeyer, S.; Jacobs, K.; Geerdens, E.; Gielen, O.; Soulier, J.; et al. Mutant JAK3 signaling is increased by loss of wild-type JAK3 or by acquisition of secondary JAK3 mutations in T-ALL. Blood 2018, 131, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Shochat, C.; Tal, N.; Bandapalli, O.R.; Palmi, C.; Ganmore, I.; te Kronnie, G.; Cario, G.; Cazzaniga, G.; Kulozik, A.E.; Stanulla, M.; et al. Gain-of-function mutations in interleukin-7 receptor-α (IL7R) in childhood acute lymphoblastic leukemias. J. Exp. Med. 2011, 208, 901–908. [Google Scholar] [CrossRef]
- Walters, D.K.; Mercher, T.; Gu, T.-L.; O’Hare, T.; Tyner, J.W.; Loriaux, M.; Goss, V.L.; Lee, K.A.; Eide, C.A.; Wong, M.J.; et al. Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer Cell 2006, 10, 65–75. [Google Scholar] [CrossRef]
- Bellanger, D.; Jacquemin, V.; Chopin, M.; Pierron, G.; Bernard, O.A.; Ghysdael, J.; Stern, M.-H. Recurrent JAK1 and JAK3 somatic mutations in T-cell prolymphocytic leukemia. Leukemia 2014, 28, 417–419. [Google Scholar] [CrossRef]
- Zhang, J.; Ding, L.; Holmfeldt, L.; Wu, G.; Heatley, S.L.; Payne-Turner, D.; Easton, J.; Chen, X.; Wang, J.; Rusch, M.; et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012, 481, 157–163. [Google Scholar] [CrossRef]
- Koo, G.C.; Tan, S.Y.; Tang, T.; Poon, S.L.; Allen, G.E.; Tan, L.; Chong, S.C.; Ong, W.S.; Tay, K.; Tao, M.; et al. Janus kinase 3-activating mutations identified in natural killer/T-cell lymphoma. Cancer Discov. 2012, 2, 591–597. [Google Scholar] [CrossRef]
- Canté-Barrett, K.; Uitdehaag, J.C.M.; Meijerink, J.P.P. Structural modeling of JAK1 mutations in T-cell acute lymphoblastic leukemia reveals a second contact site between pseudokinase and kinase domains. Haematologica 2016, 101, e189–e191. [Google Scholar] [CrossRef] [PubMed]
- Waanders, E.; Scheijen, B.; Jongmans, M.C.J.; Venselaar, H.; van Reijmersdal, S.V.; van Dijk, A.H.A.; Pastorczak, A.; Weren, R.D.A.; van der Schoot, C.E.; van de Vorst, M.; et al. Germline activating TYK2 mutations in pediatric patients with two primary acute lymphoblastic leukemia occurrences. Leukemia 2017, 31, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Kreins, A.Y.; Ciancanelli, M.J.; Okada, S.; Kong, X.-F.; Ramírez-Alejo, N.; Kilic, S.S.; El Baghdadi, J.; Nonoyama, S.; Mahdaviani, S.A.; Ailal, F.; et al. Human TYK2 deficiency: Mycobacterial and viral infections without hyper-IgE syndrome. J. Exp. Med. 2015, 212, 1641–1662. [Google Scholar] [CrossRef] [PubMed]
- Turrubiartes-Martínez, E.; Bodega-Mayor, I.; Delgado-Wicke, P.; Molina-Jiménez, F.; Casique-Aguirre, D.; González-Andrade, M.; Rapado, I.; Camós, M.; Díaz-de-Heredia, C.; Barragán, E.; et al. TYK2 Variants in B-Acute Lymphoblastic Leukaemia. Genes 2020, 11, 1434. [Google Scholar] [CrossRef] [PubMed]
