Simple Summary
Cervical cancer is the fourth leading cause of cancer-related deaths in females worldwide. Despite an active immunisation strategy in many countries, high-risk human papillomavirus infection and environmental factors may result in cervical cancer progression. Genetic factors contributing to host–pathogen interactions are incompletely understood and remain largely unknown, apart from the variants at the human leukocyte antigen (HLA) locus on 6p21.3, which replicates across populations. Novel cervical cancer susceptibility loci from recent large biobank-based genome-wide association studies and meta-analyses are likely to be more robust as compared to previous candidate gene based studies. In this review, we summarize findings from recent genome-wide association studies on cervical cancer and propose variants that may be universal susceptibility loci.
Abstract
Cervical cancer is the fourth common cancer amongst women worldwide. Infection by high-risk human papilloma virus is necessary in most cases, but not sufficient to develop invasive cervical cancer. Despite a predicted genetic heritability in the range of other gynaecological cancers, only few genomic susceptibility loci have been identified thus far. Various case-control association studies have found corroborative evidence for several independent risk variants at the 6p21.3 locus (HLA), while many reports of associations with variants outside the HLA region remain to be validated in other cohorts. Here, we review cervical cancer susceptibility variants arising from recent genome-wide association studies and meta-analysis in large cohorts and propose 2q14 (PAX8), 17q12 (GSDMB), and 5p15.33 (CLPTM1L) as consistently replicated non-HLA cervical cancer susceptibility loci. We further discuss the available evidence for these loci, knowledge gaps, future perspectives, and the potential impact of these findings on precision medicine strategies to combat cervical cancer.
Keywords:
cervical cancer; cervical dysplasia; HPV; HLA; GWAS; genetic susceptibility; meta-analysis; SNP; eQTL; papillomavirus 1. Epidemiology and Heritability of Cervical Cancer
1.1. Risk and Prevention
Cervical cancer is one of the leading causes of cancer-related deaths in women worldwide [1,2,3]. Infection by known high risk sub-types of human papillomavirus (HPV 16, 18, 31, 33, 34, 35, 39, 45, 51, 52, 56, 58, 66, 68, and 70) as well as environmental or life style factors such as multiple sexual partners, smoking, obesity, high parity, early menopause, co-infection with Chlamydia trachomatis, herpes simplex virus type-2 (HSV2), or human immunodeficiency virus (HIV) are known to increase the risk of developing cancer [4,5,6,7,8,9]. Some of the epidemiological factors may correlate with specific cervical cancer histology groups such as adenocarcinomas or squamous carcinomas [10].
As preventive measures, screening at regular intervals at clinics or self-testing at home, followed by Papanicolaou “pap” smear testing, or PCR based tests for HPV typing, help to identify the early stages of infection [11,12]. Vaccination against the most prevalent high risk HPV types (Cervarix, Gardasil, or Gardasil 9) is available in many developed nations [12,13,14]. Treatment options include surgery, irradiation, chemotherapy, targeted immunotherapy, or combinations thereof. Despite this, the mortality rate from cervical cancer is still high in developing countries.
1.2. HPV-Associated Pathogenesis
The lower part of the uterus, called the cervix uteri, contains an inner and an outer compartment (endocervix and ectocervix). The thin, flat squamous cells lining the ectocervix meet the columnar glandular cells of the inner endocervix at the transformation zone, which is prone to pre-cancerous lesions caused by HPV. The first infections in low grade squamous intraepithelial lesions (LSIL), or cervical intraepithelial neoplasia stage 1 (CIN1), often resolve on their own (in 90% cases up to two years) [6]. Re-infection may occur multiple times, without leading to cancer development in most cases. Only a small fraction of women go on to develop invasive cervical cancer after HPV infection, indicating that genetic variability may play a vital role in resolving infection and preventing progression to invasive disease [15]. A persistent HPV infection over time, gradual worsening of the lesions, viral load, tissue composition at the site of viral integration, and multiple reinfections are associated with the development of high grade squamous intraepithelial lesion (HSIL), or cervical intraepithelial neoplasia stage 2 and 3 (CIN2 and CIN3) and further, cancer in situ (CIS) [16]. These stages are together termed as high-grade dysplasia and can progress into carcinoma or invasive cervical cancer [6,17,18] (Figure 1).
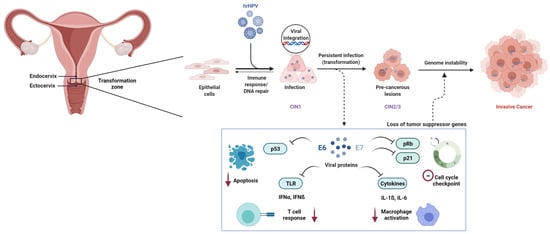
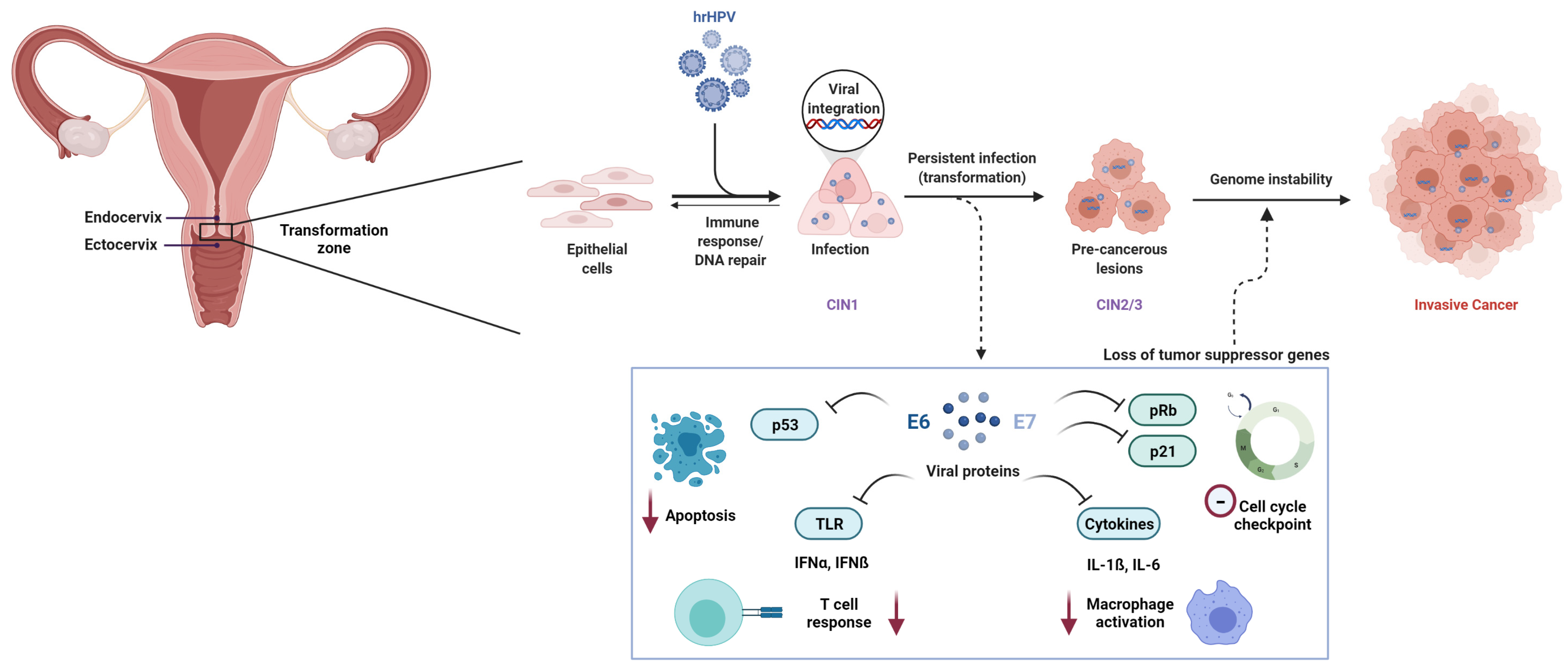
Figure 1.
Pathogenesis of cervical cancer. Epithelial cells in the transformation zone of the cervix acquire lesions upon persistent infection with high risk HPV (hrHPV). In some cases, the lesions resolve, whereas in others, upon viral integration, cells are transformed and progress from cervical intraepithelial neoplasia I to II and III (CIN1, CIN2, and CIN3). Viral proteins E6 and E7 are released and inhibit apoptosis mediated by TP53, cell cycle checkpoint by p21, T-cell response by toll-like receptors (TLR), and macrophage activation by cytokines. This leads to an insufficient immune response and viral replication, uncontrolled cell proliferation and genome instability, and further cancer in situ (CIS) or invasive cervical cancer (CC).
