Prevalence of HPV in Mexican Patients with Head and Neck Squamous Carcinoma and Identification of Potential Prognostic Biomarkers
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Selection
2.2. P16 Expression
2.3. HPV Detection and Genotypification
2.4. HTA 2.0 Expression Microarrays
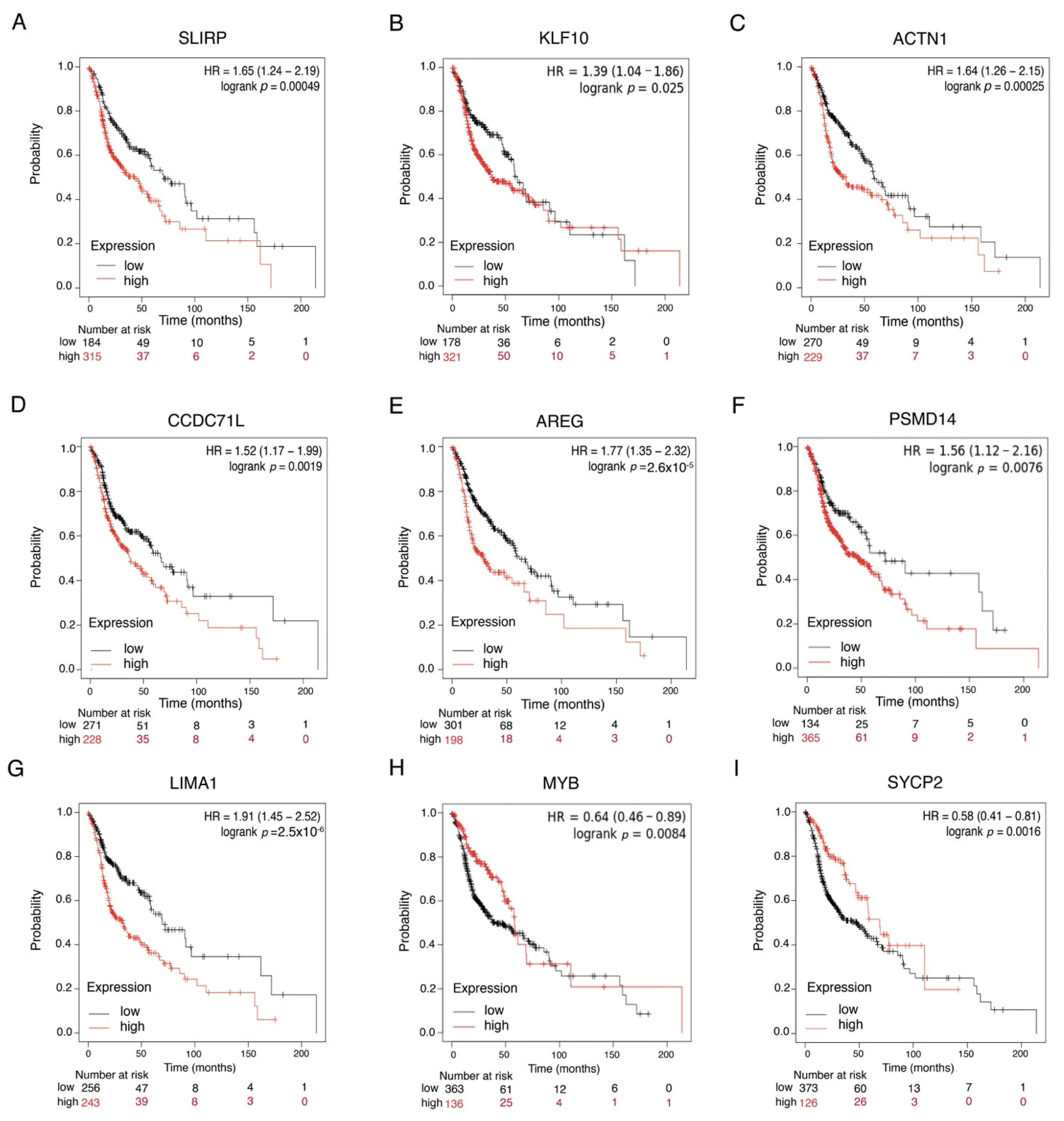
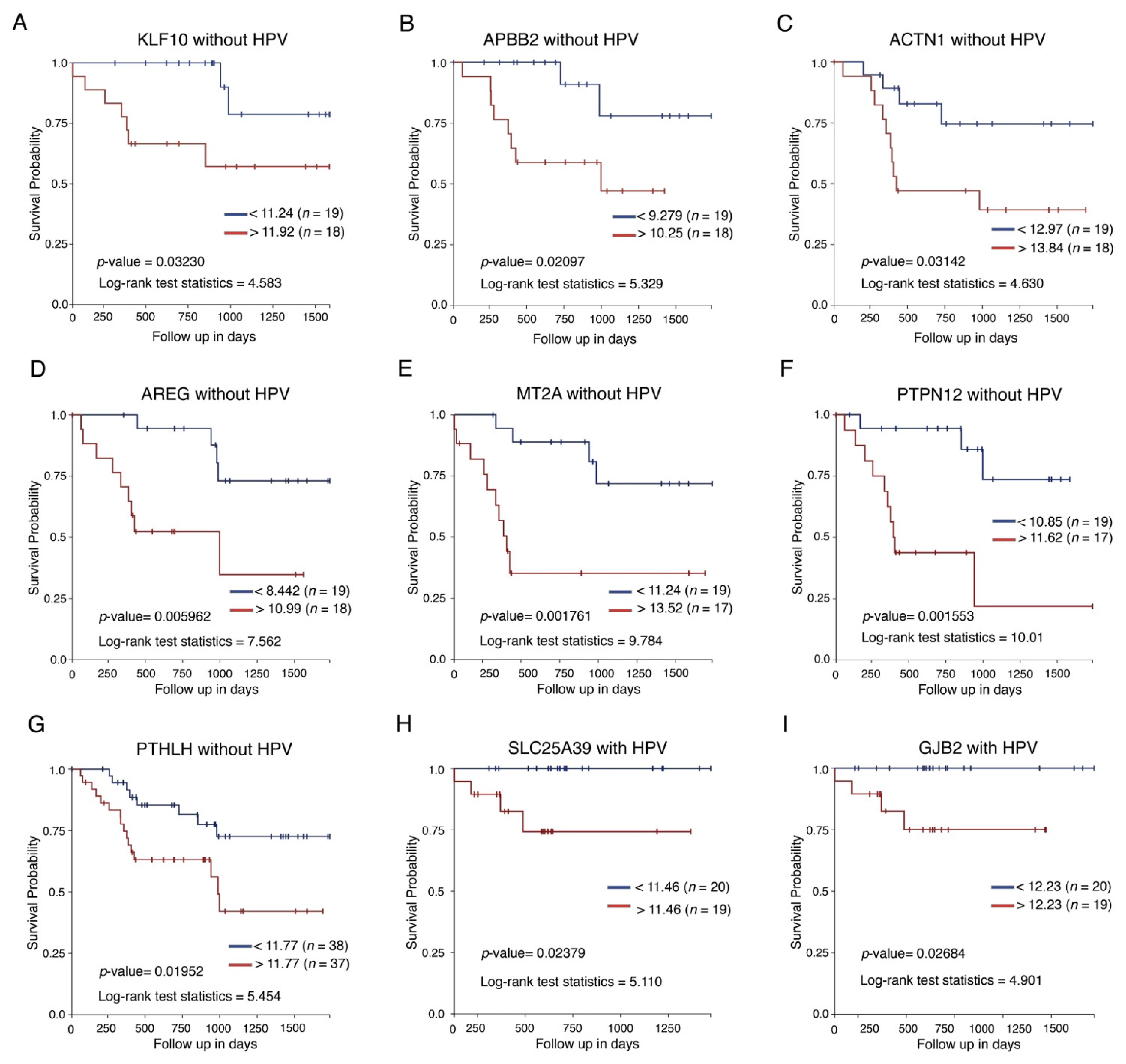
2.5. Survival Analysis
2.6. In Silico Validation
2.7. Enrichment Analysis
2.8. Heatmap
2.9. Statistical Analysis
3. Results
3.1. Clinical Data
3.2. Presence of HPV and P16
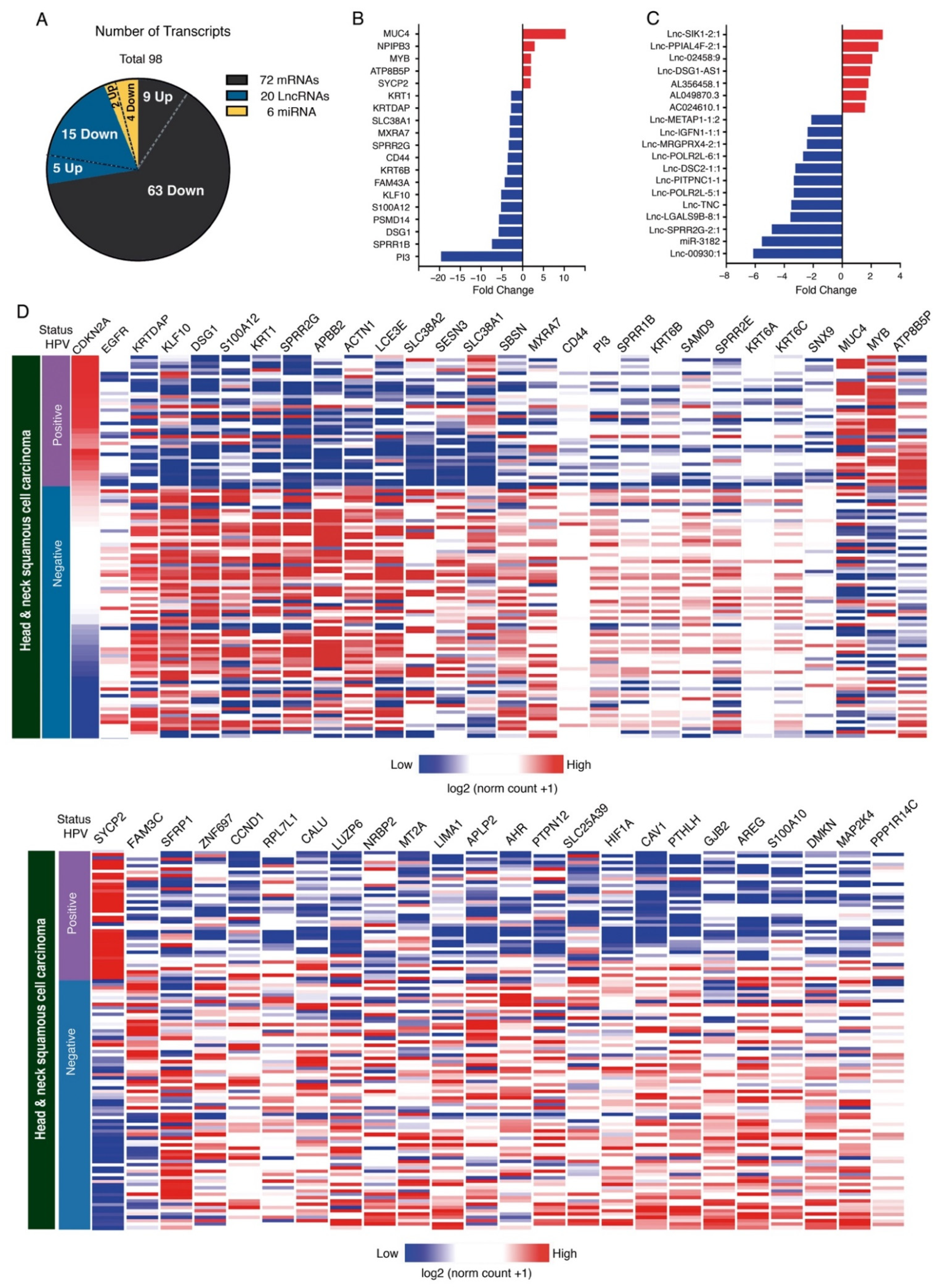
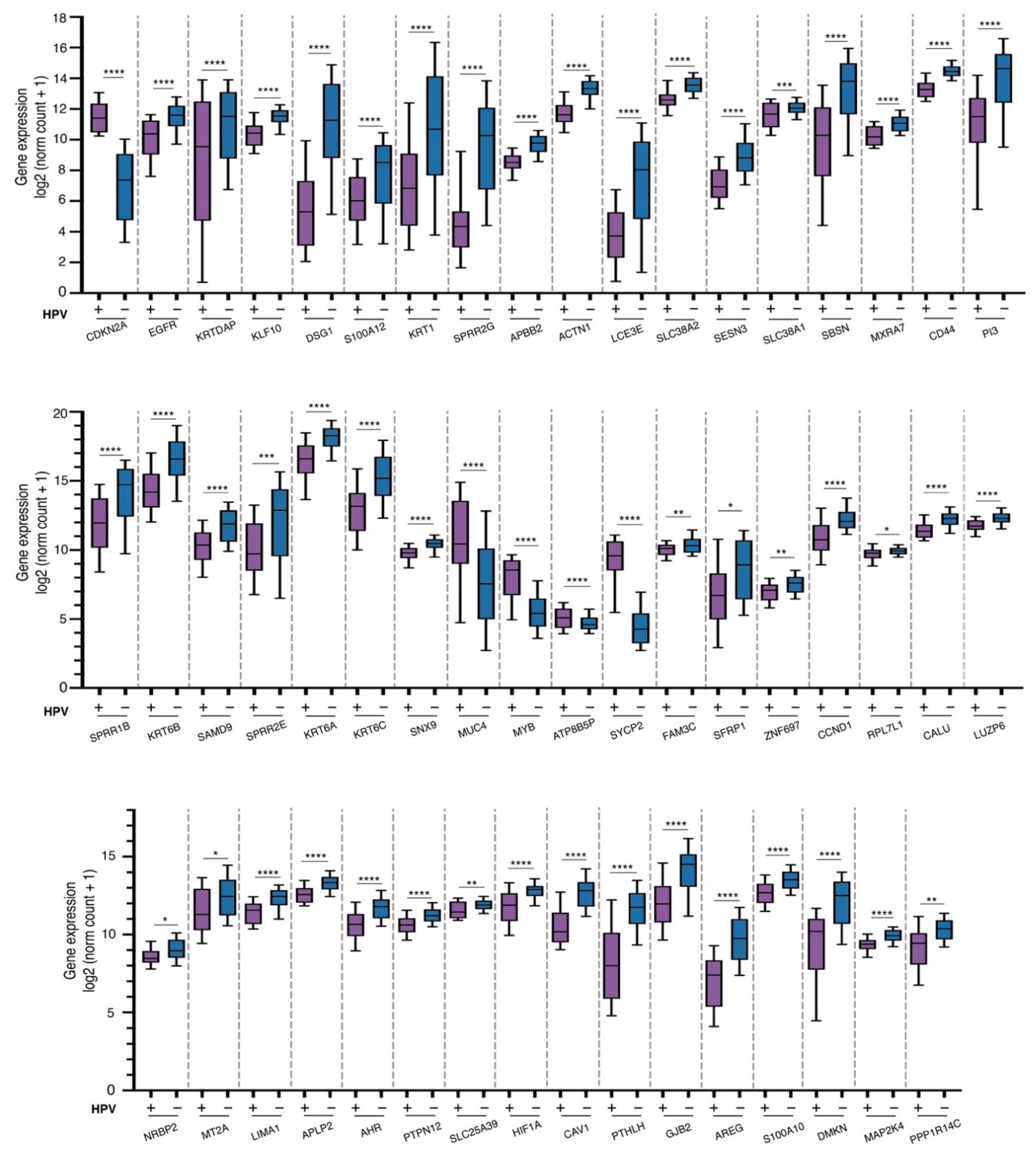
3.3. Expression Analysis
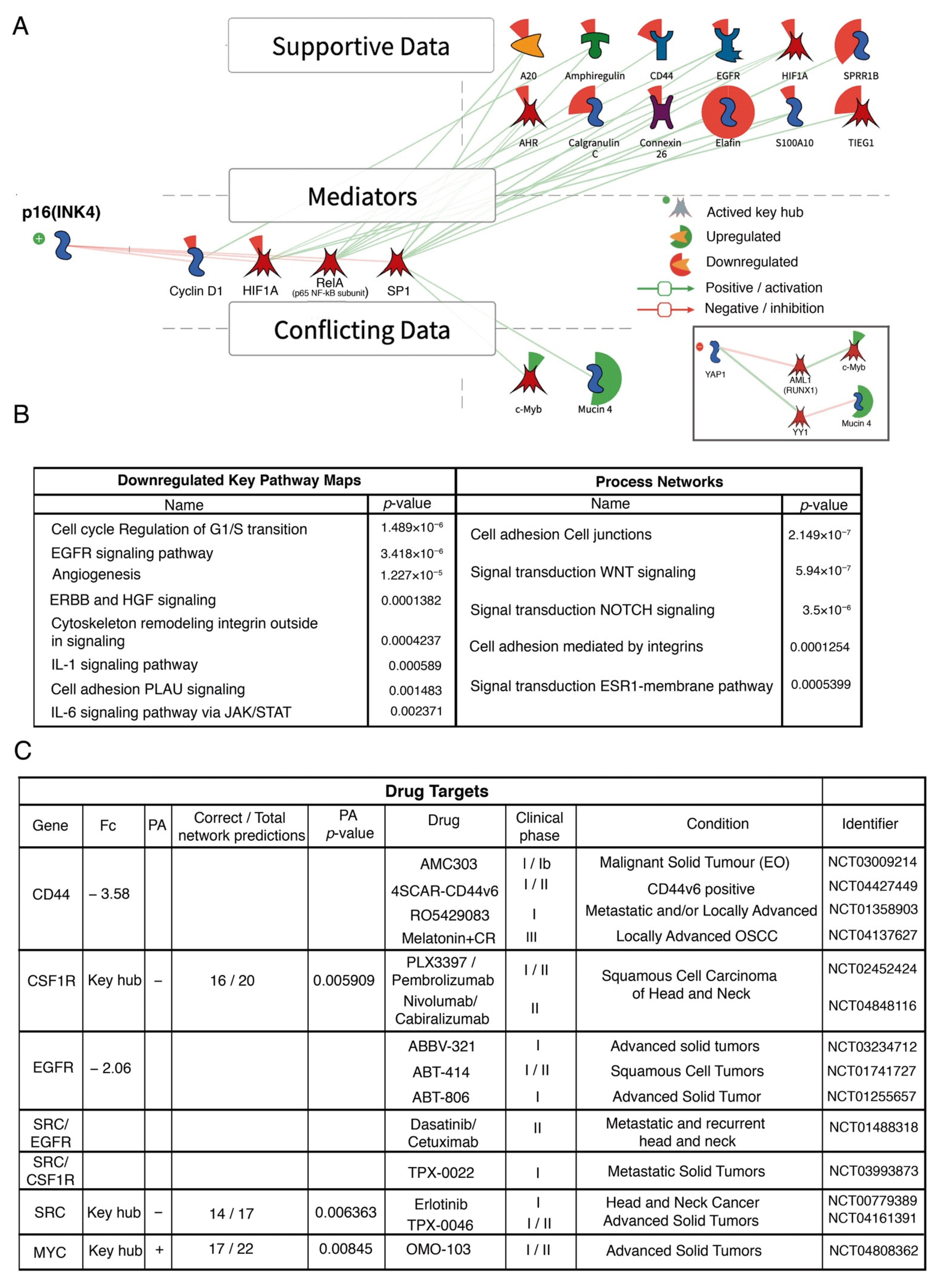
3.4. Analysis of Signaling Pathways and Molecular Processes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chow, L.Q.M. Head and Neck Cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Anantharaman, D.; Abedi-Ardekani, B.; Beachler, D.C.; Gheit, T.; Olshan, A.F.; Wisniewski, K.; Wunsch-Filho, V.; Toporcov, T.N.; Tajara, E.H.; Levi, J.E.; et al. Geographic heterogeneity in the prevalence of human papillomavirus in head and neck cancer. Int. J. Cancer 2017, 140, 1968–1975. [Google Scholar] [CrossRef] [PubMed]
- Mroz, E.; Tward, A.M.; Hammon, R.J.; Ren, Y.; Rocco, J.W. Intra-tumor Genetic Heterogeneity and Mortality in Head and Neck Cancer: Analysis of Data from The Cancer Genome Atlas. PLoS Med. 2015, 12, e1001786. [Google Scholar] [CrossRef]
- Pai, S.I.; Westra, W.H. Molecular pathology of head and neck cancer: Implications for diagnosis, prognosis, and treatment. Annu. Rev. Pathol. 2009, 4, 49–70. [Google Scholar] [CrossRef]
- Ferlay, J.E.M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. 2018. Available online: https://gco.iarc.fr/today/home (accessed on 26 June 2020).
- Syrjanen, S. The role of human papillomavirus infection in head and neck cancers. Ann. Oncol. 2010, 21 (Suppl. 7), vii243–vii245. [Google Scholar] [CrossRef]
- Mehanna, H.; Beech, T.; Nicholson, T.; El-Hariry, I.; McConkey, C.; Paleri, V.; Roberts, S. Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer-systematic review and meta-analysis of trends by time and region. Head Neck 2012, 35, 747–755. [Google Scholar] [CrossRef]
- Mahal, B.A.; Catalano, P.J.; Haddad, R.I.; Hanna, G.J.; Kass, J.I.; Schoenfeld, J.D.; Tishler, R.B.; Margalit, D.N. Incidence and Demographic Burden of HPV-Associated Oropharyngeal Head and Neck Cancers in the United States. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1660–1667. [Google Scholar] [CrossRef] [PubMed]
- Ndiaye, C.; Mena, M.; Alemany, L.; Arbyn, M.; Castellsagué, X.; Laporte, L.; Bosch, F.X.; José, F.X.B.; Trottier, H. HPV DNA, E6/E7 mRNA, and p16INK4a detection in head and neck cancers: A systematic review and meta-analysis. Lancet Oncol. 2014, 15, 1319–1331. [Google Scholar] [CrossRef]
