Kaposi’s Sarcoma-Associated Herpesvirus, the Etiological Agent of All Epidemiological Forms of Kaposi’s Sarcoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Classification
3. Structure
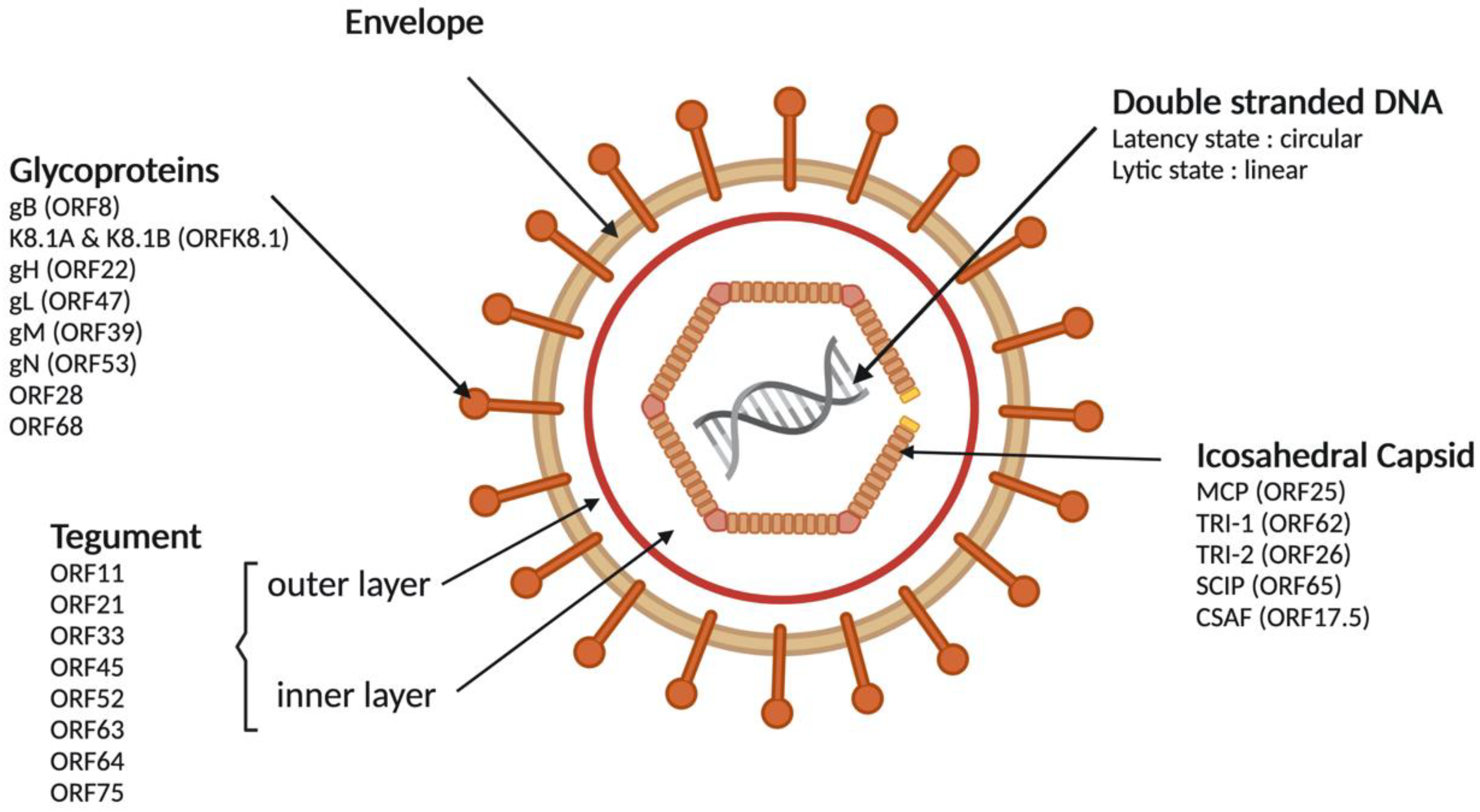
3.1. Viral Particle
3.2. Viral Genome
4. Viral Replication
4.1. Entry in the Cell
4.2. KSHV Latent Cycle
4.3. KSHV Lytic Cycle
5. Epidemiology
5.1. KSHV Seroprevalence and Transmission
5.1.1. Transmission in Countries with Low Seroprevalence
5.1.2. Transmission in Endemic Regions
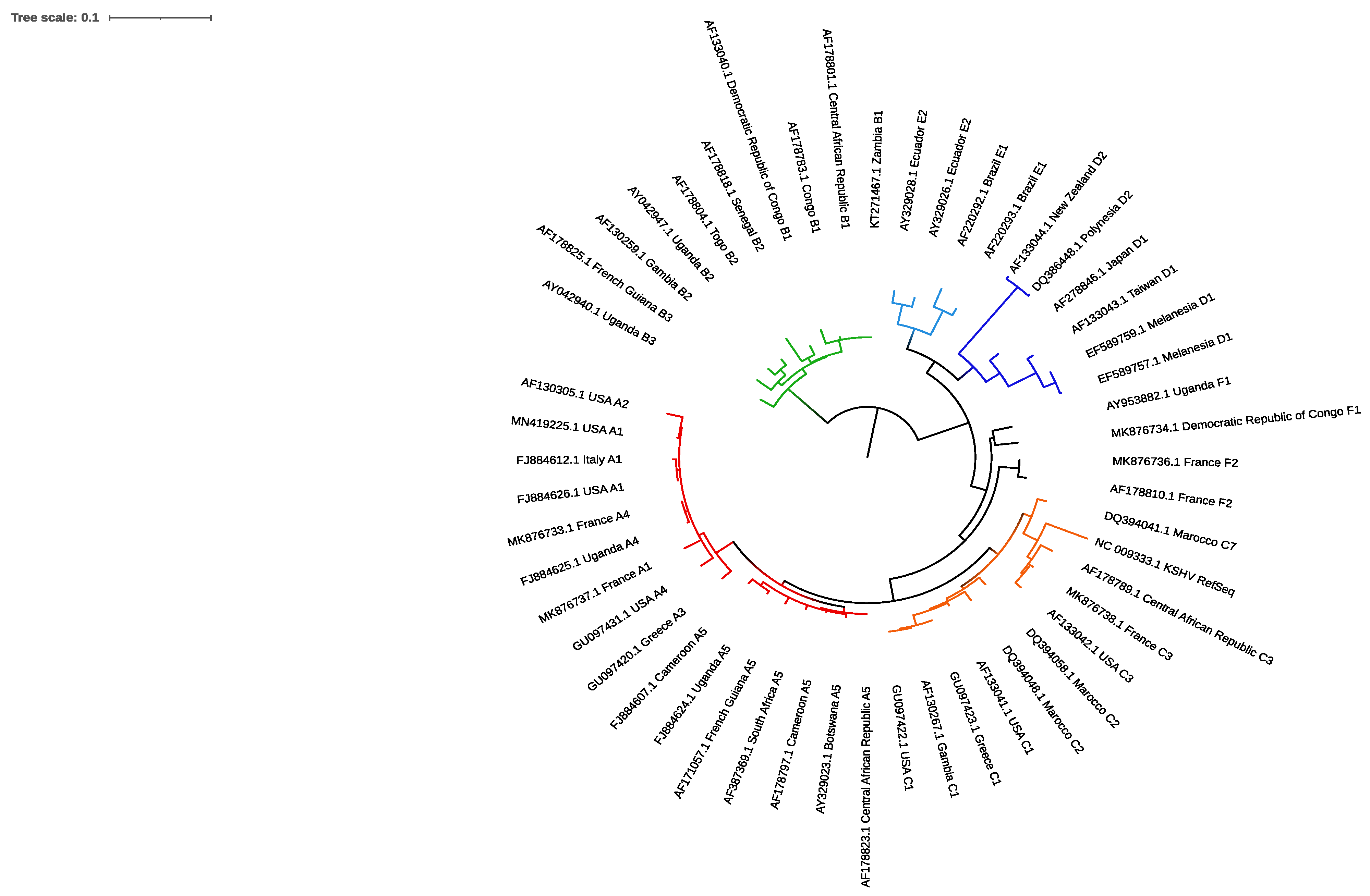
5.2. Molecular Epidemiology
5.2.1. ORF-K1
5.2.2. Other Genes
5.2.3. Molecular Diversity and Pathogenicity
6. KSHV Oncogenesis and Kaposi’s Sarcoma
6.1. KSHV, Cells Tropism and Risk Factors
6.2. KSHV Latency Proteins and Oncogenesis
6.3. Lytic Proteins and Oncogenesis
7. Rationale and Feasibility of KSHV Vaccine
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Chang, Y.; Cesarman, E.; Pessin, M.S.; Lee, F.; Culpepper, J.; Knowles, D.M.; Moore, P.S. Identification of Herpesvirus-like DNA Sequences in AIDS-Associated Kaposi’s Sarcoma. Science 1994, 266, 1865–1869. [Google Scholar] [CrossRef]
- Cesarman, E.; Chang, Y.; Moore, P.S.; Said, J.W.; Knowles, D.M. Kaposi’s Sarcoma-Associated Herpesvirus-like DNA Sequences in AIDS-Related Body-Cavity-Based Lymphomas. N. Engl. J. Med. 1995, 332, 1186–1191. [Google Scholar] [CrossRef]
- Soulier, J.; Grollet, L.; Oksenhendler, E.; Cacoub, P.; Cazals-Hatem, D.; Babinet, P.; d’Agay, M.F.; Clauvel, J.P.; Raphael, M.; Degos, L. Kaposi’s Sarcoma-Associated Herpesvirus-like DNA Sequences in Multicentric Castleman’s Disease. Blood 1995, 86, 1276–1280. [Google Scholar] [CrossRef]
- Moore, P.S.; Chang, Y. Molecular Virology of Kaposi’s Sarcoma-Associated Herpesvirus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 499–516. [Google Scholar] [CrossRef][Green Version]
- Russo, J.J.; Bohenzky, R.A.; Chien, M.C.; Chen, J.; Yan, M.; Maddalena, D.; Parry, J.P.; Peruzzi, D.; Edelman, I.S.; Chang, Y.; et al. Nucleotide Sequence of the Kaposi Sarcoma-Associated Herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 1996, 93, 14862–14867. [Google Scholar] [CrossRef]
- Melendez, L.V.; Daniel, M.D.; Hunt, R.D.; Garcia, F.G. An Apparently New Herpesvirus from Primary Kidney Cultures of the Squirrel Monkey (Saimiri Sciureus). Lab. Anim. Care 1968, 18, 374–381. [Google Scholar]
- Melendez, L.V.; Hunt, R.D.; King, N.W.; Barahona, H.H.; Daniel, M.D.; Fraser, C.E.; Garcia, F.G. Herpesvirus Ateles, a New Lymphoma Virus of Monkeys. Nat. New Biol. 1972, 235, 182–184. [Google Scholar] [CrossRef] [PubMed]
- Desrosiers, R.C.; Sasseville, V.G.; Czajak, S.C.; Zhang, X.; Mansfield, K.G.; Kaur, A.; Johnson, R.P.; Lackner, A.A.; Jung, J.U. A Herpesvirus of Rhesus Monkeys Related to the Human Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 1997, 71, 9764–9769. [Google Scholar] [CrossRef] [PubMed]
- Schultz, E.R.; Rankin, G.W.; Blanc, M.P.; Raden, B.W.; Tsai, C.C.; Rose, T.M. Characterization of Two Divergent Lineages of Macaque Rhadinoviruses Related to Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 2000, 74, 4919–4928. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bosch, M.L.; Strand, K.B.; Rose, T.M. Gammaherpesvirus Sequence Comparisons. J. Virol. 1998, 72, 8458–8459. [Google Scholar] [CrossRef] [PubMed]
- Alexander, L.; Denekamp, L.; Knapp, A.; Auerbach, M.R.; Damania, B.; Desrosiers, R.C. The Primary Sequence of Rhesus Monkey Rhadinovirus Isolate 26-95: Sequence Similarities to Kaposi’s Sarcoma-Associated Herpesvirus and Rhesus Monkey Rhadinovirus Isolate 17577. J. Virol. 2000, 74, 3388–3398. [Google Scholar] [CrossRef] [PubMed]
- Greensill, J.; Sheldon, J.A.; Renwick, N.M.; Beer, B.E.; Norley, S.; Goudsmit, J.; Schulz, T.F. Two Distinct Gamma-2 Herpesviruses in African Green Monkeys: A Second Gamma-2 Herpesvirus Lineage among Old World Primates? J. Virol. 2000, 74, 1572–1577. [Google Scholar] [CrossRef] [PubMed]
- Nealon, K.; Newcomb, W.W.; Pray, T.R.; Craik, C.S.; Brown, J.C.; Kedes, D.H. Lytic Replication of Kaposi’s Sarcoma-Associated Herpesvirus Results in the Formation of Multiple Capsid Species: Isolation and Molecular Characterization of A, B, and C Capsids from a Gammaherpesvirus. J. Virol. 2001, 75, 2866–2878. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhu, F.X.; Chong, J.M.; Wu, L.; Yuan, Y. Virion Proteins of Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 2005, 79, 800–811. [Google Scholar] [CrossRef]
- Yan, L.; Majerciak, V.; Zheng, Z.-M.; Lan, K. Towards Better Understanding of KSHV Life Cycle: From Transcription and Posttranscriptional Regulations to Pathogenesis. Virol. Sin. 2019, 34, 135–161. [Google Scholar] [CrossRef]
- Sathish, N.; Wang, X.; Yuan, Y. Tegument Proteins of Kaposi’s Sarcoma-Associated Herpesvirus and Related Gamma-Herpesviruses. Front. Microbiol. 2012, 3, 98. [Google Scholar] [CrossRef]
- Dollery, S.J. Towards Understanding KSHV Fusion and Entry. Viruses 2019, 11, 1073. [Google Scholar] [CrossRef]
- Akula, S.M.; Pramod, N.P.; Wang, F.Z.; Chandran, B. Integrin Alpha3beta1 (CD 49c/29) Is a Cellular Receptor for Kaposi’s Sarcoma-Associated Herpesvirus (KSHV/HHV-8) Entry into the Target Cells. Cell 2002, 108, 407–419. [Google Scholar] [CrossRef]
- Spear, P.G.; Longnecker, R. Herpesvirus Entry: An Update. J. Virol. 2003, 77, 10179–10185. [Google Scholar] [CrossRef]
- Birkmann, A.; Mahr, K.; Ensser, A.; Yağuboğlu, S.; Titgemeyer, F.; Fleckenstein, B.; Neipel, F. Cell Surface Heparan Sulfate Is a Receptor for Human Herpesvirus 8 and Interacts with Envelope Glycoprotein K8.1. J. Virol. 2001, 75, 11583–11593. [Google Scholar] [CrossRef]
- Muniraju, M.; Mutsvunguma, L.Z.; Foley, J.; Escalante, G.M.; Rodriguez, E.; Nabiee, R.; Totonchy, J.; Mulama, D.H.; Nyagol, J.; Wussow, F.; et al. Kaposi Sarcoma-Associated Herpesvirus Glycoprotein H Is Indispensable for Infection of Epithelial, Endothelial, and Fibroblast Cell Types. J. Virol. 2019, 93, e00630-19. [Google Scholar] [CrossRef]
- Cesarman, E.; Damania, B.; Krown, S.E.; Martin, J.; Bower, M.; Whitby, D. Kaposi Sarcoma. Nat. Rev. Dis. Primer 2019, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Mesri, E.A.; Feitelson, M.A.; Munger, K. Human Viral Oncogenesis: A Cancer Hallmarks Analysis. Cell Host Microbe 2014, 15, 266–282. [Google Scholar] [CrossRef] [PubMed]
- Abere, B.; Li, J.; Zhou, H.; Toptan, T.; Moore, P.S.; Chang, Y. Kaposi’s Sarcoma-Associated Herpesvirus-Encoded CircRNAs Are Expressed in Infected Tumor Tissues and Are Incorporated into Virions. MBio 2020, 11, e03027-19. [Google Scholar] [CrossRef]
- Chakraborty, S.; Veettil, M.V.; Chandran, B. Kaposi’s Sarcoma Associated Herpesvirus Entry into Target Cells. Front. Microbiol. 2012, 3, 6. [Google Scholar] [CrossRef]
- Akula, S.M.; Wang, F.Z.; Vieira, J.; Chandran, B. Human Herpesvirus 8 Interaction with Target Cells Involves Heparan Sulfate. Virology 2001, 282, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Akula, S.M.; Pramod, N.P.; Wang, F.Z.; Chandran, B. Human Herpesvirus 8 Envelope-Associated Glycoprotein B Interacts with Heparan Sulfate-like Moieties. Virology 2001, 284, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Dollery, S.J.; Santiago-Crespo, R.J.; Chatterjee, D.; Berger, E.A. Glycoprotein K8.1A of Kaposi’s Sarcoma-Associated Herpesvirus Is a Critical B Cell Tropism Determinant Independent of Its Heparan Sulfate Binding Activity. J. Virol. 2019, 93, e01876-18. [Google Scholar] [CrossRef]
