Phenotypic Consequences of SLC25A40-ABCB1 Fusions beyond Drug Resistance in High-Grade Serous Ovarian Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
“The law is not the survival of the “better” or the “stronger”. It is the survival of those which are constitutionally fittest to thrive under the conditions in which they are placed.”Herbert Spencer
2. Materials and Methods
2.1. Cell Line Culture
2.2. Immunodetection
2.3. IncuCyte Live Cell Imaging
2.4. MDR Efflux Assay
2.5. RNA Extraction and qRT-PCR
2.6. RNA Sequencing
2.7. Extracellular Flux Analysis
2.8. High-Throughput Drug Screening
2.9. Viability and Actin Morphology
2.10. Targeted Sequencing
2.11. Statistical Analysis
3. Results
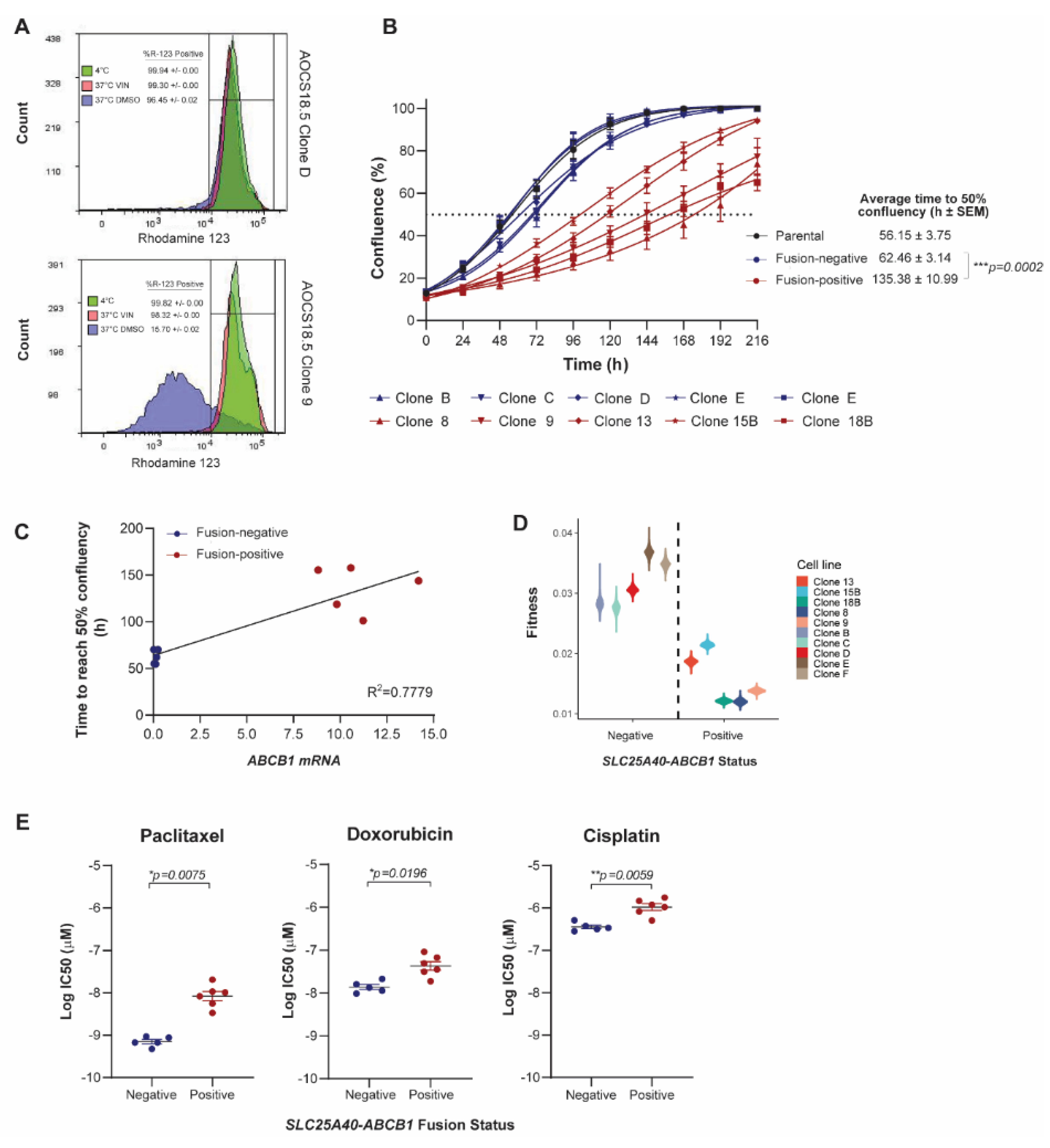
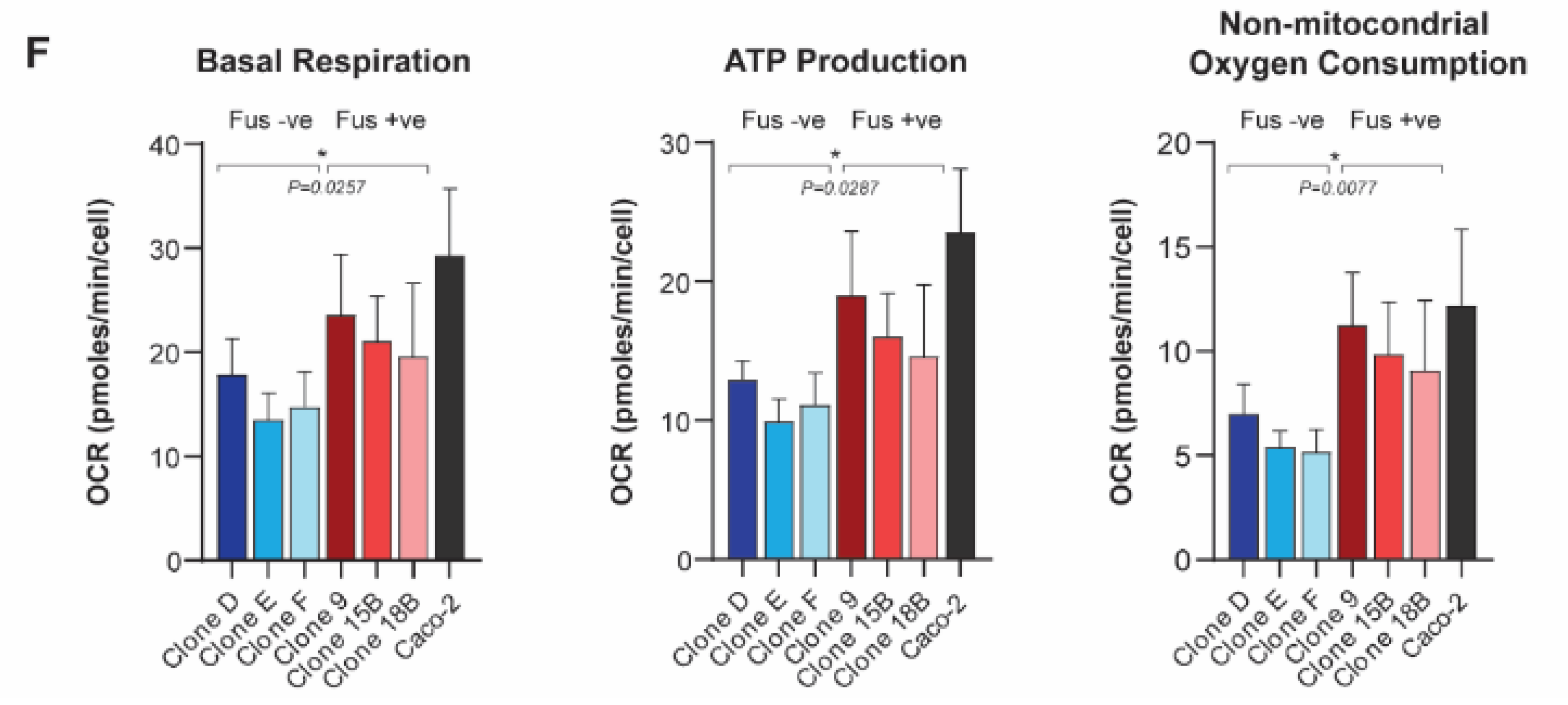
3.1. SLC25A40-ABCB1 Fusions Mediate Multi-Drug Resistance, Decreased Proliferation and Elevated Oxidative Metabolism
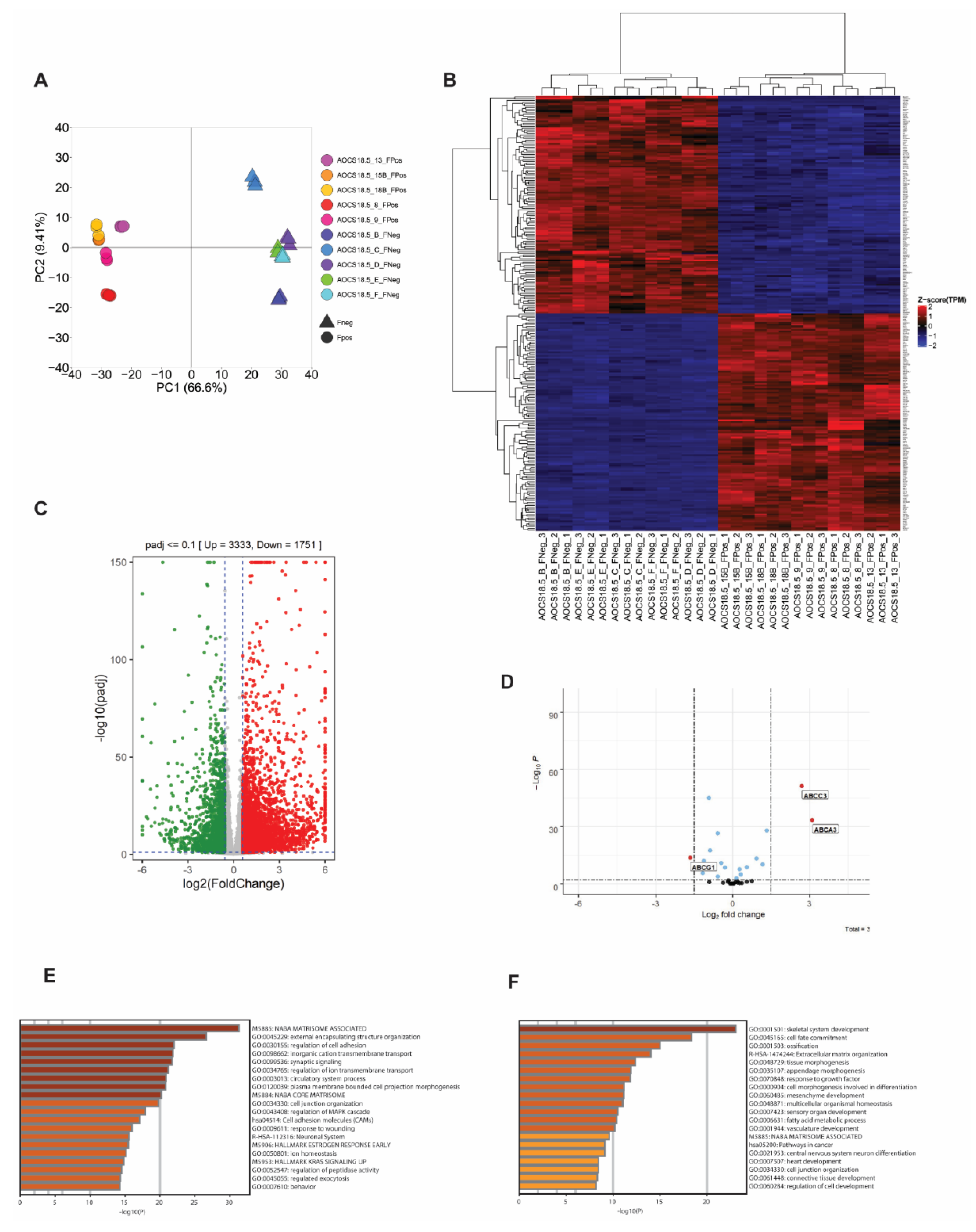
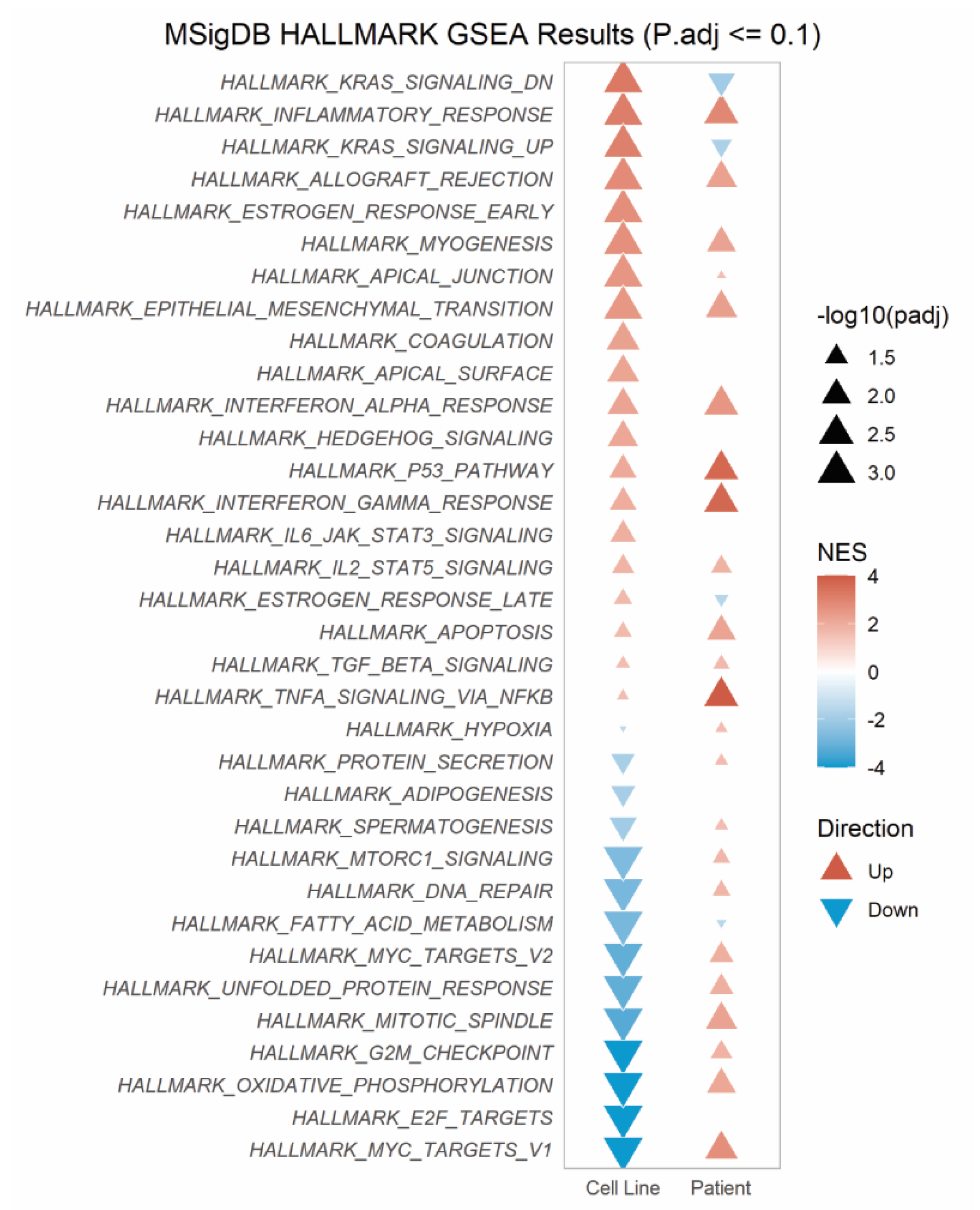
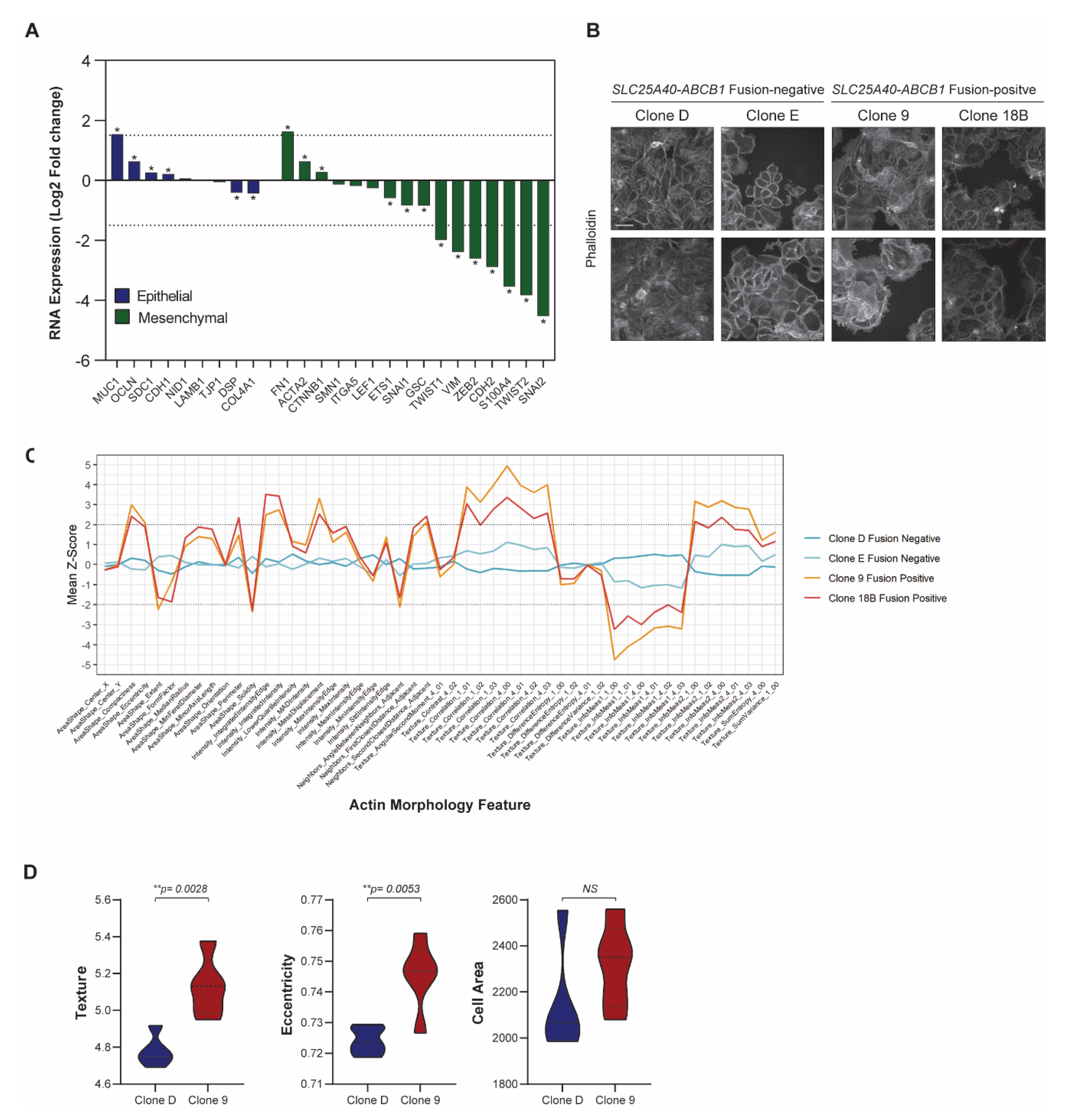
3.2. SLC25A40-ABCB1 Fusions Mediate an ECM/Inflammatory Enriched Transcriptional Profile
3.3. SLC25A40-ABCB1 Fusion-Positive Cells Display Altered Cellular Morphology
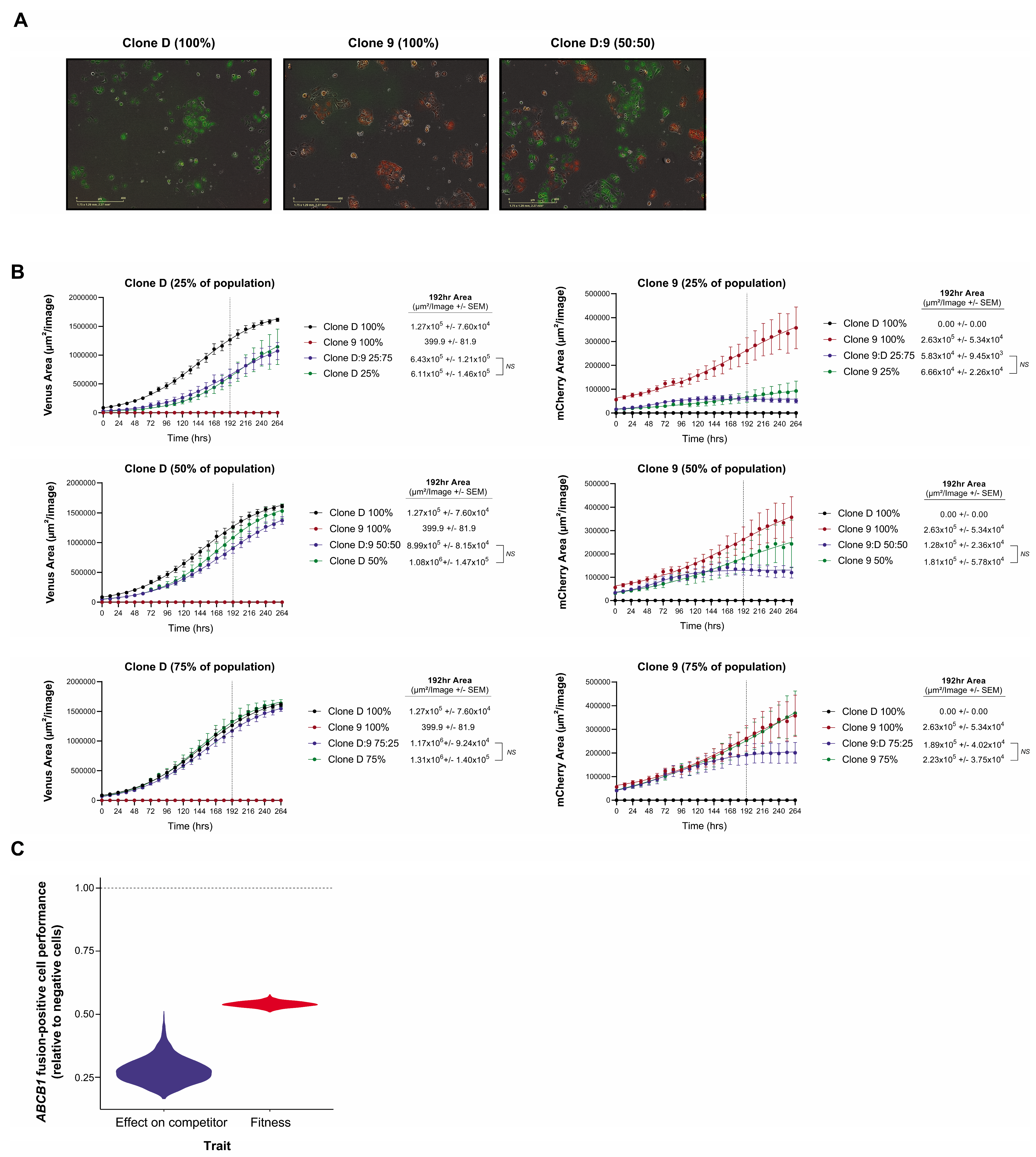
3.4. Clonal Co-Operation between Fusion-Negative and Positive Cells Does Not Promote Proliferative Fitness
3.5. High-Throughput Drug Screening Identifies FDA Agents That Induce Cytotoxic Responses Regardless of Fusion Status and Clonal Composition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gatenby, R.A.; Brown, J.S. Integrating evolutionary dynamics into cancer therapy. Nat. Rev. Clin. Oncol. 2020, 17, 675–686. [Google Scholar] [CrossRef]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2014, 136, E359–E386. [Google Scholar] [CrossRef]
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [Green Version]
- Bowtell, D.D.; Böhm, S.; Ahmed, A.A.; Aspuria, P.-J.; Robert, C.B., Jr.; Beral, V.; Berek, J.S.; Birrer, M.J.; Blagden, S.; Bookman, M.A.; et al. Rethinking ovarian cancer II: Reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer 2015, 15, 668–679. [Google Scholar] [CrossRef]
- Ahmed, A.A.; Etemadmoghadam, D.; Temple, J.; Lynch, A.; Riad, M.; Sharma, R.; Stewart, C.; Fereday, S.; Caldas, C.; DeFazio, A.; et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J. Pathol. 2010, 221, 49–56. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Patch, A.-M.; The Australian Ovarian Cancer Study Group; Christie, E.; Etemadmoghadam, D.; Garsed, D.; George, J.; Fereday, S.; Nones, K.; Cowin, P.; Alsop, K.; et al. Whole–genome characterization of chemoresistant ovarian cancer. Nature 2015, 521, 489–494. [Google Scholar] [CrossRef]
- Ledermann, J.; Harter, P.; Gourley, C.; Friedlander, M.; Vergote, I.; Rustin, G.; Scott, C.L.; Meier, W.; Shapira-Frommer, R.; Safra, T.; et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: A preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014, 15, 852–861. [Google Scholar] [CrossRef]