- Tumban, E. A Current Update on Human Papillomavirus-Associated Head and Neck Cancers. Viruses 2019, 11, 922. [Google Scholar] [CrossRef]
- de Sanjosé, S.; Serrano, B.; Tous, S.; Alejo, M.; Lloveras, B.; Quirós, B.; Clavero, O.; Vidal, A.; Ferrándiz-Pulido, C.; Pavón, M.Á.; et al. Burden of Human Papillomavirus (HPV)-Related Cancers Attributable to HPVs 6/11/16/18/31/33/45/52 and 58. JNCI Cancer Spectr. 2018, 2, pky045. [Google Scholar] [CrossRef]
- Zanoni, D.K.; Patel, S.G.; Shah, J.P. Changes in the 8th Edition of the American Joint Committee on Cancer (AJCC) Staging of Head and Neck Cancer: Rationale and Implications. Curr. Oncol. Rep. 2019, 21, 52. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.H.; Parker, J.S.; Karaca, G.; Wu, J.; Funkhouser, W.K.; Moore, D.; Butterfoss, D.; Xiang, D.; Zanation, A.; Yin, X.; et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell 2004, 5, 489–500. [Google Scholar] [CrossRef]
- Walter, V.; Yin, X.; Wilkerson, M.D.; Cabanski, C.R.; Zhao, N.; Du, Y.; Ang, M.K.; Hayward, M.C.; Salazar, A.H.; Hoadley, K.A.; et al. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS ONE 2013, 8, e56823. [Google Scholar] [CrossRef]
- De Cecco, L.; Nicolau, M.; Giannoccaro, M.; Daidone, M.G.; Bossi, P.; Locati, L.D.; Licitra, L.; Canevari, S. Head and neck cancer subtypes with biological and clinical relevance: Meta-analysis of gene-expression data. Oncotarget 2015, 6, 9627–9642. [Google Scholar] [CrossRef] [PubMed]
- De Cecco, L.; Giannoccaro, M.; Marchesi, E.; Bossi, P.; Favales, F.; Locati, L.D.; Licitra, L.; Pilotti, S.; Canevari, S. Integrative miRNA-Gene Expression Analysis Enables Refinement of Associated Biology and Prediction of Response to Cetuximab in Head and Neck Squamous Cell Cancer. Genes 2017, 8, 35. [Google Scholar] [CrossRef]
- Ibieta, B.R.; Lizano, M.; Frías-Mendivil, M.; Barrera, J.L.; Carrillo, A.; Ruíz-Godoy, L.M.; Mohar, A. Human papilloma virus in oral squamous cell carcinoma in a Mexican population. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2005, 99, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Ibieta-Zarco, B.R.; Carrillo-García, A.; Ponce-De-León-Rosales, S.; Flores-Miranda, M.M.; Mohar, A.; Lizano, M. Frequency and genotype distribution of multiple human papillomavirus infections in cancer of the head and neck in a Mexican population. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2012, 114, 350–357. [Google Scholar] [CrossRef]
