Machine Learning Models That Integrate Tumor Texture and Perfusion Characteristics Using Low-Dose Breast Computed Tomography Are Promising for Predicting Histological Biomarkers and Treatment Failure in Breast Cancer Patients
Abstract
:Simple Summary
Abstract
1. Introduction
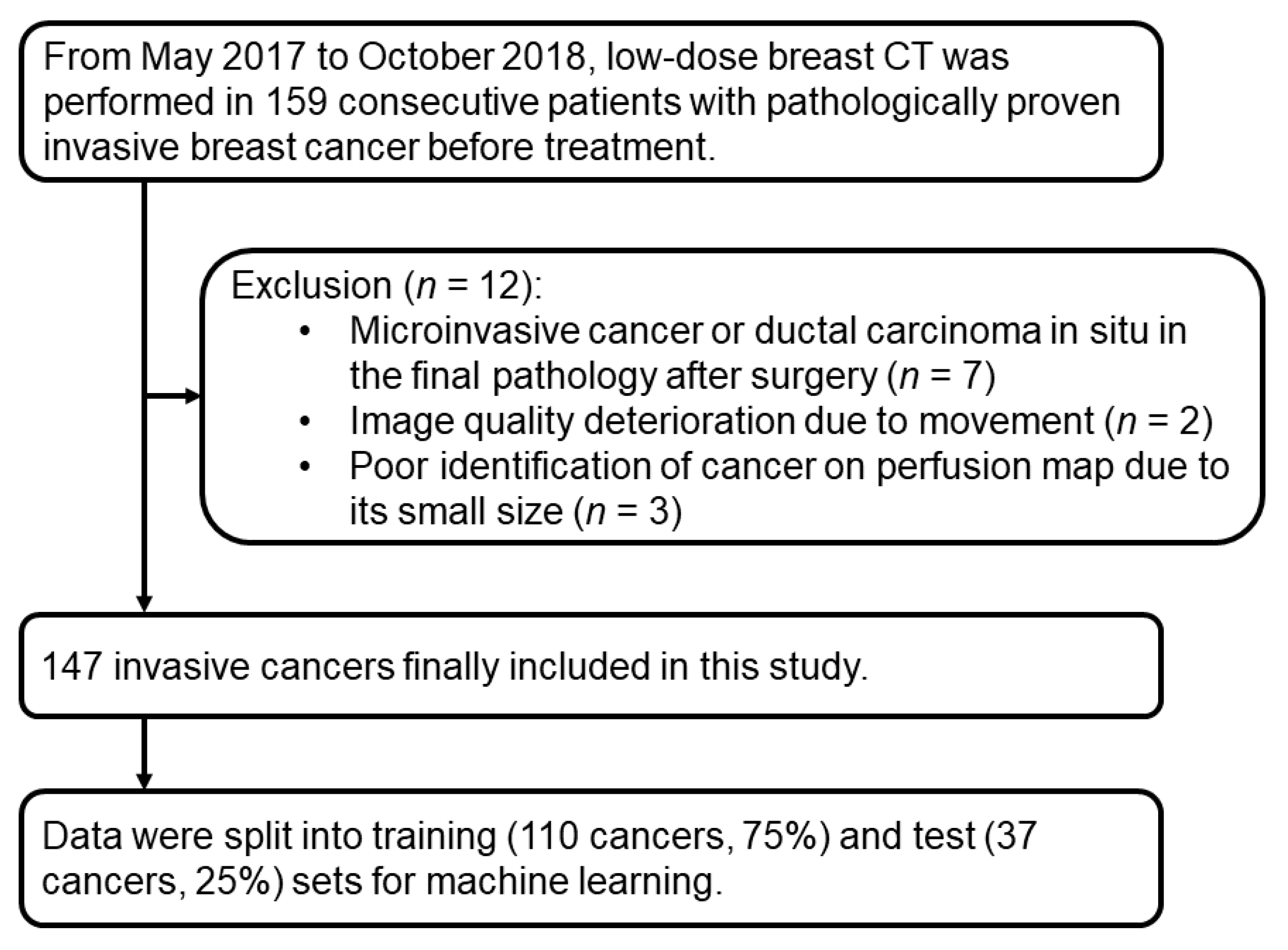
2. Materials and Methods
2.1. Patients
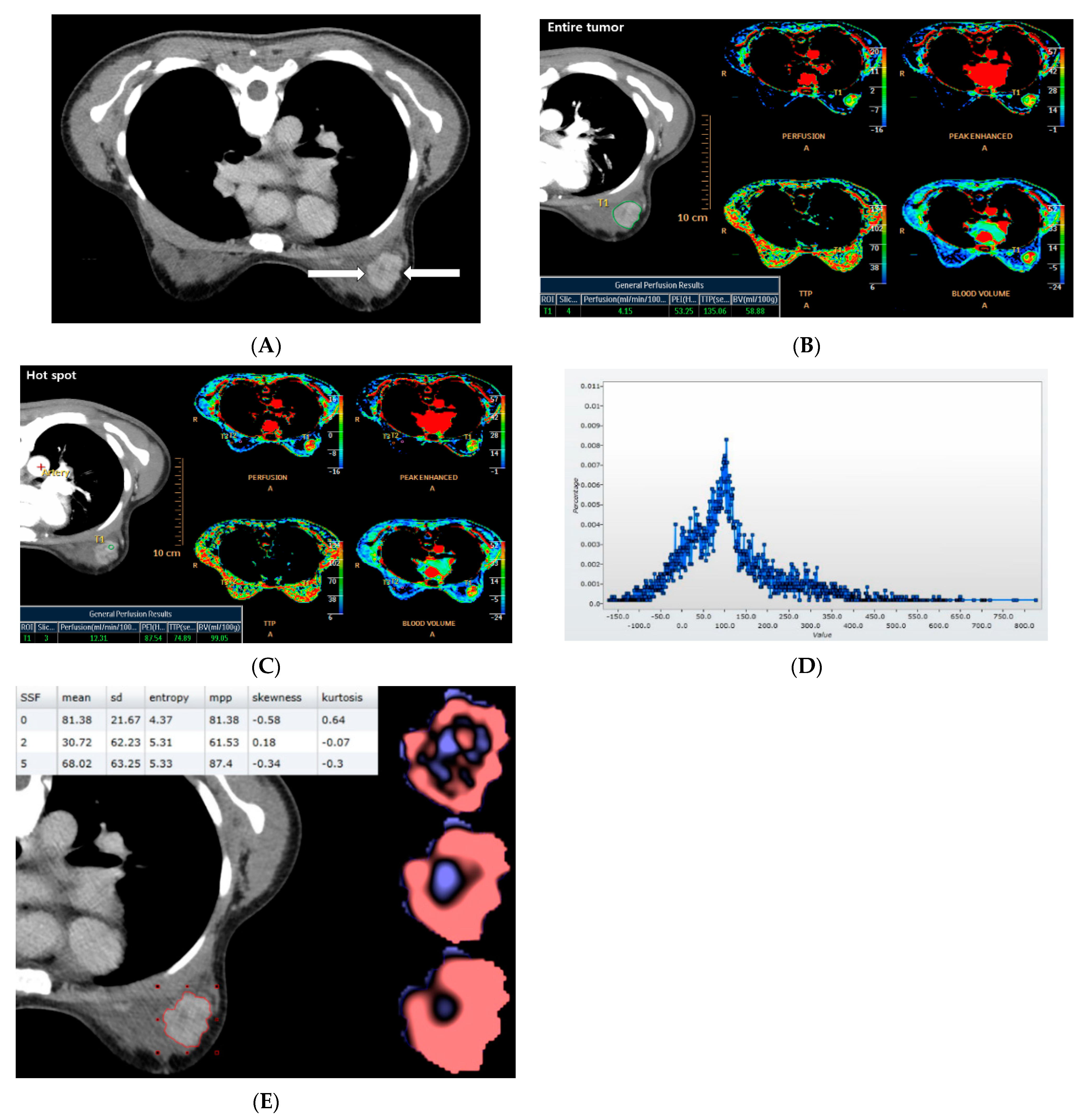
2.2. CT Acquisition and Analysis
2.3. Clinicohistological Evaluation
2.4. Statistical Analysis
3. Results
3.1. Associations between CT Parameters and Histological Biomarkers and Subtypes
3.2. Performance of the Machine Learning Models
3.3. Importance Ranking of CT Parameters for Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HER2 | Human epidermal growth factor receptor 2 |
| MRI | Magnetic resonance imaging |
| CT | Computed tomography |
| ROI | Regions of interest |
| PEI | Peak enhancement intensity |
| HU | Hounsfield units |
| TTP | Time to peak |
| SSF | Spatial scale filter |
| ER | Estrogen receptor |
| PR | Progesterone receptor |
| ANN | Artificial neural network |
| AUC | Area under the receiver-operating characteristic curve |
References
- Madu, C.O.; Wang, S.; Madu, C.O.; Lu, Y. Angiogenesis in Breast Cancer Progression, Diagnosis, and Treatment. J. Cancer 2020, 11, 4474–4494. [Google Scholar] [CrossRef]
- Folkman, J. Tumor angiogenesis: Therapeutic implications. N. Engl. J. Med. 1971, 285, 1182–1186. [Google Scholar]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. 2017, 4, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Januskeviciene, I.; Petrikaite, V. Heterogeneity of breast cancer: The importance of interaction between different tumor cell populations. Life Sci. 2019, 239, 117009. [Google Scholar] [CrossRef] [PubMed]
- Luond, F.; Tiede, S.; Christofori, G. Breast cancer as an example of tumour heterogeneity and tumour cell plasticity during malignant progression. Br. J. Cancer 2021, 125, 164–175. [Google Scholar] [CrossRef]
- Lee, J.Y.; Lee, K.S.; Seo, B.K.; Cho, K.R.; Woo, O.H.; Song, S.E.; Kim, E.K.; Lee, H.Y.; Kim, J.S.; Cha, J. Radiomic machine learning for predicting prognostic biomarkers and molecular subtypes of breast cancer using tumor heterogeneity and angiogenesis properties on MRI. Eur. Radiol. 2021. [Google Scholar] [CrossRef]
- Eun, N.L.; Kang, D.; Son, E.J.; Park, J.S.; Youk, J.H.; Kim, J.A.; Gweon, H.M. Texture Analysis with 3.0-T MRI for Association of Response to Neoadjuvant Chemotherapy in Breast Cancer. Radiology 2020, 294, 31–41. [Google Scholar] [CrossRef]
- Nagasaka, K.; Satake, H.; Ishigaki, S.; Kawai, H.; Naganawa, S. Histogram analysis of quantitative pharmacokinetic parameters on DCE-MRI: Correlations with prognostic factors and molecular subtypes in breast cancer. Breast Cancer 2019, 26, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.R.; Cho, N.; Song, I.C.; Kim, H.; Chang, J.M.; Yi, A.; Yun, B.L.; Moon, W.K. Correlation of perfusion parameters on dynamic contrast-enhanced MRI with prognostic factors and subtypes of breast cancers. J. Magn. Reson. Imaging 2012, 36, 145–151. [Google Scholar] [CrossRef]
- Li, H.; Zhu, Y.; Burnside, E.S.; Huang, E.; Drukker, K.; Hoadley, K.A.; Fan, C.; Conzen, S.D.; Zuley, M.; Net, J.M.; et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer 2016, 2. [Google Scholar] [CrossRef]