- Akula, S.M.; Naranatt, P.P.; Walia, N.-S.; Wang, F.-Z.; Fegley, B.; Chandran, B. Kaposi’s Sarcoma-Associated Herpesvirus (Human Herpesvirus 8) Infection of Human Fibroblast Cells Occurs through Endocytosis. J. Virol. 2003, 77, 7978–7990. [Google Scholar] [CrossRef]
- Inoue, N.; Winter, J.; Lal, R.B.; Offermann, M.K.; Koyano, S. Characterization of Entry Mechanisms of Human Herpesvirus 8 by Using an Rta-Dependent Reporter Cell Line. J. Virol. 2003, 77, 8147–8152. [Google Scholar] [CrossRef]
- Kerur, N.; Veettil, M.V.; Sharma-Walia, N.; Sadagopan, S.; Bottero, V.; Paul, A.G.; Chandran, B. Characterization of Entry and Infection of Monocytic THP-1 Cells by Kaposi’s Sarcoma Associated Herpesvirus (KSHV): Role of Heparan Sulfate, DC-SIGN, Integrins and Signaling. Virology 2010, 406, 103–116. [Google Scholar] [CrossRef]
- Kumar, B.; Chandran, B. KSHV Entry and Trafficking in Target Cells-Hijacking of Cell Signal Pathways, Actin and Membrane Dynamics. Viruses 2016, 8, 305. [Google Scholar] [CrossRef]
- Greene, W.; Gao, S.-J. Actin Dynamics Regulate Multiple Endosomal Steps during Kaposi’s Sarcoma-Associated Herpesvirus Entry and Trafficking in Endothelial Cells. PLoS Pathog. 2009, 5, e1000512. [Google Scholar] [CrossRef] [PubMed]
- Chandran, B. Early Events in Kaposi’s Sarcoma-Associated Herpesvirus Infection of Target Cells. J. Virol. 2010, 84, 2188–2199. [Google Scholar] [CrossRef] [PubMed]
- Uppal, T.; Banerjee, S.; Sun, Z.; Verma, S.C.; Robertson, E.S. KSHV LANA—The Master Regulator of KSHV Latency. Viruses 2014, 6, 4961–4998. [Google Scholar] [CrossRef] [PubMed]
- Gottwein, E.; Corcoran, D.L.; Mukherjee, N.; Skalsky, R.L.; Hafner, M.; Nusbaum, J.D.; Shamulailatpam, P.; Love, C.L.; Dave, S.S.; Tuschl, T.; et al. Viral MicroRNA Targetome of KSHV-Infected Primary Effusion Lymphoma Cell Lines. Cell Host Microbe 2011, 10, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Li, W.; Qin, J.; Lu, C.; Fan, W. Kaposi’s Sarcoma-Associated Herpesvirus (KSHV)-Encoded MicroRNAs Promote Matrix Metalloproteinases (MMPs) Expression and pro-Angiogenic Cytokine Secretion in Endothelial Cells. J. Med. Virol. 2017, 89, 1274–1280. [Google Scholar] [CrossRef]
- Qin, J.; Li, W.; Gao, S.-J.; Lu, C. KSHV MicroRNAs: Tricks of the Devil. Trends Microbiol. 2017, 25, 648–661. [Google Scholar] [CrossRef]
- Lin, X.; Liang, D.; He, Z.; Deng, Q.; Robertson, E.S.; Lan, K. MiR-K12-7-5p Encoded by Kaposi’s Sarcoma-Associated Herpesvirus Stabilizes the Latent State by Targeting Viral ORF50/RTA. PLoS ONE 2011, 6, e16224. [Google Scholar] [CrossRef] [PubMed]
- Hussein, H.A.M.; Alfhili, M.A.; Pakala, P.; Simon, S.; Hussain, J.; McCubrey, J.A.; Akula, S.M. MiRNAs and Their Roles in KSHV Pathogenesis. Virus Res. 2019, 266, 15–24. [Google Scholar] [CrossRef]
- Gallaher, A.M.; Das, S.; Xiao, Z.; Andresson, T.; Kieffer-Kwon, P.; Happel, C.; Ziegelbauer, J. Proteomic Screening of Human Targets of Viral MicroRNAs Reveals Functions Associated with Immune Evasion and Angiogenesis. PLoS Pathog. 2013, 9, e1003584. [Google Scholar] [CrossRef]
- Samols, M.A.; Skalsky, R.L.; Maldonado, A.M.; Riva, A.; Lopez, M.C.; Baker, H.V.; Renne, R. Identification of Cellular Genes Targeted by KSHV-Encoded MicroRNAs. PLoS Pathog. 2007, 3, e65. [Google Scholar] [CrossRef]
- Davis, D.A.; Rinderknecht, A.S.; Zoeteweij, J.P.; Aoki, Y.; Read-Connole, E.L.; Tosato, G.; Blauvelt, A.; Yarchoan, R. Hypoxia Induces Lytic Replication of Kaposi Sarcoma-Associated Herpesvirus. Blood 2001, 97, 3244–3250. [Google Scholar] [CrossRef]
- Ye, F.; Zhou, F.; Bedolla, R.G.; Jones, T.; Lei, X.; Kang, T.; Guadalupe, M.; Gao, S.-J. Reactive Oxygen Species Hydrogen Peroxide Mediates Kaposi’s Sarcoma-Associated Herpesvirus Reactivation from Latency. PLoS Pathog. 2011, 7, e1002054. [Google Scholar] [CrossRef]
- Yu, Y.; Black, J.B.; Goldsmith, C.S.; Browning, P.J.; Bhalla, K.; Offermann, M.K. Induction of Human Herpesvirus-8 DNA Replication and Transcription by Butyrate and TPA in BCBL-1 Cells. J. Gen. Virol. 1999, 80 Pt 1, 83–90. [Google Scholar] [CrossRef]
- Gradoville, L.; Gerlach, J.; Grogan, E.; Shedd, D.; Nikiforow, S.; Metroka, C.; Miller, G. Kaposi’s Sarcoma-Associated Herpesvirus Open Reading Frame 50/Rta Protein Activates the Entire Viral Lytic Cycle in the HH-B2 Primary Effusion Lymphoma Cell Line. J. Virol. 2000, 74, 6207–6212. [Google Scholar] [CrossRef] [PubMed]
- Guito, J.; Lukac, D.M. KSHV Rta Promoter Specification and Viral Reactivation. Front. Microbiol. 2012, 3, 30. [Google Scholar] [CrossRef] [PubMed]
- Aneja, K.K.; Yuan, Y. Reactivation and Lytic Replication of Kaposi’s Sarcoma-Associated Herpesvirus: An Update. Front. Microbiol. 2017, 8, 613. [Google Scholar] [CrossRef] [PubMed]
- DeCotiis, J.L.; Lukac, D.M. KSHV and the Role of Notch Receptor Dysregulation in Disease Progression. Pathogens 2017, 6, 34. [Google Scholar] [CrossRef] [PubMed]
- Conrad, N.K. New Insights into the Expression and Functions of the Kaposi’s Sarcoma-Associated Herpesvirus Long Noncoding PAN RNA. Virus Res. 2016, 212, 53–63. [Google Scholar] [CrossRef]
- Rossetto, C.C.; Tarrant-Elorza, M.; Verma, S.; Purushothaman, P.; Pari, G.S. Regulation of Viral and Cellular Gene Expression by Kaposi’s Sarcoma-Associated Herpesvirus Polyadenylated Nuclear RNA. J. Virol. 2013, 87, 5540–5553. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, S.R.; Damania, B. The Viral Interferon Regulatory Factors of KSHV: Immunosuppressors or Oncogenes? Front. Immunol. 2011, 2, 19. [Google Scholar] [CrossRef]
- Abere, B.; Mamo, T.M.; Hartmann, S.; Samarina, N.; Hage, E.; Rückert, J.; Hotop, S.-K.; Büsche, G.; Schulz, T.F. The Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) Non-Structural Membrane Protein K15 Is Required for Viral Lytic Replication and May Represent a Therapeutic Target. PLoS Pathog. 2017, 13, e1006639. [Google Scholar] [CrossRef]
- Lei, X.; Bai, Z.; Ye, F.; Xie, J.; Kim, C.-G.; Huang, Y.; Gao, S.-J. Regulation of NF-KappaB Inhibitor IkappaBalpha and Viral Replication by a KSHV MicroRNA. Nat. Cell Biol. 2010, 12, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Mesri, E.A.; Cesarman, E.; Boshoff, C. Kaposi’s Sarcoma and Its Associated Herpesvirus. Nat. Rev. Cancer 2010, 10, 707–719. [Google Scholar] [CrossRef]
- Wang, L.; Dittmer, D.P.; Tomlinson, C.C.; Fakhari, F.D.; Damania, B. Immortalization of Primary Endothelial Cells by the K1 Protein of Kaposi’s Sarcoma-Associated Herpesvirus. Cancer Res. 2006, 66, 3658–3666. [Google Scholar] [CrossRef] [PubMed]
- Ikoma, M.; Gantt, S.; Casper, C.; Ogata, Y.; Zhang, Q.; Basom, R.; Dyen, M.R.; Rose, T.M.; Barcy, S. KSHV Oral Shedding and Plasma Viremia Result in Significant Changes in the Extracellular Tumorigenic MiRNA Expression Profile in Individuals Infected with the Malaria Parasite. PLoS ONE 2018, 13, e0192659. [Google Scholar] [CrossRef]
- Nalwoga, A.; Cose, S.; Nash, S.; Miley, W.; Asiki, G.; Kusemererwa, S.; Yarchoan, R.; Labo, N.; Whitby, D.; Newton, R. Relationship Between Anemia, Malaria Coinfection, and Kaposi Sarcoma-Associated Herpesvirus Seropositivity in a Population-Based Study in Rural Uganda. J. Infect. Dis. 2018, 218, 1061–1065. [Google Scholar] [CrossRef]
- Newton, R.; Labo, N.; Wakeham, K.; Marshall, V.; Roshan, R.; Nalwoga, A.; Sebina, I.; Muhangi, L.; Webb, E.L.; Miley, W.; et al. Determinants of Gammaherpesvirus Shedding in Saliva Among Ugandan Children and Their Mothers. J. Infect. Dis. 2018, 218, 892–900. [Google Scholar] [CrossRef]
- Simonart, T. Role of Environmental Factors in the Pathogenesis of Classic and African-Endemic Kaposi Sarcoma. Cancer Lett. 2006, 244, 1–7. [Google Scholar] [CrossRef]