- Sanda, T.; Tyner, J.W.; Gutierrez, A.; Ngo, V.N.; Glover, J.; Chang, B.H.; Yost, A.; Ma, W.; Fleischman, A.G.; Zhou, W.; et al. TYK2-STAT1-BCL2 pathway dependence in T-cell acute lymphoblastic leukemia. Cancer Discov. 2013, 3, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Prutsch, N.; Gurnhofer, E.; Suske, T.; Liang, H.C.; Schlederer, M.; Roos, S.; Wu, L.C.; Simonitsch-Klupp, I.; Alvarez-Hernandez, A.; Kornauth, C.; et al. Dependency on the TYK2/STAT1/MCL1 axis in anaplastic large cell lymphoma. Leukemia 2019, 33, 696–709. [Google Scholar] [CrossRef]
- Wöss, K.; Simonović, N.; Strobl, B.; Macho-Maschler, S.; Müller, M. TYK2: An Upstream Kinase of STATs in Cancer. Cancers 2019, 11, 1728. [Google Scholar] [CrossRef]
- Elliott, N.E.; Cleveland, S.M.; Grann, V.; Janik, J.; Waldmann, T.A.; Davé, U.P. FERM domain mutations induce gain of function in JAK3 in adult T-cell leukemia/lymphoma. Blood 2011, 118, 3911–3921. [Google Scholar] [CrossRef]
- Zhao, L.; Ma, Y.; Seemann, J.; Huang, L.J. A regulating role of the JAK2 FERM domain in hyperactivation of JAK2(V617F). Biochem. J. 2010, 426, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Bandapalli, O.R.; Schuessele, S.; Kunz, J.B.; Rausch, T.; Stütz, A.M.; Tal, N.; Geron, I.; Gershman, N.; Izraeli, S.; Eilers, J.; et al. The activating STAT5B N642H mutation is a common abnormality in pediatric T-cell acute lymphoblastic leukemia and confers a higher risk of relapse. Haematologica 2014, 99, e188–e192. [Google Scholar] [CrossRef] [PubMed]
- Andersson, E.; Kuusanmäki, H.; Bortoluzzi, S.; Lagström, S.; Parsons, A.; Rajala, H.; van Adrichem, A.; Eldfors, S.; Olson, T.; Clemente, M.J.; et al. Activating somatic mutations outside the SH2-domain of STAT3 in LGL leukemia. Leukemia 2016, 30, 1204–1208. [Google Scholar] [CrossRef] [PubMed]
- Kontro, M.; Kuusanmäki, H.; Eldfors, S.; Burmeister, T.; Andersson, E.I.; Bruserud, O.; Brümmendorf, T.H.; Edgren, H.; Gjertsen, B.T.; Itälä-Remes, M.; et al. Novel activating STAT5B mutations as putative drivers of T-cell acute lymphoblastic leukemia. Leukemia 2014, 28, 1738–1742. [Google Scholar] [CrossRef] [PubMed]
- Dorritie, K.A.; McCubrey, J.A.; Johnson, D.E. STAT transcription factors in hematopoiesis and leukemogenesis: Opportunities for therapeutic intervention. Leukemia 2014, 28, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, D.; Melão, A.; van Boxtel, R.; Santos, C.I.; Silva, A.; Silva, M.C.; Cardoso, B.A.; Coffer, P.J.; Barata, J.T. STAT5 is essential for IL-7-mediated viability, growth, and proliferation of T-cell acute lymphoblastic leukemia cells. Blood Adv. 2018, 2, 2199–2213. [Google Scholar] [CrossRef] [PubMed]
- Townsend, P.A.; Scarabelli, T.M.; Davidson, S.M.; Knight, R.A.; Latchman, D.S.; Stephanou, A. STAT-1 interacts with p53 to enhance DNA damage-induced apoptosis. J. Biol. Chem. 2004, 279, 5811–5820. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.; Wright, K.L.; Ma, Y.; Wright, G.M.; Huang, M.; Irby, R.; Briggs, J.; Karras, J.; Cress, W.D.; Pardoll, D.; et al. Role of Stat3 in regulating p53 expression and function. Mol. Cell. Biol. 2005, 25, 7432–7440. [Google Scholar] [CrossRef]