- Gockley, A.; Melamed, A.; Bregar, A.J.; Clemmer, J.T.; Birrer, M.; Schorge, J.O.; Del Carmen, M.G.; Rauh-Hain, J.A. Outcomes of Women with High-Grade and Low-Grade Advanced-Stage Serous Epithelial Ovarian Cancer. Obstet. Gynecol. 2017, 129, 439–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blagden, S.P.; Nicum, S. A source of hope for platinum-resistant ovarian cancer? Lancet 2021, 397, 254–256. [Google Scholar] [CrossRef]
- Van Zyl, B.; Tang, D.; Bowden, N.A. Biomarkers of platinum resistance in ovarian cancer: What can we use to improve treatment. Endocr.-Related Cancer 2018, 25, R303–R318. [Google Scholar] [CrossRef]
- Szakacs, G.; Paterson, J.K.; Ludwig, J.A.; Booth-Genthe, C.; Gottesman, M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006, 5, 219–234. [Google Scholar] [CrossRef]
- Christie, E.; Pattnaik, S.; Beach, J.; Copeland, A.; Rashoo, N.; Fereday, S.; Hendley, J.; Alsop, K.; Brady, S.L.; Lamb, G.; et al. Multiple ABCB1 transcriptional fusions in drug resistant high-grade serous ovarian and breast cancer. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knutsen, T.; Mickley, L.A.; Ried, T.; Green, E.D.; du Manoir, S.; Schröck, E.; Macville, M.; Ning, Y.; Robey, R.; Polymeropoulos, M.; et al. Cytogenetic and molecular characterization of random chromosomal rearrangements activating the drug resistance gene, MDR1/P-glycoprotein, in drug-selected cell lines and patients with drug refractory ALL. Genes Chromosom. Cancer 1998, 23, 44–54. [Google Scholar] [CrossRef]
- Ween, M.; Armstrong, M.; Oehler, M.; Ricciardelli, C. The role of ABC transporters in ovarian cancer progression and chemoresistance. Crit. Rev. Oncol. 2015, 96, 220–256. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.I.; Haber, M.; Henderson, M.J.; Norris, M.D. ABC transporters in cancer: More than just drug efflux pumps. Nat. Rev. Cancer 2010, 10, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Kimura, Y.; Nagata, K.; Yamamoto, A.; Matsuo, M.; Ueda, K. ABC proteins: Key molecules for lipid homeostasis. Med. Mol. Morphol. 2005, 38, 2–12. [Google Scholar] [CrossRef]
- Ford, J.M.; Hait, W.N. Pharmacology of drugs that alter multidrug resistance in cancer. Pharmacol. Rev. 1990, 42, 155–199. [Google Scholar]
- Holland, I.; Blight, M.A. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol. 1999, 293, 381–399. [Google Scholar] [CrossRef]
- Hodges, L.M.; Markova, S.M.; Chinn, L.W.; Gow, J.M.; Kroetz, D.L.; Klein, T.E.; Altman, R.B. Very important pharmacogene summary: ABCB1 (MDR1, P-glycoprotein). Pharm. Genom. 2011, 21, 152–161. [Google Scholar] [CrossRef] [Green Version]
- Juliano, R.; Ling, V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim. Biophys. Acta Biomembr. 1976, 455, 152–162. [Google Scholar] [CrossRef]
- Staud, F.; Ceckova, M.; Micuda, S.; Pavek, P. Multi-Drug Resistance in Cancer; Zhou, J., Ed.; Humana Press: Totowa, NJ, USA, 2010; pp. 199–222. [Google Scholar]
- Robey, R.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Weber, K.; Thomaschewski, M.; Benten, D.; Fehse, B. RGB marking with lentiviral vectors for multicolor clonal cell tracking. Nat. Protoc. 2012, 7, 839–849. [Google Scholar] [CrossRef]