- Pelser, C.; Dazzi, C.; Graubard, B.I.; Lauria, C.; Vitale, F.; Goedert, J.J. Risk of Classic Kaposi Sarcoma with Residential Exposure to Volcanic and Related Soils in Sicily. Ann. Epidemiol. 2009, 19, 597–601. [Google Scholar] [CrossRef]
- Ziegler, J.L. Endemic Kaposi’s Sarcoma in Africa and Local Volcanic Soils. Lancet 1993, 342, 1348–1351. [Google Scholar] [CrossRef]
- Simonart, T.; Noel, J.C.; Andrei, G.; Parent, D.; Van Vooren, J.P.; Hermans, P.; Lunardi-Yskandar, Y.; Lambert, C.; Dieye, T.; Farber, C.M.; et al. Iron as a Potential Co-Factor in the Pathogenesis of Kaposi’s Sarcoma? Int. J. Cancer 1998, 78, 720–726. [Google Scholar] [CrossRef]
- Casper, C.; Wald, A.; Pauk, J.; Tabet, S.R.; Corey, L.; Celum, C.L. Correlates of Prevalent and Incident Kaposi’s Sarcoma-Associated Herpesvirus Infection in Men Who Have Sex with Men. J. Infect. Dis. 2002, 185, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Casper, C.; Carrell, D.; Miller, K.G.; Judson, F.D.; Meier, A.S.; Pauk, J.S.; Morrow, R.A.; Corey, L.; Wald, A.; Celum, C. HIV Serodiscordant Sex Partners and the Prevalence of Human Herpesvirus 8 Infection among HIV Negative Men Who Have Sex with Men: Baseline Data from the EXPLORE Study. Sex. Transm. Infect. 2006, 82, 229–235. [Google Scholar] [CrossRef]
- Giuliani, M.; Cordiali-Fei, P.; Castilletti, C.; Di Carlo, A.; Palamara, G.; Boros, S.; Rezza, G. Incidence of Human Herpesvirus 8 (HHV-8) Infection among HIV-Uninfected Individuals at High Risk for Sexually Transmitted Infections. BMC Infect. Dis. 2007, 7, 143. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, S.G.; Hirsch, H.H.; Franceschi, S.; Steffen, I.; Amari, E.B.E.; Mueller, N.J.; Magouras, I.; Biggar, R.J.; Rickenbach, M.; Clifford, G.M.; et al. Kaposi Sarcoma Herpes Virus Antibody Response and Viremia Following Highly Active Antiretroviral Therapy in the Swiss HIV Cohort Study. AIDS 2010, 24, 2245–2252. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-M.; Hung, P.-S.; Lin, C.-W. Seroepidemiology and Phylogenetic Analysis of Human Herpesvirus Type 8 in Injection Drug Users and Men Who Have Sex with Men in Northern Taiwan. J. Int. Med. Res. 2018, 48, 300060518764747. [Google Scholar] [CrossRef]
- Liu, Z.; Fang, Q.; Zuo, J.; Chen, Y.; Minhas, V.; Wood, C.; Zhang, T. Global Epidemiology of Human Herpesvirus 8 in Men Who Have Sex with Men: A Systematic Review and Meta-Analysis. J. Med. Virol. 2017, 90, 582–591. [Google Scholar] [CrossRef]
- Martin, J.N.; Ganem, D.E.; Osmond, D.H.; Page-Shafer, K.A.; Macrae, D.; Kedes, D.H. Sexual Transmission and the Natural History of Human Herpesvirus 8 Infection. N. Engl. J. Med. 1998, 338, 948–954. [Google Scholar] [CrossRef]
- Melbye, M.; Cook, P.M.; Hjalgrim, H.; Begtrup, K.; Simpson, G.R.; Biggar, R.J.; Ebbesen, P.; Schulz, T.F. Risk Factors for Kaposi’s-Sarcoma-Associated Herpesvirus (KSHV/HHV-8) Seropositivity in a Cohort of Homosexual Men, 1981–1996. Int. J. Cancer 1998, 77, 543–548. [Google Scholar] [CrossRef]
- Rohner, E.; Wyss, N.; Heg, Z.; Faralli, Z.; Mbulaiteye, S.M.; Novak, U.; Zwahlen, M.; Egger, M.; Bohlius, J. HIV and Human Herpesvirus 8 Co-Infection across the Globe: Systematic Review and Meta-Analysis. Int. J. Cancer 2016, 138, 45–54. [Google Scholar] [CrossRef]
- Smith, N.A.; Sabin, C.A.; Gopal, R.; Bourboulia, D.; Labbet, W.; Boshoff, C.; Barlow, D.; Band, B.; Peters, B.S.; de Ruiter, A.; et al. Serologic Evidence of Human Herpesvirus 8 Transmission by Homosexual but Not Heterosexual Sex. J. Infect. Dis. 1999, 180, 600–606. [Google Scholar] [CrossRef]
- Pauk, J.; Huang, M.L.; Brodie, S.J.; Wald, A.; Koelle, D.M.; Schacker, T.; Celum, C.; Selke, S.; Corey, L. Mucosal Shedding of Human Herpesvirus 8 in Men. N. Engl. J. Med. 2000, 343, 1369–1377. [Google Scholar] [CrossRef]
- Dukers, N.H.; Renwick, N.; Prins, M.; Geskus, R.B.; Schulz, T.F.; Weverling, G.J.; Coutinho, R.A.; Goudsmit, J. Risk Factors for Human Herpesvirus 8 Seropositivity and Seroconversion in a Cohort of Homosexual Men. Am. J. Epidemiol. 2000, 151, 213–224. [Google Scholar] [CrossRef]
- Casper, C.; Krantz, E.; Selke, S.; Kuntz, S.R.; Wang, J.; Huang, M.-L.; Pauk, J.S.; Corey, L.; Wald, A. Frequent and Asymptomatic Oropharyngeal Shedding of Human Herpesvirus 8 among Immunocompetent Men. J. Infect. Dis. 2007, 195, 30–36. [Google Scholar] [CrossRef]
- Corbellino, M.; Bestetti, G.; Galli, M.; Parravicini, C. Absence of HHV-8 in Prostate and Semen. N. Engl. J. Med. 1996, 335, 1237; author reply 1238–1239. [Google Scholar] [CrossRef] [PubMed]
- Kaspersen, M.D.; Höllsberg, P. Seminal Shedding of Human Herpesviruses. Virol. J. 2013, 10, 226. [Google Scholar] [CrossRef]
- Monini, P.; de Lellis, L.; Fabris, M.; Rigolin, F.; Cassai, E. Kaposi’s Sarcoma-Associated Herpesvirus DNA Sequences in Prostate Tissue and Human Semen. N. Engl. J. Med. 1996, 334, 1168–1172. [Google Scholar] [CrossRef] [PubMed]
- Pellett, P.E.; Spira, T.J.; Bagasra, O.; Boshoff, C.; Corey, L.; de Lellis, L.; Huang, M.L.; Lin, J.C.; Matthews, S.; Monini, P.; et al. Multicenter Comparison of PCR Assays for Detection of Human Herpesvirus 8 DNA in Semen. J. Clin. Microbiol. 1999, 37, 1298–1301. [Google Scholar] [CrossRef]
- Cannon, M.J.; Dollard, S.C.; Black, J.B.; Edlin, B.R.; Hannah, C.; Hogan, S.E.; Patel, M.M.; Jaffe, H.W.; Offermann, M.K.; Spira, T.J.; et al. Risk Factors for Kaposi’s Sarcoma in Men Seropositive for Both Human Herpesvirus 8 and Human Immunodeficiency Virus. AIDS 2003, 17, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Butler, L.M.; Osmond, D.H.; Jones, A.G.; Martin, J.N. Use of Saliva as a Lubricant in Anal Sexual Practices among Homosexual Men. J. Acquir. Immune Defic. Syndr. 2009, 50, 162–167. [Google Scholar] [CrossRef]
- Eltom, M.A.; Mbulaiteye, S.M.; Dada, A.J.; Whitby, D.; Biggar, R.J. Transmission of Human Herpesvirus 8 by Sexual Activity among Adults in Lagos, Nigeria. AIDS 2002, 16, 2473–2478. [Google Scholar] [CrossRef]
- Engels, E.A.; Atkinson, J.O.; Graubard, B.I.; McQuillan, G.M.; Gamache, C.; Mbisa, G.; Cohn, S.; Whitby, D.; Goedert, J.J. Risk Factors for Human Herpesvirus 8 Infection among Adults in the United States and Evidence for Sexual Transmission. J. Infect. Dis. 2007, 196, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Janier, M.; Agbalika, F.; de La Salmonière, P.; Lassau, F.; Lagrange, P.; Morel, P. Human Herpesvirus 8 Seroprevalence in an STD Clinic in Paris: A Study of 512 Patients. Sex. Transm. Dis. 2002, 29, 698–702. [Google Scholar] [CrossRef]
- Malope, B.I.; MacPhail, P.; Mbisa, G.; MacPhail, C.; Stein, L.; Ratshikhopha, E.M.; Ndhlovu, L.; Sitas, F.; Whitby, D. No Evidence of Sexual Transmission of Kaposi’s Sarcoma Herpes Virus in a Heterosexual South African Population. AIDS 2008, 22, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Crum, N.F.; Wallace, M.R.; Stephan, K.; Blazes, D.L.; Aronson, N.; Tasker, S.A.; Thomas, A.G.; Wegner, S.; Casper, C.; Wald, A.; et al. Correlates of Human Herpesvirus-8 Seropositivity among U.S. Military Members Recently Infected with Human Immunodeficiency Virus. Sex. Transm. Dis. 2003, 30, 713–718. [Google Scholar] [CrossRef]
- Renwick, N.; Dukers, N.H.T.M.; Weverling, G.J.; Sheldon, J.A.; Schulz, T.F.; Prins, M.; Coutinho, R.A.; Goudsmit, J. Risk Factors for Human Herpesvirus 8 Infection in a Cohort of Drug Users in the Netherlands, 1985–1996. J. Infect. Dis. 2002, 185, 1808–1812. [Google Scholar] [CrossRef][Green Version]