- Girardot, M.; Pecquet, C.; Chachoua, I.; Van Hees, J.; Guibert, S.; Ferrant, A.; Knoops, L.; Baxter, E.J.; Beer, P.A.; Giraudier, S.; et al. Persistent STAT5 activation in myeloid neoplasms recruits p53 into gene regulation. Oncogene 2015, 34, 1323–1332. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Aerts, J.L.; Vandenplas, H.; Wang, J.A.; Gorbenko, O.; Chen, J.P.; Giron, P.; Heirman, C.; Goyvaerts, C.; Zacksenhaus, E.; et al. Phosphorylated STAT5 regulates p53 expression via BRCA1/BARD1-NPM1 and MDM2. Cell Death Dis. 2016, 7, e2560. [Google Scholar] [CrossRef] [PubMed]
- Iacobucci, I.; Li, Y.; Roberts, K.G.; Dobson, S.M.; Kim, J.C.; Payne-Turner, D.; Harvey, R.C.; Valentine, M.; McCastlain, K.; Easton, J.; et al. Truncating Erythropoietin Receptor Rearrangements in Acute Lymphoblastic Leukemia. Cancer Cell 2016, 29, 186–200. [Google Scholar] [CrossRef] [PubMed]
- Varghese, L.N.; Defour, J.-P.; Pecquet, C.; Constantinescu, S.N. The Thrombopoietin Receptor: Structural Basis of Traffic and Activation by Ligand, Mutations, Agonists, and Mutated Calreticulin. Front. Endocrinol. 2017, 8, 59. [Google Scholar] [CrossRef] [PubMed]
- Vainchenker, W.; Plo, I.; Marty, C.; Varghese, L.N.; Constantinescu, S.N. The role of the thrombopoietin receptor MPL in myeloproliferative neoplasms: Recent findings and potential therapeutic applications. Expert Rev. Hematol. 2019, 12, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Harvey, R.C.; Mullighan, C.G.; Chen, I.-M.; Wharton, W.; Mikhail, F.M.; Carroll, A.J.; Kang, H.; Liu, W.; Dobbin, K.K.; Smith, M.A.; et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood 2010, 115, 5312–5321. [Google Scholar] [CrossRef] [PubMed]
- Roll, J.D.; Reuther, G.W. CRLF2 and JAK2 in B-Progenitor Acute Lymphoblastic Leukemia: A Novel Association in Oncogenesis. Cancer Res. 2010, 70, 7347–7352. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C.; Kiladjian, J.-J.; Al-Ali, H.K.; Gisslinger, H.; Waltzman, R.; Stalbovskaya, V.; McQuitty, M.; Hunter, D.S.; Levy, R.; Knoops, L.; et al. JAK Inhibition with Ruxolitinib versus Best Available Therapy for Myelofibrosis. N. Engl. J. Med. 2012, 366, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Verstovsek, S.; Mesa, R.A.; Gotlib, J.; Levy, R.S.; Gupta, V.; DiPersio, J.F.; Catalano, J.V.; Deininger, M.; Miller, C.; Silver, R.T.; et al. A Double-Blind, Placebo-Controlled Trial of Ruxolitinib for Myelofibrosis. N. Engl. J. Med. 2012, 366, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Kvasnicka, H.M.; Thiele, J.; Bueso-Ramos, C.E.; Sun, W.; Cortes, J.; Kantarjian, H.M.; Verstovsek, S. Long-term effects of ruxolitinib versus best available therapy on bone marrow fibrosis in patients with myelofibrosis. J. Hematol. Oncol. 