- Milon, A.; Wahab, C.A.; Kermarrec, E.; Bekhouche, A.; Taourel, P.; Thomassin-Naggara, I. Breast MRI: Is Faster Better? AJR Am. J. Roentgenol. 2020, 214, 282–295. [Google Scholar] [CrossRef] [PubMed]
- Peters, N.H.; Borel Rinkes, I.H.; Zuithoff, N.P.; Mali, W.P.; Moons, K.G.; Peeters, P.H. Meta-analysis of MR imaging in the diagnosis of breast lesions. Radiology 2008, 246, 116–124. [Google Scholar] [CrossRef]
- Gao, Y.; Heller, S.L. Abbreviated and Ultrafast Breast MRI in Clinical Practice. Radiographics 2020, 40, 1507–1527. [Google Scholar] [CrossRef] [PubMed]
- Park, E.K.; Seo, B.K.; Kwon, M.; Cho, K.R.; Woo, O.H.; Song, S.E.; Cha, J.; Lee, H.Y. Low-Dose Perfusion Computed Tomography for Breast Cancer to Quantify Tumor Vascularity: Correlation With Prognostic Biomarkers. Investig. Radiol. 2019, 54, 273–281. [Google Scholar] [CrossRef]
- Song, S.E.; Seo, B.K.; Cho, K.R.; Woo, O.H.; Ganeshan, B.; Kim, E.S.; Cha, J. Prediction of Inflammatory Breast Cancer Survival Outcomes Using Computed Tomography-Based Texture Analysis. Front. Bioeng. Biotechnol. 2021, 9, 695305. [Google Scholar] [CrossRef] [PubMed]
- Ganeshan, B.; Panayiotou, E.; Burnand, K.; Dizdarevic, S.; Miles, K. Tumour heterogeneity in non-small cell lung carcinoma assessed by CT texture analysis: A potential marker of survival. Eur. Radiol. 2012, 22, 796–802. [Google Scholar] [CrossRef]
- Lubner, M.G.; Smith, A.D.; Sandrasegaran, K.; Sahani, D.V.; Pickhardt, P.J. CT Texture Analysis: Definitions, Applications, Biologic Correlates, and Challenges. Radiographics 2017, 37, 1483–1503. [Google Scholar] [CrossRef]
- Chee, C.G.; Kim, Y.H.; Lee, K.H.; Lee, Y.J.; Park, J.H.; Lee, H.S.; Ahn, S.; Kim, B. CT texture analysis in patients with locally advanced rectal cancer treated with neoadjuvant chemoradiotherapy: A potential imaging biomarker for treatment response and prognosis. PLoS ONE 2017, 12, e0182883. [Google Scholar] [CrossRef] [Green Version]
- Mule, S.; Thiefin, G.; Costentin, C.; Durot, C.; Rahmouni, A.; Luciani, A.; Hoeffel, C. Advanced Hepatocellular Carcinoma: Pretreatment Contrast-enhanced CT Texture Parameters as Predictive Biomarkers of Survival in Patients Treated with Sorafenib. Radiology 2018, 288, 445–455. [Google Scholar] [CrossRef] [Green Version]
- Park, E.K.; Lee, K.S.; Seo, B.K.; Cho, K.R.; Woo, O.H.; Son, G.S.; Lee, H.Y.; Chang, Y.W. Machine Learning Approaches to Radiogenomics of Breast Cancer using Low-Dose Perfusion Computed Tomography: Predicting Prognostic Biomarkers and Molecular Subtypes. Sci. Rep. 2019, 9, 17847. [Google Scholar] [CrossRef]
- Saha, A.; Harowicz, M.R.; Grimm, L.J.; Kim, C.E.; Ghate, S.V.; Walsh, R.; Mazurowski, M.A. A machine learning approach to radiogenomics of breast cancer: A study of 922 subjects and 529 DCE-MRI features. Br. J. Cancer 2018, 119, 508–516. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chen, J.H.; Lin, Y.; Chan, S.; Zhou, J.; Chow, D.; Chang, P.; Kwong, T.; Yeh, D.C.; Wang, X.; et al. Prediction of breast cancer molecular subtypes on DCE-MRI using convolutional neural network with transfer learning between two centers. Eur. Radiol. 2021, 31, 2559–2567. [Google Scholar] [CrossRef] [PubMed]