- Lennette, E.T.; Blackbourn, D.J.; Levy, J.A. Antibodies to Human Herpesvirus Type 8 in the General Population and in Kaposi’s Sarcoma Patients. Lancet 1996, 348, 858–861. [Google Scholar] [CrossRef]
- Shimizu, S.; Katano, H.; Sata, T.; Chen, K.R.; Tagami, H.; Hanabusa, H.; Shimizu, H. Absence of Anti-Human Herpesvirus 8 Antibody in 32 Japanese Hemophiliacs with Advanced HIV Infection. Arch. Dermatol. Res. 2001, 293, 380–381. [Google Scholar] [CrossRef]
- Simpson, G.R.; Schulz, T.F.; Whitby, D.; Cook, P.M.; Boshoff, C.; Rainbow, L.; Howard, M.R.; Gao, S.J.; Bohenzky, R.A.; Simmonds, P.; et al. Prevalence of Kaposi’s Sarcoma Associated Herpesvirus Infection Measured by Antibodies to Recombinant Capsid Protein and Latent Immunofluorescence Antigen. Lancet 1996, 348, 1133–1138. [Google Scholar] [CrossRef]
- Chiereghin, A.; Barozzi, P.; Petrisli, E.; Piccirilli, G.; Gabrielli, L.; Riva, G.; Potenza, L.; Cappelli, G.; De Ruvo, N.; Libri, I.; et al. Multicenter Prospective Study for Laboratory Diagnosis of HHV8 Infection in Solid Organ Donors and Transplant Recipients and Evaluation of the Clinical Impact after Transplantation. Transplantation 2017, 101, 1935–1944. [Google Scholar] [CrossRef]
- Francès, C.; Marcelin, A.G.; Legendre, C.; Chevret, S.; Dussaix, E.; Lejeune, J.; Euvrard, S.; Bigorie, A.; Schulz, T.F.; Agbalika, F.; et al. The Impact of Preexisting or Acquired Kaposi Sarcoma Herpesvirus Infection in Kidney Transplant Recipients on Morbidity and Survival. Am. J. Transplant. Off. J. Am. Soc. Transplant. Am. Soc. Transpl. Surg. 2009, 9, 2580–2586. [Google Scholar] [CrossRef]
- Lebbe, C.; Porcher, R.; Marcelin, A.G.; Agbalika, F.; Dussaix, E.; Samuel, D.; Varnous, S.; Euvrard, S.; Bigorie, A.; Creusvaux, H.; et al. Human Herpesvirus 8 (HHV8) Transmission and Related Morbidity in Organ Recipients. Am. J. Transplant. Off. J. Am. Soc. Transplant. Am. Soc. Transpl. Surg. 2013, 13, 207–213. [Google Scholar] [CrossRef]
- Riva, G.; Barozzi, P.; Quadrelli, C.; Vallerini, D.; Zanetti, E.; Forghieri, F.; Chiereghin, A.; Libri, I.; Maggiore, U.; Buzio, C.; et al. Human Herpesvirus 8 (HHV8) Infection and Related Diseases in Italian Transplant Cohorts. Am. J. Transplant. Off. J. Am. Soc. Transplant. Am. Soc. Transpl. Surg. 2013, 13, 1619–1620. [Google Scholar] [CrossRef] [PubMed]
- Gessain, A.; Mauclère, P.; van Beveren, M.; Plancoulaine, S.; Ayouba, A.; Essame-Oyono, J.L.; Martin, P.M.; de Thé, G. Human Herpesvirus 8 Primary Infection Occurs during Childhood in Cameroon, Central Africa. Int. J. Cancer 1999, 81, 189–192. [Google Scholar] [CrossRef]
- Plancoulaine, S.; Abel, L.; van Beveren, M.; Trégouët, D.A.; Joubert, M.; Tortevoye, P.; de Thé, G.; Gessain, A. Human Herpesvirus 8 Transmission from Mother to Child and between Siblings in an Endemic Population. Lancet 2000, 356, 1062–1065. [Google Scholar] [CrossRef]
- Mayama, S.; Cuevas, L.E.; Sheldon, J.; Omar, O.H.; Smith, D.H.; Okong, P.; Silvel, B.; Hart, C.A.; Schulz, T.F. Prevalence and Transmission of Kaposi’s Sarcoma-Associated Herpesvirus (Human Herpesvirus 8) in Ugandan Children and Adolescents. Int. J. Cancer 1998, 77, 817–820. [Google Scholar] [CrossRef]
- Mbulaiteye, S.; Marshall, V.; Bagni, R.K.; Wang, C.-D.; Mbisa, G.; Bakaki, P.M.; Owor, A.M.; Ndugwa, C.M.; Engels, E.A.; Katongole-Mbidde, E.; et al. Molecular Evidence for Mother-to-Child Transmission of Kaposi Sarcoma-Associated Herpesvirus in Uganda and K1 Gene Evolution within the Host. J. Infect. Dis. 2006, 193, 1250–1257. [Google Scholar] [CrossRef] [PubMed]
- Dedicoat, M.; Newton, R.; Alkharsah, K.R.; Sheldon, J.; Szabados, I.; Ndlovu, B.; Page, T.; Casabonne, D.; Gilks, C.F.; Cassol, S.A.; et al. Mother-to-Child Transmission of Human Herpesvirus-8 in South Africa. J. Infect. Dis. 2004, 190, 1068–1075. [Google Scholar] [CrossRef]
- Butler, L.M.; Were, W.A.; Balinandi, S.; Downing, R.; Dollard, S.; Neilands, T.B.; Gupta, S.; Rutherford, G.W.; Mermin, J. Human Herpesvirus 8 Infection in Children and Adults in a Population-Based Study in Rural Uganda. J. Infect. Dis. 2011, 203, 625–634. [Google Scholar] [CrossRef]
- Romano, R.; Tabacchi, F.; Paganotti, G.M.; Russo, G.; Gramolelli, S.; Marinucci, F.; Ceccherini-Nelli, L.; Coluzzi, M. Evaluation of Bloodsucking Arthropod Bite as Possible Risk Co-Factor in Human Herpesvirus-8 Transmission Route. Parassitologia 2010, 52, 405–410. [Google Scholar]
- Brayfield, B.P.; Kankasa, C.; West, J.T.; Muyanga, J.; Bhat, G.; Klaskala, W.; Mitchell, C.D.; Wood, C. Distribution of Kaposi Sarcoma-Associated Herpesvirus/Human Herpesvirus 8 in Maternal Saliva and Breast Milk in Zambia: Implications for Transmission. J. Infect. Dis. 2004, 189, 2260–2270. [Google Scholar] [CrossRef] [PubMed]
- Campbell, T.B.; Borok, M.; Ndemera, B.; Fiorillo, S.; White, I.E.; Zhang, X.; Machekano, R.N.; Katzenstein, D.; Gwanzura, L. Lack of Evidence for Frequent Heterosexual Transmission of Human Herpesvirus 8 in Zimbabwe. Clin. Infect. Dis. 2009, 48, 1601–1608. [Google Scholar] [CrossRef]
- Shebl, F.M.; Dollard, S.C.; Pfeiffer, R.M.; Biryahwaho, B.; Amin, M.M.; Munuo, S.S.; Hladik, W.; Parsons, R.; Graubard, B.I.; Mbulaiteye, S.M. Human Herpesvirus 8 Seropositivity among Sexually Active Adults in Uganda. PLoS ONE 2011, 6, e21286. [Google Scholar] [CrossRef]
- Hladik, W.; Dollard, S.C.; Mermin, J.; Fowlkes, A.L.; Downing, R.; Amin, M.M.; Banage, F.; Nzaro, E.; Kataaha, P.; Dondero, T.J.; et al. Transmission of Human Herpesvirus 8 by Blood Transfusion. N. Engl. J. Med. 2006, 355, 1331–1338. [Google Scholar] [CrossRef]
- Cook, P.M.; Whitby, D.; Calabro, M.L.; Luppi, M.; Kakoola, D.N.; Hjalgrim, H.; Ariyoshi, K.; Ensoli, B.; Davison, A.J.; Schulz, T.F. Variability and Evolution of Kaposi’s Sarcoma-Associated Herpesvirus in Europe and Africa. International Collaborative Group. AIDS 1999, 13, 1165–1176. [Google Scholar] [CrossRef] [PubMed]
- Zong, J.C.; Ciufo, D.M.; Alcendor, D.J.; Wan, X.; Nicholas, J.; Browning, P.J.; Rady, P.L.; Tyring, S.K.; Orenstein, J.M.; Rabkin, C.S.; et al. High-Level Variability in the ORF-K1 Membrane Protein Gene at the Left End of the Kaposi’s Sarcoma-Associated Herpesvirus Genome Defines Four Major Virus Subtypes and Multiple Variants or Clades in Different Human Populations. J. Virol. 1999, 73, 4156–4170. [Google Scholar] [CrossRef]
- Zong, J.; Ciufo, D.M.; Viscidi, R.; Alagiozoglou, L.; Tyring, S.; Rady, P.; Orenstein, J.; Boto, W.; Kalumbuja, H.; Romano, N.; et al. Genotypic Analysis at Multiple Loci across Kaposi’s Sarcoma Herpesvirus (KSHV) DNA Molecules: Clustering Patterns, Novel Variants and Chimerism. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2002, 23, 119–148. [Google Scholar] [CrossRef]
- Bhatt, A.P.; Damania, B. AKTivation of PI3K/AKT/MTOR Signaling Pathway by KSHV. Front. Immunol. 2012, 3, 401. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-S.; Lee, S.-H.; Feng, P.; Chang, H.; Cho, N.-H.; Jung, J.U. Characterization of the Kaposi’s Sarcoma-Associated Herpesvirus K1 Signalosome. J. Virol. 2005, 79, 12173–12184. [Google Scholar] [CrossRef]
- Ötvös, R.; Juhasz, A.; Szalai, E.; Ujvari, D.; Ötvös, K.; Szabo, K.; Remenyik, E.; Szekely, L.; Gergely, L.; Konya, J. Molecular Typing of Human Herpesvirus 8 Isolates from Patients with Kaposi’s Sarcoma in Hungary. Anticancer Res. 2014, 34, 893–898. [Google Scholar] [PubMed]
- Ouyang, X.; Zeng, Y.; Fu, B.; Wang, X.; Chen, W.; Fang, Y.; Luo, M.; Wang, L. Genotypic Analysis of Kaposi’s Sarcoma-Associated Herpesvirus from Patients with Kaposi’s Sarcoma in Xinjiang, China. Viruses 2014, 6, 4800–4810. [Google Scholar] [CrossRef] [PubMed]