- Santiappillai, N.T.; Abuhammad, S.; Slater, A.; Kirby, L.; McArthur, G.A.; Sheppard, K.E.; Smith, L.K. CDK4/6 Inhibition Reprograms Mitochondrial Metabolism in BRAF(V600) Melanoma via a p53 Dependent Pathway. Cancers 2021, 13, 524. [Google Scholar] [CrossRef] [PubMed]
- Wind, N.S.; Holen, I. Multidrug Resistance in Breast Cancer: FromIn VitroModels to Clinical Studies. Int. J. Breast Cancer 2011, 2011, 967419. [Google Scholar] [CrossRef]
- Kam, Y.; Das, T.; Tian, H.; Foroutan, P.; Ruiz, E.; Martinez, G.V.; Minton, S.; Gillies, R.; Gatenby, R.A. Sweat but no gain: Inhibiting proliferation of multidrug resistant cancer cells with “ersatzdroges”. Int. J. Cancer 2014, 136, E188–E196. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-J.; Chin, J.E.; Ueda, K.; Clark, D.P.; Pastan, I.; Gottesman, M.M.; Roninson, I.B. Internal duplication and homology with bacterial transport proteins in the mdr1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell 1986, 47, 381–389. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Hynes, R.O.; Naba, A. Overview of the Matrisome—An Inventory of Extracellular Matrix Constituents and Functions. Cold Spring Harb. Perspect. Biol. 2011, 4, a004903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naba, A.; Clauser, K.; Hoersch, S.; Liu, H.; Carr, S.A.; Hynes, R.O. The Matrisome: In Silico Definition and In Vivo Characterization by Proteomics of Normal and Tumor Extracellular Matrices. Mol. Cell. Proteom. 2012, 11. [Google Scholar] [CrossRef] [Green Version]
- Peche, L.Y.; Ladelfa, M.F.; Toledo, M.F.; Mano, M.; Laiseca, J.E.; Schneider, C.; Monte, M. Human MageB2 Protein Expression Enhances E2F Transcriptional Activity, Cell Proliferation, and Resistance to Ribotoxic Stress. J. Biol. Chem. 2015, 290, 29652–29662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scanlan, M.J.; Simpson, A.J.G.; Old, L.J. The cancer/testis genes: Review, standardization, and commentary. Cancer Immun. 2004, 4, 1. [Google Scholar] [PubMed]
- Han, H.; Shim, H.; Shin, D.; Shim, J.E.; Ko, Y.; Shin, J.; Kim, H.; Cho, A.; Kim, E.; Lee, T.; et al. TRRUST: A reference database of human transcriptional regulatory interactions. Sci. Rep. 2015, 5, 11432. [Google Scholar] [CrossRef] [PubMed]
- Cornwell, M.; Smith, D. SP1 activates the MDR1 promoter through one of two distinct G-rich regions that modulate promoter activity. J. Biol. Chem. 1993, 268, 19505–19511. [Google Scholar] [CrossRef]
- Hu, Z.; Jin, S.; Scotto, K.W. Transcriptional Activation of the MDR1 Gene by UV Irradiation. J. Biol. Chem. 2000, 275, 2979–2985. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.-T. Use of arrays to investigate the contribution of ATP-binding cassette transporters to drug resistance in cancer chemotherapy and prediction of chemosensitivity. Cell Res. 2007, 17, 311–323. [Google Scholar] [CrossRef]
- Australian Institute of Health and Welfare. Cancer in Australia: In Brief 2019; Cancer Series No. 122; Cat No. CAN 126; AIHW: Canberra, Australia, 2019. [Google Scholar]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [Green Version]