2018, 11, 42. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C.N.; Schaap, N.; Vannucchi, A.M.; Kiladjian, J.-J.; Tiu, R.V.; Zachee, P.; Jourdan, E.; Winton, E.; Silver, R.T.; Schouten, H.C.; et al. Janus kinase-2 inhibitor fedratinib in patients with myelofibrosis previously treated with ruxolitinib (JAKARTA-2): A single-arm, open-label, non-randomised, phase 2, multicentre study. Lancet Haematol. 2017, 4, e317–e324. [Google Scholar] [CrossRef]
- Quintás-Cardama, A.; Vaddi, K.; Liu, P.; Manshouri, T.; Li, J.; Scherle, P.A.; Caulder, E.; Wen, X.; Li, Y.; Waeltz, P.; et al. Preclinical characterization of the selective JAK1/2 inhibitor INCB018424: Therapeutic implications for the treatment of myeloproliferative neoplasms. Blood 2010, 115, 3109–3117. [Google Scholar] [CrossRef]
- Wernig, G.; Kharas, M.G.; Okabe, R.; Moore, S.A.; Leeman, D.S.; Cullen, D.E.; Gozo, M.; McDowell, E.P.; Levine, R.L.; Doukas, J.; et al. Efficacy of TG101348, a selective JAK2 inhibitor, in treatment of a murine model of JAK2V617F-induced polycythemia vera. Cancer Cell 2008, 13, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Pardanani, A.; Lasho, T.; Smith, G.; Burns, C.J.; Fantino, E.; Tefferi, A. CYT387, a selective JAK1/JAK2 inhibitor: In vitro assessment of kinase selectivity and preclinical studies using cell lines and primary cells from polycythemia vera patients. Leukemia 2009, 23, 1441–1445. [Google Scholar] [CrossRef]
- Hart, S.; Goh, K.C.; Novotny-Diermayr, V.; Hu, C.Y.; Hentze, H.; Tan, Y.C.; Madan, B.; Amalini, C.; Loh, Y.K.; Ong, L.C.; et al. SB1518, a novel macrocyclic pyrimidine-based JAK2 inhibitor for the treatment of myeloid and lymphoid malignancies. Leukemia 2011, 25, 1751–1759. [Google Scholar] [CrossRef]
- Hexner, E.O.; Serdikoff, C.; Jan, M.; Swider, C.R.; Robinson, C.; Yang, S.; Angeles, T.; Emerson, S.G.; Carroll, M.; Ruggeri, B.; et al. Lestaurtinib (CEP701) is a JAK2 inhibitor that suppresses JAK2/STAT5 signaling and the proliferation of primary erythroid cells from patients with myeloproliferative disorders. Blood 2008, 111, 5663–5671. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Clayton, J.R.; Walgren, R.A.; Zhao, B.; Evans, R.J.; Smith, M.C.; Heinz-Taheny, K.M.; Kreklau, E.L.; Bloem, L.; Pitou, C.; et al. Discovery and characterization of LY2784544, a small-molecule tyrosine kinase inhibitor of JAK2V617F. Blood Cancer J. 2013, 3, e109. [Google Scholar] [CrossRef]
- Nakaya, Y.; Shide, K.; Niwa, T.; Homan, J.; Sugahara, S.; Horio, T.; Kuramoto, K.; Kotera, T.; Shibayama, H.; Hori, K.; et al. Efficacy of NS-018, a potent and selective JAK2/Src inhibitor, in primary cells and mouse models of myeloproliferative neoplasms. Blood Cancer J. 2011, 1, e29. [Google Scholar] [CrossRef] [PubMed]