- Varmazyar, S.; Shoja, Z.; Kakavand-Ghalehnoei, R.; Shahmahmoodi, S.; Marashi, S.M.; Jalilvand, S. Molecular Typing of Human Herpesvirus 8 among HIV Positive in Comparison to HIV-Negative Individuals in Iran. J. Med. Virol. 2017, 89, 703–709. [Google Scholar] [CrossRef]
- Isaacs, T.; Abera, A.B.; Muloiwa, R.; Katz, A.A.; Todd, G. Genetic Diversity of HHV8 Subtypes in South Africa: A5 Subtype Is Associated with Extensive Disease in AIDS-KS. J. Med. Virol. 2016, 88, 292–303. [Google Scholar] [CrossRef] [PubMed]
- Mamimandjiami, A.I.; Mouinga-Ondémé, A.; Ramassamy, J.-L.; Djuicy, D.D.; Afonso, P.V.; Mahé, A.; Lekana-Douki, J.-B.; Cassar, O.; Gessain, A. Epidemiology and Genetic Variability of HHV-8/KSHV among Rural Populations and Kaposi’s Sarcoma Patients in Gabon, Central Africa. Review of the Geographical Distribution of HHV-8 K1 Genotypes in Africa. Viruses 2021, 13, 175. [Google Scholar] [CrossRef]
- Cassar, O.; Charavay, F.; Bassot, S.; Plancoulaine, S.; Grangeon, J.-P.; Laumond-Barny, S.; Martin, P.M.V.; Chanteau, S.; Gessain, A. Divergent KSHV/HHV-8 Subtype D Strains in New Caledonia and Solomon Islands, Melanesia. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2012, 53, 214–218. [Google Scholar] [CrossRef]
- Biggar, R.J.; Whitby, D.; Marshall, V.; Linhares, A.C.; Black, F. Human Herpesvirus 8 in Brazilian Amerindians: A Hyperendemic Population with a New Subtype. J. Infect. Dis. 2000, 181, 1562–1568. [Google Scholar] [CrossRef]
- Whitby, D.; Marshall, V.A.; Bagni, R.K.; Wang, C.D.; Gamache, C.J.; Guzman, J.R.; Kron, M.; Ebbesen, P.; Biggar, R.J. Genotypic Characterization of Kaposi’s Sarcoma-Associated Herpesvirus in Asymptomatic Infected Subjects from Isolated Populations. J. Gen. Virol. 2004, 85, 155–163. [Google Scholar] [CrossRef]
- Kajumbula, H.; Wallace, R.G.; Zong, J.-C.; Hokello, J.; Sussman, N.; Simms, S.; Rockwell, R.F.; Pozos, R.; Hayward, G.S.; Boto, W. Ugandan Kaposi’s Sarcoma-Associated Herpesvirus Phylogeny: Evidence for Cross-Ethnic Transmission of Viral Subtypes. Intervirology 2006, 49, 133–143. [Google Scholar] [CrossRef]
- Jary, A.; Leducq, V.; Desire, N.; Petit, H.; Palich, R.; Joly, V.; Canestri, A.; Gothland, A.; Lambert-Niclot, S.; Surgers, L.; et al. New Kaposi’s Sarcoma-Associated Herpesvirus Variant in Men Who Have Sex with Men Associated with Severe Pathologies. J. Infect. Dis. 2020, 222, 1320–1328. [Google Scholar] [CrossRef]
- Kasolo, F.C.; Monze, M.; Obel, N.; Anderson, R.A.; French, C.; Gompels, U.A. Sequence Analyses of Human Herpesvirus-8 Strains from Both African Human Immunodeficiency Virus-Negative and -Positive Childhood Endemic Kaposi’s Sarcoma Show a Close Relationship with Strains Identified in Febrile Children and High Variation in the K1 Glycoprotein. J. Gen. Virol. 1998, 79 Pt 12, 3055–3065. [Google Scholar] [CrossRef]
- Brinkmann, M.M.; Glenn, M.; Rainbow, L.; Kieser, A.; Henke-Gendo, C.; Schulz, T.F. Activation of Mitogen-Activated Protein Kinase and NF-KappaB Pathways by a Kaposi’s Sarcoma-Associated Herpesvirus K15 Membrane Protein. J. Virol. 2003, 77, 9346–9358. [Google Scholar] [CrossRef]
- Hayward, G.S.; Zong, J.C. Modern Evolutionary History of the Human KSHV Genome. Curr. Top. Microbiol. Immunol. 2007, 312, 1–42. [Google Scholar] [PubMed]
- Poole, L.J.; Zong, J.C.; Ciufo, D.M.; Alcendor, D.J.; Cannon, J.S.; Ambinder, R.; Orenstein, J.M.; Reitz, M.S.; Hayward, G.S. Comparison of Genetic Variability at Multiple Loci across the Genomes of the Major Subtypes of Kaposi’s Sarcoma-Associated Herpesvirus Reveals Evidence for Recombination and for Two Distinct Types of Open Reading Frame K15 Alleles at the Right-Hand End. J. Virol. 1999, 73, 6646–6660. [Google Scholar] [CrossRef] [PubMed]
- Tornesello, M.L.; Biryahwaho, B.; Downing, R.; Hatzakis, A.; Alessi, E.; Cusini, M.; Ruocco, V.; Katongole-Mbidde, E.; Loquercio, G.; Buonaguro, L.; et al. Human Herpesvirus Type 8 Variants Circulating in Europe, Africa and North America in Classic, Endemic and Epidemic Kaposi’s Sarcoma Lesions during Pre-AIDS and AIDS Era. Virology 2010, 398, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Olp, L.N.; Jeanniard, A.; Marimo, C.; West, J.T.; Wood, C. Whole-Genome Sequencing of Kaposi’s Sarcoma-Associated Herpesvirus from Zambian Kaposi’s Sarcoma Biopsy Specimens Reveals Unique Viral Diversity. J. Virol. 2015, 89, 12299–12308. [Google Scholar] [CrossRef]
- Sallah, N.; Palser, A.L.; Watson, S.J.; Labo, N.; Asiki, G.; Marshall, V.; Newton, R.; Whitby, D.; Kellam, P.; Barroso, I. Genome-Wide Sequence Analysis of Kaposi Sarcoma-Associated Herpesvirus Shows Diversification Driven by Recombination. J. Infect. Dis. 2018, 218, 1700–1710. [Google Scholar] [CrossRef]
- Bellocchi, M.C.; Svicher, V.; Ceccherini-Silberstein, F. HHV-8 Genetic Diversification and Its Impact on Severe Clinical Presentation of Associated Diseases. J. Infect. Dis. 2020, 222, 1250–1253. [Google Scholar] [CrossRef]
- Mancuso, R.; Biffi, R.; Valli, M.; Bellinvia, M.; Tourlaki, A.; Athanasia, T.; Ferrucci, S.; Brambilla, L.; Delbue, S.; Ferrante, P.; et al. HHV8 a Subtype Is Associated with Rapidly Evolving Classic Kaposi’s Sarcoma. J. Med. Virol. 2008, 80, 2153–2160. [Google Scholar] [CrossRef]
- Pérez, C.L.; Tous, M.I. Diversity of Human Herpesvirus 8 Genotypes in Patients with AIDS and Non-AIDS Associated Kaposi’s Sarcoma, Castleman’s Disease and Primary Effusion Lymphoma in Argentina. J. Med. Virol. 2017, 89, 2020–2028. [Google Scholar] [CrossRef]
- White, T.; Hagen, M.; Gudza, I.; White, I.E.; Ndemera, B.; Gwanzura, L.; Borok, M.; Campbell, T.B. Genetic Diversity of the Kaposi’s Sarcoma Herpesvirus K1 Protein in AIDS-KS in Zimbabwe. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2008, 42, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Kadyrova, E.; Lacoste, V.; Duprez, R.; Pozharissky, K.; Molochkov, V.; Huerre, M.; Gurtsevitch, V.; Gessain, A. Molecular Epidemiology of Kaposi’s Sarcoma-Associated Herpesvirus/Human Herpesvirus 8 Strains from Russian Patients with Classic, Posttransplant, and AIDS-Associated Kaposi’s Sarcoma. J. Med. Virol. 2003, 71, 548–556. [Google Scholar] [CrossRef]
- Tozetto-Mendoza, T.R.; Ibrahim, K.Y.; Tateno, A.F.; Oliveira, C.M.; Sumita, L.M.; Sanchez, M.C.A.; Luna, E.J.; Pierrotti, L.C.; Drexler, J.F.; Braz-Silva, P.H.; et al. Genotypic Distribution of HHV-8 in AIDS Individuals without and with Kaposi Sarcoma: Is Genotype B Associated with Better Prognosis of AIDS-KS? Medicine 2016, 95, e5291. [Google Scholar] [CrossRef]
- Barete, S.; Calvez, V.; Mouquet, C.; Barrou, B.; Kreis, H.; Dantal, J.; Dorent, R.; Durand, F.; Dimitrov, Y.; Dupin, N.; et al. Clinical Features and Contribution of Virological Findings to the Management of Kaposi Sarcoma in Organ-Allograft Recipients. Arch. Dermatol. 2000, 136, 1452–1458. [Google Scholar] [CrossRef][Green Version]
- Tamanaha-Nakasone, A.; Uehara, K.; Tanabe, Y.; Ishikawa, H.; Yamakawa, N.; Toyoda, Z.; Kurima, K.; Kina, S.; Tsuneki, M.; Okubo, Y.; et al. K1 Gene Transformation Activities in AIDS-Related and Classic Type Kaposi’s Sarcoma: Correlation with Clinical Presentation. Sci. Rep. 2019, 9, 6416. [Google Scholar] [CrossRef]
- Bechtel, J.T.; Liang, Y.; Hvidding, J.; Ganem, D. Host Range of Kaposi’s Sarcoma-Associated Herpesvirus in Cultured Cells. J. Virol. 2003, 77, 6474–6481. [Google Scholar] [CrossRef]
