Immune Checkpoints and Innate Lymphoid Cells—New Avenues for Cancer Immunotherapy
Abstract
Simple Summary
Abstract
1. Introduction
2. Innate Lymphoid Cell Diversity within Tissues
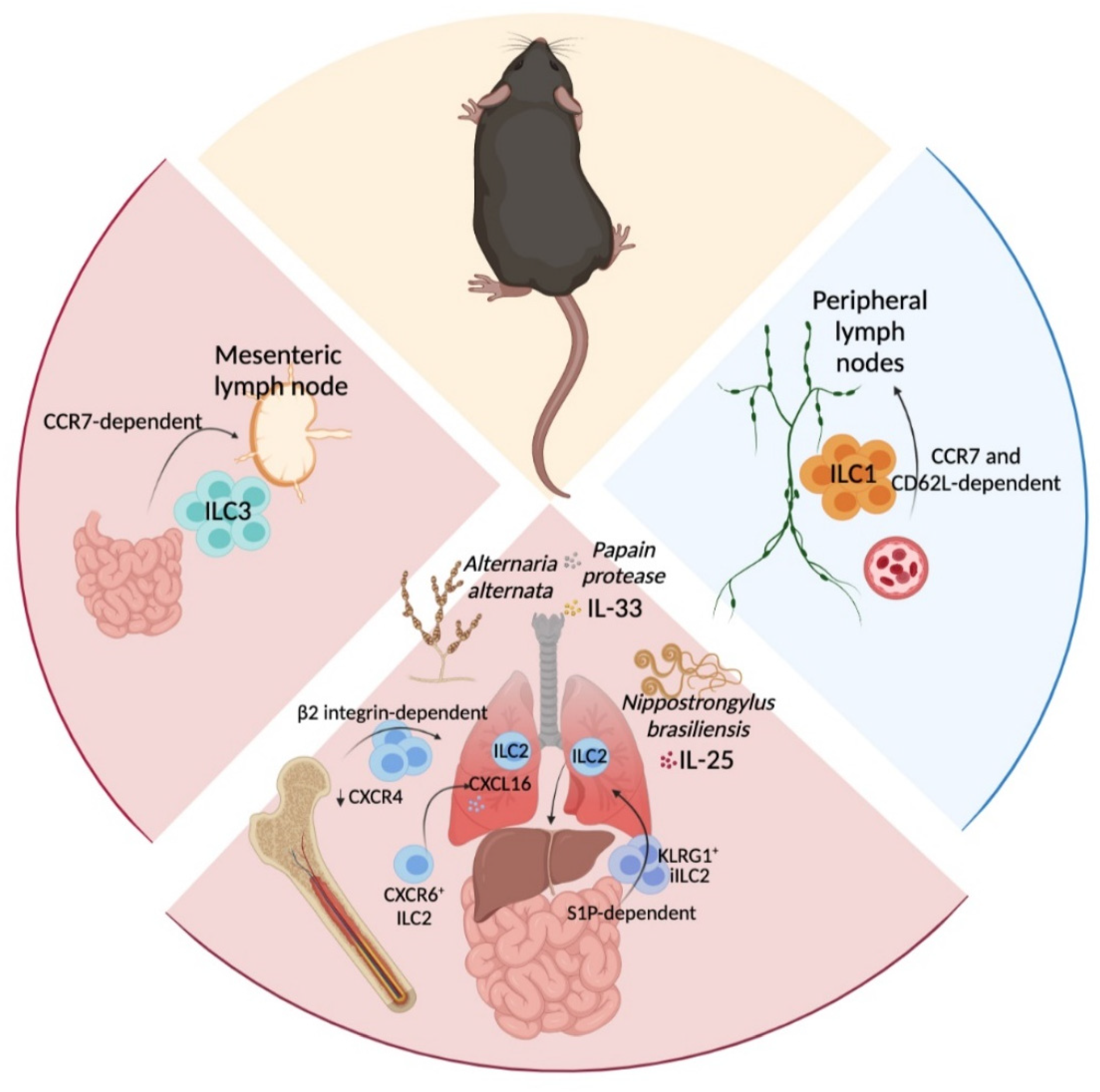
3. In Situ ILC-Poiesis and Interorgan ILC Migration
4. Peripheral Immune Tolerance—The Role of Immune Checkpoints
5. ILC and Immune Checkpoints. What Do We Know?
6. ILC Expression of IC—Parallel with Adaptive Immune Cells
6.1. PD-1 and Its Ligands
6.1.1. PD-1 Expression in ILC Development
6.1.2. PD-1 and NK Cells
6.1.3. PD-1 and ILC1
6.1.4. PD-1 and ILC2
6.1.5. PD-1 and ILC3
6.2. CTLA-4—CD80/CD86
6.3. TIM-3
6.4. TIGIT, DNAM-1 and CD96
6.5. ICOS—ICOSL
6.6. LAG3
6.7. CD137/4-1BB
6.8. KLRG1
6.9. GITR—GITRL
6.10. BTLA—HVEM
6.11. OX40—OX40L
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Artis, D.; Spits, H. The biology of innate lymphoid cells. Nature 2015, 517, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Eberl, G.; Colonna, M.; Di Santo, J.P.; McKenzie, A.N.J. Innate lymphoid cells. Innate lymphoid cells: A new paradigm in immunology. Science 2015, 348, aaa6566. [Google Scholar] [CrossRef] [PubMed]
- Vivier, E.; Artis, D.; Colonna, M.; Diefenbach, A.; Di Santo, J.P.; Eberl, G.; Koyasu, S.; Locksley, R.M.; McKenzie, A.N.J.; Mebius, R.E.; et al. Innate lymphoid cells: 10 years on. Cell 2018, 174, 1054–1066. [Google Scholar] [CrossRef] [PubMed]
- Mebius, R.E.; Rennert, P.; Weissman, I.L. Developing lymph nodes collect CD4+CD3- LTbeta+ cells that can differentiate to APC, NK Cells, and follicular cells but not T or B cells. Immunity 1997, 7, 493–504. [Google Scholar] [CrossRef]
- Sun, Z.; Unutmaz, D.; Zou, Y.R.; Sunshine, M.J.; Pierani, A.; Brenner-Morton, S.; Mebius, R.E.; Littman, D.R. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science 2000, 288, 2369–2373. [Google Scholar] [CrossRef] [PubMed]
- Eberl, G.; Marmon, S.; Sunshine, M.-J.; Rennert, P.D.; Choi, Y.; Littman, D.R. An Essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 2004, 5, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Ghaedi, M.; Takei, F. Innate Lymphoid Cell Development. J. Allergy Clin. Immunol. 2021, 147, 1549–1560. [Google Scholar] [CrossRef] [PubMed]
- Almeida, F.F.; Jacquelot, N.; Belz, G.T. Deconstructing deployment of the innate immune lymphocyte army for barrier homeostasis and protection. Immunol. Rev. 2018, 286, 6–22. [Google Scholar] [CrossRef]
- Weizman, O.-E.; Adams, N.M.; Schuster, I.S.; Krishna, C.; Pritykin, Y.; Lau, C.; Degli-Esposti, M.A.; Leslie, C.S.; Sun, J.C.; O’Sullivan, T.E. ILC1 confer early host protection at initial sites of viral infection. Cell 2017, 171, 795–808.e12. [Google Scholar] [CrossRef] [PubMed]
- Spits, H.; Bernink, J.H.; Lanier, L. NK Cells and Type 1 innate lymphoid cells: Partners in host defense. Nat. Immunol. 2016, 17, 758–764. [Google Scholar] [CrossRef] [PubMed]
- Dadi, S.; Chhangawala, S.; Whitlock, B.M.; Franklin, R.A.; Luo, C.T.; Oh, S.A.; Toure, A.; Pritykin, Y.; Huse, M.; Leslie, C.S.; et al. Cancer immunosurveillance by tissue-resident innate lymphoid cells and innate-like T cells. Cell 2016, 164, 365–377. [Google Scholar] [CrossRef]
- Krabbendam, L.; Bal, S.M.; Spits, H.; Golebski, K. New Insights into the function, development, and plasticity of Type 2 innate lymphoid cells. Immunol. Rev. 2018, 286, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Melo-Gonzalez, F.; Hepworth, M.R. Functional and phenotypic heterogeneity of group 3 innate lymphoid cells. Immunology 2017, 150, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Klose, C.S.N.; Artis, D. Innate lymphoid cells control signaling circuits to regulate tissue-specific immunity. Cell Res. 2020, 30, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Klose, C.S.N.; Artis, D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat. Immunol. 2016, 17, 765–774. [Google Scholar] [CrossRef]
- Ricardo-Gonzalez, R.R.; van Dyken, S.J.; Schneider, C.; Lee, J.; Nussbaum, J.C.; Liang, H.-E.; Vaka, D.; Eckalbar, W.L.; Molofsky, A.B.; Erle, D.J.; et al. Tissue signals imprint ILC2 identity with anticipatory function. Nat. Immunol. 2018, 19, 1093–1099. [Google Scholar] [CrossRef]
- Meininger, I.; Carrasco, A.; Rao, A.; Soini, T.; Kokkinou, E.; Mjösberg, J. Tissue-specific features of innate lymphoid cells. Trends Immunol. 2020, 41, 902–917. [Google Scholar] [CrossRef] [PubMed]
- Goc, J.; Lv, M.; Bessman, N.J.; Flamar, A.-L.; Sahota, S.; Suzuki, H.; Teng, F.; Putzel, G.G.; JRI Live Cell Bank; Eberl, G.; et al. Dysregulation of ILC3s unleashes progression and immunotherapy resistance in colon cancer. Cell 2021, 184, 5015–5030. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.K.; Colonna, M. Innate lymphoid cells in mucosal immunity. Front. Immunol. 2019, 10, 861. [Google Scholar] [CrossRef] [PubMed]
- Caputa, G.; Castoldi, A.; Pearce, E.J. Metabolic adaptations of tissue-resident immune cells. Nat. Immunol. 2019, 20, 793–801. [Google Scholar] [CrossRef]
- Di Luccia, B.; Gilfillan, S.; Cella, M.; Colonna, M.; Huang, S.C.-C. ILC3s integrate glycolysis and mitochondrial production of reactive oxygen species to fulfill activation demands. J. Exp. Med. 2019, 216, 2231–2241. [Google Scholar] [CrossRef]
- Pokrovskii, M.; Hall, J.A.; Ochayon, D.E.; Yi, R.; Chaimowitz, N.S.; Seelamneni, H.; Carriero, N.; Watters, A.; Waggoner, S.N.; Littman, D.R.; et al. Characterization of transcriptional regulatory networks that promote and restrict identities and functions of intestinal innate lymphoid cells. Immunity 2019, 51, 185–197.e6. [Google Scholar] [CrossRef]
- Bal, S.M.; Golebski, K.; Spits, H. Plasticity of innate lymphoid cell subsets. Nat. Rev. Immunol. 2020, 20, 552–565. [Google Scholar] [CrossRef]
- Bielecki, P.; Riesenfeld, S.J.; Hütter, J.-C.; Torlai Triglia, E.; Kowalczyk, M.S.; Ricardo-Gonzalez, R.R.; Lian, M.; Amezcua Vesely, M.C.; Kroehling, L.; Xu, H.; et al. Skin-resident innate lymphoid cells converge on a pathogenic effector state. Nature 2021, 592, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Vonarbourg, C.; Mortha, A.; Bui, V.L.; Hernandez, P.P.; Kiss, E.A.; Hoyler, T.; Flach, M.; Bengsch, B.; Thimme, R.; Hölscher, C.; et al. Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγt(+) innate lymphocytes. Immunity 2010, 33, 736–751. [Google Scholar] [CrossRef]
- Nussbaum, K.; Burkhard, S.H.; Ohs, I.; Mair, F.; Klose, C.S.N.; Arnold, S.J.; Diefenbach, A.; Tugues, S.; Becher, B. Tissue microenvironment dictates the fate and tumor-suppressive function of Type 3 ILCs. J. Exp. Med. 2017, 214, 2331–2347. [Google Scholar] [CrossRef] [PubMed]
- Yudanin, N.A.; Schmitz, F.; Flamar, A.-L.; Thome, J.J.C.; Tait Wojno, E.; Moeller, J.B.; Schirmer, M.; Latorre, I.J.; Xavier, R.J.; Farber, D.L.; et al. Spatial and temporal mapping of human innate lymphoid cells reveals elements of tissue specificity. Immunity 2019, 50, 505–519.e4. [Google Scholar] [CrossRef] [PubMed]
- Grégoire, C.; Chasson, L.; Luci, C.; Tomasello, E.; Geissmann, F.; Vivier, E.; Walzer, T. The trafficking of natural killer cells. Immunol. Rev. 2007, 220, 169–182. [Google Scholar] [CrossRef]
- Crinier, A.; Milpied, P.; Escalière, B.; Piperoglou, C.; Galluso, J.; Balsamo, A.; Spinelli, L.; Cervera-Marzal, I.; Ebbo, M.; Girard-Madoux, M.; et al. High-dimensional single-cell analysis identifies organ-specific signatures and conserved NK cell subsets in humans and mice. Immunity 2018, 49, 971–986.e5. [Google Scholar] [CrossRef] [PubMed]
- Dogra, P.; Rancan, C.; Ma, W.; Toth, M.; Senda, T.; Carpenter, D.J.; Kubota, M.; Matsumoto, R.; Thapa, P.; Szabo, P.A.; et al. Tissue determinants of human NK cell development, function, and residence. Cell 2020, 180, 749–763.e13. [Google Scholar] [CrossRef] [PubMed]
- Seillet, C.; Luong, K.; Tellier, J.; Jacquelot, N.; Shen, R.D.; Hickey, P.; Wimmer, V.C.; Whitehead, L.; Rogers, K.; Smyth, G.K.; et al. The neuropeptide VIP confers anticipatory mucosal immunity by regulating ILC3 activity. Nat. Immunol. 2020, 21, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Nussbaum, J.C.; Van Dyken, S.J.; von Moltke, J.; Cheng, L.E.; Mohapatra, A.; Molofsky, A.B.; Thornton, E.E.; Krummel, M.F.; Chawla, A.; Liang, H.-E.; et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 2013, 502, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Molofsky, A.B.; Nussbaum, J.C.; Liang, H.-E.; Van Dyken, S.J.; Cheng, L.E.; Mohapatra, A.; Chawla, A.; Locksley, R.M. Innate lymphoid Type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J. Exp. Med. 2013, 210, 535–549. [Google Scholar] [CrossRef] [PubMed]
- Mazzurana, L.; Czarnewski, P.; Jonsson, V.; Wigge, L.; Ringnér, M.; Williams, T.C.; Ravindran, A.; Björklund, Å.K.; Säfholm, J.; Nilsson, G.; et al. Tissue-specific transcriptional imprinting and heterogeneity in human innate lymphoid cells revealed by full-length single-cell RNA-sequencing. Cell Res. 2021, 31, 554–568. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Vienne, M.; Tang, L.; Kerdiles, Y.; Etiennot, M.; Escalière, B.; Galluso, J.; Wei, H.; Sun, R.; Vivier, E.; et al. Liver Type 1 innate lymphoid cells develop locally via an interferon-γ-dependent loop. Science 2021, 371, 6536. [Google Scholar] [CrossRef]