- Anaya-Saavedra, G.; Ramírez-Amador, V.; Irigoyen-Camacho, M.E.; Garcia-Cuellar, C.M.; Guido-Jiménez, M.; Méndez-Martínez, R.; García-Carrancá, A. High Association of Human Papillomavirus Infection with Oral Cancer: A Case-Control Study. Arch. Med Res. 2008, 39, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Peralta, R.; Garcia, P.; Valdivia, A.; Lopez, A.; Apresa, T.; Hernandez, D.M.; Gallegos, F.; Alvarado-Cabrero, I.; Vargas-De-León, C.; Davila, S.; et al. HPV Could be a Potential Factor of Survival in Laryngeal Cancer: A Preliminary Study in Mexican Patients. Asian Pac. J. Cancer Prev. 2018, 19, 1711–1716. [Google Scholar] [CrossRef]
- Lewis, J.S.; Beadle, B.; Bishop, J.A.; Chernock, R.D.; Colasacco, C.; Lacchetti, C.; Moncur, J.T.; Rocco, J.W.; Schwartz, M.R.; Seethala, R.R.; et al. Human Papillomavirus Testing in Head and Neck Carcinomas: Guideline from the College of American Pathologists. Arch. Pathol. Lab. Med. 2017, 142, 559–597. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Gillison, M.L.; Koch, W.M.; Capone, R.B.; Spafford, M.; Westra, W.H.; Wu, L.; Zahurak, M.L.; Daniel, R.W.; Viglione, M.; Symer, D.E.; et al. Evidence for a Causal Association Between Human Papillomavirus and a Subset of Head and Neck Cancers. J. Natl. Cancer Inst. 2000, 92, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Ang, K.K.; Harris, J.; Wheeler, R.; Weber, R.; Rosenthal, D.; Nguyen-Tân, P.F.; Westra, W.H.; Chung, C.H.; Jordan, R.C.; Lu, C.; et al. Human Papillomavirus and Survival of Patients with Oropharyngeal Cancer. N. Engl. J. Med. 2010, 363, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Abrahão, R.; Perdomo, S.; Pinto, L.F.R.; de Carvalho, F.N.; Dias, F.L.; de Podestá, J.R.V.; von Zeidler, S.V.; de Abreu, P.M.; Vilensky, M.; Giglio, R.E.; et al. Predictors of Survival After Head and Neck Squamous Cell Carcinoma in South America: The InterCHANGE Study. JCO Glob. Oncol. 2020, 6, 486–499. [Google Scholar] [CrossRef] [PubMed]
- De Martel, C.; Plummer, M.; Vignat, J.; Franceschi, S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int. J. Cancer 2017, 141, 664–670. [Google Scholar] [CrossRef]
- Kreimer, A.R.; Clifford, G.M.; Boyle, P.; Franceschi, S. Human Papillomavirus Types in Head and Neck Squamous Cell Carcinomas Worldwide: A Systematic Review. Cancer Epidemiol. Biomark. Prev. 2005, 14, 467–475. [Google Scholar] [CrossRef]
- Aguilar-Lemarroy, A.; Vallejo-Ruiz, V.; Cortés-Gutiérrez, E.I.; Salgado-Bernabé, M.E.; Ramos-González, N.P.; Ortega-Cervantes, L.; Arias-Flores, R.; Medina-Díaz, I.M.; Hernández-Garza, F.; Santos-López, G.; et al. Human papillomavirus infections in Mexican women with normal cytology, precancerous lesions, and cervical cancer: Type-specific prevalence and HPV coinfections. J. Med. Virol. 2015, 87, 871–884. [Google Scholar] [CrossRef]
- Tagliabue, M.; Mena, M.; Maffini, F.; Gheit, T.; Blasco, B.Q.; Holzinger, D.; Tous, S.; Scelsi, D.; Riva, D.; Grosso, E.; et al. Role of Human Papillomavirus Infection in Head and Neck Cancer in Italy: The HPV-AHEAD Study. Cancers 2020, 12, 3567. [Google Scholar] [CrossRef] [PubMed]