- Whitby, D.; Marshall, V.A.; Bagni, R.K.; Miley, W.J.; McCloud, T.G.; Hines-Boykin, R.; Goedert, J.J.; Conde, B.A.; Nagashima, K.; Mikovits, J.; et al. Reactivation of Kaposi’s Sarcoma-Associated Herpesvirus by Natural Products from Kaposi’s Sarcoma Endemic Regions. Int. J. Cancer 2007, 120, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Wakeham, K.; Webb, E.L.; Sebina, I.; Muhangi, L.; Miley, W.; Johnson, W.T.; Ndibazza, J.; Elliott, A.M.; Whitby, D.; Newton, R. Parasite Infection Is Associated with Kaposi’s Sarcoma Associated Herpesvirus (KSHV) in Ugandan Women. Infect. Agent. Cancer 2011, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Nalwoga, A.; Cose, S.; Wakeham, K.; Miley, W.; Ndibazza, J.; Drakeley, C.; Elliott, A.; Whitby, D.; Newton, R. Association between Malaria Exposure and Kaposi’s Sarcoma-Associated Herpes Virus Seropositivity in Uganda. Trop. Med. Int. Health TM IH 2015, 20, 665–672. [Google Scholar] [CrossRef]
- Simonart, T. Iron: A Target for the Management of Kaposi’s Sarcoma? BMC Cancer 2004, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- AIDS-defining Cancer Project Working Group for IeDEA and COHERE in EuroCoord. Comparison of Kaposi Sarcoma Risk in Human Immunodeficiency Virus-Positive Adults across 5 Continents: A Multiregional Multicohort Study. Clin. Infect. Dis. 2017, 65, 1316–1326. [Google Scholar] [CrossRef]
- Piselli, P.; Busnach, G.; Citterio, F.; Frigerio, M.; Arbustini, E.; Burra, P.; Pinna, A.D.; Bresadola, V.; Ettorre, G.M.; Baccarani, U.; et al. Risk of Kaposi Sarcoma after Solid-Organ Transplantation: Multicenter Study in 4767 Recipients in Italy, 1970–2006. Transplant. Proc. 2009, 41, 1227–1230. [Google Scholar] [CrossRef] [PubMed]
- Ensoli, B.; Barillari, G.; Salahuddin, S.Z.; Gallo, R.C.; Wong-Staal, F. Tat Protein of HIV-1 Stimulates Growth of Cells Derived from Kaposi’s Sarcoma Lesions of AIDS Patients. Nature 1990, 345, 84–86. [Google Scholar] [CrossRef]
- Zhu, X.; Guo, Y.; Yao, S.; Yan, Q.; Xue, M.; Hao, T.; Zhou, F.; Zhu, J.; Qin, D.; Lu, C. Synergy between Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) VIL-6 and HIV-1 Nef Protein in Promotion of Angiogenesis and Oncogenesis: Role of the AKT Signaling Pathway. Oncogene 2014, 33, 1986–1996. [Google Scholar] [CrossRef]
- Aoki, Y.; Tosato, G. Interactions between HIV-1 Tat and KSHV. Curr. Top. Microbiol. Immunol. 2007, 312, 309–326. [Google Scholar] [CrossRef]
- Zeng, Y.; Zhang, X.; Huang, Z.; Cheng, L.; Yao, S.; Qin, D.; Chen, X.; Tang, Q.; Lv, Z.; Zhang, L.; et al. Intracellular Tat of Human Immunodeficiency Virus Type 1 Activates Lytic Cycle Replication of Kaposi’s Sarcoma-Associated Herpesvirus: Role of JAK/STAT Signaling. J. Virol. 2007, 81, 2401–2417. [Google Scholar] [CrossRef]
- Ensoli, B.; Gendelman, R.; Markham, P.; Fiorelli, V.; Colombini, S.; Raffeld, M.; Cafaro, A.; Chang, H.K.; Brady, J.N.; Gallo, R.C. Synergy between Basic Fibroblast Growth Factor and HIV-1 Tat Protein in Induction of Kaposi’s Sarcoma. Nature 1994, 371, 674–680. [Google Scholar] [CrossRef] [PubMed]
- Rusnati, M.; Presta, M. HIV-1 Tat Protein and Endothelium: From Protein/Cell Interaction to AIDS-Associated Pathologies. Angiogenesis 2002, 5, 141–151. [Google Scholar] [CrossRef]
- Wood, N.H.; Feller, L. The Malignant Potential of HIV-Associated Kaposi Sarcoma. Cancer Cell Int. 2008, 8, 14. [Google Scholar] [CrossRef]
- Cancer Project Working Group for the Collaboration of Observational HIV Epidemiological Research Europe (COHERE) Study in EuroCoord. Changing Incidence and Risk Factors for Kaposi Sarcoma by Time since Starting Antiretroviral Therapy: Collaborative Analysis of 21 European Cohort Studies. Clin. Infect. Dis. 2016, 63, 1373–1379. [Google Scholar] [CrossRef] [PubMed]
- Boshoff, C.; Chang, Y. Kaposi’s Sarcoma-Associated Herpesvirus: A New DNA Tumor Virus. Annu. Rev. Med. 2001, 52, 453–470. [Google Scholar] [CrossRef]
- Asahi-Ozaki, Y.; Sato, Y.; Kanno, T.; Sata, T.; Katano, H. Quantitative Analysis of Kaposi Sarcoma-Associated Herpesvirus (KSHV) in KSHV-Associated Diseases. J. Infect. Dis. 2006, 193, 773–782. [Google Scholar] [CrossRef]
- Alkharsah, K.R.; Singh, V.V.; Bosco, R.; Santag, S.; Grundhoff, A.; Konrad, A.; Stürzl, M.; Wirth, D.; Dittrich-Breiholz, O.; Kracht, M.; et al. Deletion of Kaposi’s Sarcoma-Associated Herpesvirus FLICE Inhibitory Protein, VFLIP, from the Viral Genome Compromises the Activation of STAT1-Responsive Cellular Genes and Spindle Cell Formation in Endothelial Cells. J. Virol. 2011, 85, 10375–10388. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, C.; Podgrabinska, S.; Skobe, M.; Ganem, D. Activation of NF-KappaB by the Latent VFLIP Gene of Kaposi’s Sarcoma-Associated Herpesvirus Is Required for the Spindle Shape of Virus-Infected Endothelial Cells and Contributes to Their Proinflammatory Phenotype. J. Virol. 2006, 80, 7179–7185. [Google Scholar] [CrossRef]
- Jha, H.C.; Banerjee, S.; Robertson, E.S. The Role of Gammaherpesviruses in Cancer Pathogenesis. Pathogens 2016, 5, 18. [Google Scholar] [CrossRef]
- Gasperini, P.; Espigol-Frigole, G.; McCormick, P.J.; Salvucci, O.; Maric, D.; Uldrick, T.S.; Polizzotto, M.N.; Yarchoan, R.; Tosato, G. Kaposi Sarcoma Herpesvirus Promotes Endothelial-to-Mesenchymal Transition through Notch-Dependent Signaling. Cancer Res. 2012, 72, 1157–1169. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-W.; Trotter, M.W.B.; Lagos, D.; Bourboulia, D.; Henderson, S.; Mäkinen, T.; Elliman, S.; Flanagan, A.M.; Alitalo, K.; Boshoff, C. Kaposi Sarcoma Herpesvirus-Induced Cellular Reprogramming Contributes to the Lymphatic Endothelial Gene Expression in Kaposi Sarcoma. Nat. Genet. 2004, 36, 687–693. [Google Scholar] [CrossRef]
- Hong, Y.-K.; Foreman, K.; Shin, J.W.; Hirakawa, S.; Curry, C.L.; Sage, D.R.; Libermann, T.; Dezube, B.J.; Fingeroth, J.D.; Detmar, M. Lymphatic Reprogramming of Blood Vascular Endothelium by Kaposi Sarcoma-Associated Herpesvirus. Nat. Genet. 2004, 36, 683–685. [Google Scholar] [CrossRef] [PubMed]
- Cancian, L.; Hansen, A.; Boshoff, C. Cellular Origin of Kaposi’s Sarcoma and Kaposi’s Sarcoma-Associated Herpesvirus-Induced Cell Reprogramming. Trends Cell Biol. 2013, 23, 421–432. [Google Scholar] [CrossRef]
- Katano, H.; Sato, Y.; Kurata, T.; Mori, S.; Sata, T. Expression and Localization of Human Herpesvirus 8-Encoded Proteins in Primary Effusion Lymphoma, Kaposi’s Sarcoma, and Multicentric Castleman’s Disease. Virology 2000, 269, 335–344. [Google Scholar] [CrossRef]
- Decker, L.L.; Shankar, P.; Khan, G.; Freeman, R.B.; Dezube, B.J.; Lieberman, J.; Thorley-Lawson, D.A. The Kaposi Sarcoma-Associated Herpesvirus (KSHV) Is Present as an Intact Latent Genome in KS Tissue but Replicates in the Peripheral Blood Mononuclear Cells of KS Patients. J. Exp. Med. 1996, 184, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Lin, S.F.; Staskus, K.; Gradoville, L.; Grogan, E.; Haase, A.; Miller, G. Kinetics of Kaposi’s Sarcoma-Associated Herpesvirus Gene Expression. J. Virol. 1999, 73, 2232–2242. [Google Scholar] [CrossRef]
- Grundhoff, A.; Ganem, D. Inefficient Establishment of KSHV Latency Suggests an Additional Role for Continued Lytic Replication in Kaposi Sarcoma Pathogenesis. J. Clin. Investig. 2004, 113, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Parravicini, C.; Chandran, B.; Corbellino, M.; Berti, E.; Paulli, M.; Moore, P.S.; Chang, Y. Differential Viral Protein Expression in Kaposi’s Sarcoma-Associated Herpesvirus-Infected Diseases: Kaposi’s Sarcoma, Primary Effusion Lymphoma, and Multicentric Castleman’s Disease. Am. J. Pathol. 2000, 156, 743–749. [Google Scholar] [CrossRef]