- Mackay, L.K.; Minnich, M.; Kragten, N.A.M.; Liao, Y.; Nota, B.; Seillet, C.; Zaid, A.; Man, K.; Preston, S.; Freestone, D.; et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 2016, 352, 459–463. [Google Scholar] [CrossRef]
- Robinette, M.L.; Fuchs, A.; Cortez, V.S.; Lee, J.S.; Wang, Y.; Durum, S.K.; Gilfillan, S.; Colonna, M. Immunological genome consortium transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 2015, 16, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Cretney, E.; Hayakawa, Y.; Ota, T.; Akiba, H.; Ogasawara, K.; Yagita, H.; Kinoshita, K.; Okumura, K.; Smyth, M.J. TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood 2005, 105, 2082–2089. [Google Scholar] [CrossRef] [PubMed]
- Cortez, V.S.; Cervantes-Barragan, L.; Robinette, M.L.; Bando, J.K.; Wang, Y.; Geiger, T.L.; Gilfillan, S.; Fuchs, A.; Vivier, E.; Sun, J.C.; et al. Transforming growth factor-β signaling guides the differentiation of innate lymphoid cells in salivary glands. Immunity 2016, 44, 1127–1139. [Google Scholar] [CrossRef] [PubMed]
- Simoni, Y.; Fehlings, M.; Kløverpris, H.N.; McGovern, N.; Koo, S.-L.; Loh, C.Y.; Lim, S.; Kurioka, A.; Fergusson, J.R.; Tang, C.-L.; et al. Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity 2017, 46, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Roan, F.; Stoklasek, T.A.; Whalen, E.; Molitor, J.A.; Bluestone, J.A.; Buckner, J.H.; Ziegler, S.F. CD4+ Group 1 innate lymphoid cells form a functionally distinct ILC subset that is increased in systemic sclerosis. J. Immunol. 2016, 196, 2051–2062. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Zhang, J.; Ji, Y.; Sun, H.; Gu, Z.; Sun, Q.; Bai, M.; Gong, J.; Tang, J.; Zhang, Y.; et al. Tissue signals imprint aiolos expression in ILC2s to modulate Type 2 immunity. Mucosal Immunol. 2021, 14, 1306–1322. [Google Scholar] [CrossRef]
- Camelo, A.; Rosignoli, G.; Ohne, Y.; Stewart, R.A.; Overed-Sayer, C.; Sleeman, M.A.; May, R.D. IL-33, IL-25, and TSLP induce a distinct phenotypic and activation profile in human type 2 innate lymphoid cells. Blood Adv. 2017, 1, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Moral, J.A.; Leung, J.; Rojas, L.A.; Ruan, J.; Zhao, J.; Sethna, Z.; Ramnarain, A.; Gasmi, B.; Gururajan, M.; Redmond, D.; et al. ILC2s amplify PD-1 blockade by activating tissue-specific cancer immunity. Nature 2020, 579, 130–135. [Google Scholar] [CrossRef]
- Halim, T.Y.F.; Krauss, R.H.; Sun, A.C.; Takei, F. Lung natural helper cells are a critical source of Th2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity 2012, 36, 451–463. [Google Scholar] [CrossRef]
- Jacquelot, N.; Seillet, C.; Wang, M.; Pizzolla, A.; Liao, Y.; Hediyeh-zadeh, S.; Grisaru-Tal, S.; Louis, C.; Huang, Q.; Schreuder, J.; et al. Blockade of the co-inhibitory molecule PD-1 unleashes ILC2-dependent antitumor immunity in melanoma. Nat. Immunol. 2021, 22, 851–864. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, M.F.; Trabanelli, S.; Racle, J.; Salomé, B.; Cesson, V.; Gharbi, D.; Bohner, P.; Domingos-Pereira, S.; Dartiguenave, F.; Fritschi, A.-S.; et al. ILC2-modulated T Cell-to-MDSC balance is associated with bladder cancer recurrence. J. Clin. Investig. 2017, 127, 2916–2929. [Google Scholar] [CrossRef] [PubMed]
- Trabanelli, S.; Chevalier, M.F.; Martinez-Usatorre, A.; Gomez-Cadena, A.; Salomé, B.; Lecciso, M.; Salvestrini, V.; Verdeil, G.; Racle, J.; Papayannidis, C.; et al. Tumour-derived PGD2 and NKp30-B7H6 engagement drives an immunosuppressive ILC2-MDSC axis. Nat. Commun. 2017, 8, 593. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, B.; Gertz, E.M.; Schäffer, A.A.; Craig, A.; Ruf, B.; Subramanyam, V.; McVey, J.C.; Diggs, L.P.; Heinrich, S.; Rosato, U.; et al. The tumour microenvironment shapes innate lymphoid cells in patients with Hepatocellular Carcinoma. Gut 2021, gutjnl-2021-325288, Online ahead of print. [Google Scholar] [CrossRef]
- Klose, C.S.N.; Kiss, E.A.; Schwierzeck, V.; Ebert, K.; Hoyler, T.; d’Hargues, Y.; Göppert, N.; Croxford, A.L.; Waisman, A.; Tanriver, Y.; et al. A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature 2013, 494, 261–265. [Google Scholar] [CrossRef]
- Rankin, L.C.; Groom, J.R.; Chopin, M.; Herold, M.J.; Walker, J.A.; Mielke, L.A.; McKenzie, A.N.J.; Carotta, S.; Nutt, S.L.; Belz, G.T. The transcription factor T-Bet is essential for the development of NKp46+ innate lymphocytes via the Notch Pathway. Nat. Immunol. 2013, 14, 389–395. [Google Scholar] [CrossRef]
- Tizian, C.; Lahmann, A.; Hölsken, O.; Cosovanu, C.; Kofoed-Branzk, M.; Heinrich, F.; Mashreghi, M.-F.; Kruglov, A.; Diefenbach, A.; Neumann, C. C-Maf restrains T-Bet-driven programming of CCR6-negative group 3 innate lymphoid cells. eLife 2020, 9, e52549. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.E.; Barrera, A.; Wheaton, J.D.; Zuberbuehler, M.K.; Allan, D.S.J.; Carlyle, J.R.; Reddy, T.E.; Ciofani, M. C-Maf regulates the plasticity of group 3 innate lymphoid cells by restraining the type 1 program. J. Exp. Med. 2020, 217, e20191030. [Google Scholar] [CrossRef]
- Chea, S.; Perchet, T.; Petit, M.; Verrier, T.; Guy-Grand, D.; Banchi, E.-G.; Vosshenrich, C.A.J.; Di Santo, J.P.; Cumano, A.; Golub, R. Notch signaling in group 3 innate lymphoid cells modulates their plasticity. Sci. Signal. 2016, 9, ra45. [Google Scholar] [CrossRef] [PubMed]
- Viant, C.; Rankin, L.C.; Girard-Madoux, M.J.H.; Seillet, C.; Shi, W.; Smyth, M.J.; Bartholin, L.; Walzer, T.; Huntington, N.D.; Vivier, E.; et al. Transforming growth factor-β and notch ligands act as opposing environmental cues in regulating the plasticity of type 3 innate lymphoid cells. Sci. Signal. 2016, 9, ra46. [Google Scholar] [CrossRef] [PubMed]
- Cella, M.; Gamini, R.; Sécca, C.; Collins, P.L.; Zhao, S.; Peng, V.; Robinette, M.L.; Schettini, J.; Zaitsev, K.; Gordon, W.; et al. Subsets of ILC3−ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat. Immunol. 2019, 20, 980–991. [Google Scholar] [CrossRef] [PubMed]
- Lim, A.I.; Li, Y.; Lopez-Lastra, S.; Stadhouders, R.; Paul, F.; Casrouge, A.; Serafini, N.; Puel, A.; Bustamante, J.; Surace, L.; et al. Systemic human ILC precursors provide a substrate for tissue ILC differentiation. Cell 2017, 168, 1086–1100.e10. [Google Scholar] [CrossRef]
- Nagasawa, M.; Heesters, B.A.; Kradolfer, C.M.A.; Krabbendam, L.; Martinez-Gonzalez, I.; de Bruijn, M.J.W.; Golebski, K.; Hendriks, R.W.; Stadhouders, R.; Spits, H.; et al. KLRG1 and NKp46 Discriminate subpopulations of human CD117+CRTH2− ILCs biased toward ILC2 or ILC3. J. Exp. Med. 2019, 216, 1762–1776. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, G.; Fan, X.; Dikiy, S.; Lee, S.Y.; Rudensky, A.Y. Tissue residency of innate lymphoid cells in lymphoid and non-lymphoid organs. Science 2015, 350, 981–985. [Google Scholar] [CrossRef]
- Moro, K.; Kabata, H.; Tanabe, M.; Koga, S.; Takeno, N.; Mochizuki, M.; Fukunaga, K.; Asano, K.; Betsuyaku, T.; Koyasu, S. Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat. Immunol. 2016, 17, 76–86. [Google Scholar] [CrossRef]
- Kim, M.H.; Taparowsky, E.J.; Kim, C.H. Retinoic acid differentially regulates the migration of innate lymphoid cell subsets to the gut. Immunity 2015, 43, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Dutton, E.E.; Gajdasik, D.W.; Willis, C.; Fiancette, R.; Bishop, E.L.; Camelo, A.; Sleeman, M.A.; Coccia, M.; Didierlaurent, A.M.; Tomura, M.; et al. Peripheral lymph nodes contain migratory and resident innate lymphoid cell populations. Sci. Immunol. 2019, 4. [Google Scholar] [CrossRef]
- Kobayashi, T.; Voisin, B.; Kim, D.Y.; Kennedy, E.A.; Jo, J.-H.; Shih, H.-Y.; Truong, A.; Doebel, T.; Sakamoto, K.; Cui, C.-Y.; et al. Homeostatic control of sebaceous glands by innate lymphoid cells regulates commensal bacteria equilibrium. Cell 2019, 176, 982–997.e16. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Mao, K.; Chen, X.; Sun, M.-A.; Kawabe, T.; Li, W.; Usher, N.; Zhu, J.; Urban, J.F.; Paul, W.E.; et al. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science 2018, 359, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Stier, M.T.; Zhang, J.; Goleniewska, K.; Cephus, J.Y.; Rusznak, M.; Wu, L.; Van Kaer, L.; Zhou, B.; Newcomb, D.C.; Peebles, R.S., Jr. IL-33 Promotes the Egress of Group 2 Innate Lymphoid Cells from the Bone Marrow. J. Exp. Med. 2017, 215, 263–281. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, S.; Chi, Y.; Yang, Y.; Chen, X.; Wang, H.; Lv, Z.; Wang, J.; Yuan, L.; Huang, P.; et al. Kinetics of the accumulation of group 2 innate lymphoid cells in IL-33-induced and IL-25-induced murine models of asthma: A potential role for the chemokine CXCL16. Cell Mol. Immunol. 2019, 16, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Ricardo-Gonzalez, R.R.; Schneider, C.; Liao, C.; Lee, J.; Liang, H.-E.; Locksley, R.M. Tissue-specific pathways extrude activated ILC2s to disseminate Type 2 immunity. J. Exp. Med. 2020, 217, e20191172. [Google Scholar] [CrossRef]