- Castellsague, X.; Mena, M.; Alemany, L. Epidemiology of HPV-Positive Tumors in Europe and in the World. Recent Results Cancer Res. 2017, 206, 27–35. [Google Scholar]
- Albers, A.E.; Qian, X.; Kaufmann, A.M.; Coordes, A. Meta analysis: HPV and p16 pattern determines survival in patients with HNSCC and identifies potential new biologic subtype. Sci. Rep. 2017, 7, 16715. [Google Scholar] [CrossRef]
- Chakravarthy, A.; Henderson, S.; Thirdborough, S.M.; Ottensmeier, C.; Su, X.; Lechner, M.; Feber, A.; Thomas, G.J.; Fenton, T.R. Human Papillomavirus Drives Tumor Development Throughout the Head and Neck: Improved Prognosis Is Associated with an Immune Response Largely Restricted to the Oropharynx. J. Clin. Oncol. 2016, 34, 4132–4141. [Google Scholar] [CrossRef] [PubMed]
- Blahak, J.; Zelinka, J.; Gumulec, J.; Machacek, C.; Danek, Z.; Bulik, O. HPV, protein p16 and squamous cell carcinoma of the oral cavity. Biomed. Pap. 2020, 164, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Guillen, J.M.; Palacios-Saucedo, G.C.; Rivera-Morales, L.G.; Alonzo-Morado, M.V.; Burciaga-Bernal, S.B.; Montufar-Martinez, M.; Ortiz-Lopez, R.; Gonzalez-Villasana, V.; Martinez-Torres, A.C.; Serna-Hernandez, J.C.; et al. Infection and coinfection by human papillomavirus, Epstein-Barr virus and Merkel cell polyomavirus in patients with squamous cell carcinoma of the larynx: A retrospective study. PeerJ 2018, 6, e5834. [Google Scholar] [CrossRef] [PubMed]
- Vidal Loustau, A.C.; Dulguerov, N.; Curvoisier, D.; McKee, T.; Lombardi, T. Low prevalence of HPV-induced oral squamous cell carcinoma in Geneva, Switzerland. Oral Dis. 2019, 25, 1283–1290. [Google Scholar] [CrossRef]
- Padhi, S.S.; Roy, S.; Kar, M.; Saha, A.; Roy, S.; Adhya, A.; Baisakh, M.; Banerjee, B. Role of CDKN2A/p16 expression in the prognostication of oral squamous cell carcinoma. Oral Oncol. 2017, 73, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.; Chakravarthy, A.R.; Walter, V.; Masterson, L.; Feber, A.; Jay, A.; Weinberger, P.M.; McIndoe, R.; Forde, C.; Chester, K.; et al. Frequent HPV-independent p16/INK4A overexpression in head and neck cancer. Oral Oncol. 2018, 83, 32–37. [Google Scholar] [CrossRef]
- Li, Y.; Nichols, M.A.; Shay, J.W.; Xiong, Y. Transcriptional repression of the D-type cyclin-dependent kinase inhibitor p16 by the retinoblastoma susceptibility gene product pRb. Cancer Res. 1994, 54, 6078–6082. [Google Scholar]
- Bryant, A.K.; Sojourner, E.J.; Vitzthum, L.K.; Zakeri, K.; Shen, H.; Nguyen, C.; Murphy, J.D.; Califano, J.A.; Cohen, E.E.W.; Mell, L.K. Prognostic Role of p16 in Nonoropharyngeal Head and Neck Cancer. J. Natl. Cancer Inst. 2018, 110, 1393–1399. [Google Scholar] [CrossRef] [PubMed]