- Coffey, G.; Betz, A.; DeGuzman, F.; Pak, Y.; Inagaki, M.; Baker, D.C.; Hollenbach, S.J.; Pandey, A.; Sinha, U. The Novel Kinase Inhibitor PRT062070 (Cerdulatinib) Demonstrates Efficacy in Models of Autoimmunity and B Cell Cancer. J. Pharmacol. Exp. Ther. 2014. [Google Scholar] [CrossRef]
- Koppikar, P.; Bhagwat, N.; Kilpivaara, O.; Manshouri, T.; Adli, M.; Hricik, T.; Liu, F.; Saunders, L.M.; Mullally, A.; Abdel-Wahab, O.; et al. Heterodimeric JAK-STAT activation as a mechanism of persistence to JAK2 inhibitor therapy. Nature 2012, 489, 155–159. [Google Scholar] [CrossRef]
- Tvorogov, D.; Thomas, D.; Liau, N.P.D.; Dottore, M.; Barry, E.F.; Lathi, M.; Kan, W.L.; Hercus, T.R.; Stomski, F.; Hughes, T.P.; et al. Accumulation of JAK activation loop phosphorylation is linked to type I JAK inhibitor withdrawal syndrome in myelofibrosis. Sci. Adv. 2018, 4. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-C.; Li, L.S.; Kopp, N.; Montero, J.; Chapuy, B.; Yoda, A.; Christie, A.L.; Liu, H.; Christodoulou, A.; van Bodegom, D.; et al. Activity of the Type II JAK2 Inhibitor CHZ868 in B Cell Acute Lymphoblastic Leukemia. Cancer Cell 2015, 28, 29–41. [Google Scholar] [CrossRef]
- Andraos, R.; Qian, Z.; Bonenfant, D.; Rubert, J.; Vangrevelinghe, E.; Scheufler, C.; Marque, F.; Régnier, C.H.; De Pover, A.; Ryckelynck, H.; et al. Modulation of activation-loop phosphorylation by JAK inhibitors is binding mode dependent. Cancer Discov. 2012. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.C.; Keller, M.D.; Chiu, S.; Koppikar, P.; Guryanova, O.A.; Rapaport, F.; Xu, K.; Manova, K.; Pankov, D.; O’Reilly, R.J.; et al. CHZ868, a Type II JAK2 Inhibitor, Reverses Type I JAK Inhibitor Persistence and Demonstrates Efficacy in Myeloproliferative Neoplasms. Cancer Cell 2015, 28, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Jatiani, S.S.; Cosenza, S.C.; Reddy, M.V.R.; Ha, J.H.; Baker, S.J.; Samanta, A.K.; Olnes, M.J.; Pfannes, L.; Sloand, E.M.; Arlinghaus, R.B.; et al. A Non-ATP-Competitive Dual Inhibitor of JAK2 and BCR-ABL Kinases: Elucidation of a Novel Therapeutic Spectrum Based on Substrate Competitive Inhibition. Genes Cancer 2010, 1, 331–345. [Google Scholar] [CrossRef]
- Lipka, D.B.; Hoffmann, L.S.; Heidel, F.; Markova, B.; Blum, M.-C.; Breitenbuecher, F.; Kasper, S.; Kindler, T.; Levine, R.L.; Huber, C.; et al. LS104, a non-ATP-competitive small-molecule inhibitor of JAK2, is potently inducing apoptosis in JAK2V617F-positive cells. Mol. Cancer Ther. 2008, 7, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Liosi, M.-E.; Krimmer, S.G.; Newton, A.S.; Dawson, T.K.; Puleo, D.E.; Cutrona, K.J.; Suzuki, Y.; Schlessinger, J.; Jorgensen, W.L. Selective Janus Kinase 2 (JAK2) Pseudokinase Ligands with a Diaminotriazole Core. J. Med. Chem. 2020, 63, 5324–5340. [Google Scholar] [CrossRef]
- Puleo, D.E.; Kucera, K.; Hammarén, H.M.; Ungureanu, D.; Newton, A.S.; Silvennoinen, O.; Jorgensen, W.L.; Schlessinger, J. Identification and Characterization of JAK2 Pseudokinase Domain Small Molecule Binders. ACS Med. Chem. Lett. 2017, 8, 618–621. [Google Scholar] [CrossRef]