- Kästele, V.; Mayer, J.; Lee, E.S.; Papazian, N.; Cole, J.J.; Cerovic, V.; Belz, G.; Tomura, M.; Eberl, G.; Goodyear, C.; et al. Intestinal-derived ILCs migrating in lymph increase IFNγ production in response to salmonella typhimurium infection. Mucosal Immunol. 2021, 14, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Zeis, P.; Lian, M.; Fan, X.; Herman, J.S.; Hernandez, D.C.; Gentek, R.; Elias, S.; Symowski, C.; Knöpper, K.; Peltokangas, N.; et al. In situ maturation and tissue adaptation of Type 2 innate lymphoid cell progenitors. Immunity 2020, 53, 775–792.e9. [Google Scholar] [CrossRef]
- Mathä, L.; Romera-Hernández, M.; Steer, C.A.; Yin, Y.H.; Orangi, M.; Shim, H.; Chang, C.; Rossi, F.M.; Takei, F. Migration of lung resident group 2 innate lymphoid cells link allergic lung inflammation and liver immunity. Front. Immunol. 2021, 12, 679509. [Google Scholar] [CrossRef]
- Ghaedi, M.; Shen, Z.Y.; Orangi, M.; Martinez-Gonzalez, I.; Wei, L.; Lu, X.; Das, A.; Heravi-Moussavi, A.; Marra, M.A.; Bhandoola, A.; et al. Single-cell analysis of RORα tracer mouse lung reveals ILC progenitors and effector ILC2 subsets. J. Exp. Med. 2020, 217, e20182293. [Google Scholar] [CrossRef]
- Schneider, C.; Lee, J.; Koga, S.; Ricardo-Gonzalez, R.R.; Nussbaum, J.C.; Smith, L.K.; Villeda, S.A.; Liang, H.-E.; Locksley, R.M. Tissue-resident group 2 innate lymphoid cells differentiate by layered ontogeny and in situ perinatal priming. Immunity 2019, 50, 1425–1438.e5. [Google Scholar] [CrossRef]
- Ali, A.; Canaday, L.M.; Feldman, H.A.; Cevik, H.; Moran, M.T.; Rajaram, S.; Lakes, N.; Tuazon, J.A.; Seelamneni, H.; Krishnamurthy, D.; et al. Natural killer cell immunosuppressive function requires CXCR3-dependent redistribution within lymphoid tissues. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Carrega, P.; Bonaccorsi, I.; Carlo, E.D.; Morandi, B.; Paul, P.; Rizzello, V.; Cipollone, G.; Navarra, G.; Mingari, M.C.; Moretta, L.; et al. CD56brightPerforinlow Noncytotoxic human NK cells are abundant in both healthy and neoplastic solid tissues and recirculate to secondary lymphoid organs via afferent lymph. J. Immunol. 2014, 192, 3805–3815. [Google Scholar] [CrossRef]
- Walzer, T.; Vivier, E. G-Protein-coupled receptors in control of natural killer cell migration. Trends Immunol. 2011, 32, 486–492. [Google Scholar] [CrossRef]
- Salimi, M.; Barlow, J.L.; Saunders, S.P.; Xue, L.; Gutowska-Owsiak, D.; Wang, X.; Huang, L.-C.; Johnson, D.; Scanlon, S.T.; McKenzie, A.N.J.; et al. A role for IL-25 and IL-33-driven Type-2 innate lymphoid cells in atopic dermatitis. J. Exp. Med. 2013, 210, 2939–2950. [Google Scholar] [CrossRef]
- Karta, M.R.; Rosenthal, P.S.; Beppu, A.; Vuong, C.Y.; Miller, M.; Das, S.; Kurten, R.C.; Doherty, T.A.; Broide, D.H. B2 Integrins rather than B1 integrins mediate alternaria-induced group 2 innate lymphoid cell trafficking to the lung. J. Allergy Clin. Immunol. 2018, 141, 329–338.e12. [Google Scholar] [CrossRef]
- Mackley, E.C.; Houston, S.; Marriott, C.L.; Halford, E.E.; Lucas, B.; Cerovic, V.; Filbey, K.J.; Maizels, R.M.; Hepworth, M.R.; Sonnenberg, G.F.; et al. CCR7-dependent trafficking of RORγ+ ILCs creates a unique microenvironment within mucosal draining lymph nodes. Nat. Commun. 2015, 6, 5862. [Google Scholar] [CrossRef]
- Lim, A.I.; Di Santo, J.P. ILC-Poiesis: Ensuring tissue ILC differentiation at the right place and time. Eur. J. Immunol. 2018, 49, 11–18. [Google Scholar] [CrossRef]
- Mjösberg, J.M.; Trifari, S.; Crellin, N.K.; Peters, C.P.; van Drunen, C.M.; Piet, B.; Fokkens, W.J.; Cupedo, T.; Spits, H. Human IL-25- and IL-33-responsive Type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat. Immunol. 2011, 12, 1055–1062. [Google Scholar] [CrossRef]
- Maric, J.; Ravindran, A.; Mazzurana, L.; Björklund, Å.K.; Van Acker, A.; Rao, A.; Friberg, D.; Dahlén, S.-E.; Heinemann, A.; Konya, V.; et al. Prostaglandin E2 suppresses human group 2 innate lymphoid cell function. J. Allergy Clin. Immunol. 2018, 141, 1761–1773.e6. [Google Scholar] [CrossRef]
- Chang, J.E.; Doherty, T.A.; Baum, R.; Broide, D. Prostaglandin D2 regulates human type 2 innate lymphoid cell chemotaxis. J. Allergy Clin. Immunol. 2014, 133, 899–901.e3. [Google Scholar] [CrossRef]
- Soriani, A.; Stabile, H.; Gismondi, A.; Santoni, A.; Bernardini, G. Chemokine regulation of innate lymphoid cell tissue distribution and function. Cytokine Growth Factor Rev. 2018, 42, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Laurent, P.; Allard, B.; Manicki, P.; Jolivel, V.; Levionnois, E.; Jeljeli, M.; Henrot, P.; Izotte, J.; Leleu, D.; Groppi, A.; et al. TGFβ Promotes low IL10-producing ILC2 with profibrotic ability involved in skin fibrosis in systemic sclerosis. Ann. Rheum. Dis. 2021, 80, 1594–1603. [Google Scholar] [CrossRef] [PubMed]
- Gronski, M.A.; Boulter, J.M.; Moskophidis, D.; Nguyen, L.T.; Holmberg, K.; Elford, A.R.; Deenick, E.K.; Kim, H.O.; Penninger, J.M.; Odermatt, B.; et al. TCR affinity and negative regulation limit autoimmunity. Nat. Med. 2004, 10, 1234–1239. [Google Scholar] [CrossRef] [PubMed]
- Probst, H.C.; McCoy, K.; Okazaki, T.; Honjo, T.; van den Broek, M. Resting dendritic cells induce peripheral CD8+ T cell tolerance through PD-1 and CTLA-4. Nat. Immunol. 2005, 6, 280–286. [Google Scholar] [CrossRef]
- Walker, L.S.K. Treg and CTLA-4: Two intertwining pathways to immune tolerance. J. Autoimmun. 2013, 45, 49–57. [Google Scholar] [CrossRef]
- Latchman, Y.E.; Liang, S.C.; Wu, Y.; Chernova, T.; Sobel, R.A.; Klemm, M.; Kuchroo, V.K.; Freeman, G.J.; Sharpe, A.H. PD-L1-Deficient mice show that PD-L1 on T cells, antigen-presenting cells, and host tissues negatively regulates T cells. Proc. Natl. Acad. Sci. USA 2004, 101, 10691–10696. [Google Scholar] [CrossRef]
- Peggs, K.S.; Quezada, S.A.; Chambers, C.A.; Korman, A.J.; Allison, J.P. Blockade of CTLA-4 on both effector and regulatory T cell compartments contributes to the antitumor activity of anti-CTLA-4 antibodies. J. Exp. Med. 2009, 206, 1717–1725. [Google Scholar] [CrossRef]
- Wang, J.; Yoshida, T.; Nakaki, F.; Hiai, H.; Okazaki, T.; Honjo, T. Establishment of NOD-Pdcd1-/- mice as an efficient animal model of Type I diabetes. Proc. Natl. Acad. Sci. USA 2005, 102, 11823–11828. [Google Scholar] [CrossRef]
- Nishimura, H.; Nose, M.; Hiai, H.; Minato, N.; Honjo, T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity 1999, 11, 141–151. [Google Scholar] [CrossRef]
- Nishimura, H.; Okazaki, T.; Tanaka, Y.; Nakatani, K.; Hara, M.; Matsumori, A.; Sasayama, S.; Mizoguchi, A.; Hiai, H.; Minato, N.; et al. Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science 2001, 291, 319–322. [Google Scholar] [CrossRef]
- Okazaki, T.; Tanaka, Y.; Nishio, R.; Mitsuiye, T.; Mizoguchi, A.; Wang, J.; Ishida, M.; Hiai, H.; Matsumori, A.; Minato, N.; et al. Autoantibodies against Cardiac Troponin I are responsible for dilated cardiomyopathy in PD-1-deficient mice. Nat. Med. 2003, 9, 1477–1483. [Google Scholar] [CrossRef]
- Waterhouse, P.; Penninger, J.M.; Timms, E.; Wakeham, A.; Shahinian, A.; Lee, K.P.; Thompson, C.B.; Griesser, H.; Mak, T.W. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science 1995, 270, 985–988. [Google Scholar] [CrossRef]
- Tivol, E.A.; Borriello, F.; Schweitzer, A.N.; Lynch, W.P.; Bluestone, J.A.; Sharpe, A.H. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity 1995, 3, 541–547. [Google Scholar] [CrossRef]
- Ogishi, M.; Yang, R.; Aytekin, C.; Langlais, D.; Bourgey, M.; Khan, T.; Ali, F.A.; Rahman, M.; Delmonte, O.M.; Chrabieh, M.; et al. Inherited PD-1 deficiency underlies tuberculosis and autoimmunity in a child. Nat. Med. 2021, 27, 1646–1654. [Google Scholar] [CrossRef]
- Delamain, M.T.; Gomez, G.V.B.; Lourenço, G.J.; de Souza, C.A.; Lima, C.S.P. Increased Risk of Hodgkin Lymphoma in Males with Inherited T Lymphocyte Receptor Programed Death-1 Deficiency. Leukemia Lymphoma 2019, 60, 3552–3556. [Google Scholar] [CrossRef]
- Schubert, D.; Bode, C.; Kenefeck, R.; Hou, T.Z.; Wing, J.B.; Kennedy, A.; Bulashevska, A.; Petersen, B.-S.; Schäffer, A.A.; Grüning, B.A.; et al. Autosomal dominant immune dysregulation syndrome in humans with CTLA4 mutations. Nat. Med. 2014, 20, 1410–1416. [Google Scholar] [CrossRef]
- Kuehn, H.S.; Ouyang, W.; Lo, B.; Deenick, E.K.; Niemela, J.E.; Avery, D.T.; Schickel, J.-N.; Tran, D.Q.; Stoddard, J.; Zhang, Y.; et al. Immune dysregulation in human subjects with heterozygous germline mutations in CTLA4. Science 2014, 345, 1623–1627. [Google Scholar] [CrossRef]