- Larque, A.B.; Conde, L.; Hakim, S.; Alos, L.; Jares, P.; Vilaseca, I.; Cardesa, A.; Nadal, A. p16INK4a overexpression is associated with CDKN2A mutation and worse prognosis in HPV-negative laryngeal squamous cell carcinomas. Virchows Archiv. 2015, 466, 375–382. [Google Scholar] [CrossRef]
- Slebos, R.J.; Yi, Y.; Ely, K.; Carter, J.; Evjen, A.; Zhang, X.; Shyr, Y.; Murphy, B.M.; Cmelak, A.J.; Burkey, B.B.; et al. Gene Expression Differences Associated with Human Papillomavirus Status in Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. 2006, 12, 701–709. [Google Scholar] [CrossRef]
- Costa, R.L.; Boroni, M.; Soares, M.A. Distinct co-expression networks using multi-omic data reveal novel interventional targets in HPV-positive and negative head-and-neck squamous cell cancer. Sci. Rep. 2018, 8, 15254. [Google Scholar] [CrossRef]
- Pyeon, D.; Newton, M.A.; Lambert, P.F.; Boon, J.A.D.; Sengupta, S.; Marsit, C.; Woodworth, C.D.; Connor, J.P.; Haugen, T.; Smith, E.M.; et al. Fundamental Differences in Cell Cycle Deregulation in Human Papillomavirus–Positive and Human Papillomavirus–Negative Head/Neck and Cervical Cancers. Cancer Res. 2007, 67, 4605–4619. [Google Scholar] [CrossRef]
- Masterson, L.; Sorgeloos, F.; Winder, D.; Lechner, M.; Marker, A.; Malhotra, S.; Sudhoff, H.; Jani, P.; Goon, P.; Sterling, J. Deregulation of SYCP 2 predicts early stage human papillomavirus-positive oropharyngeal carcinoma: A prospective whole transcriptome analysis. Cancer Sci. 2015, 106, 1568–1575. [Google Scholar] [CrossRef]
- Hosoya, N.; Miyagawa, K. Synaptonemal complex proteins modulate the level of genome integrity in cancers. Cancer Sci. 2021, 112, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.L.; Campos, S.K.; Wandinger-Ness, A.; Ozbun, M.A. Caveolin-1-dependent infectious entry of human papillomavirus type 31 in human keratinocytes proceeds to the endosomal pathway for pH-dependent uncoating. J. Virol. 2008, 82, 9505–9512. [Google Scholar] [CrossRef]
- Razani, B.; Altschuler, Y.; Zhu, L.; Pestell, R.G.; Mostov, K.E.; Lisanti, M.P. Caveolin-1 Expression Is Down-Regulated in Cells Transformed by the Human Papilloma Virus in a p53-Dependent Manner. Replacement of Caveolin-1 Expression Suppresses HPV-Mediated Cell Transformation. Biochemistry 2000, 39, 13916–13924. [Google Scholar] [CrossRef]
- Nohata, N.; Abba, M.C.; Gutkind, J.S. Unraveling the oral cancer lncRNAome: Identification of novel lncRNAs associated with malignant progression and HPV infection. Oral Oncol. 2016, 59, 58–66. [Google Scholar] [CrossRef]
- Lavik, M.; Shatokhina, T.; Sana, J.; Ahmad, P.; Kazda, T.; Selingerova, I.; Hermanova, M.; Cervena, R.; Novotny, T.; Burkon, P.; et al. Expression of CD44, EGFR, p16, and their mutual combinations in patients with head and neck cancer: Impact on outcomes of intensity-modulated radiation therapy. Head Neck 2019, 41, 940–949. [Google Scholar]