- Bashir, U.; Siddique, M.M.; McLean, E.; Goh, V.; Cook, G.J. Imaging Heterogeneity in Lung Cancer: Techniques, Applications, and Challenges. AJR Am. J. Roentgenol. 2016, 207, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Genestie, C.; Zafrani, B.; Asselain, B.; Fourquet, A.; Rozan, S.; Validire, P.; Vincent-Salomon, A.; Sastre-Garau, X. Comparison of the prognostic value of Scarff-Bloom-Richardson and Nottingham histological grades in a series of 825 cases of breast cancer: Major importance of the mitotic count as a component of both grading systems. Anticancer Res. 1998, 18, 571–576. [Google Scholar]
- Rakha, E.A.; El-Sayed, M.E.; Lee, A.H.; Elston, C.W.; Grainge, M.J.; Hodi, Z.; Blamey, R.W.; Ellis, I.O. Prognostic significance of Nottingham histologic grade in invasive breast carcinoma. J. Clin. Oncol. 2008, 26, 3153–3158. [Google Scholar] [CrossRef] [PubMed]
- Hammond, M.E.; Hayes, D.F.; Dowsett, M.; Allred, D.C.; Hagerty, K.L.; Badve, S.; Fitzgibbons, P.L.; Francis, G.; Goldstein, N.S.; Hayes, M.; et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch. Pathol. Lab. Med. 2010, 134, 48–72. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 2013, 31, 3997–4013. [Google Scholar] [CrossRef]
- Bustreo, S.; Osella-Abate, S.; Cassoni, P.; Donadio, M.; Airoldi, M.; Pedani, F.; Papotti, M.; Sapino, A.; Castellano, I. Optimal Ki67 cut-off for luminal breast cancer prognostic evaluation: A large case series study with a long-term follow-up. Breast Cancer Res. Treat 2016, 157, 363–371. [Google Scholar] [CrossRef] [Green Version]
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thurlimann, B.; Senn, H.J.; Panel, M. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer. Ann. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef]
- Danish Breast Cancer Cooperative, G.; Nielsen, H.M.; Overgaard, M.; Grau, C.; Jensen, A.R.; Overgaard, J. Study of failure pattern among high-risk breast cancer patients with or without postmastectomy radiotherapy in addition to adjuvant systemic therapy: Long-term results from the Danish Breast Cancer Cooperative Group DBCG 82 b and c randomized studies. J. Clin. Oncol. 2006, 24, 2268–2275. [Google Scholar]
- Kim, J.Y.; Kim, J.J.; Hwangbo, L.; Suh, H.B.; Kim, S.; Choo, K.S.; Nam, K.J.; Kang, T. Kinetic Heterogeneity of Breast Cancer Determined Using Computer-aided Diagnosis of Preoperative MRI Scans: Relationship to Distant Metastasis-Free Survival. Radiology 2020, 295, 517–526. [Google Scholar] [CrossRef]
- Ho, T.K. The Random Subspace Method for Constructing Decision Forests. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20, 832–844. [Google Scholar]
- Tan, P.-N.; Steinbach, M.; Karpatne, A.; Kumar, V. Introduction to Data Mining, 2nd ed.; Pearson: Hoboken, NJ, USA, 2018. [Google Scholar]
- Mettler, F.A., Jr.; Bhargavan, M.; Faulkner, K.; Gilley, D.B.; Gray, J.E.; Ibbott, G.S.; Lipoti, J.A.; Mahesh, M.; McCrohan, J.L.; Stabin, M.G.; et al. Radiologic and nuclear medicine studies in the United States and worldwide: Frequency, radiation dose, and comparison with other radiation sources—1950-2007. Radiology 2009, 253, 520–531. [Google Scholar] [CrossRef]