- Okazaki, T.; Chikuma, S.; Iwai, Y.; Fagarasan, S.; Honjo, T. A rheostat for immune responses: The unique properties of PD-1 and their advantages for clinical application. Nat. Immunol. 2013, 14, 1212–1218. [Google Scholar] [CrossRef]
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 2008, 26, 677–704. [Google Scholar] [CrossRef]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Mellman, I.; Coukos, G.; Dranoff, G. Cancer immunotherapy comes of age. Nature 2011, 480, 480–489. [Google Scholar] [CrossRef]
- Ferris, R.L.; Blumenschein, G.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef]
- Tawbi, H.A.; Forsyth, P.A.; Algazi, A.; Hamid, O.; Hodi, F.S.; Moschos, S.J.; Khushalani, N.I.; Lewis, K.; Lao, C.D.; Postow, M.A.; et al. Combined nivolumab and ipilimumab in melanoma metastatic to the brain. N. Engl. J. Med. 2018, 379, 722–730. [Google Scholar] [CrossRef]
- Robert, C.; Long, G.V.; Brady, B.; Dutriaux, C.; Maio, M.; Mortier, L.; Hassel, J.C.; Rutkowski, P.; McNeil, C.; Kalinka-Warzocha, E.; et al. Nivolumab in previously untreated melanoma without BRAF Mutation. N. Engl. J. Med. 2015, 372, 320–330. [Google Scholar] [CrossRef]
- Motzer, R.J.; Escudier, B.; McDermott, D.F.; George, S.; Hammers, H.J.; Srinivas, S.; Tykodi, S.S.; Sosman, J.A.; Procopio, G.; Plimack, E.R.; et al. Nivolumab versus Everolimus in advanced renal-cell carcinoma. N. Engl. J. Med. 2015, 373, 1803–1813. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Cowey, C.L.; Lao, C.D.; Schadendorf, D.; Dummer, R.; Smylie, M.; Rutkowski, P.; et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 2015, 373, 23–34. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Paz-Ares, L.; Bernabe Caro, R.; Zurawski, B.; Kim, S.-W.; Carcereny Costa, E.; Park, K.; Alexandru, A.; Lupinacci, L.; de la Mora Jimenez, E.; et al. Nivolumab plus ipilimumab in advanced non–small-cell lung cancer. N. Engl. J. Med. 2019, 381, 2020–2031. [Google Scholar] [CrossRef]
- Ansell, S.M.; Lesokhin, A.M.; Borrello, I.; Halwani, A.; Scott, E.C.; Gutierrez, M.; Schuster, S.J.; Millenson, M.M.; Cattry, D.; Freeman, G.J.; et al. PD-1 Blockade with nivolumab in relapsed or refractory Hodgkin’s Lymphoma. N. Engl. J. Med. 2015, 372, 311–319. [Google Scholar] [CrossRef]
- Ercolano, G.; Wyss, T.; Salomé, B.; Romero, P.; Trabanelli, S.; Jandus, C. Distinct and shared gene expression for human innate versus adaptive helper lymphoid cells. J. Leukoc. Biol. 2020, 108, 723–737. [Google Scholar] [CrossRef] [PubMed]
- Andrews, L.P.; Yano, H.; Vignali, D.A.A. Inhibitory Receptors and Ligands beyond PD-1, PD-L1 and CTLA-4: Breakthroughs or backups. Nat. Immunol. 2019, 20, 1425–1434. [Google Scholar] [CrossRef]
- Mariotti, F.R.; Quatrini, L.; Munari, E.; Vacca, P.; Moretta, L. Innate lymphoid cells: Expression of PD-1 and other checkpoints in normal and pathological conditions. Front. Immunol. 2019, 10, 910. [Google Scholar] [CrossRef]
- Quatrini, L.; Wieduwild, E.; Escaliere, B.; Filtjens, J.; Chasson, L.; Laprie, C.; Vivier, E.; Ugolini, S. Endogenous glucocorticoids control host resistance to viral infection through the tissue-specific regulation of PD-1 expression on NK cells. Nat. Immunol. 2018, 19, 954–962. [Google Scholar] [CrossRef]
- Hsu, J.; Hodgins, J.J.; Marathe, M.; Nicolai, C.J.; Bourgeois-Daigneault, M.-C.; Trevino, T.N.; Azimi, C.S.; Scheer, A.K.; Randolph, H.E.; Thompson, T.W.; et al. Contribution of NK Cells To Immunotherapy Mediated by PD-1/PD-L1 Blockade. Available online: https://www.jci.org/articles/view/99317/pdf (accessed on 9 August 2021).
- Quatrini, L.; Vacca, P.; Tumino, N.; Besi, F.; Pace, A.L.D.; Scordamaglia, F.; Martini, S.; Munari, E.; Mingari, M.C.; Ugolini, S.; et al. Glucocorticoids and the Cytokines IL-12, IL-15, and IL-18 present in the tumor microenvironment induce PD-1 expression on human natural killer cells. J. Allergy Clin. Immunol. 2021, 147, 349–360. [Google Scholar] [CrossRef]
- Pesce, S.; Greppi, M.; Tabellini, G.; Rampinelli, F.; Parolini, S.; Olive, D.; Moretta, L.; Moretta, A.; Marcenaro, E. Identification of a subset of human natural killer cells expressing high levels of programmed Death 1: A phenotypic and functional characterization. J. Allergy Clin. Immunol. 2017, 139, 335–346.e3. [Google Scholar] [CrossRef]
- Davis, Z.; Felices, M.; Lenvik, T.; Badal, S.; Walker, J.T.; Hinderlie, P.; Riley, J.L.; Vallera, D.A.; Blazar, B.R.; Miller, J.S. Low-density PD-1 expression on resting human natural killer cells is functional and upregulated after transplantation. Blood Advances 2021, 5, 1069–1080. [Google Scholar] [CrossRef]
- Beldi-Ferchiou, A.; Lambert, M.; Dogniaux, S.; Vély, F.; Vivier, E.; Olive, D.; Dupuy, S.; Levasseur, F.; Zucman, D.; Lebbé, C.; et al. PD-1 mediates functional exhaustion of activated NK cells in patients with Kaposi Sarcoma. Oncotarget 2016, 7, 72961–72977. [Google Scholar] [CrossRef]
- Vari, F.; Arpon, D.; Keane, C.; Hertzberg, M.S.; Talaulikar, D.; Jain, S.; Cui, Q.; Han, E.; Tobin, J.; Bird, R.; et al. Immune evasion via PD-1/PD-L1 on NK cells and monocyte/macrophages is more prominent in hodgkin lymphoma than DLBCL. Blood 2018, 131, 1809–1819. [Google Scholar] [CrossRef]
- Tumino, N.; Martini, S.; Munari, E.; Scordamaglia, F.; Besi, F.; Mariotti, F.R.; Bogina, G.; Mingari, M.C.; Vacca, P.; Moretta, L. Presence of innate lymphoid cells in pleural effusions of primary and metastatic tumors: Functional analysis and expression of PD-1 receptor. Int. J. Cancer 2019, 145, 1660–1668. [Google Scholar] [CrossRef]
- Niu, C.; Li, M.; Zhu, S.; Chen, Y.; Zhou, L.; Xu, D.; Xu, J.; Li, Z.; Li, W.; Cui, J. PD-1-positive natural killer cells have a weaker antitumor function than that of PD-1-negative natural killer cells in lung cancer. Int. J. Med. Sci. 2020, 17, 1964–1973. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, Y.; Xu, Y.; Wang, Z.; Du, X.; Li, C.; Peng, J.; Gao, L.; Liang, X.; Ma, C. Increased expression of programmed cell death Protein 1 on NK cells inhibits NK-cell-mediated anti-tumor function and indicates poor prognosis in digestive cancers. Oncogene 2017, 36, 6143–6153. [Google Scholar] [CrossRef]
- Salimi, M.; Wang, R.; Yao, X.; Li, X.; Wang, X.; Hu, Y.; Chang, X.; Fan, P.; Dong, T.; Ogg, G. Activated innate lymphoid cell populations accumulate in human tumour tissues. BMC Cancer 2018, 18, 341. [Google Scholar] [CrossRef]
- Ndhlovu, L.C.; Lopez-Vergès, S.; Barbour, J.D.; Jones, R.B.; Jha, A.R.; Long, B.R.; Schoeffler, E.C.; Fujita, T.; Nixon, D.F.; Lanier, L.L. Tim-3 marks human natural killer cell maturation and suppresses cell-mediated cytotoxicity. Blood 2012, 119, 3734–3743. [Google Scholar] [CrossRef] [PubMed]
- Gleason, M.K.; Lenvik, T.R.; McCullar, V.; Felices, M.; O’Brien, M.S.; Cooley, S.A.; Verneris, M.R.; Cichocki, F.; Holman, C.J.; Panoskaltsis-Mortari, A.; et al. Tim-3 is an inducible human natural killer cell receptor that enhances interferon gamma production in response to Galectin-9. Blood 2012, 119, 3064–3072. [Google Scholar] [CrossRef] [PubMed]
- da Silva, I.P.; Gallois, A.; Jimenez-Baranda, S.; Khan, S.; Anderson, A.C.; Kuchroo, V.K.; Osman, I.; Bhardwaj, N. Reversal of NK-cell exhaustion in advanced melanoma by Tim-3 blockade. Cancer Immunol. Res. 2014, 2, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Komita, H.; Koido, S.; Hayashi, K.; Kan, S.; Ito, M.; Kamata, Y.; Suzuki, M.; Homma, S. Expression of immune checkpoint molecules of T cell immunoglobulin and mucin Protein 3/Galectin-9 for NK cell suppression in human gastrointestinal stromal tumors. Oncol. Rep. 2015, 34, 2099–2105. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, J.; Gu, H.; Yuan, Y.; Zhang, B.; Zhu, D.; Zhou, J.; Zhu, Y.; Chen, W. The clinical significance of abnormal Tim-3 expression on NK cells from patients with gastric cancer. Immunol. Investig. 2015, 44, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, J.; Gao, W.; Song, B.; Shao, Q.; Zhao, L.; Zhang, Y.; Wang, Q.; Zhang, Y.; Qu, X. Preoperative Tim-3 Expression on peripheral NK cells is correlated with pathologic TNM staging in colorectal cancer. Mol. Med. Rep. 2017, 15, 3810–3818. [Google Scholar] [CrossRef][Green Version]