- Yu, S.S.; Cirillo, N. The molecular markers of cancer stem cells in head and neck tumors. J. Cell. Physiol. 2020, 235, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Kokko, L.-L.; Hurme, S.; Maula, S.-M.; Alanen, K.; Grénman, R.; Kinnunen, I.; Ventelä, S. Significance of site-specific prognosis of cancer stem cell marker CD44 in head and neck squamous-cell carcinoma. Oral Oncol. 2011, 47, 510–516. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Y.; Chen, X.; Zhou, Y.; Jiang, F.; Chen, J.; Wang, L.; Zhang, W.-F. MiR-34a suppresses amphiregulin and tumor metastatic potential of head and neck squamous cell carcinoma (HNSCC). Oncotarget 2015, 6, 7454–7469. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kimple, R.J.; Smith, M.; Blitzer, G.C.; Torres, A.D.; Martin, J.A.; Yang, R.Z.; Peet, C.R.; Lorenz, L.D.; Nickel, K.P.; Klingelhutz, A.; et al. Enhanced Radiation Sensitivity in HPV-Positive Head and Neck Cancer. Cancer Res. 2013, 73, 4791–4800. [Google Scholar] [CrossRef]
- Kogashiwa, Y.; Inoue, H.; Kuba, K.; Araki, R.; Yasuda, M.; Nakahira, M.; Sugasawa, M. Prognostic role of epiregulin/amphiregulin expression in recurrent/metastatic head and neck cancer treated with cetuximab. Head Neck 2018, 40, 2424–2431. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Yeh, C.-M.; Lee, Y.-J.; Ko, P.-Y.; Lin, Y.-M.; Sung, W.-W. High Expression of KLF10 Is Associated with Favorable Survival in Patients with Oral Squamous Cell Carcinoma. Medicina 2020, 57, 17. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, M.; Hawse, J.R.; Rajamannan, N.M.; Ingle, J.N.; Spelsberg, T.C. Functional role of KLF10 in multiple disease processes. BioFactors 2010, 36, 8–18. [Google Scholar] [CrossRef]
- Schmidt, S.; Linge, A.; Zwanenburg, A.; Leger, S.; Lohaus, F.; Krenn, C.; Appold, S.; Gudziol, V.; Nowak, A.; von Neubeck, C.; et al. Development and Validation of a Gene Signature for Patients with Head and Neck Carcinomas Treated by Postoperative Radio(chemo)therapy. Clin. Cancer Res. 2018, 24, 1364–1374. [Google Scholar] [CrossRef]
- Billard-Sandu, C.; Tao, Y.G.; Sablin, M.P.; Dumitrescu, G.; Billard, D.; Deutsch, E. CDK4/6 inhibitors in P16/HPV16-negative squamous cell carcinoma of the head and neck. Eur. Arch. Otorhinolaryngol. 2020, 277, 1273–1280. [Google Scholar] [CrossRef]
- Paternot, S.; Bockstaele, L.; Bisteau, X.; Kooken, H.; Coulonval, K.; Roger, P. Rb inactivation in cell cycle and cancer: The puzzle of highly regulated activating phosphorylation of CDK4 versus constitutively active CDK-activating kinase. Cell Cycle 2010, 9, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Meyer, H.J.; Leifels, L.; Hamerla, G.; Höhn, A.K.; Surov, A. Associations between Histogram Analysis Parameters Derived from DCE-MRI and Histopathological Features including Expression of EGFR, p16, VEGF, Hif1-alpha, and p53 in HNSCC. Contrast Media Mol. Imaging 2019, 2019, 1–10. [Google Scholar] [CrossRef]