- Brayman, M.; Thathiah, A.; Carson, D.D. MUC1: A multifunctional cell surface component of reproductive tissue epithelia. Reprod. Biol. Endocrinol. 2004, 2, 4–14. [Google Scholar] [CrossRef] [Green Version]
- Hafner, M.; Heiser, L.M.; Williams, E.H.; Niepel, M.; Wang, N.J.; Korkola, J.E.; Gray, J.W.; Sorger, P.K. Quantification of sensitivity and resistance of breast cancer cell lines to anti-cancer drugs using GR metrics. Sci. Data 2017, 4, 170166. [Google Scholar] [CrossRef] [Green Version]
- Bukowski, K.; Kciuk, M.; Kontek, R. Mechanisms of Multidrug Resistance in Cancer Chemotherapy. Int. J. Mol. Sci. 2020, 21, 3233. [Google Scholar] [CrossRef]
- Avendaño, C.; Menéndez, J.C. Chapter 14—Drugs That Modulate Resistance to Antitumor Agents. In Medicinal Chemistry of Anticancer Drugs, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2015; pp. 655–700. [Google Scholar]
- Kadioglu, O.; Saeed, M.E.M.; Munder, M.; Spuller, A.; Greten, H.J.; Efferth, T. Effect of ABC transporter expression and mutational status on survival rates of cancer patients. Biomed. Pharmacother. 2020, 131, 110718. [Google Scholar] [CrossRef]
- Sun, S.; Cai, J.; Yang, Q.; Zhu, Y.; Zhao, S.; Wang, Z. Prognostic Value and Implication for Chemotherapy Treatment of ABCB1 in Epithelial Ovarian Cancer: A Meta-Analysis. PLoS ONE 2016, 11, e0166058. [Google Scholar] [CrossRef]
- Johnatty, S.E.; Beesley, J.; Gao, B.; Chen, X.; Lu, Y.; Law, M.H.; Henderson, M.J.; Russell, A.J.; Hedditch, E.L.; Emmanuel, C.; et al. ABCB1 (MDR1) polymorphisms and ovarian cancer progression and survival: A comprehensive analysis from the Ovarian Cancer Association Consortium and The Cancer Genome Atlas. Gynecol. Oncol. 2013, 131, 8–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, S.-Y.; Ueno, M.; Takakura, N. Involvement of MDR1 Function in Proliferation of Tumour Cells. J. Biochem. 2007, 143, 517–524. [Google Scholar] [CrossRef]
- Yamada, T.; Mori, Y.; Hayashi, R.; Takada, M.; Ino, Y.; Naishiro, Y.; Kondo, T.; Hirohashi, S. Suppression of intestinal polyposis in Mdr1-deficient ApcMin/+ mice. Cancer Res. 2003, 63, 895–901. [Google Scholar] [PubMed]
- Schinkel, A.H.; Mayer, U.; Wagenaar, E.; Mol, C.A.A.M.; van Deemter, L.; Smit, J.J.M.; van der Valk, M.A.; Voordouw, A.C.; Spits, H.; van Tellingen, O.; et al. Normal viability and altered pharmacokinetics in mice lacking mdr1-type (drug-transporting) P-glycoproteins. Proc. Natl. Acad. Sci. USA 1997, 94, 4028–4033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gatenby, R.; Brown, J. The Evolution and Ecology of Resistance in Cancer Therapy. Cold Spring Harb. Perspect. Med. 2017, 8, a033415. [Google Scholar] [CrossRef]
- Tabassum, D.P.; Polyak, K. Tumorigenesis: It takes a village. Nat. Rev. Cancer 2015, 15, 473–483. [Google Scholar] [CrossRef]
- Hötte, K.; Smyrek, I.; Starzinski-Powitz, A.; Stelzer, E.H.K. Endogenous AJAP1 associates with the cytoskeleton and attenuates angiogenesis in endothelial cells. Biol. Open 2017, 6, 723–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanekura, T.; Chen, X.; Kanzaki, T. Basigin (cd147) is expressed on melanoma cells and induces tumor cell invasion by stimulating production of matrix metalloproteinases by fibroblasts. Int. J. Cancer 2002, 99, 520–528. [Google Scholar] [CrossRef] [PubMed]