- Schauer, D.A.; Linton, O.W. National Council on Radiation Protection and Measurements report shows substantial medical exposure increase. Radiology 2009, 253, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Mettler, F.A., Jr.; Huda, W.; Yoshizumi, T.T.; Mahesh, M. Effective doses in radiology and diagnostic nuclear medicine: A catalog. Radiology 2008, 248, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Ytre-Hauge, S.; Dybvik, J.A.; Lundervold, A.; Salvesen, O.O.; Krakstad, C.; Fasmer, K.E.; Werner, H.M.; Ganeshan, B.; Hoivik, E.; Bjorge, L.; et al. Preoperative tumor texture analysis on MRI predicts high-risk disease and reduced survival in endometrial cancer. J. Magn. Reson. Imaging 2018, 48, 1637–1647. [Google Scholar] [CrossRef]
- Ahn, S.Y.; Park, C.M.; Park, S.J.; Kim, H.J.; Song, C.; Lee, S.M.; McAdams, H.P.; Goo, J.M. Prognostic value of computed tomography texture features in non-small cell lung cancers treated with definitive concomitant chemoradiotherapy. Investig. Radiol. 2015, 50, 719–725. [Google Scholar] [CrossRef]
- Kim, J.H.; Ko, E.S.; Lim, Y.; Lee, K.S.; Han, B.K.; Ko, E.Y.; Hahn, S.Y.; Nam, S.J. Breast Cancer Heterogeneity: MR Imaging Texture Analysis and Survival Outcomes. Radiology 2017, 282, 665–675. [Google Scholar] [CrossRef]
- Park, C.M.; Goo, J.M.; Lee, H.J.; Kim, M.A.; Kim, H.C.; Kim, K.G.; Lee, C.H.; Im, J.G. FN13762 murine breast cancer: Region-by-region correlation of first-pass perfusion CT indexes with histologic vascular parameters. Radiology 2009, 251, 721–730. [Google Scholar] [CrossRef]
- Kino, A.; Shaffer, J.; Maturen, K.E.; Schmiedeskamp, H.; Koong, A.C.; Chang, D.T.; Fleischmann, D.; Kamaya, A. Perfusion CT measurements predict tumor response in rectal carcinoma. Abdom. Radiol. 2017, 42, 1132–1140. [Google Scholar] [CrossRef]
- Miles, K.A.; Lee, T.Y.; Goh, V.; Klotz, E.; Cuenod, C.; Bisdas, S.; Groves, A.M.; Hayball, M.P.; Alonzi, R.; Brunner, T.; et al. Current status and guidelines for the assessment of tumour vascular support with dynamic contrast-enhanced computed tomography. Eur. Radiol. 2012, 22, 1430–1441. [Google Scholar] [CrossRef]
- Cuenod, C.A.; Fournier, L.; Balvay, D.; Guinebretiere, J.M. Tumor angiogenesis: Pathophysiology and implications for contrast-enhanced MRI and CT assessment. Abdom. Imaging 2006, 31, 188–193. [Google Scholar] [CrossRef]
- Ko, E.S.; Kim, J.H.; Lim, Y.; Han, B.K.; Cho, E.Y.; Nam, S.J. Assessment of Invasive Breast Cancer Heterogeneity Using Whole-Tumor Magnetic Resonance Imaging Texture Analysis: Correlations With Detailed Pathological Findings. Medicine 2016, 95, e2453. [Google Scholar] [CrossRef] [PubMed]
- Yip, C.; Landau, D.; Kozarski, R.; Ganeshan, B.; Thomas, R.; Michaelidou, A.; Goh, V. Primary esophageal cancer: Heterogeneity as potential prognostic biomarker in patients treated with definitive chemotherapy and radiation therapy. Radiology 2014, 270, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, K.; Sun, X.; Wang, K.; Sun, Y.; Zhang, G.; Shen, B. Parameters of dynamic contrast-enhanced MRI as imaging markers for angiogenesis and proliferation in human breast cancer. Med. Sci. Monit. 2015, 21, 376–382. [Google Scholar] [PubMed] [Green Version]
- Liu, F.; Wang, M.; Li, H. Role of perfusion parameters on DCE-MRI and ADC values on DWMRI for invasive ductal carcinoma at 3.0 Tesla. World J. Surg. Oncol. 2018, 16, 239. [Google Scholar] [CrossRef] [PubMed]
- La Forgia, D.; Fanizzi, A.; Campobasso, F.; Bellotti, R.; Didonna, V.; Lorusso, V.; Moschetta, M.; Massafra, R.; Tamborra, P.; Tangaro, S.; et al. Radiomic Analysis in Contrast-Enhanced Spectral Mammography for Predicting Breast Cancer Histological Outcome. Diagnostics 2020, 10, 708. [Google Scholar] [CrossRef] [PubMed]