- Omori, H.; Nishio, M.; Masuda, M.; Miyachi, Y.; Ueda, F.; Nakano, T.; Sato, K.; Mimori, K.; Taguchi, K.; Hikasa, H.; et al. YAP1 is a potent driver of the onset and progression of oral squamous cell carcinoma. Sci. Adv. 2020, 6, eaay3324. [Google Scholar] [CrossRef] [PubMed]
- Rocha-Lima, C.M.; Soares, H.P.; Raez, L.E.; Singal, R. EGFR Targeting of Solid Tumors. Cancer Control. 2007, 14, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Fasano, M.; Della Corte, C.M.; Viscardi, G.; Di Liello, R.; Paragliola, F.; Sparano, F.; Iacovino, M.L.; Castrichino, A.; Doria, F.; Sica, A.; et al. Head and neck cancer: The role of anti-EGFR agents in the era of immunotherapy. Ther. Adv. Med. Oncol. 2021, 13, 1758835920949418. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Cui, J.; Xu, N.; Wang, M.; Xu, T.; Tian, H.; Han, F. p16(INK4a) status and survival benefit of EGFR inhibitors in head and neck squamous cell cancer: A systematic review and meta-analysis. Crit. Rev. Oncol. 2018, 124, 11–20. [Google Scholar] [CrossRef]
- Yang, Z.; Liao, J.; Cullen, K.J.; Dan, H. Inhibition of IKKbeta/NF-kappaB signaling pathway to improve Dasatinib efficacy in suppression of cisplatin-resistant head and neck squamous cell carcinoma. Cell Death Discov. 2020, 6, 36. [Google Scholar] [CrossRef]
- Duffy, M.J.; O’Grady, S.; Tang, M.; Crown, J. MYC as a target for cancer treatment. Cancer Treat. Rev. 2021, 94, 102154. [Google Scholar] [CrossRef]

| Characteristics | Total | HPV+ | P16+ | |||
|---|---|---|---|---|---|---|
| % | n = 414 | % | n = 368 | % | n = 412 | |
| Age | ||||||
| ≤55 | 24.1 | 88 | 35.4 | 29 | 38.6 | 34 |
| 56–75 | 53.7 | 196 | 20.1 | 35 | 33.8 | 66 |
| ≥76 | 22.2 | 81 | 12.5 | 9 | 21.3 | 17 |
| p = 0.002 | p = 0.043 | |||||
| Gender | ||||||
| Men | 71.3 | 286 | 22.5 | 57 | 32.7 | 93 |
| Woman | 28.7 | 115 | 22.5 | 23 | 31.3 | 36 |
| Anatomic site | ||||||
| Larynx | 39.1 | 162 | 18.2 | 25 | 30.4 | 49 |
| Oropharynx | 19.6 | 81 | 40.5 | 30 | 54.3 | 44 |
| Oral cavity | 41.3 | 171 | 17.2 | 27 | 23.5 | 40 |
| p = 0.000 | p = 0.000 | |||||
| Stage | ||||||
| I–II | 36.5 | 91 | 15.6 | 14 | 18.7 | 17 |
| III–IV | 63.5 | 158 | 20.1 | 30 | 32.3 | 51 |
| p = 0.26 | ||||||
| Histological Grade | ||||||
| I | 22.2 | 75 | 18.5 | 12 | 34.7 | 26 |
| II | 66.6 | 225 | 17.3 | 35 | 25.0 | 56 |
| III | 11.2 | 38 | 24.2 | 8 | 36.8 | 14 |
| Smoke | ||||||
| Yes | 64.4 | 125 | 18.9 | 23 | 30.4 | 38 |
| No | 35.6 | 69 | 23.8 | 15 | 37.7 | 26 |
| Alcohol | ||||||
| Yes | 61.5 | 112 | 19.1 | 21 | 30.4 | 34 |
| No | 38.5 | 70 | 14.3 | 9 | 31.4 | 22 |
| Characteristics | HPV+/P16+ | HPV−/P16− | Others | p | |||
|---|---|---|---|---|---|---|---|
| % | n | % | n | % | n | ||