- Gross, J.C.; Schreiner, A.; Engels, K.; Starzinski-Powitz, A. E-cadherin Surface Levels in Epithelial Growth Factor-stimulated Cells Depend on Adherens Junction Protein Shrew-1. Mol. Biol. Cell 2009, 20, 3598–3607. [Google Scholar] [CrossRef] [Green Version]
- McDonald, J.M.; Dunlap, S.; Cogdell, D.; Dunmire, V.; Wei, Q.; Starzinski-Powitz, A.; Sawaya, R.; Bruner, J.; Fuller, G.N.; Aldape, K.; et al. The SHREW1 gene, frequently deleted in oligodendrogliomas, functions to inhibit cell adhesion and migration. Cancer Biol. Ther. 2006, 5, 300–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivas, J.D.L.; Brozovic, A.; Izraely, S.; Casas-Pais, A.; Witz, I.P.; Figueroa, A. Cancer drug resistance induced by EMT: Novel therapeutic strategies. Arch. Toxicol. 2021, 95, 2279–2297. [Google Scholar] [CrossRef]
- Saxena, M.; A Stephens, M.; Pathak, H.; Rangarajan, A. Transcription factors that mediate epithelial–mesenchymal transition lead to multidrug resistance by upregulating ABC transporters. Cell Death Dis. 2011, 2, e179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miletti-González, K.E.; Chen, S.; Muthukumaran, N.; Saglimbeni, G.N.; Wu, X.; Yang, J.; Apolito, K.; Shih, W.J.; Hait, W.N.; Rodríguez-Rodríguez, L. The CD44 Receptor Interacts with P-Glycoprotein to Promote Cell Migration and Invasion in Cancer. Cancer Res. 2005, 65, 6660–6667. [Google Scholar] [CrossRef] [Green Version]
- Barakat, S.; Turcotte, S.; Demeule, M.; Lachambre, M.-P.; Régina, A.; Baggetto, L.G.; Béliveau, R. Regulation of brain endothelial cells migration and angiogenesis by P-glycoprotein/caveolin-1 interaction. Biochem. Biophys. Res. Commun. 2008, 372, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Tsou, S.-H.; Chen, T.-M.; Hsiao, H.-T.; Chen, Y.-H. A Critical Dose of Doxorubicin Is Required to Alter the Gene Expression Profiles in MCF-7 Cells Acquiring Multidrug Resistance. PLoS ONE 2015, 10, e0116747. [Google Scholar] [CrossRef]
- Robbiani, D.F.; Finch, R.A.; Jäger, D.; Muller, W.A.; Sartorelli, A.C.; Randolph, G.J. The leukotriene C(4) transporter MRP1 regulates CCL19 (MIP-3beta, ELC)-dependent mobilization of dendritic cells to lymph nodes. Cell 2000, 103, 757–768. [Google Scholar] [CrossRef] [Green Version]
| Cisplatin | Clone D (Fus − ve) and Clone 15B (Fus + ve) | Clone E (Fus − ve) and Clone 9 (Fus + ve) | Clone E (Fus − ve) and Clone 18B (Fus + ve) | ||||||
| 72 h IC50 (µM) | ^ p-Value | # p-Value | 72 h IC50 (µM) | ^ p-Value | # p-Value | 72 h IC50 (µM) | ^ p-Value | # p-Value | |
| 100% Fusion-negative | 0.298 ± 0.087 | - | - | 0.357 ± 0.099 | - | - | 0.288 ± 0.068 | - | - |
| 100% Fusion-positive | 1.051 ± 0.245 | 0.022 | - | 1.296 ± 0.368 | 0.036 | - | 1.158 ± 0.211 | 0.008 | - |
| 25:75% Fusion-negative/positive | 0.387 ± 0.057 | NS | 0.027 | 0.464 ± 0.073 | NS | 0.048 | 0.343 ± 0.084 | NS | 0.012 |