Although all high-risk human papillomaviruses predispose a woman to cervical cancer, there are notable differences between them. The most prevalent anogenital HPV type, HPV16, is also the most carcinogenic. Variations within the viral genome appear to modulate the pathogenicity of the respective lineages of HPV16, HPV33, or HPV45 [19,20,21]. Such genomic evolution of the virus may be partially driven by host cell DNA-editing enzymes of the APOBEC family [22]. The mechanisms underlying the different pathogenicity of diverse lineages are largely unknown, but may relate to differential expression levels or alternate splicing of viral oncogenes [23]. Further differences have been noted with regard to the tumour characteristics. The second most carcinogenic HPV type, HPV18, accounts for approximately 12% of squamous cell carcinoma, but 37% of adenocarcinoma of the cervix worldwide [24], suggesting etiological differences in tumour development after HPV16 or HPV18 infections. Patterns of integration within the host genome can also vary between different HPV types. HPV18 appears to integrate fully in cervical intraepithelial neoplasia grade 3 (CIN3) or invasive cancer whereas HPV16 can be either completely integrated or remain as episomes [25,26,27]. HPV integration sites within the human genome have been closely scrutinised with reported changes in gene and protein expression near integration hot spots as well as differential pathway activation [27,28,29]. Possible mechanisms include differences in promoter methylation, which have also been reported in HPV positive versus negative lesions [30,31,32], direct disruption of genes, or activation of retroelements [33].
The viral DNA replicates inside the host cell and starts expressing early genes E6 and E7, among others [7]. The E6 protein is known to bind and initiate the degradation of p53, a well-characterised tumour suppressor, blocking apoptosis and accumulating DNA damage within the cells, which can lead to uncontrolled proliferation [34,35,36]. The E7 protein binds and inactivates the retinoblastoma (Rb) protein, which otherwise sequesters the transcription factor E2F [37,38,39]. Upon the inactivation of Rb, E2F lifts the cell cycle checkpoint inhibition, and induces unchecked cell proliferation. E7 via E2F also upregulates cyclin-dependent kinase inhibitor 2A (p16INK4A, or CDKN2A) expression, which acts as a prognostic biomarker for cervical cancer [40,41,42]. CDKN2A is then gets hypermethylated [43,44,45], however, this inhibition of CDKN2A cannot effectively arrest the cell cycle while Rb is blocked and p53 is degraded. E7 also suppresses p53 effector and tumour suppressor protein cyclin-dependent kinase inhibitor 1A (p21 or CDKN1A) [37,38,39].
E6 and E7 overexpression appears to downregulate the toll-like receptor (TLR) mediated type I interferon (IFN- α and ß) response [46]. Additionally, once the HPV integrates inside the cells, this interferon mediated response is ineffective [47]. Keratinocytes express cytokines, which are crucial for the activation of macrophages and Langerhans cells and for T-cell activation, but HPV episomes have been shown to downregulate the cytokines IL-1ß and IL-6 [48,49]. In some women, due to a combination of such mechanisms, immune cells are not able to constrain viral replication, and the unrestrained cell proliferation due to the E6 and E7 downstream activities results in progression to HSIL and ultimately, cervical cancer (Figure 1).
HPV-induced inactivation of p53 and unscheduled replication and cell proliferation make the host cell prone to mutagenesis. As with all cancers, the host genome acquires selective advantage through the accumulation of somatic mutations and epigenetic changes. Some 537 genes associated with cervical cancer progression have been included in an early manually curated cervical cancer gene database [50], while genes with an experimentally validated role in cervical cancer have been listed in the Disease-Gene Association database DisGeNET [51]. More recently, large sequencing efforts such as those of the TCGA and the BioRAIDs consortia have established key genes and pathways for cervical cancer [27,52]. It is possible, but yet under-investigated, that differential regulation of some of them are also relevant in the germline predisposition of cervical cancer risk.
1.3. Heritability of Cervical Cancer
Genetic factors contributing to the development of HSIL and invasive cervical cancer are largely unknown. However, some evidence for familial clustering has been noted for more than 60 years [53], thus previous studies have examined whether there is a hereditary component to acquiring cervical cancer. There is clear evidence for a high familial relative risk (FRR), which has been deduced from the Swedish cancer registry and indicated towards disease clustering in families [54,55,56]. These studies reported that female offspring and siblings have a relative risk (RR) of 1.5–2.3 for developing cervical cancer. This risk is substantial and comes close to the familial relative risk for breast cancer, which has a strong genetic component.
However, in contrast to breast and ovarian cancer, there have not been many large multiple-case families described with cervical cancer [53,55,57,58,59], suggesting that high-penetrance germline variants are rare in this malignancy and many of the heritable factors may be low- or intermediate penetrant and may act in synergy with HPV infection. With regard to the outstanding role of HPV infection, cervical cancer has sometimes been called a unique human neoplasia that has one single necessary cause [60]. Nevertheless, there has been accumulating evidence that cervical cancer can occur in the absence of HPV [61,62,63] and these tumours with undetected HPV have specific molecular pathology [27,64]. It is important to note that some of the HPV negative cervical cancer cases may arise from false diagnoses or outdated genotyping methods [65]. However, with the advancements in HPV genotyping and enhanced detection of more HPV sub-types, it has become possible to identify true HPV-negative cervical cancer cases. Genome-wide sequencing of a large series of such cases may reveal whether germline variants in certain genes are enriched in cancers with undetected HPV.
Further studies towards the hypothesis of germline predisposition suggested that heritability via genetic factors might contribute some 27–36% of cervical cancer risk variation [56,66], and a large proportion of this heritability was attributed to unidentified autosomal common SNPs with low penetrance [67]. A pan-cancer heritability study using UK Biobank and US Kaiser Permanente Genetic Epidemiology Research on Adult Health and Aging (GERA) data found an array heritability estimate of 7% for cervical cancer, which was similar to the estimates for ovarian or colon cancer [68]. As will be discussed in Section 2.2 below, several genomic loci have been identified by recent GWAS at genome-wide significance. However, the contribution of these variants is small, and the major fraction of the estimated heritability still remains to be defined.
1.4. Candidate Gene Based Studies
There have been many candidate-gene based studies performed for cervical cancer, but the findings have been restricted to specific populations. Since host genetic factors are thought to play a major role in the response to cancer and HPV infection, most cervical cancer candidate gene based studies have focused on genes with relevant roles in immunity or carcinogenesis.
Candidate cervical cancer susceptibility gene variants have been reported in the tumour-suppressor gene TP53 [69,70,71] or the p53 regulating ubiquitin ligase gene MDM2 [70,72,73], and in further DNA damage response or cell cycle genes such as ATM [74], BRIP1 [75], CDKN1A [76,77,78], CDKN2A [79], FANCA, FANCC, and FANCL [80], XRCC1 [81,82,83], or XRCC3 [84]. Variants in immune response genes, which may confer immune advantage to the virus or to the host, in genes such as T-cell surface molecules CD83 [85,86] and CTLA4 [87], CARD8 [88], or secreted factors such as tumour necrosis factor alpha (TNFA) [89,90,91,92], interleukins [93,94,95,96], transforming-growth factor beta (TGFB1) [97], interferon-gamma (IFNG) [76,98] have also been studied, among many others.
Despite these considerable efforts, the vast majority of proposed risk variants from candidate gene studies have not been replicated (e.g., a debated ArgR72Pro variant in p53 [99]) and have not reached statistical significance in large case-control studies or meta-analyses (except for certain HLA alleles, e.g., [67]). With technological advancements over the past decade, stronger evidence for additional risk variants has come from the massively parallel analysis of millions of variants throughout the whole genome. In the following section, we will discuss the progress made through these genome-wide association studies.