| Age | 0.019 | ||||||
| ≤55 | 26.8 | 22 | 53.7 | 44 | 19.5 | 16 | |
| 56–75 | 8.5 | 32 | 68.8 | 119 | 12.7 | 22 | |
| ≥76 | 8.5 | 6 | 76.1 | 54 | 15.5 | 11 | |
| Gender | 0.210 | ||||||
| Men | 18.7 | 47 | 67.3 | 169 | 13.9 | 35 | |
| Woman | 16.7 | 17 | 61.8 | 63 | 21.6 | 22 | |
| Anatomic site | 0.000 | ||||||
| Larynx | 14.7 | 20 | 69.1 | 94 | 16.2 | 22 | |
| Oropharynx | 39.2 | 29 | 48.6 | 36 | 12.2 | 9 | |
| Oral cavity | 9.6 | 15 | 69.9 | 109 | 20.5 | 32 | |
| Stage | 0.238 | ||||||
| I–II | 11.1 | 10 | 76.7 | 69 | 12.2 | 11 | |
| III–IV | 19.5 | 29 | 69.8 | 104 | 10.7 | 16 | |
| Histological Grade | 0.485 | ||||||
| I | 16.9 | 11 | 64.6 | 42 | 18.5 | 12 | |
| II | 11.9 | 24 | 72.6 | 146 | 15.4 | 31 | |
| III | 21.2 | 7 | 60.6 | 20 | 18.2 | 6 | |
| Smoke | 0.514 | ||||||
| Yes | 18.9. | 23 | 71.3 | 87 | 9.8 | 12 | |
| No | 22.2 | 14 | 63.5 | 40 | 14.3 | 9 | |
| Alcohol | 0.803 | ||||||
| Yes | 18.2 | 20 | 70.0 | 77 | 11.8 | 13 | |
| No | 14.2 | 9 | 73.0 | 46 | 12.7 | 8 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Méndez-Matías, G.; Velázquez-Velázquez, C.; Castro-Oropeza, R.; Mantilla-Morales, A.; Ocampo-Sandoval, D.; Burgos-González, A.; Heredia-Gutiérrez, C.; Alvarado-Cabrero, I.; Sánchez-Sandoval, R.; Barco-Bazán, A.; et al. Prevalence of HPV in Mexican Patients with Head and Neck Squamous Carcinoma and Identification of Potential Prognostic Biomarkers. Cancers 2021, 13, 5602. https://doi.org/10.3390/cancers13225602
Méndez-Matías G, Velázquez-Velázquez C, Castro-Oropeza R, Mantilla-Morales A, Ocampo-Sandoval D, Burgos-González A, Heredia-Gutiérrez C, Alvarado-Cabrero I, Sánchez-Sandoval R, Barco-Bazán A, et al. Prevalence of HPV in Mexican Patients with Head and Neck Squamous Carcinoma and Identification of Potential Prognostic Biomarkers. Cancers. 2021; 13(22):5602. https://doi.org/10.3390/cancers13225602
Chicago/Turabian StyleMéndez-Matías, Galo, Cindy Velázquez-Velázquez, Rosario Castro-Oropeza, Alejandra Mantilla-Morales, Diana Ocampo-Sandoval, Ana Burgos-González, Carlos Heredia-Gutiérrez, Isabel Alvarado-Cabrero, Rosa Sánchez-Sandoval, Abigail Barco-Bazán, and et al. 2021. "Prevalence of HPV in Mexican Patients with Head and Neck Squamous Carcinoma and Identification of Potential Prognostic Biomarkers" Cancers 13, no. 22: 5602. https://doi.org/10.3390/cancers13225602
APA StyleMéndez-Matías, G., Velázquez-Velázquez, C., Castro-Oropeza, R., Mantilla-Morales, A., Ocampo-Sandoval, D., Burgos-González, A., Heredia-Gutiérrez, C., Alvarado-Cabrero, I., Sánchez-Sandoval, R., Barco-Bazán, A., Chilaca-Rosas, F., & Piña-Sánchez, P. (2021). Prevalence of HPV in Mexican Patients with Head and Neck Squamous Carcinoma and Identification of Potential Prognostic Biomarkers. Cancers, 13(22), 5602. https://doi.org/10.3390/cancers13225602