- Yin, M.; Di, G.; Bian, M. Dysfunction of natural killer cells mediated by PD-1 and Tim-3 pathway in anaplastic thyroid cancer. Int. Immunopharmacol. 2018, 64, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Feng, H.; Chen, Q.; Lu, X.; Ge, J. The functional potency of natural killer cells in response to IL-2/IL-15/IL-21 stimulation is limited by a concurrent upregulation of Tim-3 in bladder cancer. Exp. Cell Res. 2018, 372, 92–98. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, Y.; Lian, J.; Yang, H.; Li, F.; Zhao, S.; Qi, Y.; Zhang, Y.; Huang, L. TNF-α-Induced Tim-3 expression marks the dysfunction of infiltrating natural killer cells in human esophageal cancer. J. Transl. Med. 2019, 17, 165. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Xu, Y.; Wang, Z.; Wang, T.; Du, X.; Song, X.; Guo, X.; Peng, J.; Zhang, J.; Liang, Y.; et al. Tim-3 hampers tumor surveillance of liver-resident and conventional NK cells by disrupting PI3K signaling. Cancer Res. 2020, 80, 1130–1142. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Huang, Y.; Tan, L.; Yu, W.; Chen, D.; Lu, C.; He, J.; Wu, G.; Liu, X.; Zhang, Y. Increased Tim-3 expression in peripheral NK cells predicts a poorer prognosis and Tim-3 blockade improves NK cell-mediated cytotoxicity in human lung adenocarcinoma. Int. Immunopharmacol. 2015, 29, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.; Simonetta, F.; Baker, J.; Pierini, A.; Wenokur, A.S.; Morrison, A.R.; Murphy, W.J.; Negrin, R.S. Regulation of murine NK cell exhaustion through the activation of the DNA damage repair pathway. JCI Insight 2019, 4, e127729. [Google Scholar] [CrossRef]
- Judge, S.J.; Dunai, C.; Aguilar, E.G.; Vick, S.C.; Sturgill, I.R.; Khuat, L.T.; Stoffel, K.M.; Van Dyke, J.; Longo, D.L.; Darrow, M.A.; et al. Minimal PD-1 Expression in Mouse and Human NK Cells under Diverse Conditions. J. Clin. Investig. 2020, 130, 3051–3068. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, A.; Campbell, D.; Hannum, C.; Yssel, H.; Franz-Bacon, K.; McClanahan, T.; Kitamura, T.; Nicholl, J.; Sutherland, G.R.; Lanier, L.L.; et al. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity 1996, 4, 573–581. [Google Scholar] [CrossRef]
- Dougall, W.C.; Kurtulus, S.; Smyth, M.J.; Anderson, A.C. TIGIT and CD96: New checkpoint receptor targets for cancer immunotherapy. Immunol. Rev. 2017, 276, 112–120. [Google Scholar] [CrossRef]
- Chan, C.J.; Martinet, L.; Gilfillan, S.; Souza-Fonseca-Guimaraes, F.; Chow, M.T.; Town, L.; Ritchie, D.S.; Colonna, M.; Andrews, D.M.; Smyth, M.J. The Receptors CD96 and CD226 Oppose Each Other in the Regulation of Natural Killer Cell Functions. Nat. Immunol. 2014, 15, 431–438. [Google Scholar] [CrossRef]
- O’Donnell, J.S.; Madore, J.; Li, X.-Y.; Smyth, M.J. Tumor Intrinsic and Extrinsic Immune Functions of CD155. Semin. Cancer Biol. 2020, 65, 189–196. [Google Scholar] [CrossRef]
- Zhang, Q.; Bi, J.; Zheng, X.; Chen, Y.; Wang, H.; Wu, W.; Wang, Z.; Wu, Q.; Peng, H.; Wei, H.; et al. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat. Immunol. 2018, 19, 723–732. [Google Scholar] [CrossRef]
- Workman, C.J.; Rice, D.S.; Dugger, K.J.; Kurschner, C.; Vignali, D.A.A. Phenotypic analysis of the murine CD4-related glycoprotein, CD223 (LAG-3). Eur. J. Immunol. 2002, 32, 2255–2263. [Google Scholar] [CrossRef]
- Merino, A.; Zhang, B.; Dougherty, P.; Luo, X.; Wang, J.; Blazar, B.R.; Miller, J.S.; Cichocki, F. Chronic stimulation drives human NK cell dysfunction and epigenetic reprograming. J. Clin. Investig. 2019, 129, 3770–3785. [Google Scholar] [CrossRef]
- Miyazaki, T.; Dierich, A.; Benoist, C.; Mathis, D. Independent modes of natural killing distinguished in mice lacking Lag3. Science 1996, 272, 405–408. [Google Scholar] [CrossRef]
- Huard, B.; Tournier, M.; Triebel, F. LAG-3 does not define a specific mode of natural killing in human. Immunol. Lett. 1998, 61, 109–112. [Google Scholar] [CrossRef]
- Narayanan, S.; Ahl, P.J.; Bijin, V.A.; Kaliaperumal, N.; Lim, S.G.; Wang, C.-I.; Fairhurst, A.-M.; Connolly, J.E. LAG3 Is a Central Regulator of NK Cell Cytokine Production. Available online: https://www.biorxiv.org/content/10.1101/2020.01.31.928200v1.abstract (accessed on 13 October 2021).
- Sordo-Bahamonde, C.; Lorenzo-Herrero, S.; González-Rodríguez, A.P.; Payer, Á.R.; González-García, E.; López-Soto, A.; Gonzalez, S. LAG-3 blockade with relatlimab (BMS-986016) restores anti-leukemic responses in chronic lymphocytic leukemia. Cancers 2021, 13, 2112. [Google Scholar] [CrossRef]
- Ito, M.; Maruyama, T.; Saito, N.; Koganei, S.; Yamamoto, K.; Matsumoto, N. Killer cell lectin-like receptor G1 binds three members of the classical cadherin family to inhibit NK cell cytotoxicity. J. Exp. Med. 2006, 203, 289–295. [Google Scholar] [CrossRef]
- Huntington, N.D.; Tabarias, H.; Fairfax, K.; Brady, J.; Hayakawa, Y.; Degli-Esposti, M.A.; Smyth, M.J.; Tarlinton, D.M.; Nutt, S.L. NK Cell Maturation and Peripheral Homeostasis Is Associated with KLRG1 Up-Regulation. J. Immunol. 2007, 178, 4764–4770. [Google Scholar] [CrossRef]
- Wang, J.M.; Cheng, Y.Q.; Shi, L.; Ying, R.S.; Wu, X.Y.; Li, G.Y.; Moorman, J.P.; Yao, Z.Q. KLRG1 negatively regulates natural killer cell functions through the Akt pathway in individuals with chronic Hepatitis C virus infection. J. Virol. 2013, 87, 11626–11636. [Google Scholar] [CrossRef]
- Tata, A.; Dodard, G.; Fugère, C.; Leget, C.; Ors, M.; Rossi, B.; Vivier, E.; Brossay, L. Combination blockade of KLRG1 and PD-1 promotes immune control of local and disseminated cancers. OncoImmunology 2021, 10, 1933808. [Google Scholar] [CrossRef]
- Sakurai, T.; Okuyama, Y.; Kobayashi, S.; Phung, H.T.; Asao, A.; Kawabe, T.; Ndhlovu, L.C.; Riccardi, C.; Kudo, H.; Wada, M.; et al. GITR controls intestinal inflammation by suppressing IL-15-dependent NK cell activity. FASEB J. 2020, 34, 14820–14831. [Google Scholar] [CrossRef]
- Placke, T.; Salih, H.R.; Kopp, H.-G. GITR ligand provided by thrombopoietic cells inhibits NK cell antitumor activity. J. Immunol. 2012, 189, 154–160. [Google Scholar] [CrossRef]
- Buechele, C.; Baessler, T.; Wirths, S.; Schmohl, J.U.; Schmiedel, B.J.; Salih, H.R. Glucocorticoid-induced TNFR-related protein (GITR) ligand modulates cytokine release and NK cell reactivity in chronic lymphocytic leukemia (CLL). Leukemia 2012, 26, 991–1000. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, Z.; Mahesh, S.P.; Pantanelli, S.; Hwang, F.S.; Siu, W.O.; Nussenblatt, R.B. Glucocorticoid-induced tumor necrosis factor receptor negatively regulates activation of human primary natural killer (NK) cells by blocking proliferative signals and increasing NK cell apoptosis. J. Biol. Chem. 2008, 283, 8202–8210. [Google Scholar] [CrossRef]
- Baltz, K.M.; Krusch, M.; Baessler, T.; Schmiedel, B.J.; Bringmann, A.; Brossart, P.; Salih, H.R. Neutralization of tumor-derived soluble glucocorticoid-induced tnfr-related protein ligand increases NK cell anti-tumor reactivity. Blood 2008, 112, 3735–3743. [Google Scholar] [CrossRef]
- Montgomery, R.I.; Warner, M.S.; Lum, B.J.; Spear, P.G. Herpes simplex Virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 1996, 87, 427–436. [Google Scholar] [CrossRef]
- Sordo-Bahamonde, C.; Lorenzo-Herrero, S.; Gonzalez-Rodriguez, A.P.; R Payer, Á.; González-García, E.; López-Soto, A.; Gonzalez, S. BTLA/HVEM axis induces NK cell immunosuppression and poor outcome in chronic lymphocytic Leukemia. Cancers 2021, 13, 1766. [Google Scholar] [CrossRef] [PubMed]
- Rethacker, L.; Roelens, M.; Bejar, C.; Maubec, E.; Moins-Teisserenc, H.; Caignard, A. Specific patterns of blood ILCs in metastatic melanoma patients and their modulations in response to immunotherapy. Cancers 2021, 13, 1446. [Google Scholar] [CrossRef]
- Gao, Y.; Souza-Fonseca-Guimaraes, F.; Bald, T.; Ng, S.S.; Young, A.; Ngiow, S.F.; Rautela, J.; Straube, J.; Waddell, N.; Blake, S.J.; et al. Tumor immunoevasion by the conversion of effector NK Cells into Type 1 innate lymphoid cells. Nat. Immunol. 2017, 18, 1004–1015. [Google Scholar] [CrossRef]
- Yu, Y.; Tsang, J.C.H.; Wang, C.; Clare, S.; Wang, J.; Chen, X.; Brandt, C.; Kane, L.; Campos, L.S.; Lu, L.; et al. Single-cell RNA-seq identifies a PD-1hi ILC progenitor and defines its development pathway. Nature 2016, 539, 102–106. [Google Scholar] [CrossRef]
- Shen, C.; Liu, C.; Zhang, Z.; Ping, Y.; Shao, J.; Tian, Y.; Yu, W.; Qin, G.; Liu, S.; Wang, L.; et al. PD-1 Affects the immunosuppressive function of group 2 innate lymphoid cells in human non-small cell lung cancer. Front. Immunol. 2021, 12, 2318. [Google Scholar] [CrossRef]
- Azuma, M.; Ito, D.; Yagita, H.; Okumura, K.; Phillips, J.H.; Lanier, L.L.; Somoza, C. B70 antigen is a second ligand for CTLA-4 and CD28. Nature 1993, 366, 76–79. [Google Scholar] [CrossRef]
- Freeman, G.J.; Gribben, J.G.; Boussiotis, V.A.; Ng, J.W.; Restivo, V.A.; Lombard, L.A.; Gray, G.S.; Nadler, L.M. Cloning of B7-2: A CTLA-4 counter-receptor that costimulates human T cell proliferation. Science 1993, 262, 909–911. [Google Scholar] [CrossRef]
- Linsley, P.S.; Brady, W.; Urnes, M.; Grosmaire, L.S.; Damle, N.K.; Ledbetter, J.A. CTLA-4 is a second receptor for the B cell activation antigen B7. J. Exp. Med. 1991, 174, 561–569. [Google Scholar] [CrossRef]
- Linsley, P.S.; Greene, J.L.; Brady, W.; Bajorath, J.; Ledbetter, J.A.; Peach, R. Human B7-1 (CD80) and B7-2 (CD86) bind with similar avidities but distinct kinetics to CD28 and CTLA-4 receptors. Immunity 1994, 1, 793–801. [Google Scholar] [CrossRef]
- Vashist, N.; Trittel, S.; Ebensen, T.; Chambers, B.J.; Guzmán, C.A.; Riese, P. Influenza-activated ILC1s contribute to antiviral immunity partially influenced by differential GITR expression. Front. Immunol. 2018, 9, 505. [Google Scholar] [CrossRef]