| 50:50% Fusion-negative/positive | 0.399 ± 0.096 | NS | NS | 0.348 ± 0.075 | NS | 0.032 | 0.329 ± 0.033 | NS | 0.008 |
| 75:25% Fusion-negative/positive | 0.313 ± 0.119 | NS | 0.031 | 0.285 ± 0.078 | NS | 0.025 | 0.201 ± 0.050 | NS | 0.005 |
| Doxorubicin | Clone D (Fus − ve) and Clone 15B (Fus + ve) | Clone E (Fus − ve) and Clone 9 (Fus + ve) | Clone E (Fus − ve) and Clone 18B (Fus + ve) | ||||||
| 72 h IC50 (nM) | ^ p-Value | # p-Value | 72 h IC50 (nM) | ^ p-Value | # p-Value | 72 h IC50 (nM) | ^ p-Value | # p-Value | |
| 100% Fusion-negative | 16.174 ± 5.165 | - | - | 31.373 ± 6.600 | - | - | 43.532 ± 6.878 | - | - |
| 100% Fusion-positive | 44.062 ± 4.500 | 0.007 | - | 182.293 ± 57.578 | 0.001 | - | 112.613 ± 28.684 | 0.034 | - |
| 25:75% Fusion-negative/positive | 27.061 ± 3.916 | NS | 0.029 | 36.423 ± 7.845 | NS | 0.046 | 46.339 ± 6.600 | NS | 0.040 |
| 50:50% Fusion-negative/positive | 21.580 ± 2.115 | NS | 0.004 | 41.369 ± 6.141 | NS | 0.002 | 53.862 ± 10.862 | NS | NS |
| 75:25% Fusion-negative/positive | 16.203 ± 3.854 | NS | 0.003 | 23.714 ± 9.745 | NS | 0.035 | 42.472 ± 10.916 | NS | 0.039 |
| Paclitaxel | Clone D (Fus − ve) and Clone 15B (Fus + ve) | Clone E (Fus − ve) and Clone 9 (Fus + ve) | Clone E (Fus − ve) and Clone 18B (Fus + ve) | ||||||
| 72 h IC50 (nM) | ^ p-Value | # p-Value | 72 h IC50 (nM) | ^ p-Value | # p-Value | 72 h IC50 (nM) | ^ p-Value | # p-Value | |
| 100% Fusion-negative | 0.769 ± 0.243 | - | - | 0.663 ± 0.203 | - | - | 1.001 ± 0.149 | - | - |
| 100% Fusion-positive | 7.902 ± 0.028 | 0.028 | - | 86.189 ± 33.774 | 0.045 | 44.035 ± 16.859 | 0.028 | - | |
| 25:75% Fusion-negative/positive | 2.316 ± 0.537 | 0.039 | NS | 1.403 ± 0.407 | NS | 0.024 | 2.799 ± 0.525 | 0.017 | 0.033 |
| 50:50% Fusion-negative/positive | 0.995 ± 0.285 | NS | 0.032 | 1.026 ± 0.231 | NS | 0.024 | 1.297 ± 0.345 | NS | 0.029 |
| 75:25% Fusion-negative/positive | 1.096 ± 0.396 | NS | 0.034 | 1.025 ± 0.385 | NS | 0.024 | 1.157 ± 0.570 | NS | 0.029 |
| Compound ID | Class | Mechanism of Action | Transporter * | GR50 (µM) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clone D | Clone E | Clone 9 | Clone 18B | Clone D:9 (50:50 Mix) | Fusion Positive Average | Fusion Negative Average | # p-Value | ^ p-Value | ||||
| 4-demethyl-epicro-podophyllotoxin | Lignan | Antimitotic, binds α- and β-tubulin | ABCB1 | 0.156 | 0.179 | 0.136 | 0.100 | 0.170 | 0.167 | 0.118 | NS | NS |
| 7-ethyl-10-hydroxycamptothecin | Alkaloid | Inhibition of DNA topoisomerase I | ABCB1, ABCG2 | 0.066 | 0.076 | 0.006 | 0.043 | 0.053 | 0.071 | 0.024 | NS | NS |
| amsacrine | Acridine | DNA intercalation and inhibition of topoisomerase II | ABCB1 | 0.057 | 0.134 | 0.027 | 0.048 | 0.056 | 0.095 | 0.037 | NS | NS |
| bleomycin | Antibiotic | Inhibition of DNA synthesis | 0.298 | 0.219 | 0.505 | 0.553 | 0.353 | 0.258 | 0.529 | 0.028 | NS | |
| camptothecin | Alkaloid | Inhibition of DNA topoisomerase I | ABCB1, ABCG2 | 0.108 | 0.109 | 0.047 | 0.070 | 0.089 | 0.108 | 0.059 | NS | NS |
| cedrelone | Limonoid | 1.147 | 1.269 | 1.833 | 1.012 | 0.973 | 1.208 | 1.422 | NS | NS | ||