- McNally, R.; Li, Q.; Li, K.; Dekker, C.; Vangrevelinghe, E.; Jones, M.; Chène, P.; Machauer, R.; Radimerski, T.; Eck, M.J. Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain. ACS Chem. Biol. 2019, 14, 587–593. [Google Scholar] [CrossRef]
- Wrobleski, S.T.; Moslin, R.; Lin, S.; Zhang, Y.; Spergel, S.; Kempson, J.; Tokarski, J.S.; Strnad, J.; Zupa-Fernandez, A.; Cheng, L.; et al. Highly Selective Inhibition of Tyrosine Kinase 2 (TYK2) for the Treatment of Autoimmune Diseases: Discovery of the Allosteric Inhibitor BMS-986165. J. Med. Chem. 2019, 62, 8973–8995. [Google Scholar] [CrossRef]
- Papp, K.; Gordon, K.; Thaçi, D.; Morita, A.; Gooderham, M.; Foley, P.; Girgis, I.G.; Kundu, S.; Banerjee, S. Phase 2 Trial of Selective Tyrosine Kinase 2 Inhibition in Psoriasis. N. Engl. J. Med. 2018, 379, 1313–1321. [Google Scholar] [CrossRef]
- Akahane, K.; Li, Z.; Etchin, J.; Berezovskaya, A.; Gjini, E.; Masse, C.E.; Miao, W.; Rocnik, J.; Kapeller, R.; Greenwood, J.R.; et al. Anti-leukaemic activity of the TYK2 selective inhibitor NDI-031301 in T-cell acute lymphoblastic leukaemia. Br. J. Haematol. 2017, 177, 271–282. [Google Scholar] [CrossRef]
- Degryse, S.; de Bock, C.E.; Cox, L.; Demeyer, S.; Gielen, O.; Mentens, N.; Jacobs, K.; Geerdens, E.; Gianfelici, V.; Hulselmans, G.; et al. JAK3 mutants transform hematopoietic cells through JAK1 activation, causing T-cell acute lymphoblastic leukemia in a mouse model. Blood 2014, 124, 3092–3100. [Google Scholar] [CrossRef]
- Crisà, E.; Cilloni, D.; Elli, E.M.; Martinelli, V.; Palumbo, G.A.; Pugliese, N.; Beggiato, E.; Frairia, C.; Cerrano, M.; Lanzarone, G.; et al. The use of erythropoiesis-stimulating agents is safe and effective in the management of anaemia in myelofibrosis patients treated with ruxolitinib. Br. J. Haematol. 2018, 182, 701–704. [Google Scholar] [CrossRef] [PubMed]
- McMullin, M.F.; Harrison, C.N.; Niederwieser, D.; Demuynck, H.; Jäkel, N.; Gopalakrishna, P.; McQuitty, M.; Stalbovskaya, V.; Recher, C.; Theunissen, K.; et al. The use of erythropoiesis-stimulating agents with ruxolitinib in patients with myelofibrosis in COMFORT-II: An open-label, phase 3 study assessing efficacy and safety of ruxolitinib versus best available therapy in the treatment of myelofibrosis. Exp. Hematol. Oncol. 2015, 4, 26. [Google Scholar] [CrossRef] [PubMed]
- Gowin, K.; Kosiorek, H.; Dueck, A.; Mascarenhas, J.; Hoffman, R.; Reeder, C.; Camoriano, J.; Tibes, R.; Gano, K.; Palmer, J.; et al. Multicenter phase 2 study of combination therapy with ruxolitinib and danazol in patients with myelofibrosis. Leuk. Res. 2017, 60, 31–35. [Google Scholar] [CrossRef]