- Hoyler, T.; Klose, C.S.N.; Souabni, A.; Turqueti-Neves, A.; Pfeifer, D.; Rawlins, E.L.; Voehringer, D.; Busslinger, M.; Diefenbach, A. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity 2012, 37, 634–648. [Google Scholar] [CrossRef]
- Taylor, S.; Huang, Y.; Mallett, G.; Stathopoulou, C.; Felizardo, T.C.; Sun, M.-A.; Martin, E.L.; Zhu, N.; Woodward, E.L.; Elias, M.S.; et al. PD-1 regulates KLRG1+ group 2 innate lymphoid cells. J. Exp. Med. 2017, 214, 1663–1678. [Google Scholar] [CrossRef]
- Gründemann, C.; Bauer, M.; Schweier, O.; von Oppen, N.; Lässing, U.; Saudan, P.; Becker, K.-F.; Karp, K.; Hanke, T.; Bachmann, M.F.; et al. Cutting edge: Identification of E-cadherin as a ligand for the murine killer cell lectin-like receptor G1. J. Immunol. 2006, 176, 1311–1315. [Google Scholar] [CrossRef]
- Tessmer, M.S.; Fugere, C.; Stevenaert, F.; Naidenko, O.V.; Chong, H.J.; Leclercq, G.; Brossay, L. KLRG1 binds cadherins and preferentially associates with SHIP-1. Int Immunol. 2007, 19, 391–400. [Google Scholar] [CrossRef]
- Huang, Y.; Guo, L.; Qiu, J.; Chen, X.; Hu-Li, J.; Siebenlist, U.; Williamson, P.R.; Urban, J.F.; Paul, W.E. IL-25-Responsive, lineage-negative KLRG1(Hi) cells are multipotential “inflammatory” type 2 innate lymphoid cells. Nat. Immunol. 2015, 16, 161–169. [Google Scholar] [CrossRef]
- Blaser, C.; Kaufmann, M.; Pircher, H. Cutting edge: Virus-activated CD8 T cells and lymphokine-activated NK cells express the mast cell function-associated antigen, an inhibitory c-type lectin. J. Immunol. 1998, 161, 6451–6454. [Google Scholar]
- Beyersdorf, N.B.; Ding, X.; Karp, K.; Hanke, T. Expression of inhibitory “killer cell lectin-like receptor g1” identifies unique subpopulations of effector and memory CD8 T cells. Eur. J. Immunol. 2001, 31, 3443–3452. [Google Scholar] [CrossRef]
- Hanke, T.; Corral, L.; Vance, R.E.; Raulet, D.H. 2F1 antigen, the mouse homolog of the rat “mast cell function-associated antigen”, is a lectin-like type II transmembrane receptor expressed by natural killer cells. Eur. J. Immunol. 1998, 28, 4409–4417. [Google Scholar] [CrossRef]
- Blanquart, E.; Mandonnet, A.; Mars, M.; Cenac, C.; Anesi, N.; Mercier, P.; Audouard, C.; Roga, S.; Serrano de Almeida, G.; Bevan, C.L.; et al. Targeting androgen signaling in ILC2s protects from IL-33–driven lung inflammation, independently of KLRG1. J. Allergy Clin. Immunol. 2021. [Google Scholar] [CrossRef]
- Wang, S.; Qu, Y.; Xia, P.; Chen, Y.; Zhu, X.; Zhang, J.; Wang, G.; Tian, Y.; Ying, J.; Fan, Z. Transdifferentiation of tumor infiltrating innate lymphoid cells during progression of colorectal cancer. Cell Res. 2020, 30, 610–622. [Google Scholar] [CrossRef] [PubMed]
- Helou, D.G.; Shafiei-Jahani, P.; Lo, R.; Howard, E.; Hurrell, B.P.; Galle-Treger, L.; Painter, J.D.; Lewis, G.; Soroosh, P.; Sharpe, A.H.; et al. PD-1 pathway regulates ILC2 metabolism and PD-1 agonist treatment ameliorates airway hyperreactivity. Nat. Commun. 2020, 11, 3998. [Google Scholar] [CrossRef]
- Falquet, M.; Ercolano, G.; Jandus, P.; Jandus, C.; Trabanelli, S. Healthy and patient Type 2 innate lymphoid cells are differently affected by in vitro culture conditions. J. Asthma Allergy 2021, 14, 773–783. [Google Scholar] [CrossRef]
- Akama, Y.; Park, E.J.; Satoh-Takayama, N.; Gaowa, A.; Ito, A.; Kawamoto, E.; Darkwah, S.; Appiah, M.G.; Myint, P.K.; Ohno, H.; et al. Sepsis induces deregulation of IL-13 production and PD-1 expression in lung group 2 innate lymphoid cells. Shock 2021, 55, 357–370. [Google Scholar] [CrossRef]
- Schwartz, C.; Khan, A.R.; Floudas, A.; Saunders, S.P.; Hams, E.; Rodewald, H.-R.; McKenzie, A.N.J.; Fallon, P.G. ILC2s Regulate adaptive Th2 cell functions via PD-L1 checkpoint control. J. Exp. Med. 2017, 214, 2507–2521. [Google Scholar] [CrossRef]
- Morita, H.; Kubo, T.; Rückert, B.; Ravindran, A.; Soyka, M.B.; Rinaldi, A.O.; Sugita, K.; Wawrzyniak, M.; Wawrzyniak, P.; Motomura, K.; et al. Induction of human regulatory innate lymphoid cells from group 2 innate lymphoid cells by retinoic acid. J. Allergy Clin. Immunol. 2019, 143, 2190–2201.e9. [Google Scholar] [CrossRef]
- Dutton, E.E.; Camelo, A.; Sleeman, M.; Herbst, R.; Carlesso, G.; Belz, G.T.; Withers, D.R. Characterisation of innate lymphoid cell populations at different sites in mice with defective T cell immunity. Wellcome Open Res. 2017, 2, 117. [Google Scholar] [CrossRef]
- Roediger, B.; Kyle, R.; Yip, K.H.; Sumaria, N.; Guy, T.V.; Kim, B.S.; Mitchell, A.J.; Tay, S.S.; Jain, R.; Forbes-Blom, E.; et al. Cutaneous immunosurveillance and regulation of inflammation by group 2 innate lymphoid cells. Nat. Immunol. 2013, 14, 564–573. [Google Scholar] [CrossRef]
- Molofsky, A.B.; Van Gool, F.; Liang, H.-E.; Van Dyken, S.J.; Nussbaum, J.C.; Lee, J.; Bluestone, J.A.; Locksley, R.M. Interleukin-33 and interferon-γ counter-regulate group 2 innate lymphoid cell activation during immune perturbation. Immunity 2015, 43, 161–174. [Google Scholar] [CrossRef]
- Maazi, H.; Patel, N.; Sankaranarayanan, I.; Suzuki, Y.; Rigas, D.; Soroosh, P.; Freeman, G.J.; Sharpe, A.H.; Akbari, O. ICOS:ICOS-ligand interaction is required for type 2 innate lymphoid cell function, homeostasis, and induction of airway hyperreactivity. Immunity 2015, 42, 538–551. [Google Scholar] [CrossRef]
- Paclik, D.; Stehle, C.; Lahmann, A.; Hutloff, A.; Romagnani, C. ICOS Regulates the pool of group 2 innate lymphoid cells under homeostatic and inflammatory conditions in mice. Eur. J. Immunol. 2015, 45, 2766–2772. [Google Scholar] [CrossRef] [PubMed]
- Kamachi, F.; Isshiki, T.; Harada, N.; Akiba, H.; Miyake, S. ICOS promotes group 2 innate lymphoid cell activation in lungs. Biochem. Biophys. Res. Commun. 2015, 463, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Winkler, C.; Hochdörfer, T.; Israelsson, E.; Hasselberg, A.; Cavallin, A.; Thörn, K.; Muthas, D.; Shojaee, S.; Lüer, K.; Müller, M.; et al. Activation of Group 2 Innate Lymphoid Cells after Allergen Challenge in Asthmatic Patients. J. Allergy Clin. Immunol. 2019, 144, 61–69.e7. [Google Scholar] [CrossRef]
- Cavagnero, K.J.; Badrani, J.H.; Naji, L.H.; Amadeo, M.B.; Shah, V.S.; Gasparian, S.; Pham, A.; Wang, A.W.; Seumois, G.; Croft, M.; et al. Unconventional ST2- and CD127-negative lung ILC2 populations are induced by the fungal allergen alternaria alternata. J. Allergy Clin. Immunol. 2019, 144, 1432–1435.e9. [Google Scholar] [CrossRef]
- Galle-Treger, L.; Sankaranarayanan, I.; Hurrell, B.P.; Howard, E.; Lo, R.; Maazi, H.; Lewis, G.; Banie, H.; Epstein, A.L.; Hu, P.; et al. Costimulation of type-2 innate lymphoid cells by GITR promotes effector function and ameliorates type 2 diabetes. Nat. Commun. 2019, 10, 713. [Google Scholar] [CrossRef]
- Halim, T.Y.F.; Rana, B.M.J.; Walker, J.A.; Kerscher, B.; Knolle, M.D.; Jolin, H.E.; Serrao, E.M.; Haim-Vilmovsky, L.; Teichmann, S.A.; Rodewald, H.-R.; et al. Tissue-restricted adaptive type 2 immunity is orchestrated by expression of the costimulatory molecule OX40L on group 2 innate lymphoid cells. Immunity 2018, 48, 1195–1207.e6. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Cui, Y.; Zhu, W.; Bai, S.; Zhao, N.; Liu, B. Critical role of OX40/OX40L in ILC2-mediated activation of CD4+T cells during respiratory syncytial virus infection in mice. Int. Immunopharmacol. 2019, 76, 105784. [Google Scholar] [CrossRef]
- Drake, L.Y.; Iijima, K.; Kita, H. Group 2 innate lymphoid cells and CD4+ T cells cooperate to mediate type 2 immune response in mice. Allergy 2014, 69, 1300–1307. [Google Scholar] [CrossRef]
- Deng, T.; Suo, C.; Chang, J.; Yang, R.; Li, J.; Cai, T.; Qiu, J. ILC3-Derived OX40L is essential for homeostasis of intestinal tregs in immunodeficient mice. Cell Mol. Immunol. 2020, 17, 163–177. [Google Scholar] [CrossRef]
- Kim, M.-Y.; Gaspal, F.M.C.; Wiggett, H.E.; McConnell, F.M.; Gulbranson-Judge, A.; Raykundalia, C.; Walker, L.S.K.; Goodall, M.D.; Lane, P.J.L. CD4+CD3−Accessory cells costimulate primed CD4 T cells through OX40 and CD30 at sites where T cells collaborate with B cells. Immunity 2003, 18, 643–654. [Google Scholar] [CrossRef]
- Castellanos, J.G.; Woo, V.; Viladomiu, M.; Putzel, G.; Lima, S.; Diehl, G.E.; Marderstein, A.R.; Gandara, J.; Perez, A.R.; Withers, D.R.; et al. Microbiota-induced TNF-like ligand 1A drives group 3 innate lymphoid cell-mediated barrier protection and intestinal T cell activation during colitis. Immunity 2018, 49, 1077–1089.e5. [Google Scholar] [CrossRef] [PubMed]
- Vacca, P.; Pesce, S.; Greppi, M.; Fulcheri, E.; Munari, E.; Olive, D.; Mingari, M.C.; Moretta, A.; Moretta, L.; Marcenaro, E. PD-1 is expressed by and regulates human group 3 innate lymphoid cells in human decidua. Mucosal Immunol. 2019, 12, 624–631. [Google Scholar] [CrossRef]