2. Genomic Susceptibility Variants for Cervical Cancer
2.1. Genome-Wide Association Studies
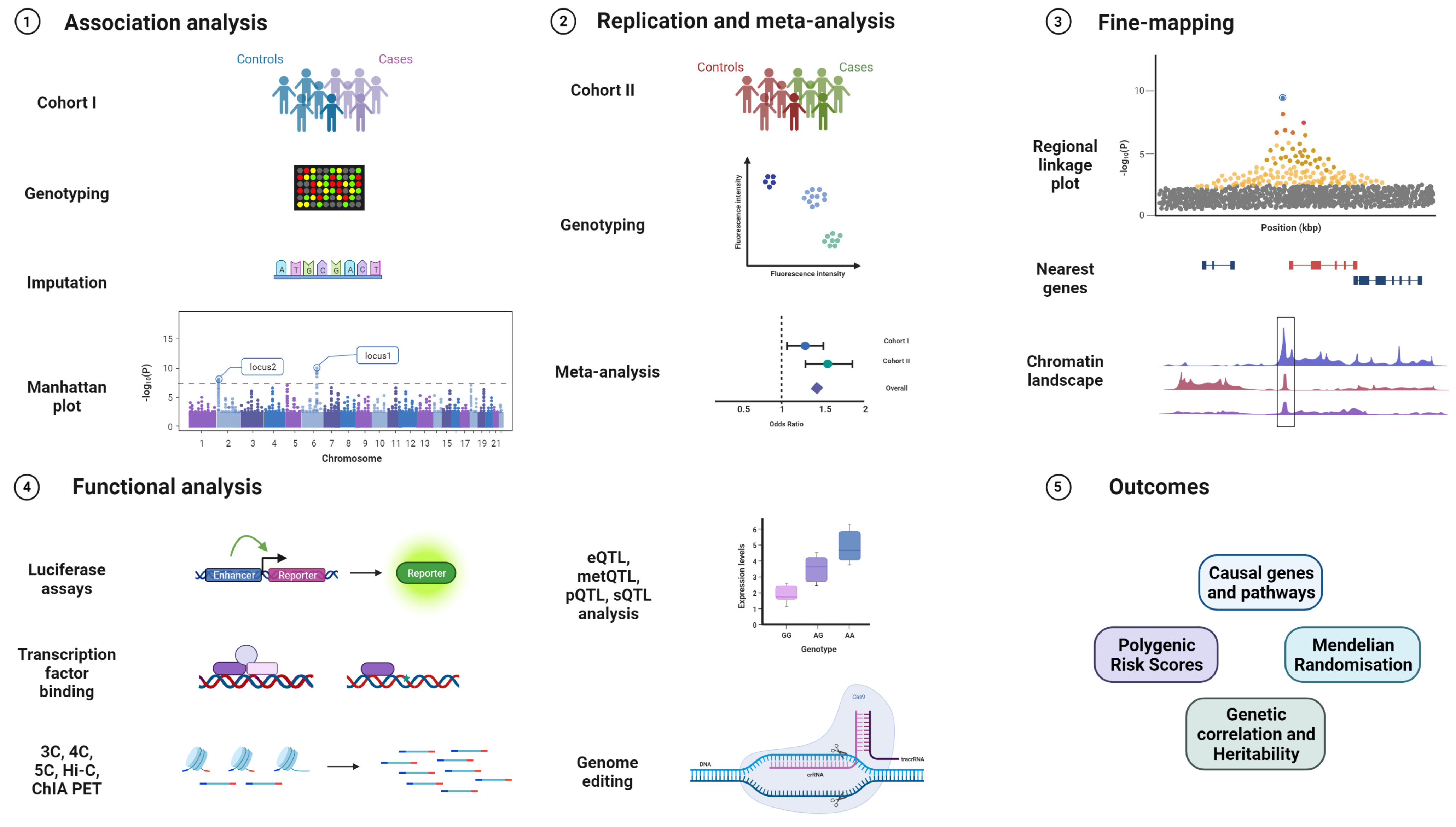
GWAS are powerful tools to identify common susceptibility variants in the population and have very successfully been applied to cancer research [100]. After genotyping and imputation, association analysis is performed using software such as PLINK or Regenie [101,102]. After associated variants are identified, replication studies in additional cohorts and meta-analysis are performed to validate new loci. Fine-mapping approaches along with bioinformatic annotations and colocalisation help to identify the causal SNP from independent sets of correlated, highly associated variants (iCHAVs). In silico predictions are used to annotate variants for known chromatin marks, genes in the vicinity, and for testing gene and pathway enrichment. These predictions become crucial in understanding the role of identified susceptibility variants since a majority of them are from the non-coding genome [103,104]. Furthermore, functional assays are designed to assess biological functions of the lead variants in the form of luciferase reporter assays, quantitative trait loci for expression, methylation, splicing, and protein levels (eQTL, metQTL, sQTL, and pQTL), chromatin immunoprecipitation (ChIP), chromosome conformation capture and related technologies (3C, 4C, 5C, Hi-C, ChIA-PET), or functional studies after genome editing of the sequences containing the variant by CRISPR/Cas or related techniques [105,106] (Figure 2).
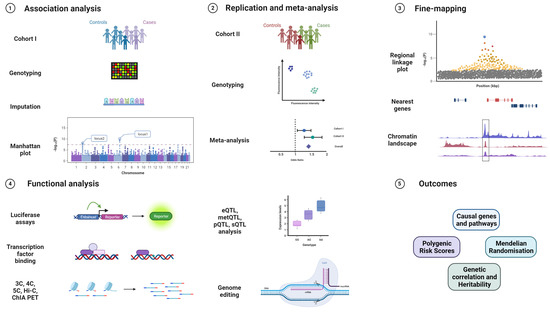
Figure 2.
GWAS workflow from replication, validation, fine-mapping, and identifying biological mechanisms to clinically relevant outcomes. The various stages of a genome-wide association study, starting from genotyping on custom arrays, imputation on reference genomes, association analysis, and visualisation, followed by replication in an independent cohort, validation genotyping, and meta-analysis. The top loci are then fine-mapped and integrated with bioinformatic annotations before proceeding to functional experiments in relevant cell and tissue types such as promoter and enhancer luciferase assays, ChIP, 3C, 4C, 5C, Hi-C, ChIA-PET, eQTL analysis, and genome editing via the CRISPR/Cas system. Expected outcomes are the identification of relevant genes and pathways affected by the variant, and extraction of polygenic risk scores (PRS), Mendelian randomisation (MR), and genetic correlation with other traits.
GWAS are conducted to identify common trait-associated variants above the genome-wide significance (GWS) threshold of p < 5 × 10−8, however, sub-significant variants may contribute small effects towards traits and are worthy of scrutiny [106]. The availability of rich data sources such as GTEx, HaploReg, SNPNexus, SNiPA, among many others, enable lookup of known eQTL and chromatin status, motif changes, linked variants, and previous known associations [107,108,109,110]. Similarly, the availability of summary statistics from published GWAS studies enables meta-analyses and validation of proposed trait-associated variants in different populations. These may eventually aid previously sub-significant variants to now cross the GWS threshold since combined studies have an increased power to detect common variants with small effects.
GWAS in large cohorts derived from biobanks (UK, FinnGen, Japan, Estonia, IARC, among others) are increasingly being used to quantify disease risk, derive polygenic risk scores (PRS), determine the genetic correlation between traits having shared environmental factors, and test causality between exposures and outcomes (Mendelian randomisation) [111,112,113]. These will drive future decisions in precision medicine and preventive screening [109,114].
2.2. Results from Cervical Cancer GWAS
The power of GWAS for the detection of cervical cancer susceptibility has been increasingly exploited over the past decade [115]. There have been a handful of cervical cancer-specific GWAS worldwide, which, however, have been complemented with recent studies from large biobank based cohorts (Table 1). Some studies have focused on invasive cervical cancer while others combined dysplasia and invasive cancer or have analysed dysplasia separately. The features and main findings of these GWAS are sequentially summarised in Table 1.

Table 1.
List of cervical cancer GWAS performed so far, with study population, genome-wide significant variants, and references. Signals that are correlated at r2 > 0.3 [110] with other GWAS variants listed above are indicated with a–e. Replication results are indicated with an asterisk (*) before the rsID. LoF: loss of function.
Although the GWAS addressed different populations, there has been some overlap between the results. We will consider this shared evidence across two or more GWAS as successful replication. Most of the consistent genome-wide significant variants arose from the human leukocyte antigen (HLA) locus in the chromosome 6p21.3 region. However, three non-HLA signals on chromosomes 2q13 (PAX8), 5p15.33 (TERT-CLPTM1L), and 17q12 (GSDMB) have also been replicated in different study populations. In the following sub-sections, we highlight these consistent loci, but also pay attention to those that still need replication in independent cohorts.
2.2.1. 6p21.3 (HLA)
The human leukocyte antigen (HLA) locus on chromosome 6p21.3 is the human equivalent of the major histocompatibility complex (MHC) and contains several genes that encode membrane proteins responsible for the regulation of the immune system [125,126,127]. The MHC is divided into three subclasses: the class I region, which includes the classical, highly polymorphic HLA-A, HLA-B, and HLA-C genes, and the less polymorphic HLA-E, HLA-F, and HLA-G genes as well as the non-canonical MHC class I-related chain (MIC) genes MICA and MICB that encode ligands for the activating natural killer cell receptor NKG2D; the class II region, which includes the HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQA2, HLA-DQB1, HLA-DQB2, HLA-DRA, HLA-DRB1, HLA-DRB2, HLA-DRB3, HLA-DRB4, and HLA-DRB5 genes as well as less variable genes involved in antigen processing and presentation; and the class III region, which contains genes implicated in inflammatory responses, leukocyte maturation, and the complement cascade [127]. Class I and Class II MHCs form two types of peptide presenting complexes, and differential representation or peptide binding of these may be associated with differential disease susceptibility.
Although we highlight the HLA region as the first consistent cervical cancer susceptibility locus, it is, in fact, made up of several separate signals. The first cervical cancer GWAS, performed in the Swedish population, identified multiple variants at the HLA locus [116]. It confirmed allelic associations with HLA-B*07:02, HLA-DRB1*13:01-DQA1*01:03-DQB1*06:03, and HLA-DRB1*15:01-DQB1*06:02, which had previously been reported in candidate gene studies, and further identified three novel loci for CIN3 in the MHC region: rs9272143 between HLA-DRB1 and HLA-DQA1; rs2516448 adjacent to MICA; and rs3117027 at HLA-DPB2 [115,116]. Interestingly, the risk allele rs2516448 was in perfect linkage disequilibrium with a frameshift mutation (the A5.1 allele) in exon 5 of MICA, resulting in a truncated MICA protein and less membrane-detectable MICA in cervical lesions, which may compromise the immune response towards HPV infection or neoplastic change [115,116,128,129]. Other polymorphisms at the same MICA exon 5 microsatellite sequence were also associated with cervical cancer [128]. Further SNPs in the vicinity (rs9271898, rs3130196, and rs73730372) were identified by follow-up investigations by combining cohorts and via pathway analysis by the same group [115,118,130].