- Hunte, R.; Alonso, P.; Thomas, R.; Bazile, C.A.; Ramos, J.C.; van der Weyden, L.; Dominguez-Bendala, J.; Khan, W.N.; Shembade, N. CADM1 Is Essential for KSHV-Encoded VGPCR-and VFLIP-Mediated Chronic NF-ΚB Activation. PLoS Pathog. 2018, 14, e1006968. [Google Scholar] [CrossRef]
- Kim, H.; Jang, J.H.; Song, Y.E.; Seo, T. Kaposi’s Sarcoma-Associated Herpesvirus Viral Protein Kinase Phosphorylates Extracellular Signal-Regulated Kinase and Activates MAPK/ERK Signaling Pathway. Biochem. Biophys. Res. Commun. 2020, 521, 1083–1088. [Google Scholar] [CrossRef]
- Broussard, G.; Damania, B. Regulation of KSHV Latency and Lytic Reactivation. Viruses 2020, 12, 1034. [Google Scholar] [CrossRef]
- Uppal, T.; Jha, H.C.; Verma, S.C.; Robertson, E.S. Chromatinization of the KSHV Genome during the KSHV Life Cycle. Cancers 2015, 7, 112–142. [Google Scholar] [CrossRef]
- Fuentes-González, A.M.; Contreras-Paredes, A.; Manzo-Merino, J.; Lizano, M. The Modulation of Apoptosis by Oncogenic Viruses. Virol. J. 2013, 10, 182. [Google Scholar] [CrossRef]
- Verma, S.C.; Borah, S.; Robertson, E.S. Latency-Associated Nuclear Antigen of Kaposi’s Sarcoma-Associated Herpesvirus up-Regulates Transcription of Human Telomerase Reverse Transcriptase Promoter through Interaction with Transcription Factor Sp1. J. Virol. 2004, 78, 10348–10359. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, A.J.; Chugh, P.; Wang, L.; Netto, E.M.; Luz, E.; Harrington, W.J.; Dezube, B.J.; Damania, B.; Dittmer, D.P. Pre-Micro RNA Signatures Delineate Stages of Endothelial Cell Transformation in Kaposi Sarcoma. PLoS Pathog. 2009, 5, e1000389. [Google Scholar] [CrossRef]
- Matta, H.; Chaudhary, P.M. Activation of Alternative NF-Kappa B Pathway by Human Herpes Virus 8-Encoded Fas-Associated Death Domain-like IL-1 Beta-Converting Enzyme Inhibitory Protein (VFLIP). Proc. Natl. Acad. Sci. USA 2004, 101, 9399–9404. [Google Scholar] [CrossRef]
- Ballon, G.; Akar, G.; Cesarman, E. Systemic Expression of Kaposi Sarcoma Herpesvirus (KSHV) Vflip in Endothelial Cells Leads to a Profound Proinflammatory Phenotype and Myeloid Lineage Remodeling in Vivo. PLoS Pathog. 2015, 11, e1004581. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, P.M.; Jasmin, A.; Eby, M.T.; Hood, L. Modulation of the NF-Kappa B Pathway by Virally Encoded Death Effector Domains-Containing Proteins. Oncogene 1999, 18, 5738–5746. [Google Scholar] [CrossRef]
- Ojala, P.M.; Tiainen, M.; Salven, P.; Veikkola, T.; Castaños-Vélez, E.; Sarid, R.; Biberfeld, P.; Mäkelä, T.P. Kaposi’s Sarcoma-Associated Herpesvirus-Encoded v-Cyclin Triggers Apoptosis in Cells with High Levels of Cyclin-Dependent Kinase 6. Cancer Res. 1999, 59, 4984–4989. [Google Scholar]
- Lan, K.; Kuppers, D.A.; Verma, S.C.; Sharma, N.; Murakami, M.; Robertson, E.S. Induction of Kaposi’s Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen by the Lytic Transactivator RTA: A Novel Mechanism for Establishment of Latency. J. Virol. 2005, 79, 7453–7465. [Google Scholar] [CrossRef]
- Shamay, M.; Krithivas, A.; Zhang, J.; Hayward, S.D. Recruitment of the de Novo DNA Methyltransferase Dnmt3a by Kaposi’s Sarcoma-Associated Herpesvirus LANA. Proc. Natl. Acad. Sci. USA 2006, 103, 14554–14559. [Google Scholar] [CrossRef] [PubMed]
- Toth, Z.; Papp, B.; Brulois, K.; Choi, Y.J.; Gao, S.-J.; Jung, J.U. LANA-Mediated Recruitment of Host Polycomb Repressive Complexes onto the KSHV Genome during De Novo Infection. PLoS Pathog. 2016, 12, e1005878. [Google Scholar] [CrossRef]
- Sun, R.; Liang, D.; Gao, Y.; Lan, K. Kaposi’s Sarcoma-Associated Herpesvirus-Encoded LANA Interacts with Host KAP1 to Facilitate Establishment of Viral Latency. J. Virol. 2014, 88, 7331–7344. [Google Scholar] [CrossRef]
- Zhang, G.; Chan, B.; Samarina, N.; Abere, B.; Weidner-Glunde, M.; Buch, A.; Pich, A.; Brinkmann, M.M.; Schulz, T.F. Cytoplasmic Isoforms of Kaposi Sarcoma Herpesvirus LANA Recruit and Antagonize the Innate Immune DNA Sensor CGAS. Proc. Natl. Acad. Sci. USA 2016, 113, E1034–E1043. [Google Scholar] [CrossRef]
- Tolani, B.; Matta, H.; Gopalakrishnan, R.; Punj, V.; Chaudhary, P.M. NEMO Is Essential for Kaposi’s Sarcoma-Associated Herpesvirus-Encoded VFLIP K13-Induced Gene Expression and Protection against Death Receptor-Induced Cell Death, and Its N-Terminal 251 Residues Are Sufficient for This Process. J. Virol. 2014, 88, 6345–6354. [Google Scholar] [CrossRef]
- Rosario, S.A.; Santiago, G.E.; Mesri, E.A.; Verdun, R.E. Kaposi’s Sarcoma-Associated Herpesvirus-Encoded Viral IL-6 (VIL-6) Enhances Immunoglobulin Class-Switch Recombination. Front. Microbiol. 2018, 9, 3119. [Google Scholar] [CrossRef]
- Hiura, K.; Strahan, R.; Uppal, T.; Prince, B.; Rossetto, C.C.; Verma, S.C. KSHV ORF59 and PAN RNA Recruit Histone Demethylases to the Viral Chromatin during Lytic Reactivation. Viruses 2020, 12, 420. [Google Scholar] [CrossRef]
- Yamin, R.; Kaynan, N.S.; Glasner, A.; Vitenshtein, A.; Tsukerman, P.; Bauman, Y.; Ophir, Y.; Elias, S.; Bar-On, Y.; Gur, C.; et al. The Viral KSHV Chemokine VMIP-II Inhibits the Migration of Naive and Activated Human NK Cells by Antagonizing Two Distinct Chemokine Receptors. PLoS Pathog. 2013, 9, e1003568. [Google Scholar] [CrossRef] [PubMed]
- Manners, O.; Murphy, J.C.; Coleman, A.; Hughes, D.J.; Whitehouse, A. Contribution of the KSHV and EBV Lytic Cycles to Tumourigenesis. Curr. Opin. Virol. 2018, 32, 60–70. [Google Scholar] [CrossRef]
- Azzi, S.; Smith, S.S.; Dwyer, J.; Leclair, H.M.; Alexia, C.; Hebda, J.K.; Dupin, N.; Bidère, N.; Gavard, J. YGLF Motif in the Kaposi Sarcoma Herpes Virus G-Protein-Coupled Receptor Adjusts NF-ΚB Activation and Paracrine Actions. Oncogene 2014, 33, 5609–5618. [Google Scholar] [CrossRef]
- Martin, D.; Galisteo, R.; Ji, Y.; Montaner, S.; Gutkind, J.S. An NF-KappaB Gene Expression Signature Contributes to Kaposi’s Sarcoma Virus VGPCR-Induced Direct and Paracrine Neoplasia. Oncogene 2008, 27, 1844–1852. [Google Scholar] [CrossRef] [PubMed]
- Gramolelli, S.; Schulz, T.F. The Role of Kaposi Sarcoma-Associated Herpesvirus in the Pathogenesis of Kaposi Sarcoma. J. Pathol. 2015, 235, 368–380. [Google Scholar] [CrossRef]
- Cousins, E.; Nicholas, J. Molecular Biology of Human Herpesvirus 8: Novel Functions and Virus-Host Interactions Implicated in Viral Pathogenesis and Replication. Viruses Hum. Cancer 2014, 193, 227–268. [Google Scholar] [CrossRef]
- Bala, K.; Bosco, R.; Gramolelli, S.; Haas, D.A.; Kati, S.; Pietrek, M.; Hävemeier, A.; Yakushko, Y.; Singh, V.V.; Dittrich-Breiholz, O.; et al. Kaposi’s Sarcoma Herpesvirus K15 Protein Contributes to Virus-Induced Angiogenesis by Recruiting PLCγ1 and Activating NFAT1-Dependent RCAN1 Expression. PLoS Pathog. 2012, 8, e1002927. [Google Scholar] [CrossRef]
- Choi, Y.B.; Nicholas, J. Autocrine and Paracrine Promotion of Cell Survival and Virus Replication by Human Herpesvirus 8 Chemokines. J. Virol. 2008, 82, 6501–6513. [Google Scholar] [CrossRef]
- Golas, G.; Jang, S.J.; Naik, N.G.; Alonso, J.D.; Papp, B.; Toth, Z. Comparative Analysis of the Viral Interferon Regulatory Factors of KSHV for Their Requisite for Virus Production and Inhibition of the Type I Interferon Pathway. Virology 2020, 541, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Palich, R.; Veyri, M.; Valantin, M.-A.; Marcelin, A.-G.; Guihot, A.; Pourcher, V.; Jary, A.; Solas, C.; Makinson, A.; Poizot-Martin, I.; et al. Recurrence and Occurrence of Kaposi’s Sarcoma in Patients Living With Human Immunodeficiency Virus (HIV) and on Antiretroviral Therapy, Despite Suppressed HIV Viremia. Clin. Infect. Dis. 2020, 70, 2435–2438. [Google Scholar] [CrossRef]