- Duan, M.; Zhou, D. Improvement of the hematologic toxicities of ruxolitinib in patients with MPN-associated myelofibrosis using a combination of thalidomide, stanozolol and prednisone. Hematology 2019, 24, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.J. Azacitidine: A Review in Myelodysplastic Syndromes and Acute Myeloid Leukaemia. Drugs 2016, 76, 889–900. [Google Scholar] [CrossRef] [PubMed]
- Masarova, L.; Verstovsek, S.; Hidalgo-Lopez, J.E.; Pemmaraju, N.; Bose, P.; Estrov, Z.; Jabbour, E.J.; Ravandi-Kashani, F.; Takahashi, K.; Cortes, J.E.; et al. A phase 2 study of ruxolitinib in combination with azacitidine in patients with myelofibrosis. Blood 2018, 132, 1664–1674. [Google Scholar] [CrossRef]
- DeAngelo, D.J.; Mesa, R.A.; Fiskus, W.; Tefferi, A.; Paley, C.; Wadleigh, M.; Ritchie, E.K.; Snyder, D.S.; Begna, K.; Ganguly, S.; et al. Phase II trial of panobinostat, an oral pan-deacetylase inhibitor in patients with primary myelofibrosis, post–essential thrombocythaemia, and post–polycythaemia vera myelofibrosis. Br. J. Haematol. 2013, 162, 326–335. [Google Scholar] [CrossRef]
- Evrot, E.; Ebel, N.; Romanet, V.; Roelli, C.; Andraos, R.; Qian, Z.; Dölemeyer, A.; Dammassa, E.; Sterker, D.; Cozens, R.; et al. JAK1/2 and Pan-Deacetylase Inhibitor Combination Therapy Yields Improved Efficacy in Preclinical Mouse Models of JAK2V617F-Driven Disease. Clin. Cancer Res. 2013, 19, 6230–6241. [Google Scholar] [CrossRef]
- Mascarenhas, J.; Marcellino, B.K.; Lu, M.; Kremyanskaya, M.; Fabris, F.; Sandy, L.; Mehrotra, M.; Houldsworth, J.; Najfeld, V.; El Jamal, S.; et al. A phase I study of panobinostat and ruxolitinib in patients with primary myelofibrosis (PMF) and post--polycythemia vera/essential thrombocythemia myelofibrosis (post--PV/ET MF). Leuk. Res. 2020, 88, 106272. [Google Scholar] [CrossRef]
- t Ribrag, V.; Harrison, C.N.; Heidel, F.H.; Kiladjian, J.-J.; Acharyya, S.; Mu, S.; Liu, T.; Williams, D.; Giles, F.J.; Conneally, E.; et al. A Phase 1b, Dose-Finding Study of Ruxolitinib Plus Panobinostat in Patients with Primary Myelofibrosis (PMF), Post–Polycythemia Vera MF (PPV-MF), Or Post–Essential Thrombocythemia MF (PET-MF): Identification of the Recommended Phase 2 Dose. Blood 2013, 122, 4045. [Google Scholar] [CrossRef]
- Hamilton, A.; Helgason, G.V.; Schemionek, M.; Zhang, B.; Myssina, S.; Allan, E.K.; Nicolini, F.E.; Müller-Tidow, C.; Bhatia, R.; Brunton, V.G.; et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood 2012, 119, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Corbin, A.S.; Agarwal, A.; Loriaux, M.; Cortes, J.; Deininger, M.W.; Druker, B.J. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J. Clin. Investig. 2011, 121, 396–409. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Wolff, N.; Li, L.; Zhang, S.; Ilaria, R.L., Jr. STAT5 signaling is required for the efficient induction and maintenance of CML in mice. Blood 2006, 107, 4917–4925. [Google Scholar] [CrossRef] [PubMed]