| cerivastatin lactone | Statin | Competitive HMG-CoA reductase inhibitor | ABCB1, ABCC2, ABCG2, ABC11, SLCO1B1 | 0.299 | 0.265 | 0.160 | 0.209 | 0.251 | 0.282 | 0.185 | NS | NS |
| colchicine | Alkaloid | Inhibition of inflammation caused by tubulin disruption | ABCB1 | UTBD | UTBD | UTBD | UTBD | UTBD | - | - | - | - |
| convallatoxin | Cardiac glycoside | Inhibition of Na+/K+- ATPase | ABCB1 | 0.017 | 0.018 | 0.011 | 0.029 | 0.017 | 0.018 | 0.020 | NS | NS |
| cytarabine | Antimetabolite | Pyrimidine nucleoside | ABCC10, SLC22A4, SLC29A1 | 0.036 | 0.036 | 0.018 | 0.067 | 0.044 | 0.036 | 0.043 | NS | NS |
| dactinomycin | Antibiotic | DNA intercalation | ABCB1, ABCC6, ABCC1, ABCG2, SLC22A5, | 1.207 | 1.490 | 0.589 | 1.086 | 1.032 | 1.348 | 0.837 | NS | NS |
| dasatinib | Tyrosine kinase | BCR/ABL and Src family tyrosine kinase inhibitor | ABCB1, ABCG2 | 0.060 | 0.117 | 0.090 | 0.035 | 0.081 | 0.089 | 0.062 | NS | NS |
| digitoxin | Cardiac glycoside | Inhibition of Na+/K+- ATPase | SLCO1A2, SLCO4C1 | 0.403 | 0.400 | 0.388 | 0.373 | 0.413 | 0.402 | 0.380 | NS | NS |
| docetaxel | Taxoid | Antimitotic, binds tubulin beta-1 chain | ABCB1, ABCC10, ABCG2, ABCC1, ABCC2, SLCO1B3, SLC22A7, | UTBD | UTBD | UTBD | UTBD | UTBD | - | - | - | - |
| emetine dihydrochloride | Antiprotozoal agent and emetic | Inhibition of protein synthesis | 0.009 | 0.036 | 0.036 | 0.043 | 0.012 | 0.023 | 0.040 | NS | NS | |
| gentian_violet | Antifungal | Mitotic poison | SLC22A1 | 0.170 | 0.319 | 0.311 | 0.350 | 0.235 | 0.245 | 0.331 | NS | NS |
| gramicidin | Antibiotic | Membrane disruption and permeabilization (Gram-positive bacteria) | ABCB1 | 0.382 | 0.396 | 0.444 | 0.487 | 0.405 | 0.389 | 0.465 | NS | NS |
| harringtonine | Cephalotaxine alkaloid | Inhibition of protein synthesis | 0.042 | 0.049 | 0.026 | 0.050 | 0.035 | 0.046 | 0.038 | NS | NS | |
| hexachlorophene | Chlorinated bisphenol antiseptic | Inhibition of respiratory D-lactate dehydrogenase | 3.855 | 2.758 | 1.658 | 1.911 | 3.330 | 3.306 | 1.784 | NS | NS | |
| irinotecan_hcl trihydrate | Antineoplastic | Inhibition of DNA topoisomerase I | ABCB1, ABCC1, ABCG2, ABCC2, SLC22A3, SLCO1B1 | 0.234 | 0.125 | 0.006 | 0.024 | 0.113 | 0.179 | 0.015 | NS | NS |
| irinotecan_hydrochloride | Antineoplastic | Inhibition of DNA topoisomerase I | ABCB1, ABCC1, ABCG2, ABCC2, SLC22A3, SLCO1B1 | 0.604 | 0.227 | 0.012 | 0.060 | 0.246 | 0.415 | 0.036 | NS | NS |
| lanatoside c | Cardiac glycoside | 0.133 | 0.124 | 0.116 | 0.125 | 0.129 | 0.128 | 0.121 | NS | NS | ||
| mitoxanthrone hcl | Anthracenediones | DNA Intercalation | ABCB1, ABCC1, ABCG2 | 0.000 | 0.001 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | NS | NS |
| nocodazole | Antineoplastic | Inhibition of microtubule polymerization | 0.087 | 0.111 | 0.100 | 0.079 | 0.081 | 0.099 | 0.090 | NS | NS | |
| ouabain_octahydrate | Cardioactive glycoside | Inhibition of Na+/K+- ATPase | SLCO1A2, SLC22A8, SLCO4C1, SLCO1B3, SLCO1C1, SLCO1B1 | 0.058 | 0.043 | 0.060 | 0.062 | 0.053 | 0.050 | 0.061 | NS | NS |
| parthenolide | Sesquiterpene lactone | Inhibition of IkB kinase (IKK) and IKKβ | 0.332 | 0.583 | 0.220 | 0.343 | 0.228 | 0.457 | 0.281 | NS | NS | |