- Losurdo, L.; Fanizzi, A.; Basile, T.M.A.; Bellotti, R.; Bottigli, U.; Dentamaro, R.; Didonna, V.; Lorusso, V.; Massafra, R.; Tamborra, P.; et al. Radiomics Analysis on Contrast-Enhanced Spectral Mammography Images for Breast Cancer Diagnosis: A Pilot Study. Entropy 2019, 21, 1110. [Google Scholar] [CrossRef] [Green Version]
- Koh, J.; Kim, M.J. Introduction of a New Staging System of Breast Cancer for Radiologists: An Emphasis on the Prognostic Stage. Korean J. Radiol. 2019, 20, 69–82. [Google Scholar] [CrossRef] [Green Version]
- Cortazar, P.; Zhang, L.; Untch, M.; Mehta, K.; Costantino, J.P.; Wolmark, N.; Bonnefoi, H.; Cameron, D.; Gianni, L.; Valagussa, P.; et al. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet 2014, 384, 164–172. [Google Scholar] [CrossRef] [Green Version]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Lee, W.J.; Seo, B.K.; Cho, P.K.; Yie, A.; Cho, K.R.; Woo, O.H.; Cha, S.H.; Son, G.S.; Lee, G.Y. The Clinical Use of Low-Dose Multidetector Row Computed Tomography for Breast Cancer Patients in the Prone Position. J. Breast Cancer 2010, 13, 357–365. [Google Scholar] [CrossRef]
- Pizzolon, F.; Horvath, E.; Wenzel, H.; Cádiz, F.; Serrano, G.; Aburto, M.; Morgado, O.; Baina, J.; Duran, M.P. Etapificación pre-operatoria con TC de pacientes con cáncer mamario: Alternativa a la RM en un hospital público. Rev. Chil. Radiol. 2014, 20, 135–140. [Google Scholar] [CrossRef] [Green Version]
- James, J.; Teo, M.; Ramachandran, V.; Law, M.; Stoney, D.; Cheng, M. A critical review of the chest CT scans performed to detect asymptomatic synchronous metastasis in new and recurrent breast cancers. World J. Surg. Oncol. 2019, 17, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papanikolaou, N.; Matos, C.; Koh, D.M. How to develop a meaningful radiomic signature for clinical use in oncologic patients. Cancer Imaging 2020, 20, 33. [Google Scholar] [CrossRef]
- Herold, C.J.; Lewin, J.S.; Wibmer, A.G.; Thrall, J.H.; Krestin, G.P.; Dixon, A.K.; Schoenberg, S.O.; Geckle, R.J.; Muellner, A.; Hricak, H. Imaging in the Age of Precision Medicine: Summary of the Proceedings of the 10th Biannual Symposium of the International Society for Strategic Studies in Radiology. Radiology 2016, 279, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Giardino, A.; Gupta, S.; Olson, E.; Sepulveda, K.; Lenchik, L.; Ivanidze, J.; Rakow-Penner, R.; Patel, M.J.; Subramaniam, R.M.; Ganeshan, D. Role of Imaging in the Era of Precision Medicine. Acad. Radiol. 2017, 24, 639–649. [Google Scholar] [CrossRef]
- Lubner, M.G.; Stabo, N.; Lubner, S.J.; del Rio, A.M.; Song, C.; Halberg, R.B.; Pickhardt, P.J. CT textural analysis of hepatic metastatic colorectal cancer: Pre-treatment tumor heterogeneity correlates with pathology and clinical outcomes. Abdom. Imaging 2015, 40, 2331–2337. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | Training Set (n = 110) | Test Set (n = 37) |
|---|---|---|
| Age | 52.2 ± 9 years | 52.5 ± 10 years |
| Tumor size | 25.3 ± 16 mm | 23.5 ± 14 mm |
| ER status | ||
| Negative | 39 (35%) | 9 (24%) |
| Positive | 71 (65%) | 28 (76%) |
| PR status | ||
| Negative | 41 (37%) | 9 (24%) |
| Positive | 69 (63%) | 28 (76%) |
| HER2 status | ||