There were several replications of these findings. The first Asian cervical cancer GWAS replicated the HLA locus in identifying another signal (rs4282438, HLA-DPB2) in the Chinese population [117]. Apart from a multi-centric study on Caucasians, which corroborated variants at the HLA locus (esp. rs9271858) [131], a cervical cancer GWAS meta-analysis combining >400,000 samples from the UK Biobank and Kaiser Permanente GERA cohorts also confirmed previously known variants at the HLA locus and identified a novel HLA signal, rs2856437 at PBX2 [68]. The UK Biobank cervical cancer GWAS, which combined CIN3 and invasive cervical cancer, confirmed variants at HLA-DQA1 (rs9272050), MICA (rs6938453), and HLA-DQB1 (rs55986091), of which HLA-DQA1 (rs9272050) was also replicated at a genome-wide significance in the FinnGen biobank cohort [121]. The MICA variant rs6938453 is only very weakly correlated with the initially reported variant rs2516448 (r2 = 0.22). This study furthermore identified a novel association with rs9266183 in HLA-B that encodes a rare missense substitution, p.Asp54Gly [121].
Although some studies have focused on invasive cervical cancer or did not distinguish between invasive cancers and dysplasia, there is solid evidence that HLA variants already affect the risk at the dysplasia stage [116,121,122,132]. The first Swedish GWAS was performed on high-grade dysplasia, CIN3 [116]. The UK Biobank and FinnGen study also performed a GWAS restricted to cervical dysplasia and reported rs9272245 (HLA-DQA1) as a signal for dysplasia alone [121]. A recent trans-ethnic GWAS meta-analysis including the Estonian population proposed two signals at the HLA locus, rs1053726 (HLA-B) and rs36214159 (HLA-DQA1), from a specific analysis that only included dysplasia cases [122]. This indicates that the risk conferred by at least some HLA alleles manifests early in the process of cervical carcinogenesis.
Chen et al. investigated HLA alleles specifically and identified significant associations with cervical dysplasia and cancer for HLA-B*07:02, HLA-B*15:01, HLA-DRB1*13:01, HLA-DRB1*15:01, HLA-DQA1*01:03, HLA-DQB1*06:03, HLA-DQB1*06:02, and HLA-C*07:02 in the Swedish population [133]. Further studies in different populations have reported, amongst others, associations with HLA-DQB1*05:01 and HLA-DRB3*99:01, which have recently been recovered in the UK Biobank [134,135,136]. More work will be required to reduce the reported associations in the HLA region to conditionally independent alleles with high-confidence associations.
From the known cervical cancer GWAS thus far, ten apparently independent HLA signals have emerged that are not in linkage disequilibrium at r2 > 0.3, and another five HLA signals have been suggested from the cross-cancer analyses (Table 1). As the pattern of linkage disequilibrium at the HLA region is complex and varies according to ethnic background, there could be hidden correlation for some of the GWAS hits. Of the clearly independent hits, the two signals initially represented by rs2516448 and rs9272143 have been replicated by three or four other GWAS, respectively, as well as through direct genotyping in case-control studies [131,132]. The genes underlying these consistent signals in the HLA region are unknown, although plausible candidates including MICA and HLA-DRB1 have been proposed [128,132]. A study of 278 affected sib pairs revealed significant excess genetic sharing for all three HLA class II loci studied: HLA-DPB1, HLA-DQB1, and HLA-DRB1 (with the strongest evidence for DQB1 and DRB1) whereas no evidence of excess sharing was observed for the HLA class I HLA-A and HLA-B loci [137]. On the other hand, HLA-B has also been repeatedly identified as a separate GWAS locus [121,122]. As the HLA region is very large and gene-dense, several genes may be affected and the causal genes may not necessarily be those in the closest vicinity. Some evidence has indicated that there may be long-range transcriptional regulation of HLA gene expression in cervical specimens [132]. Adding to the complexity, such regulations might be dependent on HPV status and on tissue constitution.
2.2.2. 17q12 (GSDMB)
The first GWA study in the Chinese population identified two novel risk loci at 4q12 (rs13117307, EXOC1) and 17q12 (rs8067378, GSDMB) [117]. While the first one has not been fully replicated, the signal on 17q12 also came up in a transethnic meta-analysis GWAS from the Estonian Biobank, the FinnGen study, the UK Biobank, and Biobank Japan (rs12603332, [122]). The closest gene is GSDMB, which encodes a member of the gasdermin family that forms pores in the membrane to aid the release of cytokines. Gasdermin B participates in the regulation of cell pyroptosis, a pro-inflammatory form of regulated cell death that is designed to attract a nonspecific innate response to the site of infection [138,139]. It also appears to activate STAT3 signalling and thereby tumour growth [140]. Interestingly, higher levels of Gasdermin B (also known as Gasdermin-like, GSDML) had previously been associated with tumour progression in cervical cancer [141]. Although it is an attractive candidate for cervical cancer risk, experimental evidence for GSDMB as the causal gene underlying the signal on 17q12 is still lacking.
2.2.3. 2q13 (PAX8)
A cervical cancer GWAS meta-analysis combining >460,000 samples from the UK Biobank and the Kaiser Permanente Genetic Epidemiology Research on Adult Health and Ageing investigated heritability and pleiotropy across 18 cancer types [68]. This study not only confirmed previously known variants at the HLA locus, but also identified a novel variant at genome-wide significance on 2q13 (rs10175462) near the PAX8 gene that encodes the transcription factor Paired box 8 [68]. This signal replicated in the UK Biobank and in the FinnGen Biobank [121] as well as in a German case-control study [142] and in the transethnic meta-analysis GWAS from the Estonian Biobank, the FinnGen study, the UK Biobank and Biobank Japan (rs4849177, [122]). Two weakly correlated variants, rs4848320 and rs1110839, had previously been associated with cervical cancer in a candidate gene study of PAX8-AS1 haplotypes [143]. In gene expression analyses, the variant rs10175462 presented as a cis-eQTL for the PAX8-AS1 non-coding RNA and also appeared to modulate PAX8 levels in response to HPV [142]. PAX8 is also emerging as an important transcription factor for other gynaecological cancers [144] but more work will be required to elucidate its role in cervical cancer pathogenesis. It is possible that the PAX8/PAX8-AS1 locus has a central role in cervical biology and pathology, as it has also been associated with cervical ectropion and cervicitis [122].
2.2.4. 5p15.33 (CLPTM1L)
The UK Biobank cervical cancer GWAS identified a genome-wide significant locus at CLPTM1L (rs27069) [121]. This finding was replicated in an independent series from the FinnGen biobank cohort at p = 2.5 × 10−7 [121] and remained genome-wide significant in a trans-ethnic meta-analysis that included the UK, Finnish, Japanese, and Estonian Biobanks [122]. rs27069 is located 10 kbp upstream of CLPTM1L, which encodes a membrane protein involved in apoptosis. Some 50 kbp apart lies TERT, the gene encoding human telomerase reverse transcriptase. The CLPTM1L-TERT locus is relevant for several gynaecological cancers [121,145,146,147,148], however, the functional significance of the identified variant and its contribution to cervical cancer pathogenesis via the CLPTM1L-TERT locus remain to be elucidated.
2.3. Other GWAS Loci for Cervical Cancer or Dysplasia
The first cervical cancer GWAS in the Japanese population failed to identify any SNPs at genome-wide significance, but a second, larger GWAS in the East Asian population (with cohorts from China and Japan), identified and linked their top novel variant rs59661306 on chromosome 5q to the ARRDC3 (Arrestin domain-containing 3) gene, encoding a known tumour suppressor and regulator of insulin sensitivity [119,149,150,151]. However, these results have not yet been replicated in European populations.
Another study on cross-trait analysis in gynaecological cancers with some overlap with the Japanese Biobank identified two novel cervical cancer variants at INS-IGF2 (rs150806792) and SOX9 (rs140991990), requiring further validation [152]. The most recent Japanese Biobank cervical cancer GWAS did not identify any new variants at GWS, although it confirmed previous findings [120] (Table 1).
Cross-trait analyses have also been performed in the pan-cancer meta-analysis of the UK Biobank and Kaiser Permanente GERA cohorts [68]. Although this study did not find evidence for a genetic correlation of cervical cancer with any other cancer at p < 0.05, it identified some loci with pleiotropic variants that reached genome-wide significance after combining cervical cancer with other cancer cases. Among those were seven HLA variants but also two signals at 4q24 (rs10007915, TET2) and 8q24.21 (rs117952826, CASC8) (Table 1). Further work will be required to validate whether these associations were significantly driven by cervical cancer.