| patulin | Polyketide mycotoxin | 0.147 | 0.236 | 0.095 | 0.219 | 0.091 | 0.191 | 0.157 | NS | NS | ||
| podofilox | Lignan | Inhibition of DNA topoisomerase II | UTBD | UTBD | UTBD | UTBD | UTBD | - | - | - | - | |
| podophyllin_acetate | Keratolytic | Binds to tubulin to prevent formation of microtubules | 0.065 | 0.079 | 0.067 | 0.053 | 0.066 | 0.072 | 0.060 | NS | NS | |
| proscillaridin a | Cardioactive glycoside | Inhibition of Na+/K+- ATPase | 0.008 | 0.008 | 0.006 | 0.011 | 0.012 | 0.008 | 0.009 | NS | NS | |
| pyrithione_zinc | Antimicrobial | Copper-mediated loss of function of iron–sulphur proteins | 0.440 | 0.423 | 0.731 | 1.056 | 0.382 | 0.432 | 0.893 | NS | NS | |
| strophanthidin acetate | Cardiac glycoside | Inhibition of Na+/K+- ATPase | 0.349 | 0.202 | 0.250 | 0.302 | 0.232 | 0.275 | 0.276 | NS | NS | |
| teniposide | Antineoplastic | Inhibition of DNA topoisomerase II | ABCC6, ABCG2 | 0.039 | 0.065 | 0.005 | 0.032 | 0.018 | 0.052 | 0.019 | NS | NS |
| topotecan_hydrochloride | Antineoplastic | Inhibition of DNA topoisomerase I | ABCB1, ABCG2, SLC47A1, SLC47A2, | 0.093 | 0.105 | 0.023 | 0.073 | 0.084 | 0.099 | 0.048 | NS | NS |
| triptolide | Diterpenoid epoxide | Inhibits transcription and nucleotide excision repair activity of RNA polymerase II | 0.012 | 0.007 | 0.006 | 0.006 | 0.008 | 0.010 | 0.006 | NS | NS | |
| vinblastine sulfate | Vinca alkaloid | Inhibition of microtubule polymerization | ABCB1, ABCC1, ABCC2, ABCC6, ABCB11, SLCO1B1, | 0.011 | 0.021 | 0.013 | 0.011 | 0.012 | 0.016 | 0.012 | NS | NS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pishas, K.I.; Cowley, K.J.; Pandey, A.; Hoang, T.; Beach, J.A.; Luu, J.; Vary, R.; Smith, L.K.; Shembrey, C.E.; Rashoo, N.; et al. Phenotypic Consequences of SLC25A40-ABCB1 Fusions beyond Drug Resistance in High-Grade Serous Ovarian Cancer. Cancers 2021, 13, 5644. https://doi.org/10.3390/cancers13225644
Pishas KI, Cowley KJ, Pandey A, Hoang T, Beach JA, Luu J, Vary R, Smith LK, Shembrey CE, Rashoo N, et al. Phenotypic Consequences of SLC25A40-ABCB1 Fusions beyond Drug Resistance in High-Grade Serous Ovarian Cancer. Cancers. 2021; 13(22):5644. https://doi.org/10.3390/cancers13225644
Chicago/Turabian StylePishas, Kathleen I., Karla J. Cowley, Ahwan Pandey, Therese Hoang, Jessica A. Beach, Jennii Luu, Robert Vary, Lorey K. Smith, Carolyn E. Shembrey, Nineveh Rashoo, and et al. 2021. "Phenotypic Consequences of SLC25A40-ABCB1 Fusions beyond Drug Resistance in High-Grade Serous Ovarian Cancer" Cancers 13, no. 22: 5644. https://doi.org/10.3390/cancers13225644
APA StylePishas, K. I., Cowley, K. J., Pandey, A., Hoang, T., Beach, J. A., Luu, J., Vary, R., Smith, L. K., Shembrey, C. E., Rashoo, N., White, M. O., Simpson, K. J., Bild, A., Griffiths, J. I., Cheasley, D., Campbell, I., Bowtell, D. D. L., & Christie, E. L. (2021). Phenotypic Consequences of SLC25A40-ABCB1 Fusions beyond Drug Resistance in High-Grade Serous Ovarian Cancer. Cancers, 13(22), 5644. https://doi.org/10.3390/cancers13225644