| Negative | 93 (85%) | 30 (81%) |
| Positive | 17 (15%) | 7 (19%) |
| Ki67 status | ||
| Negative | 50 (45%) | 23 (62%) |
| Positive | 60 (55%) | 14 (38%) |
| Tumor grade | ||
| Low | 68 (62%) | 28 (76%) |
| High | 42 (38%) | 9 (24%) |
| Molecular subtype | ||
| Luminal | 75 (68%) | 28 (76%) |
| Non-luminal | 35(32%) | 9 (24%) |
| CT Perfusion Parameter | ER | PR | HER2 | Ki67 | Grade | Subtype |
|---|---|---|---|---|---|---|
| Hot spot | ||||||
| Perfusion | <0.001 | 0.001 | 0.01 | <0.001 | <0.001 | <0.001 |
| PEI | 0.001 | 0.06 | 0.01 | <0.001 | 0.001 | 0.001 |
| TTP | 0.001 | 0.02 | 0.04 | 0.02 | 0.01 | <0.001 |
| Blood volume | 0.06 | 0.23 | 0.09 | 0.001 | 0.004 | 0.001 |
| Whole tumor | ||||||
| Perfusion | 0.001 | 0.10 | 0.11 | 0.01 | 0.001 | 0.01 |
| PEI | 0.01 | 0.17 | 0.08 | 0.01 | 0.04 | 0.01 |
| TTP | 0.02 | 0.28 | 0.43 | 0.37 | 0.01 | 0.23 |
| Blood volume | 0.04 | 0.14 | 0.35 | 0.001 | 0.01 | 0.001 |
| CT Texture Parameter | ER | PR | HER2 | Ki67 | Grade | Subtype |
|---|---|---|---|---|---|---|
| SSF 0 | ||||||
| Mean_precontrast | 0.11 | 0.41 | 0.003 | 0.02 | <0.001 | 0.01 |
| Standard deviation_precontrast | 0.55 | 0.18 | 0.15 | 0.07 | 0.30 | 0.01 |
| Entropy_precontrast | 0.002 | <0.001 | 0.09 | 0.33 | 0.07 | 0.01 |
| Mean of positive pixels_precontrast | 0.05 | 0.26 | 0.01 | 0.18 | <0.001 | 0.09 |
| Skewness_precontrast | 0.048 | 0.14 | 0.07 | 0.15 | 0.09 | 0.21 |
| Kurtosis_precontrast | 0.05 | 0.11 | 0.12 | 0.06 | 0.02 | 0.18 |
| Mean_postccontrast | 0.05 | 0.38 | 0.01 | 0.02 | 0.01 | 0.03 |
| Standard deviation_postcontrast | 0.56 | 0.41 | 0.16 | 0.24 | 0.51 | 0.05 |
| Entropy_postcontrast | <0.001 | <0.001 | 0.003 | 0.02 | 0.002 | <0.001 |
| Mean of positive pixels_postcontrast | 0.049 | 0.43 | 0.02 | 0.05 | 0.02 | 0.07 |
| Skewness_postcontrast | 0.01 | 0.15 | 0.06 | 0.09 | 0.03 | 0.05 |
| Kurtosis_postcontrast | 0.01 | 0.18 | 0.22 | 0.048 | 0.06 | 0.08 |
| SSF 2 | ||||||
| Mean_precontrast | 0.046 | 0.12 | 0.12 | 0.01 | 0.06 | 0.01 |
| Standard deviation_precontrast | 0.82 | 0.19 | 0.19 | 0.92 | 0.68 | 0.32 |
| Entropy_precontrast | <0.001 | <0.001 | <0.001 | 0.01 | 0.002 | <0.001 |
| Mean of positive pixels_precontrast | 0.21 | 0.55 | 0.55 | 0.04 | 0.18 | 0.01 |
| Skewness_precontrast | 0.13 | 0.46 | 0.46 | 0.70 | 0.23 | 0.23 |
| Kurtosis_precontrast | 0.77 | 0.37 | 0.37 | 0.79 | 0.68 | 0.85 |
| Mean_postccontrast | 0.04 | 0.06 | 0.06 | 0.03 | 0.046 | 0.03 |
| Standard deviation_postcontrast | 0.63 | 0.72 | 0.72 | 0.37 | 0.85 | 0.78 |
| Entropy_postcontrast | <0.001 | <0.001 | <0.001 | 0.001 | <0.001 | <0.001 |
| Mean of positive pixels_postcontrast | 0.16 | 0.26 | 0.26 | 0.15 | 0.21 | 0.08 |
| Skewness_postcontrast | 0.97 | 0.50 | 0.50 | 0.18 | 0.35 | 0.02 |
| Kurtosis_postcontrast | 0.70 | 0.55 | 0.55 | 0.20 | 0.18 | 0.50 |
| SSF 5 | ||||||
| Mean_precontrast | 0.18 | 0.29 | 0.21 | 0.02 | 0.27 | 0.04 |
| Standard deviation_precontrast | 0.88 | 0.55 | 0.10 | 0.84 | 0.86 | 0.79 |
| Entropy_precontrast | <0.001 | <0.001 | <0.001 | 0.004 | 0.003 | <0.001 |
| Mean of positive pixels_precontrast | 0.21 | 0.34 | 0.22 | 0.05 | 0.33 | 0.11 |
| Skewness_precontrast | 0.57 | 0.93 | 0.35 | 0.80 | 0.97 | 0.63 |
| Kurtosis_precontrast | 0.01 | 0.04 | 0.01 | 0.11 | 0.04 | 0.02 |
| Mean_postccontrast | 0.14 | 0.30 | 0.14 | 0.07 | 0.19 | 0.10 |