Bowden et al. performed a separate GWAS for invasive cervical cancer in the UK and FinnGen Biobanks and reported two novel signals for invasive cancer only, rs138446575 (TTC34) and rs117960705 (ACACB) [121]. Polymorphisms in the latter gene have also been associated with obesity and diabetes [153]. However, these results would need to be replicated and the causal genes identified.
The first GWAS in the Chinese population identified a risk locus on chromosome 4q12 (rs13117307, EXOC1) [117], which has not been replicated by subsequent GWAS results, although a multicentric case-control study reported borderline evidence of association [131]. A second GWAS designed to detect association with neoadjuvant chemotherapy response discovered four loci for prognosis at 4q (rs6812281), 10q (rs4590782), 14q (rs1742101), and 16q (rs1364121) [154]. These loci might affect treatment response rather than the primary risk and have not been validated thus far.
Koel et al. performed a GWAS trans-ethnic meta-analyses that included Estonian, UK, Finnish, and Japanese Biobanks, and reported genome-wide significant signals for five loci associated with invasive cervical cancer including one novel signal at LINC00339/CDC42 (rs2268177) on chromosome 1p36 [122]. Furthermore, this study reported a novel locus for cervical dysplasia at DAPL1 (rs12611652) on chromosome 2q24. In a joint analysis of cervical dysplasia and invasive cancer, there was another novel locus on chromosome 19p13 (rs425787) near CD70 [122]. This large GWAS had not yet been fully peer-reviewed at the time of this review but will increase the number of potential cervical cancer risk loci and candidate genes that merit validation.
In a genome-wide sequencing study from Iceland, rare loss-of-function variants in PTPN14 were associated with high risk of cervical cancer (OR, 12.7, p = 1.6 × 10−4) [123]. These overall variants were also associated with earlier age at diagnosis. PTPN14 is a p53-regulated gene that encodes a protein tyrosine phosphatase targeted by high-risk HPV E7 proteins, and the authors speculate that its germline mutation may enhance oncogenic activation of the Hippo–YAP pathway [123]. This finding indicates a potential high-penetrance germline predisposition for cervical cancer, however, the role of PTPN14 needs replication in other populations and corroboration in functional studies.
2.4. Other GWAS Loci for Viral Infection
Since not all women infected with high-risk HPV (hrHPV) go on to develop cancer and it is known that viral diversity correlates with an excess of genomic variants in immune response loci [155], there has been obvious interest to understand genetic factors that contribute to a persistent HPV infection. Such an attempt to identify host genetic factors that influence HPV seropositivity found a variant at 6p21.32, rs9357152, to be associated with seropositivity for HPV8, a β-cutaneous HPV type [156]. Although this is not among the high-risk types for cervical cancer, the result provided supportive evidence that genomic variants in the HLA region can influence immune response against HPV. Another GWAS in a Nigerian population reported further variants associated with cervical hrHPV infections at sub-genome-wide significance [157]. Increasing study sizes in the future may allow genomic factors associated with viral exposure to be established that could help to identify women at increased risk at an early stage [158].
3. Follow-Up Studies of Cervical Cancer GWAS Results
As mentioned in the previous sections, the identification of significantly associated variants is just the first step. Apart from GWAS, gene-based and pathway analysis thus contribute towards the understanding of cervical cancer aetiology. At present, the genes underlying cervical cancer GWAS signals are largely unknown, although prediction tools are established to prioritise genes by the use of established data on large-scale chromatin conformation or tissue-specific gene expression. A transcriptome-wide association study (TWAS) based on the GWA studies by Leo et al. [66] and Takeuchi et al. [119] identified 20 genes to be associated with cervical cancer in using transcriptome databases for six different tissues [159]. These genes were mainly expressed at the HLA locus, however, four non-HLA genes were also identified. However, the tissues used in this study did not include cervical epithelial cells or cervical cancer lines, and these findings need further replication [159].
There are well-characterised methylation changes in cervical cancer prognosis, and an integrative analysis combining multi-omics approaches may help to further assign functional roles to susceptibility variants and understand the mechanisms underlying cervical cancer. Recent multi-omics approaches in tumours found that HPV related squamous carcinomas have defined molecular and genetic signatures [160]. However, the genomic germline factors determining hereditary cervical cancer risk and the somatic epigenetic and genetic variations do not necessarily share a large overlap. Nevertheless, the integration of methylome, proteome, and metabolome data could aid to narrow down causal genes and eventually identify novel risk factors.
While these processes of gene identification and functional follow-up are ongoing, parallel work will aim to make use of the identified genomic risk factors to define the individual risk of cancer in an unaffected woman with higher precision. Biobank-based large cohorts provide the possibility of testing the correlation between traits and draw polygenic risk scores (PRS) that can eventually help to design preventive measures and personalise treatment methods. In correlation studies, cervical cancer was not strongly correlated with other gynaecological cancers [68], although it has been found to be correlated with bladder cancer in one analysis [112]. In attempts to define polygenic risk scores, it has not been possible thus far to predict a strong PRS for cervical cancer due to the low number of known susceptibility variants given as input [161]. Nevertheless, polygenic risk scores can be a powerful instrument when more genomic risk loci become identified, as was shown for breast cancer [162], and this also bears great potential for cervical cancer [163].
In addition, Mendelian randomisation studies can be very useful for the robust identification of associated traits and will become more powerful with the increasing size of cervical cancer GWAS data. In this type of analysis, genetic variants replace exposure measures as instrumental variables to infer whether a risk factor affects a clinical outcome. The assumption is that the genetic variant is associated with the risk factor and influences the outcome only through the risk factor, independent of confounders. Thus far, Mendelian randomisation studies did not detect causal relationships between cervical cancer and obesity [164] or cervical cancer and Alzheimer’s disease [165], but they suggested a possible link between cervical cancer and type II diabetes mellitus [166], and they strongly supported the complementary role of smoking, older age at first pregnancy, and number of sexual partners in the risk of developing cervical cancer [121].
4. Conclusions
There is now ample evidence for a strong genetic component of cervical cancer risk that adds to known risks posed by epidemiological factors and HPV infection. There have been few cervical cancer GWA studies so far, with the HLA region emerging as a consistently associated locus across different populations. Cervical cancer thus adds to the large number of autoimmune and immune-mediated diseases associated with this genetic region [127,167,168]. As for most other HLA-associated cancers with infectious aetiology, the associations were driven by multiple HLA regions, suggesting that both cytotoxic and helper T-cell responses might be important [169]. This may open a perspective for personalised antigen-specific disease prevention through harnessing HLA–ligand interactions for clinical benefit [127]. More recent biobanking and consortia efforts have also led to the identification of non-HLA susceptibility loci such as rs8067378 (GSDMB), rs10175462 (PAX8), and rs27069 (CLPTM1L-TERT), which warrant further investigation. Such association results from large-scale case-control series will become increasingly useful to inform risk assessment strategies as well as biological and pharmacological studies.
Cohort size, apart from effect allele frequency and the magnitude of risk, determines the power of the study and is therefore a crucial factor in identifying novel susceptibility loci. There are only a limited number of consistent cervical cancer susceptibility variants thus far, compared to other traits, and even fewer follow-up studies to identify the causal variant via fine-mapping. Even where the causal variants were determined, it still remains a challenge to assign biological function to the variants and identify the causal gene(s). Furthermore, only a handful of results so far has been attempted for fine-mapping or functional validation via chromatin conformation or eQTL analysis [119,128,132,142,152,170].
Although there are rare coding variants in MICA, HLA-B, or PTPN14 that would directly affect the function of the gene product [121,123,128], the majority of genome-wide significant signals identified at susceptibility regions seem to be regulatory variants. For the HLA region, specific amino acids in the MHC-peptide binding groove may also explain some of the SNP associations, as is known for HLA-B and HIV infection [171,172]. Additional work will be required to trace down target genes at every locus and to understand their molecular mechanisms. It is highly likely that at least some of these genomic factors play a role in viral infection and host immunity. The identification of these genes and their function(s) could provide important insights into the aetiology of cervical cancer and fuel further pharmacological research [173].
Thus far, the low number of known cervical cancer susceptibility variants and genes has restricted further research towards precision medicine approaches. It is therefore the need of the hour to perform large-cohort based cervical cancer GWAS and meta-analysis in order to identify further susceptibility loci, and national biobanking has proven to be a very fruitful resource for genomic research in this regard. With ongoing efforts, we may eventually be able to screen for disease-associated risk factors early on as part of preventive cancer surveillance strategies.
Author Contributions
Conceptualisation, writing, review, editing, D.R. and T.D.; Visualisation, D.R.; Supervision, funding acquisition, T.D. Both authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Bruno and Helene Jöster Foundation.