- Zhu, C.; Anderson, A.C.; Schubart, A.; Xiong, H.; Imitola, J.; Khoury, S.J.; Zheng, X.X.; Strom, T.B.; Kuchroo, V.K. The Tim-3 Ligand galectin-9 negatively regulates T helper type 1 immunity. Nat. Immunol. 2005, 6, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- DeKruyff, R.H.; Bu, X.; Ballesteros, A.; Santiago, C.; Chim, Y.-L.E.; Lee, H.-H.; Karisola, P.; Pichavant, M.; Kaplan, G.G.; Umetsu, D.T.; et al. T cell/transmembrane, Ig, and Mucin-3 Allelic variants differentially recognize phosphatidylserine and mediate phagocytosis of apoptotic cells. J. Immunol. 2010, 184, 1918–1930. [Google Scholar] [CrossRef]
- Chiba, S.; Baghdadi, M.; Akiba, H.; Yoshiyama, H.; Kinoshita, I.; Dosaka-Akita, H.; Fujioka, Y.; Ohba, Y.; Gorman, J.V.; Colgan, J.D.; et al. Tumor-Infiltrating DCs suppress nucleic acid-mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1. Nat. Immunol. 2012, 13, 832–842. [Google Scholar] [CrossRef]
- Huang, Y.-H.; Zhu, C.; Kondo, Y.; Anderson, A.C.; Gandhi, A.; Russell, A.; Dougan, S.K.; Petersen, B.-S.; Melum, E.; Pertel, T.; et al. CEACAM1 regulates TIM-3-mediated tolerance and exhaustion. Nature 2015, 517, 386–390. [Google Scholar] [CrossRef]
- Aspeslagh, S.; Postel-Vinay, S.; Rusakiewicz, S.; Soria, J.-C.; Zitvogel, L.; Marabelle, A. Rationale for anti-OX40 cancer immunotherapy. Eur. J. Cancer 2016, 52, 50–66. [Google Scholar] [CrossRef]
- Ishida, Y.; Agata, Y.; Shibahara, K.; Honjo, T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992, 11, 3887–3895. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Zhu, G.; Tamada, K.; Chen, L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat. Med. 1999, 5, 1365–1369. [Google Scholar] [CrossRef] [PubMed]
- Freeman, G.J.; Long, A.J.; Iwai, Y.; Bourque, K.; Chernova, T.; Nishimura, H.; Fitz, L.J.; Malenkovich, N.; Okazaki, T.; Byrne, M.C.; et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med. 2000, 192, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Latchman, Y.; Wood, C.R.; Chernova, T.; Chaudhary, D.; Borde, M.; Chernova, I.; Iwai, Y.; Long, A.J.; Brown, J.A.; Nunes, R.; et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol. 2001, 2, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Tseng, S.Y.; Otsuji, M.; Gorski, K.; Huang, X.; Slansky, J.E.; Pai, S.I.; Shalabi, A.; Shin, T.; Pardoll, D.M.; Tsuchiya, H. B7-DC, a new dendritic cell molecule with potent costimulatory properties for T cells. J. Exp. Med. 2001, 193, 839–846. [Google Scholar] [CrossRef]
- Day, C.L.; Kaufmann, D.E.; Kiepiela, P.; Brown, J.A.; Moodley, E.S.; Reddy, S.; Mackey, E.W.; Miller, J.D.; Leslie, A.J.; DePierres, C.; et al. PD-1 expression on HIV-specific T cells is associated with t-cell exhaustion and disease progression. Nature 2006, 443, 350–354. [Google Scholar] [CrossRef]
- Francisco, L.M.; Sage, P.T.; Sharpe, A.H. The PD-1 pathway in tolerance and autoimmunity. Immunol. Rev. 2010, 236, 219–242. [Google Scholar] [CrossRef]
- Seillet, C.; Mielke, L.A.; Amann-Zalcenstein, D.B.; Su, S.; Gao, J.; Almeida, F.F.; Shi, W.; Ritchie, M.E.; Naik, S.H.; Huntington, N.D.; et al. Deciphering the innate lymphoid cell transcriptional program. Cell Rep. 2016, 17, 436–447. [Google Scholar] [CrossRef]
- Hasim, M.S.; Marotel, M.; Hodgins, J.J.; Vulpis, E.; Shih, H.-Y.; Scheer, A.; MacMillan, O.; Alonso, F.G.; Burke, K.P.; Cook, D.P.; et al. When Killers Become Thieves: Trogocytosed PD-1 Inhibits NK Cells in Cancer. Available online: https://www.biorxiv.org/content/10.1101/2020.06.26.174342v2.abstract (accessed on 13 October 2021).
- Oldenhove, G.; Boucquey, E.; Taquin, A.; Acolty, V.; Bonetti, L.; Ryffel, B.; Le Bert, M.; Englebert, K.; Boon, L.; Moser, M. PD-1 is involved in the dysregulation of type 2 innate lymphoid cells in a murine model of obesity. Cell Rep. 2018, 25, 2053–2060.e4. [Google Scholar] [CrossRef]
- Myklebust, J.H.; Irish, J.M.; Brody, J.; Czerwinski, D.K.; Houot, R.; Kohrt, H.E.; Timmerman, J.; Said, J.; Green, M.R.; Delabie, J.; et al. High PD-1 expression and suppressed cytokine signaling distinguish T cells infiltrating follicular lymphoma tumors from peripheral T cells. Blood 2013, 121, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Kavousanaki, M.; Ioannou, M.; Boumpas, D.; Verginis, P. The Negative Costimulatory Molecule PD-1 Modulates the Balance between Immunity and Tolerance via MiR-21. Eur. J. Immunol. 2011, 41, 1754–1763. [Google Scholar] [CrossRef]
- Linsley, P.S.; Ledbetter, J.A. The role of the CD28 receptor during T cell responses to antigen. Annu. Rev. Immunol. 1993, 11, 191–212. [Google Scholar] [CrossRef] [PubMed]
- Linsley, P.S.; Clark, E.A.; Ledbetter, J.A. T-cell antigen CD28 mediates adhesion with B cells by interacting with activation antigen B7/BB-1. Proc. Natl. Acad. Sci. USA 1990, 87, 5031–5035. [Google Scholar] [CrossRef] [PubMed]
- Linsley, P.S.; Bradshaw, J.; Greene, J.; Peach, R.; Bennett, K.L.; Mittler, R.S. Intracellular Trafficking of CTLA-4 and Focal Localization towards Sites of TCR Engagement. Immunity 1996, 4, 535–543. [Google Scholar] [CrossRef]
- Jago, C.B.; Yates, J.; Olsen Saraiva Câmara, N.; Lechler, R.I.; Lombardi, G. Differential expression of CTLA-4 among T cell subsets. Clin. Exp. Immunol. 2004, 136, 463–471. [Google Scholar] [CrossRef]
- Takahashi, T.; Tagami, T.; Yamazaki, S.; Uede, T.; Shimizu, J.; Sakaguchi, N.; Mak, T.W.; Sakaguchi, S. Immunologic self-tolerance maintained by CD25(+)CD4(+) regulatory T cells constitutively expressing cytotoxic T lymphocyte-associated antigen 4. J. Exp. Med. 2000, 192, 303–310. [Google Scholar] [CrossRef]
- Wing, K.; Onishi, Y.; Prieto-Martin, P.; Yamaguchi, T.; Miyara, M.; Fehervari, Z.; Nomura, T.; Sakaguchi, S. CTLA-4 Control over Foxp3+ regulatory T cell function. Science 2008, 322, 271–275. [Google Scholar] [CrossRef]
- Wolf, Y.; Anderson, A.C.; Kuchroo, V.K. TIM3 comes of age as an inhibitory receptor. Nat. Rev. Immunol. 2020, 20, 173–185. [Google Scholar] [CrossRef]
- Blake, S.J.; Stannard, K.; Liu, J.; Allen, S.; Yong, M.C.R.; Mittal, D.; Aguilera, A.R.; Miles, J.J.; Lutzky, V.P.; de Andrade, L.F.; et al. Suppression of metastases using a new lymphocyte checkpoint target for cancer immunotherapy. Cancer Discov. 2016, 6, 446–459. [Google Scholar] [CrossRef] [PubMed]
- Hutloff, A.; Dittrich, A.M.; Beier, K.C.; Eljaschewitsch, B.; Kraft, R.; Anagnostopoulos, I.; Kroczek, R.A. ICOS Is an inducible t-cell co-stimulator structurally and functionally related to CD28. Nature 1999, 397, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Coyle, A.J.; Lehar, S.; Lloyd, C.; Tian, J.; Delaney, T.; Manning, S.; Nguyen, T.; Burwell, T.; Schneider, H.; Gonzalo, J.A.; et al. The CD28-related molecule ICOS is required for effective T cell-dependent immune responses. Immunity 2000, 13, 95–105. [Google Scholar] [CrossRef]
- Toker, A.; Nguyen, L.T.; Stone, S.C.; Yang, S.Y.C.; Katz, S.R.; Shaw, P.A.; Clarke, B.A.; Ghazarian, D.; Al-Habeeb, A.; Easson, A.; et al. Regulatory T cells in ovarian cancer are characterized by a highly activated phenotype distinct from that in melanoma. Clin. Cancer Res. 2018, 24, 5685–5696. [Google Scholar] [CrossRef] [PubMed]
- Yoshinaga, S.K.; Zhang, M.; Pistillo, J.; Horan, T.; Khare, S.D.; Miner, K.; Sonnenberg, M.; Boone, T.; Brankow, D.; Dai, T.; et al. Characterization of a new human B7-related protein: B7RP-1 is the ligand to the co-stimulatory protein ICOS. Int. Immunol. 2000, 12, 1439–1447. [Google Scholar] [CrossRef] [PubMed]
- Swallow, M.M.; Wallin, J.J.; Sha, W.C. B7h, a novel costimulatory homolog of B7.1 and B7.2, is induced by TNFα. Immunity 1999, 11, 423–432. [Google Scholar] [CrossRef]
- Khayyamian, S.; Hutloff, A.; Büchner, K.; Gräfe, M.; Henn, V.; Kroczek, R.A.; Mages, H.W. ICOS-ligand, expressed on human endothelial cells, costimulates Th1 and Th2 cytokine secretion by memory CD4+ T cells. Proc. Natl. Acad. Sci. USA 2002, 99, 6198–6203. [Google Scholar] [CrossRef]
- Dong, C.; Juedes, A.E.; Temann, U.A.; Shresta, S.; Allison, J.P.; Ruddle, N.H.; Flavell, R.A. ICOS Co-stimulatory receptor is essential for T-cell activation and function. Nature 2001, 409, 97–101. [Google Scholar] [CrossRef]
- Gonzalo, J.A.; Tian, J.; Delaney, T.; Corcoran, J.; Rottman, J.B.; Lora, J.; Al-Garawi, A.; Kroczek, R.; Gutierrez-Ramos, J.C.; Coyle, A.J. ICOS is critical for t helper cell-mediated lung mucosal inflammatory responses. Nat. Immunol. 2001, 2, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Nurieva, R.I.; Duong, J.; Kishikawa, H.; Dianzani, U.; Rojo, J.M.; Ho, I.C.; Flavell, R.A.; Dong, C. Transcriptional regulation of Th2 differentiation by inducible costimulator. Immunity 2003, 18, 801–811. [Google Scholar] [CrossRef]
- Rigas, D.; Lewis, G.; Aron, J.L.; Wang, B.; Banie, H.; Sankaranarayanan, I.; Galle-Treger, L.; Maazi, H.; Lo, R.; Freeman, G.J.; et al. ILC2 suppression by regulatory T cells attenuates airway hyperreactivity and requires ICOS:ICOS-ligand interaction. J. Allergy Clin. Immunol. 2017, 139, 1468–1477.e2. [Google Scholar] [CrossRef]