| Standard deviation_postcontrast | 0.67 | 0.99 | 0.63 | 0.87 | 0.71 | 0.73 |
| Entropy_postcontrast | <0.001 | <0.001 | <0.001 | 0.001 | 0.001 | <0.001 |
| Mean of positive pixels_postcontrast | 0.20 | 0.41 | 0.16 | 0.10 | 0.24 | 0.12 |
| Skewness_postcontrast | 0.74 | 0.97 | 0.26 | 0.10 | 0.96 | 0.58 |
| Kurtosis_postcontrast | 0.03 | 0.12 | 0.07 | 0.15 | 0.59 | 0.12 |
| Machine Learning Model | Diagnostic Performance | Perfusion Features * | Texture Features † | Integrating Perfusion and Texture Features ‡ | p Value § |
|---|---|---|---|---|---|
| Decision tree | AUC median | 0.55 | 0.59 | 0.65 | 0.04 |
| AUC mean | 0.55 | 0.58 | 0.62 | ||
| AUC SD | 0.25 | 0.34 | 0.28 | ||
| AUC 95% CI | 0.35, 0.75 | 0.42, 0.74 | 0.40, 0.84 | ||
| accuracy | 59% | 66% | 73% | ||
| sensitivity | 68% | 38% | 51% | ||
| specificity | 47% | 64% | 73% | ||
| NPV | 42% | 49% | 61% | ||
| PPV | 73% | 57% | 58% | ||
| Naïve Bayes | AUC median | 0.69 | 0.54 | 0.73 | 0.63 |
| AUC mean | 0.69 | 0.59 | 0.71 | ||
| AUC SD | 0.31 | 0.18 | 0.32 | ||
| AUC 95% CI | 0.44, 0.94 | 0.51, 0.67 | 0.45, 0.97 | ||
| accuracy | 67% | 51% | 65% | ||
| sensitivity | 80% | 60% | 60% | ||
| specificity | 52% | 67% | 64% | ||
| NPV | 56% | 63% | 78% | ||
| PPV | 77% | 69% | 61% | ||
| Logistic regression | AUC median | 0.65 | 0.50 | 0.71 | 0.41 |
| AUC mean | 0.63 | 0.53 | 0.70 | ||
| AUC SD | 0.29 | 0.42 | 0.32 | ||
| AUC 95% CI | 0.40, 0.86 | 0.34, 0.72 | 0.44, 0.96 | ||
| accuracy | 63% | 62% | 73% | ||
| sensitivity | 80% | 24% | 46% | ||
| specificity | 43% | 68% | 71% | ||
| NPV | 53% | 45% | 64% | ||
| PPV | 74% | 25% | 27% | ||
| ANN | AUC median | 0.60 | 0.56 | 0.66 | 0.17 |
| AUC mean | 0.60 | 0.57 | 0.68 | ||
| AUC SD | 0.27 | 0.28 | 0.31 | ||
| AUC 95% CI | 0.38, 0.82 | 0.44, 0.70 | 0.43, 0.93 | ||
| accuracy | 61% | 59% | 68% | ||
| sensitivity | 70% | 39% | 55% | ||
| specificity | 43% | 67% | 72% | ||
| NPV | 38% | 47% | 71% | ||
| PPV | 74% | 55% | 61% | ||
| Random forest | AUC median | 0.65 | 0.61 | 0.76 | … |
| AUC mean | 0.66 | 0.61 | 0.75 | ||
| AUC SD | 0.30 | 0.32 | 0.34 | ||
| AUC 95% CI | 0.42, 0.90 | 0.46, 0.76 | 0.48, 1.00 | ||
| accuracy | 65% | 65% | 74% | ||
| sensitivity | 81% | 27% | 50% | ||
| specificity | 36% | 72% | 76% | ||
| NPV | 48% | 48% | 69% | ||
| PPV | 72% | 64% | 70% |
| Machine Learning Model | Diagnostic Performance | ER | PR | HER2 | Ki67 | Grade | Subtype |
|---|---|---|---|---|---|---|---|
| Decision tree | AUC median | 0.65 | 0.55 | 0.61 | 0.53 | 0.69 | 0.68 |
| accuracy | 77% | 59% | 83% | 52% | 73% | 73% | |
| Naïve Bayes | AUC median | 0.76 | 0.60 | 0.75 | 0.72 | 0.73 | 0.68 |
| accuracy | 65% | 59% | 62% | 68% | 73% | 65% | |
| Logistic regression | AUC median | 0.76 | 072 | 0.67 | 0.55 | 0.69 | 0.79 |
| accuracy | 70% | 73% | 89% | 49% | 73% | 81% | |
| ANN | AUC median | 0.66 | 0.60 | 0.67 | 0.66 | 0.73 | 0.75 |
| accuracy | 68% | 54% | 84% | 68% | 68% | 65% | |
| Random forest | AUC median | 0.76 | 0.69 | 0.86 | 0.65 | 0.75 | 0.79 |
| accuracy | 76% | 74% | 92% | 65% | 67% | 75% |
| Rank | Important CT Parameters |
|---|---|
| 1 | Entropy_postcontrast |
| 2 | Perfusion_hot spot |
| 3 | TTP_hot spot |
| 4 | PEI_hot spot |
| 5 | Entropy_precontrast |
| Histological Facor | Entropy_ Postcontrast | Perfusion_ Hot Spot | TTP_ Hot Spot | PEI_ Hot Spot | Entropy_Precontrast |