Data Availability Statement
The GWAS summary statistics for most of the studies described in this text are available from the following online repositories, in addition to the respective cited research articles. Leo et al. (https://www.ebi.ac.uk/gwas/efotraits/EFO_0001061GWASCatalog, Accession ID GCST004833), Rashkin et al. (https://www.ebi.ac.uk/gwas/efotraits/EFO_0001061GWASCatalog, Accession ID GCST90011816), UK Biobank (CC GWAS with female controls only, https://github.com/Nealelab/UK_Biobank_GWAS, file: 20001_1041.gwas.imputed_v3.female), and FinnGen freeze 5 (https://r5.finngen.fi/), Japan Biobank (https://pheweb.jp/).
Acknowledgments
The authors would like to acknowledge the diligent scientists who have conducted large scale genomic studies on cervical cancer and made their datasets available for public use. We furthermore thank Professor Peter Hillemanns for his continuous support. The images were created on Biorender.com.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
HPV human papillomavirus; GWAS genome-wide association study; HLA human leukocyte antigen; HIV human immunodeficiency virus; PCR polymerase chain reaction; LSIL low grade squamous intraepithelial lesions; CIN cervical intraepithelial neoplasia stage; HSIL high grade squamous intraepithelial lesions; CIS carcinoma in situ; hrHPV high risk HPV; RR relative risk; FRR familial RR; iCHAVs independent sets of correlated highly associated variants; QTL quantitative trait loci; eQTL expression QTL; metQTL methylation QTL; sQTL splicing QTL; pQTL protein QTL; PRS polygenic risk score; MR Mendelian randomisation; ChIP chromatin immunoprecipitation; 3C chromatin conformation capture; 4C chromatin conformation capture on chip; 5C chromatin conformation capture carbon copy; Hi-C high throughput chromatin conformation capture; ChIA-PET chromatin interaction analysis by paired-end tag sequencing; CRISPR clustered regularly interspaced short palindromic repeats; MHC major histocompatibility complex; LoF loss of function.
References
- Bruni, L.; Albero, G.; Serrano, B.; Mena, M.; Gómez, D.; Muñoz, J.; Bosch, F.; de Sanjosé, S. ICO HPV Information Centre Human Papillomavirus and Related Diseases Report-Germany. Summary Report. ICO/IARC Information Centre HPV Cancer. 2019. Available online: https://hpvcentre.net/statistics/reports/DEU.pdf?t=1575294458729 (accessed on 12 October 2021).
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Arbyn, M.; Weiderpass, E.; Bruni, L.; de Sanjosé, S.; Saraiya, M.; Ferlay, J.; Bray, F. Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob. Health 2020, 8, e191–e203. [Google Scholar] [CrossRef]
- zur Hausen, H. Papillomaviruses in the causation of human cancers—A brief historical account. Virology 2009, 384, 260–265. [Google Scholar] [CrossRef]
- Burd, E. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 2003, 16, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Burd, E.M.; Dean, C.L. Human Papillomavirus. Microbiol. Spectr. 2016, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Zur Hausen, H. Papillomaviruses and cancer: From basic studies to clinical application. Nat. Rev. Cancer 2002, 2, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, N.; Franceschi, S.; Bosetti, C.; Moreno, V.; Herrero, R.; Smith, J.S.; Shah, K.V.; Meijer, C.J.; Bosch, F.X. Role of parity and human papillomavirus in cervical cancer: The IARC multicentric case-control study. Lancet 2002, 359, 1093–1101. [Google Scholar] [CrossRef]
- Franceschi, S.; Rajkumar, T.; Vaccarella, S.; Gajalakshmi, V.; Sharmila, A.; Snijders, P.J.F.; Muñoz, N.; Meijer, C.J.L.M.; Herrero, R. Human papillomavirus and risk factors for cervical cancer in Chennai, India: A case-control study. Int. J. Cancer 2003, 107, 127–133. [Google Scholar] [CrossRef]
- Brinton, L.A.; Tashima, K.T.; Lehman, H.F.; Levine, R.S.; Mallin, K.; Savitz, D.A.; Stolley, P.D.; Fraumeni, J.F. Epidemiology of cervical cancer by cell type. Cancer Res. 1987, 47, 1706–1711. [Google Scholar]
- Bruni, L.; Albero, G.; Serrano, B.; Mena, M.; Gómez, D.; Muñoz, J.; Bosch, F.; de Sanjosé, S. ICO HPV Information Centre Human Papillomavirus and Related Diseases Report-World Summary Report. ICO/IARC Information Centre HPV Cancer. 2019. Available online: https://hpvcentre.net/statistics/reports/XWX.pdf (accessed on 12 October 2021).
- Bujan Rivera, J.; Klug, S.J. Cervical cancer screening in Germany. Bundesgesundheitsblatt Gesundh. Gesundh. 2018, 61, 1528–1535. [Google Scholar] [CrossRef]
- Hillemanns, P.; Soergel, P.; Hertel, H.; Jentschke, M. Epidemiology and Early Detection of Cervical Cancer. Oncol. Res. Treat. 2016, 39, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Starker, A.; Buttmann-Schweiger, N.; Krause, L.; Barnes, B.; Kraywinkel, K.; Holmberg, C. Cancer screening in Germany: Availability and participation. Bundesgesundheitsblatt Gesundh. Gesundh. 2018, 61, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- de Araujo Souza, P.S.; Sichero, L.; Maciag, P.C. HPV variants and HLA polymorphisms: The role of variability on the risk of cervical cancer. Futur. Oncol. 2009, 5, 359–370. [Google Scholar] [CrossRef] [PubMed]
- De Brot, L.; Pellegrini, B.; Moretti, S.T.; Carraro, D.M.; Soares, F.A.; Rocha, R.M.; Baiocchi, G.; da Cunha, I.W.; de Andrade, V.P. Infections with multiple high-risk HPV types are associated with high-grade and persistent low-grade intraepithelial lesions of the cervix. Cancer Cytopathol. 2017, 125, 138–143. [Google Scholar] [CrossRef]
- Feyrter, F. Über das Oberflächenkarzinom im Bereich des Collum uteri TT-Surface carcinoma of the uterine cervix. Dtsch. Med. Wochenschr. 1955, 80, 1686–1691. [Google Scholar] [CrossRef]
- Gray, L.A.; Barnes, M.L.; Lee, J.J. Carcinoma-in-situ and dysplasia of the cervix. Ann. Surg. 1960, 151, 951–960. [Google Scholar] [CrossRef]
- Cornet, I.; Gheit, T.; Franceschi, S.; Vignat, J.; Burk, R.D.; Sylla, B.S.; Tommasino, M.; Clifford, G.M. Human Papillomavirus Type 16 Genetic Variants: Phylogeny and Classification Based on E6 and LCR. J. Virol. 2012, 86, 6855–6861. [Google Scholar] [CrossRef]
- Chen, A.A.; Heideman, D.A.M.; Boon, D.; Chen, Z.; Burk, R.D.; De Vuyst, H.; Gheit, T.; Snijders, P.J.F.; Tommasino, M.; Franceschi, S.; et al. Human papillomavirus 33 worldwide genetic variation and associated risk of cervical cancer. Virology 2014, 448, 356–362. [Google Scholar] [CrossRef]
- Chen, A.A.; Heideman, D.A.M.; Boon, D.; Gheit, T.; Snijders, P.J.F.; Tommasino, M.; Franceschi, S.; Clifford, G.M. Human Papillomavirus 45 Genetic Variation and Cervical Cancer Risk Worldwide. J. Virol. 2014, 88, 4514–4521. [Google Scholar] [CrossRef]
- Hirose, Y.; Onuki, M.; Tenjimbayashi, Y.; Mori, S.; Ishii, Y.; Takeuchi, T.; Tasaka, N.; Satoh, T.; Morisada, T.; Iwata, T.; et al. Within-Host Variations of Human Papillomavirus Reveal APOBEC Signature Mutagenesis in the Viral Genome. J. Virol. 2018, 92, 1–14. [Google Scholar] [CrossRef]
- Mirabello, L.; Clarke, M.A.; Nelson, C.W.; Dean, M.; Wentzensen, N.; Yeager, M.; Cullen, M.; Boland, J.F.; Alemany, L.; Banks, L.; et al. The intersection of HPV epidemiology, genomics and mechanistic studies of HPV-mediated carcinogenesis. Viruses 2018, 10, 80. [Google Scholar] [CrossRef]
- Chen, A.A.; Gheit, T.; Franceschi, S.; Tommasino, M.; Clifford, G.M. Human Papillomavirus 18 Genetic Variation and Cervical Cancer Risk Worldwide. J. Virol. 2015, 89, 10680–10687. [Google Scholar] [CrossRef]