- Makkouk, A.; Chester, C.; Kohrt, H.E. Rationale for Anti-CD137 cancer immunotherapy. Eur. J. Cancer 2016, 54, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Golebski, K.; Layhadi, J.A.; Sahiner, U.; Steveling-Klein, E.H.; Lenormand, M.M.; Li, R.C.Y.; Bal, S.M.; Heesters, B.A.; Vilà-Nadal, G.; Hunewald, O.; et al. Induction of IL-10-Producing type 2 innate lymphoid cells by allergen immunotherapy is associated with clinical response. Immunity 2021, 54, 291–307.e7. [Google Scholar] [CrossRef] [PubMed]
- Clouthier, D.L.; Watts, T.H. Cell-specific and context-dependent effects of GITR in cancer, autoimmunity, and infection. Cytokine Growth Factor Rev. 2014, 25, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Rauber, S.; Luber, M.; Weber, S.; Maul, L.; Soare, A.; Wohlfahrt, T.; Lin, N.-Y.; Dietel, K.; Bozec, A.; Herrmann, M.; et al. Resolution of inflammation by interleukin-9-producing type 2 innate lymphoid cells. Nat. Med. 2017, 23, 938–944. [Google Scholar] [CrossRef]
- Murphy, K.M.; Nelson, C.A.; Šedý, J.R. Balancing co-stimulation and inhibition with BTLA and HVEM. Nat. Rev. Immunol. 2006, 6, 671–681. [Google Scholar] [CrossRef]
- Sugamura, K.; Ishii, N.; Weinberg, A.D. Therapeutic targeting of the effector T-cell co-stimulatory molecule OX40. Nat. Rev. Immunol. 2004, 4, 420–431. [Google Scholar] [CrossRef]

| ILC Subset | Checkpoint Marker | Ligands | Mouse | Human | Function | Context of Expression | References |
|---|---|---|---|---|---|---|---|
| NK cells | PD-1 | PD-L1/2 | Yes | Yes | Negative regulation. Reduced NK cell cytokine production and cytotoxicity resulting in tumor escape. | Induced upon activation by viral infections including MCMV, HCMV, HIV and HCV as well as tumor microenvironment including ovarian carcinoma, Kaposi sarcoma, Hodgkin lymphoma, pleural effusions of primary and metastatic tumors, digestive, lung and breast cancers. | [114,115,116,117,118,119,120,121,122,123,124] |
| TIM-3 | Galectin-9, phosphatidylserine, HMGB1, Caecam-1 | Yes | Yes | Negative regulation. Impaired NK cell-derived IFN-γ and TNF-α expression as well as perforin production and cytotoxicity. | Expressed at steady state (CD56dim NK cells). TIM-3 is expressed in NK cells upon stimulation with IL-2, IL-12+IL-18 or IL-15 and cancers including gastrointestinal stromal tumors, gastric cancer, oesophageal cancer, melanoma, colorectal cancer, anaplastic thyroid cancer, bladder cancer, hepatocellular carcinoma and lung adenocarcinoma. | [125,126,127,128,129,130,131,132,133,134,135,136] | |
| TIGIT | CD155 CD112 | Yes | Yes | Negative regulation. Impaired NK cell activity. | Expressed at steady state in circulation and tumor microenvironment including in melanoma and colorectal cancer. | [137,138,139,140,141,142] | |
| LAG-3 | MHC-II, galectin-3, LSECtin, FGL-1 | Yes | Negative regulation. Reduced IFN-γ expression and may inhibit NK cell function. | Chronic stimulation through NKG2C, NKp30 or NKG2D and stimulation with IFN-α as well as chronic lymphocytic leukaemia. | [143,144,145,146,147,148] | ||
| KLRG1 | E-, N, R-Cadherin | Yes | Yes | Negative regulation. Reduced NK cell cytotoxicity, proliferation and increased apoptosis. | Klrg1−/− NK cells have increased capacities to produce pro-inflammatory cytokines in tumor microenvironment | [149,150,151,152] | |
| GITR | GITRL | Yes | Yes | Negative regulation. Dampened NK cell function, proliferation, and survival. | Reduced anti-tumor activities | [153,154,155,156,157] | |
| BTLA | HVEM | Yes | Negative regulation. Reduced NK cell IFN-γ expression and killing capacity. | Chronic lymphocytic leukaemia | [158,159] | ||
| CTLA-4 | CD80, CD86 | Yes | Yes | Not determined | A fraction of circulating NK cells in melanoma patients. | [160] | |
| ILC1 | PD-1 | PD-L1/2 | Yes | Not determined | Tumor microenvironment including non-small cell lung cancer, breast and gastrointestinal tumors. | [124,161,162,163] | |
| CTLA-4 | CD80, CD86 | Yes | Yes | Not determined | Tumor microenvironment including melanoma, hepatocellular carcinoma, breast and gastrointestinal cancer. | [49,111,124,160,161,164,165,166,167] | |
| TIGIT | CD155 CD112 | Yes | May negatively regulate ILC1 function. | Tumor microenvironment | [161] | ||
| LAG-3 | MHC-II, galectin-3, LSECtin, FGL-1 | Yes | Yes | Not determined | Tumor microenvironment | [111,161] | |
| KLRG1 | E-, N, R-Cadherin | Yes | Not determined | Tumor microenvironment including non-small cell lung cancer, breast and gastrointestinal tumors. | [124] | ||
| GITR | GITRL | Yes | Negative regulation. Inhibited ILC1 IFN-γ production and activity. | Expressed at steady state and enhanced upon activation following influenza infection. | [168] | ||
| ILC2 | KLRG1 | E-, N, R-Cadherin | Yes | Yes | Negative regulation. May negatively regulate ILC2 accumulation and function in the lungs following IL-33 injection. | During steady state on iILC2 and upon IL-25 or IL-33 activation on nILC2. | [46,149,150,169,170,171,172,173,174,175,176,177] |
| PD-1 | PD-L1/2 | Yes | Yes | Negative regulation. Regulation of ILC2 metabolism and STAT5 phosphorylation resulting in decreased cytokine expression and proliferation. | Lowly expressed at steady state and is further increased upon IL-33 and γc cytokines stimulation in contexts including influenza infection, papain challenge and cancer including breast, gastrointestinal, colorectal, melanoma, non-small lung and pancreatic adenocarcinoma cancers. | [44,46,49,124,162,163,170,178,179,180,181] | |
| PD-L1 | PD-1 | Yes | Promoted Th2 polarization and type 2 inflammatory responses. | Nippostrongylus brasiliensis infection and retinoic acid stimulation. | [111,170,182] [183] | ||
| CTLA-4 | CD80, CD86 | Yes | Yes | Negative regulation. May negatively regulate ILC2 maintenance and may bestow immunosuppressive properties on ILC2. | Retinoic acid stimulation and tumor microenvironment including hepatocellular carcinoma, breast, and gastrointestinal cancer. | [49,124,183,184] | |
| ICOS | ICOS-L | Yes | Yes | Positive regulation. Promoted lung and small intestine ILC2 accumulation, cytokine expression, survival, and proliferation. | Notably expressed on lung and skin ILC2. | [185,186,187,188,189] | |
| CD137/ 4-1BB | CD137L/ 4-1BBL | Yes | Yes | May modulate ILC2 effector function. | Expressed in mouse large and small intestine ILC2. | [34,42,111,190] | |
| GITR | GITRL | Yes | Yes | Positive regulation. Promoted ILC2 cytokine expression and function. | Expressed at steady state and has an increased expression upon activation including Alternia-induced lung inflammation. | [191,192] | |
| OX40L | OX40 | Yes | Promoted Th2 and Treg cell responses critical to anti-helminth and allergic type 2 immunity. | Upon papain or IL-33 stimulation, helminth infection, allergic reactions, and respiratory syncytial virus infection. | [193,194,195,196,197,198] | ||
| ILC3 | PD-1 | PD-L1/2 | Yes | Yes | Negative regulation. Inhibited ILC3 function. | Mouse intestine, human decidua and tumors including breast and gastrointestinal tumors. | [49,121,124,162,199] |
| TIM-3 | Galectin-9, phosphatidylserine, HMGB1, Caecam-1 | Yes | Negative regulation. May inhibit ILC3 function. | Human decidua | [199,200,201,202,203] | ||
| OX40L | OX40 | Yes | Yes | LTi cell expression is required for CD4+ T cell memory maintenance within secondary lymphoid organs. ILC3 maintains intestinal barrier homeostasis and promotes conventional and regulatory CD4+ T cell maintenance, proliferation, and function. | Microbiota and intestinal inflammation promote mononuclear macrophages TL1A secretion, which drives ILC3 OX40L expression. Hence, intestinal ILC3 constitutively express OX40L and the level of expression is increased upon inflammation such as Crohn’s disease. | [196,197,198,204] | |
| CTLA-4 | CD80, CD86 | Yes | May negatively regulate ILC3 maintenance. | Hepatocellular carcinoma | [49,184] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jacquelot, N.; Ghaedi, M.; Warner, K.; Chung, D.C.; Crome, S.Q.; Ohashi, P.S. Immune Checkpoints and Innate Lymphoid Cells—New Avenues for Cancer Immunotherapy. Cancers 2021, 13, 5967. https://doi.org/10.3390/cancers13235967
Jacquelot N, Ghaedi M, Warner K, Chung DC, Crome SQ, Ohashi PS. Immune Checkpoints and Innate Lymphoid Cells—New Avenues for Cancer Immunotherapy. Cancers. 2021; 13(23):5967. https://doi.org/10.3390/cancers13235967
Chicago/Turabian StyleJacquelot, Nicolas, Maryam Ghaedi, Kathrin Warner, Douglas C. Chung, Sarah Q. Crome, and Pamela S. Ohashi. 2021. "Immune Checkpoints and Innate Lymphoid Cells—New Avenues for Cancer Immunotherapy" Cancers 13, no. 23: 5967. https://doi.org/10.3390/cancers13235967
APA StyleJacquelot, N., Ghaedi, M., Warner, K., Chung, D. C., Crome, S. Q., & Ohashi, P. S. (2021). Immune Checkpoints and Innate Lymphoid Cells—New Avenues for Cancer Immunotherapy. Cancers, 13(23), 5967. https://doi.org/10.3390/cancers13235967