|---|---|---|---|---|---|
| ER | |||||
| − | 4.75 | 46.98 4.34 | 34.68 | 82.22 | 4.54 |
| + | 4.50 | 26.94 | 51.92 | 65.54 | 4.35 |
| HER2 | |||||
| − | 4.55 | 31.33 | 47.63 | 68.69 | 4.40 |
| + | 4.76 | 48.88 | 35.82 | 86.61 | 4.54 |
| Grade | |||||
| low | 4.52 | 27.67 | 51.01 | 64.33 | 4.36 |
| high | 4.70 | 44.36 | 37.38 | 83.00 | 4.51 |
| Subtype | |||||
| luminal A | 4.48 | 20.60 | 53.20 | 58.03 | 4.35 |
| luminal B | 4.54 | 35.46 | 54.59 | 80.88 | 4.35 0.05 |
| HER2-enriched | 4.78 | 59.22 | 21.06 | 86.98 | 4.58 0.04 |
| Triple-negative | 4.75 | 44.71 | 33.32 | 76.89 | 4.56 0.05 |
| CT Parameter | AUC | p Value * |
|---|---|---|
| All parameters for predicting ER, HER2, and molecular subtype | ||
| Decision tree | 0.59 | 0.002 |
| Naïve Bayes | 0.75 | 0.52 |
| Logistic regression | 0.76 | 0.63 |
| ANN | 0.67 | 0.04 |
| Random forest | 0.79 | … |
| Top five important parameters for predicting ER, HER2, and molecular subtype | ||
| Decision tree | 0.62 | 0.03 |
| Naïve Bayes | 0.76 | 0.95 |
| Logistic regression | 0.76 | 0.97 |
| ANN | 0.70 | 0.34 |
| Random forest | 0.76 | … |
| All parameters for predicting ER, HER2, molecular subtype, and treatment failure | ||
| Decision tree | 0.52 | 0.001 |
| Naïve Bayes | 0.81 | 0.29 |
| Logistic regression | 0.69 | 0.26 |
| ANN | 0.70 | 0.36 |
| Random forest | 0.76 | … |
| Top five parameters for predicting ER, HER2, molecular subtype, and treatment failure | ||
| Decision tree | 0.52 | 0.001 |
| Naïve Bayes | 0.83 | 0.06 |
| Logistic regression | 0.72 | 0.82 |
| ANN | 0.68 | 0.64 |
| Random forest | 0.74 | … |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, H.-S.; Lee, K.-s.; Seo, B.-K.; Kim, E.-S.; Cho, K.-R.; Woo, O.-H.; Song, S.-E.; Lee, J.-Y.; Cha, J. Machine Learning Models That Integrate Tumor Texture and Perfusion Characteristics Using Low-Dose Breast Computed Tomography Are Promising for Predicting Histological Biomarkers and Treatment Failure in Breast Cancer Patients. Cancers 2021, 13, 6013. https://doi.org/10.3390/cancers13236013
Park H-S, Lee K-s, Seo B-K, Kim E-S, Cho K-R, Woo O-H, Song S-E, Lee J-Y, Cha J. Machine Learning Models That Integrate Tumor Texture and Perfusion Characteristics Using Low-Dose Breast Computed Tomography Are Promising for Predicting Histological Biomarkers and Treatment Failure in Breast Cancer Patients. Cancers. 2021; 13(23):6013. https://doi.org/10.3390/cancers13236013
Chicago/Turabian StylePark, Hyun-Soo, Kwang-sig Lee, Bo-Kyoung Seo, Eun-Sil Kim, Kyu-Ran Cho, Ok-Hee Woo, Sung-Eun Song, Ji-Young Lee, and Jaehyung Cha. 2021. "Machine Learning Models That Integrate Tumor Texture and Perfusion Characteristics Using Low-Dose Breast Computed Tomography Are Promising for Predicting Histological Biomarkers and Treatment Failure in Breast Cancer Patients" Cancers 13, no. 23: 6013. https://doi.org/10.3390/cancers13236013
APA StylePark, H.-S., Lee, K.-s., Seo, B.-K., Kim, E.-S., Cho, K.-R., Woo, O.-H., Song, S.-E., Lee, J.-Y., & Cha, J. (2021). Machine Learning Models That Integrate Tumor Texture and Perfusion Characteristics Using Low-Dose Breast Computed Tomography Are Promising for Predicting Histological Biomarkers and Treatment Failure in Breast Cancer Patients. Cancers, 13(23), 6013. https://doi.org/10.3390/cancers13236013