- Badaracco, G.; Venuti, A.; Sedati, A.; Marcante, M.L. HPV16 and HPV18 in genital tumors: Significantly different levels of viral integration and correlation to tumor invasiveness. J. Med. Virol. 2002, 67, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Pirami, L.; Giache, V.; Becciolini, A. Pre-Invasive and Invasive Lesions of the Uterine Cervix. Obstet. Gynecol. 1997, 600–604. [Google Scholar]
- Burk, R.D.; Chen, Z.Z.; Saller, C.; Tarvin, K.; Carvalho, A.L.; Scapulatempo-Neto, C.; Silveira, H.C.; Fregnani, J.H.; Creighton, C.J.; Anderson, M.L.; et al. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [Google Scholar] [CrossRef]
- Hu, Z.; Zhu, D.; Wang, W.; Li, W.; Jia, W.; Zeng, X.; Ding, W.; Yu, L.; Wang, X.; Wang, L.; et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat. Genet. 2015, 47, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Ojesina, A.I.; Lichtenstein, L.; Freeman, S.S.; Pedamallu, C.S.; Imaz-Rosshandler, I.; Pugh, T.J.; Cherniack, A.D.; Ambrogio, L.; Cibulskis, K.; Bertelsen, B.; et al. Landscape of genomic alterations in cervical carcinomas. Nature 2014, 506, 371–375. [Google Scholar] [CrossRef]
- Henken, F.E.; Wilting, S.M.; Overmeer, R.M.; Van Rietschoten, J.G.I.; Nygren, A.O.H.; Errami, A.; Schouten, J.P.; Meijer, C.J.L.M.; Snijders, P.J.F.; Steenbergen, R.D.M. Sequential gene promoter methylation during HPV-induced cervical carcinogenesis. Br. J. Cancer 2007, 97, 1457–1464. [Google Scholar] [CrossRef]
- Dong, S.M.; Kim, H.S.; Rha, S.H.; Sidransky, D. Promoter hypermethylation of multiple genes in carcinoma of the uterine cervix. Clin. Cancer Res. 2001, 7, 1982–1986. [Google Scholar]
- Sartor, M.A.; Dolinoy, D.C.; Jones, T.R.; Colacino, J.A.; Prince, M.E.P.; Carey, T.E.; Rozek, L.S. Genome-wide methylation and expression differences in HPV(+) and HPV(-) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics 2011, 6, 777–787. [Google Scholar] [CrossRef]
- Curty, G.; Menezes, A.N.; Brant, A.C.; de Mulder Rougvie, M.; Moreira, M.Â.M.; Soares, M.A. Expression of Retroelements in Cervical Cancer and Their Interplay with HPV Infection and Host Gene Expression. Cancers 2021, 13, 3513. [Google Scholar] [CrossRef] [PubMed]
- Scheffner, M.; Werness, B.A.; Huibregtse, J.M.; Levine, A.J.; Howley, P.M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 1990, 63, 1129–1136. [Google Scholar] [CrossRef]
- Werness, B.A.; Levine, A.J.; Howley, P.M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 1990, 248, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; David, P.; Banks, L. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene 1999, 18, 7690–7700. [Google Scholar] [CrossRef] [PubMed]
- Helt, A.-M.; Funk, J.O.; Galloway, D.A. Inactivation of both the Retinoblastoma Tumor Suppressor and p21 by the Human Papillomavirus Type 16 E7 Oncoprotein Is Necessary To Inhibit Cell Cycle Arrest in Human Epithelial Cells. J. Virol. 2002, 76, 10559–10568. [Google Scholar] [CrossRef]
- Shin, M.K.; Balsitis, S.; Brake, T.; Lambert, P.F. Human papillomavirus E7 oncoprotein overrides the tumor suppressor activity of p21Cip1 in cervical carcinogenesis. Cancer Res. 2009, 69, 5656–5663. [Google Scholar] [CrossRef]
- Jones, D.L.; Alani, R.M.; Münger, K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21(Cip1)-mediated inhibition of cdk2. Genes Dev. 1997, 11, 2101–2111. [Google Scholar] [CrossRef]
- Sano, T.; Oyama, T.; Kashiwabara, K.; Fukuda, T.; Nakajima, T. Expression Status of p16 Protein Is Associated with Human Papillomavirus Oncogenic Potential in Cervical and Genital Lesions. Am. J. Pathol. 1998, 153, 1741–1748. [Google Scholar] [CrossRef]
- Keating, J.T.; Cviko, A.; Riethdorf, S.; Riethdorf, L.; Quade, B.J.; Sun, D.; Duensing, S.; Sheets, E.E.; Munger, K.; Crum, C.P. Ki-67, Cyclin E, and p16 INK4 Are Complimentary Surrogate Biomarkers for Human Papilloma Virus-Related Cervical Neoplasia. Am. J. Surg. Pathol. 2001, 25, 884–891. [Google Scholar] [CrossRef]
- Von Knebel Doeberitz, M. New markers for cervical dysplasia to visualise the genomic chaos created by aberrant oncogenic papillomavirus infections. Eur. J. Cancer 2002, 38, 2229–2242. [Google Scholar] [CrossRef]
- Nuovo, G.J.; Plaia, T.W.; Belinsky, S.A.; Baylin, S.B.; Herman, J.G. In situ detection of the hypermethylation-induced inactivation of the p16 gene as an early event in oncogenesis. Proc. Natl. Acad. Sci. USA 1999, 96, 12754–12759. [Google Scholar] [CrossRef]
- Wijetunga, N.A.; Belbin, T.J.; Burk, R.D.; Whitney, K.; Abadi, M.; Greally, J.M.; Einstein, M.H.; Schlecht, N.F. Novel epigenetic changes in CDKN2A are associated with progression of cervical intraepithelial neoplasia. Gynecol. Oncol. 2016, 142, 566–573. [Google Scholar] [CrossRef]
- McLaughlin-Drubin, M.E.; Park, D.; Munger, K. Tumor suppressor p16INK4A is necessary for survival of cervical carcinoma cell lines. Proc. Natl. Acad. Sci. USA 2013, 110, 16175–16180. [Google Scholar] [CrossRef]
- Le Bon, A.; Tough, D.F. Links between innate and adaptive immunity via type I interferon. Curr. Opin. Immunol. 2002, 14, 432–436. [Google Scholar] [CrossRef]
- Herdman, T.M.; Pett, M.R.; Roberts, I.; Alazawi, W.O.F.; Teschendorff, A.E.; Zhang, X.Y.; Stanley, M.A.; Coleman, N. Interferon-β treatment of cervical keratinocytes naturally infected with human papillomavirus 16 episomes promotes rapid reduction in episome numbers and emergence of latent integrants. Carcinogenesis 2006, 27, 2341–2353. [Google Scholar] [CrossRef] [PubMed]
- Black, A.P.B.; Ardern-Jones, M.R.; Kasprowicz, V.; Bowness, P.; Jones, L.; Bailey, A.S.; Ogg, G.S. Human keratinocyte induction of rapid effector function in antigen-specific memory CD4+ and CD8+ T cells. Eur. J. Immunol. 2007, 37, 1485–1493. [Google Scholar] [CrossRef]
- Karim, R.; Meyers, C.; Backendorf, C.; Ludigs, K.; Offringa, R.; van Ommen, G.J.B.; Melief, C.J.M.; van der Burg, S.H.; Boer, J.M. Human papillomavirus deregulates the response of a cellular network comprising of chemotactic and proinflammatory genes. PLoS ONE 2011, 6, e17848. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.M.; Raghav, D.; Singh, H.; Raghava, G.P.S. CCDB: A curated database of genes involved in cervix cancer. Nucleic Acids Res. 2011, 39, 975–979. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Bravo, Á.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017, 45, D833–D839. [Google Scholar] [CrossRef] [PubMed]
- Ngo, C.; Samuels, S.; Bagrintseva, K.; Slocker, A.; Hupé, P.; Kenter, G.; Popovic, M.; Samet, N.; Tresca, P.; von der Leyen, H.; et al. From prospective biobanking to precision medicine: BIO-RAIDs—An EU study protocol in cervical cancer. BMC Cancer 2015, 15, 842. [Google Scholar] [CrossRef]
- Way, S.; Hetherington, J.; Galloway, D.C. Simultaneous cytological diagnosis of cervical cancer in three sisters. Lancet 1959, 2, 890–891. [Google Scholar] [CrossRef]
- Ahlbom, A.; Lichtenstein, P.; Malmström, H.; Feychting, M.; Hemminki, K.; Pedersen, N.L. Cancer in twins: Genetic and nongenetic familial risk factors. J. Natl. Cancer Inst. 1997, 89, 287–293. [Google Scholar] [CrossRef]
- Andrews, F.J.; Linehan, J.J.; Melcher, D.H. Cervical carcinoma in both mother and daughter. Acta Cytol. 1981, 25, 3–4. [Google Scholar] [PubMed]
- Magnusson, P.K.E.; Lichtenstein, P.; Gyllensten, U.B. Heritability of cervical tumours. Int. J. Cancer 2000, 88, 698–701. [Google Scholar] [CrossRef]
- Bruinse, H.W.; te Velde, E.R.; de Gast, B.C. Human leukocyte antigen patterns in a family with cervical cancer. Gynecol. Oncol. 1981, 12, 249–252. [Google Scholar] [CrossRef]