- Yanik, E.L.; Achenbach, C.J.; Gopal, S.; Coghill, A.E.; Cole, S.R.; Eron, J.J.; Moore, R.D.; Mathews, W.C.; Drozd, D.R.; Hamdan, A.; et al. Changes in Clinical Context for Kaposi’s Sarcoma and Non-Hodgkin Lymphoma Among People With HIV Infection in the United States. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2016, 34, 3276–3283. [Google Scholar] [CrossRef]
- Reid, E.; Suneja, G.; Ambinder, R.F.; Ard, K.; Baiocchi, R.; Barta, S.K.; Carchman, E.; Cohen, A.; Crysler, O.V.; Gupta, N.; et al. AIDS-Related Kaposi Sarcoma, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2019, 17, 171–189. [Google Scholar] [CrossRef]
- Beldi-Ferchiou, A.; Lambert, M.; Dogniaux, S.; Vély, F.; Vivier, E.; Olive, D.; Dupuy, S.; Levasseur, F.; Zucman, D.; Lebbé, C.; et al. PD-1 Mediates Functional Exhaustion of Activated NK Cells in Patients with Kaposi Sarcoma. Oncotarget 2016, 7, 72961–72977. [Google Scholar] [CrossRef] [PubMed]
- Dupuy, S.; Lambert, M.; Zucman, D.; Choukem, S.-P.; Tognarelli, S.; Pages, C.; Lebbé, C.; Caillat-Zucman, S. Human Herpesvirus 8 (HHV8) Sequentially Shapes the NK Cell Repertoire during the Course of Asymptomatic Infection and Kaposi Sarcoma. PLoS Pathog. 2012, 8, e1002486. [Google Scholar] [CrossRef]
- Guihot, A.; Dupin, N.; Marcelin, A.-G.; Gorin, I.; Bedin, A.-S.; Bossi, P.; Galicier, L.; Oksenhendler, E.; Autran, B.; Carcelain, G. Low T Cell Responses to Human Herpesvirus 8 in Patients with AIDS-Related and Classic Kaposi Sarcoma. J. Infect. Dis. 2006, 194, 1078–1088. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.; Gannagé, M.; Karras, A.; Abel, M.; Legendre, C.; Kerob, D.; Agbalika, F.; Girard, P.-M.; Lebbe, C.; Caillat-Zucman, S. Differences in the Frequency and Function of HHV8-Specific CD8 T Cells between Asymptomatic HHV8 Infection and Kaposi Sarcoma. Blood 2006, 108, 3871–3880. [Google Scholar] [CrossRef]
- Wu, T.-T.; Qian, J.; Ang, J.; Sun, R. Vaccine Prospect of Kaposi Sarcoma-Associated Herpesvirus. Curr. Opin. Virol. 2012, 2, 482–488. [Google Scholar] [CrossRef]
- Kumar, P.; Kuwa, N.Y.; Minhas, V.; Marimo, C.; Shea, D.M.; Kankasa, C.; Wood, C. Higher Levels of Neutralizing Antibodies against KSHV in KS Patients Compared to Asymptomatic Individuals from Zambia. PLoS ONE 2013, 8, e71254. [Google Scholar] [CrossRef] [PubMed]
- Dialyna, I.A.; Graham, D.; Rezaee, R.; Blue, C.E.; Stavrianeas, N.G.; Neisters, H.G.M.; Spandidos, D.A.; Blackbourn, D.J. Anti-HHV-8/KSHV Antibodies in Infected Individuals Inhibit Infection in Vitro. AIDS 2004, 18, 1263–1270. [Google Scholar] [CrossRef]
- Mortazavi, Y.; Lidenge, S.J.; Tran, T.; West, J.T.; Wood, C.; Tso, F.Y. The Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) GH/GL Complex Is the Predominant Neutralizing Antigenic Determinant in KSHV-Infected Individuals. Viruses 2020, 12, 256. [Google Scholar] [CrossRef] [PubMed]


| Low | Intermediate | High | |
|---|---|---|---|
| World region | North America Western Europe Eastern Asia | Mediterranean basin Eastern Europe Southern America Western Africa | Central Africa Eastern Africa |
| KSHV Seroprevalence | <10% | 10–30% | >50% |
| Transmission | Sexual Iatrogenic | Sexual Iatrogenic Nonsexual | Childhood Sexual |
| Risk factors | Risky sexual behavior: STIs including HIV, number of different sexual partners Use of poppers? | Infectious agents (malaria)? Soils rich in metals? Chemical substances from plants? | Infectious agents (malaria)? Soils rich in metals? Chemical substances from plants? Transfusion |
| At-risk population | MSM Organ transplant patients | MSM Organ transplant patients Elderly men | Children Elderly men Low socio-economic level |
| Proteins or RNA | Gene | Viral Cycle | Cellular Homologs | Essential Functions |
|---|---|---|---|---|
| LANA-1 | ORF73 | Latent | - | Inhibition of lytic cycle by inhibiting the expression of RTA [177] Sequestration of KSHV episome in nucleus and transmission to daughters’ cell during mitosis Inhibition of apoptosis by interacting with p53 and pRB Recruitment of DNA methyltransferase [178] Recruitment of host PRC and KAP1 to suppress lytic gene expression [179,180] Cytoplasmic isoform of LANA-1 inhibits cGAS to promote KSHV reactivation [181] |
| v-cyclin | ORF72 | Latent | Cyclin D2 | Cell proliferation: v-cyclin-CDK6 => inhibition of p21 and p27 Cell transformation |
| v-FLIP | ORF71 | Latent | FLICE | Inhibition of apoptosis Activation of NF-kB pathway by interaction with NEMO [182] Inhibition of RBP-Jk (co-activator of RTA) |
| miRNA miRNA-K7-5p miRNA-K9-5p miRNA K1 | - - - miR-K11 | Latent | - | Immunomodulation, immune escape, inhibition of apoptosis Post transcriptional inhibition of RTA expression Post transcriptional inhibition of RTA expression Activation of NF-kB pathway |
| Kaposin | ORF-K12 | Latent | - | Cell transformation Activation of p18/MK2 pathways |
| LANA-2 /v-IRF3 | ORF 10.5 | Latent | - | Inhibition of p53 pathway Immune escape |
| RTA | ORF50 | Lytic | - | Activation of lytic cycle Stimulation of human IL-6 production Inhibition of p53 Activation of LANA-1 expression [177] |
| K-bZIP /RAP | ORF K8 | Lytic | Zta (ZEBRA) in EBV | Bind to RTA protein and suppression of its transactivation Interaction with CREB binding protein Stimulation of p53 and p21 and promotion of cell cycle arrest |
| v-IL-6 | ORF K2 | Lytic | IL-6 | Inhibition of apoptosis Interaction with cellular cycle Activation of JAK-STAT, MAPK-ERK and PI3-AKT pathways => Cell survival, and pro-inflammatory and pro-angiogenic environment Functional modulation of B cell by promoting CSR [183] |
| v-GPCR | ORF 74 | Lytic | IL8 receptor, CXCR2 | Interaction with MAPKs, PI3K-AKT and NF-kB pathways Cell transformation Stimulation of angiogenesis |
| v-bcl-2 | ORF 16 | Lytic | Bcl-2 | Inhibition of apoptosis |
| K1 (KIST) | ORF K1 | Lytic | STP in SaHV | Inhibition of NF-kB pathway Cell survival by activating PI3K-AKT pathway Immune escape by the ITAM motif |
| v-IRF1 | ORF K9 | Lytic | IRF1 | Inhibition of NF-kB pathway Cell proliferation Inhibition of genes expression induced by INF |
| K14 | ORF K14 | Lytic | - | Inhibition of NF-kB pathway |
| PAN RNA | ORF K7 | Lytic | - | LANA-1 sequestration [50] Recruitment of histone demethylase to the viral chromosome [184] Generated circRNA potentially involved in early infection [24] |
| v-MIP-1 v-MIP-2 v-MIP-3 | ORF K6 ORF K4 ORF K4.1 | Lytic | MIP-1alpha - MIP-1béta | Stimulation of angiogenesis Inhibition of naïve and active human NK cells [185] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jary, A.; Veyri, M.; Gothland, A.; Leducq, V.; Calvez, V.; Marcelin, A.-G. Kaposi’s Sarcoma-Associated Herpesvirus, the Etiological Agent of All Epidemiological Forms of Kaposi’s Sarcoma. Cancers 2021, 13, 6208. https://doi.org/10.3390/cancers13246208
Jary A, Veyri M, Gothland A, Leducq V, Calvez V, Marcelin A-G. Kaposi’s Sarcoma-Associated Herpesvirus, the Etiological Agent of All Epidemiological Forms of Kaposi’s Sarcoma. Cancers. 2021; 13(24):6208. https://doi.org/10.3390/cancers13246208
Chicago/Turabian StyleJary, Aude, Marianne Veyri, Adélie Gothland, Valentin Leducq, Vincent Calvez, and Anne-Geneviève Marcelin. 2021. "Kaposi’s Sarcoma-Associated Herpesvirus, the Etiological Agent of All Epidemiological Forms of Kaposi’s Sarcoma" Cancers 13, no. 24: 6208. https://doi.org/10.3390/cancers13246208
APA StyleJary, A., Veyri, M., Gothland, A., Leducq, V., Calvez, V., & Marcelin, A.-G. (2021). Kaposi’s Sarcoma-Associated Herpesvirus, the Etiological Agent of All Epidemiological Forms of Kaposi’s Sarcoma. Cancers, 13(24), 6208. https://doi.org/10.3390/cancers13246208