- Kuepper, M.K.; Bütow, M.; Herrmann, O.; Ziemons, J.; Chatain, N.; Maurer, A.; Kirschner, M.; Maié, T.; Costa, I.G.; Eschweiler, J.; et al. Stem cell persistence in CML is mediated by extrinsically activated JAK1-STAT3 signaling. Leukemia 2019, 33, 1964–1977. [Google Scholar] [CrossRef]
- Gallipoli, P.; Cook, A.; Rhodes, S.; Hopcroft, L.; Wheadon, H.; Whetton, A.D.; Jørgensen, H.G.; Bhatia, R.; Holyoake, T.L. JAK2/STAT5 inhibition by nilotinib with ruxolitinib contributes to the elimination of CML CD34+ cells in vitro and in vivo. Blood 2014, 124, 1492–1501. [Google Scholar] [CrossRef]
- Yokoyama, N.; Reich, N.C.; Todd Miller, W. Determinants for the interaction between Janus kinase 2 and protein phosphatase 2A. Arch. Biochem. Biophys. 2003, 417, 87–95. [Google Scholar] [CrossRef]
- Mascarenhas, J.; Kremyanskaya, M.; Hoffman, R.; Bose, P.; Talpaz, M.; Harrison, C.N.; Gupta, V.; Leber, B.; Sirhan, S.; Kabir, S.; et al. MANIFEST, a Phase 2 Study of CPI-0610, a Bromodomain and Extraterminal Domain Inhibitor (BETi), As Monotherapy or “Add-on” to Ruxolitinib, in Patients with Refractory or Intolerant Advanced Myelofibrosis. Blood 2019, 134, 670. [Google Scholar] [CrossRef]
- Harrison, C.N.; Garcia, J.S.; Mesa, R.A.; Somervaille, T.C.P.; Komrokji, R.S.; Pemmaraju, N.; Jamieson, C.; Papadantonakis, N.; Foran, J.M.; O’Connell, C.L.; et al. Results from a Phase 2 Study of Navitoclax in Combination with Ruxolitinib in Patients with Primary or Secondary Myelofibrosis. Blood 2019, 134, 671. [Google Scholar] [CrossRef]


| Inhibitor | IC50 (nM) | Condition | Phase | Ref. | |||
|---|---|---|---|---|---|---|---|
| JAK1 | JAK2 | JAK3 | TYK2 | ||||
| Ruxolitinib | 3.3 | 2.8 | 428 | 19 | MF, PV | Approved | [114] |
| CMML, CLL, SLL, ATL, AML, | Phase 2 | ||||||
| Fedratinib | 105 | 3 | 1002 | 405 | MF | Approved | [115] |
| Momelotinib | 11 | 18 | 155 | nd | MF | Phase 3 | [116] |
| Pacritinib | 1280 | 23 | 520 | 50 | MF | Phase 3 | [117] |
| Lestaurtinib | nd | 1 | 3 | nd | AML, MF, PV, ET | Phase 2 * | [118] |
| Gandotinib | 19.8 | 2.5 | 48 | 44 | PV, ET, MF | Phase 2 * | [119] |
| Ilginatinib | 33 | 0.72 | 39 | 22 | MF | Phase 2 * | [120] |
| Cerdulatinib | 12 | 6 | 8 | 0.5 | CLL, SLL, NHS | Phase 2 | [121] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raivola, J.; Haikarainen, T.; Abraham, B.G.; Silvennoinen, O. Janus Kinases in Leukemia. Cancers 2021, 13, 800. https://doi.org/10.3390/cancers13040800
Raivola J, Haikarainen T, Abraham BG, Silvennoinen O. Janus Kinases in Leukemia. Cancers. 2021; 13(4):800. https://doi.org/10.3390/cancers13040800
Chicago/Turabian StyleRaivola, Juuli, Teemu Haikarainen, Bobin George Abraham, and Olli Silvennoinen. 2021. "Janus Kinases in Leukemia" Cancers 13, no. 4: 800. https://doi.org/10.3390/cancers13040800
APA StyleRaivola, J., Haikarainen, T., Abraham, B. G., & Silvennoinen, O. (2021). Janus Kinases in Leukemia. Cancers, 13(4), 800. https://doi.org/10.3390/cancers13040800