- Horn, L.-C.; Raptis, G.; Fischer, U. Familial cancer history in patients with carcinoma of the cervix uteri. Eur. J. Obstet. Gynecol. Reprod. Biol. 2002, 101, 54–57. [Google Scholar] [CrossRef]
- Zoodsma, M.; Sijmons, R.H.; de Vries, E.G.; Zee, A. Familial Cervical Cancer: Case Reports, Review and Clinical Implications. Hered. Cancer Clin. Pract. 2004, 2, 99. [Google Scholar] [CrossRef][Green Version]
- De Oliveira, C.M.; Levi, J.E. The Biological Impact of Genomic Diversity in Cervical Cancer Development. Acta Cytol. 2016, 60, 513–517. [Google Scholar] [CrossRef]
- Rodríguez-Carunchio, L.; Soveral, I.; Steenbergen, R.D.M.; Torné, A.; Martinez, S.; Fusté, P.; Pahisa, J.; Marimon, L.; Ordi, J.; del Pino, M. HPV-negative carcinoma of the uterine cervix: A distinct type of cervical cancer with poor prognosis. BJOG Int. J. Obstet. Gynaecol. 2015, 122, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Tjalma, W. HPV negative cervical cancers and primary HPV screening. Facts Views Vis. ObGyn 2018, 10, 107–113. [Google Scholar]
- Tjalma, W.A.A.; Trinh, X.B.; Rosenlund, M.; Makar, A.P.; Kridelka, F.; Rosillon, D.; Van Dam, P.A.; Collas De Souza, S.; Holl, K.; Simon, P.; et al. A cross-sectional, multicentre, epidemiological study on human papillomavirus (HPV) type distribution in adult women diagnosed with invasive cervical cancer in Belgium. Facts Views Vis. ObGyn 2015, 7, 101–108. [Google Scholar]
- Ruiz, F.J.; Sundaresan, A.; Zhang, J.; Pedamallu, C.S.; Halle, M.K.; Srinivasasainagendra, V.; Zhang, J.; Muhammad, N.; Stanley, J.; Markovina, S.; et al. Genomic Characterization and Therapeutic Targeting of HPV Undetected Cervical Carcinomas. Cancers 2021, 13, 4551. [Google Scholar] [CrossRef] [PubMed]
- Petry, K.U.; Liebrich, C.; Luyten, A.; Zander, M.; Iftner, T. Surgical staging identified false HPV-negative cases in a large series of invasive cervical cancers. Papillomavirus Res. 2017, 4, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Leo, P.J.; Madeleine, M.M.; Wang, S.; Schwartz, S.M.; Newell, F.; Pettersson-Kymmer, U.; Hemminki, K.; Hallmans, G.; Tiews, S.; Steinberg, W.; et al. Defining the genetic susceptibility to cervical neoplasia—A genome-wide association study. PLoS Genet. 2017, 13, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Cui, T.; Ek, W.E.; Liu, H.; Wang, H.; Gyllensten, U. Analysis of the genetic architecture of susceptibility to cervical cancer indicates that common SNPs explain a large proportion of the heritability. Carcinogenesis 2015, 36, 992–998. [Google Scholar] [CrossRef] [PubMed]
- Rashkin, S.R.; Graff, R.E.; Kachuri, L.; Thai, K.K.; Alexeeff, S.E.; Blatchins, M.A.; Cavazos, T.B.; Corley, D.A.; Emami, N.C.; Hoffman, J.D.; et al. Pan-cancer study detects genetic risk variants and shared genetic basis in two large cohorts. Nat. Commun. 2020, 11, 4423. [Google Scholar] [CrossRef]
- Von Keyserling, H.; Bergmann, T.; Schuetz, M.; Schiller, U.; Stanke, J.; Hoffmann, C.; Schneider, A.; Lehrach, H.; Dahl, A.; Kaufmann, A.M. Analysis of 4 single-nucleotide polymorphisms in relation to cervical dysplasia and cancer development using a high-throughput ligation-detection reaction procedure. Int. J. Gynecol. Cancer 2011, 21, 1664–1671. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, Z.; Ma, D.; Huettner, P.C.; Massad, L.S.; Nguyen, L.; Borecki, I.; Rader, J.S. TP53, MDM2, NQO1, and susceptibility to cervical cancer. Cancer Epidemiol. Biomarkers Prev. 2010, 19, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.A.; Kim, J.W.; Roh, J.W.; Choi, J.Y.; Lee, K.M.; Yoo, K.Y.; Song, Y.S.; Kang, D. Genetic polymorphisms of GSTM1, p21, p53 and HPV infection with cervical cancer in Korean women. Gynecol. Oncol. 2004, 93, 14–18. [Google Scholar] [CrossRef]
- Nunobiki, O.; Ueda, M.; Yamamoto, M.; Toji, E.; Sato, N.; Izuma, S.; Okamoto, Y.; Torii, K.; Noda, S. MDM2 SNP 309 human papillomavirus infection in cervical carcinogenesis. Gynecol. Oncol. 2010, 118, 258–261. [Google Scholar] [CrossRef]
- Roszak, A.; Misztal, M.; Sowińska, A.; Jagodziński, P.P. Murine Double-Minute 2 Homolog Single Nucleotide Polymorphisms 285 and 309 in Cervical Carcinogenesis. Mol. Diagnosis Ther. 2015, 19, 235–244. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oliveira, S.; Ribeiro, J.; Sousa, H.; Pinto, D.; Baldaque, I.; Medeiros, R. Genetic polymorphisms and cervical cancer development: ATM G5557A and p53bp1 C1236G. Oncol. Rep. 2012, 27, 1188–1192. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.D.; Cai, G.Q.; Zou, W.; Huang, Y.H.; Zhang, J.R.; Wang, D.T.; Chen, B.L. BRIP1 variations analysis reveals their relative importance as genetic susceptibility factor for cervical cancer. Biochem. Biophys. Res. Commun. 2013, 433, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Nava, G.A.; Fernández-Niño, J.A.; Madrid-Marina, V.; Torres-Poveda, K. Cervical cancer genetic susceptibility: A systematic review and meta-analyses of recent evidence. PLoS ONE 2016, 11, e0157344. [Google Scholar] [CrossRef]
- Wang, N.; Wang, S.; Zhang, Q.; Lu, Y.; Wei, H.; Li, W.; Zhang, S.; Yin, D.; Ou, Y. Association of p21 SNPs and risk of cervical cancer among Chinese women. BMC Cancer 2012, 12, 589. [Google Scholar] [CrossRef]
- Lima, G.; Santos, E.; Angelo, H.; Oliveira, M.; Heráclio, S.; Leite, F.; de Melo, C.; Crovella, S.; Maia, M.; Souza, P. Association between p21 Ser31Arg polymorphism and the development of cervical lesion in women infected with high risk HPV. Tumor Biol. 2016, 37, 10935–10941. [Google Scholar] [CrossRef] [PubMed]
- Thakur, N.; Hussain, S.; Nasare, V.; Das, B.C.; Basir, S.F.; Bharadwaj, M. Association analysis of p16 (CDKN2A) and RB1 polymorphisms with susceptibility to cervical cancer in Indian population. Mol. Biol. Rep. 2012, 39, 407–414. [Google Scholar] [CrossRef]
- Juko-Pecirep, I.; Ivansson, E.L.; Gyllensten, U.B. Evaluation of Fanconi anaemia genes FANCA, FANCC and FANCL in cervical cancer susceptibility. Gynecol. Oncol. 2011, 122, 377–381. [Google Scholar] [CrossRef]
- Chung, H.H.; Kim, M.K.; Kim, J.W.; Park, N.H.; Song, Y.S.; Kang, S.B.; Lee, H.P. XRCC1 R399Q polymorphism is associated with response to platinum-based neoadjuvant chemotherapy in bulky cervical cancer. Gynecol. Oncol. 2006, 103, 1031–1037. [Google Scholar] [CrossRef]
- Kim, K.; Kang, S.B.; Chung, H.H.; Kim, J.W.; Park, N.H.; Song, Y.S. XRCC1 Arginine194Tryptophan and GGH-401Cytosine/Thymine polymorphisms are associated with response to platinum-based neoadjuvant chemotherapy in cervical cancer. Gynecol. Oncol. 2008, 111, 509–515. [Google Scholar] [CrossRef]
- Cheng, X.-D.; Lu, W.-G.; Ye, F.; Wan, X.-Y.; Xie, X. The association of XRCC1 gene single nucleotide polymorphisms with response to neoadjuvant chemotherapy in locally advanced cervical carcinoma. J. Exp. Clin. Cancer Res. 2009, 28, 91. [Google Scholar] [CrossRef] [PubMed]
- Alsbeih, G.; Al-Harbi, N.; El-Sebaie, M.; Al-Badawi, I. HPV prevalence and genetic predisposition to cervical cancer in Saudi Arabia. Infect. Agent. Cancer 2013, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.J.; Rader, J.S.; Borecki, I.; Zhang, Z.; Hildesheim, A. CD83 polymorphisms and cervical cancer risk. Gynecol. Oncol. 2009, 114, 319–322. [Google Scholar] [CrossRef]
- Zhang, Z.; Borecki, I.; Nguyen, L.; Ma, D.; Smith, K.; Huettner, P.C.; Mutch, D.G.; Herzog, T.J.; Gibb, R.K.; Powell, M.A.; et al. CD83 gene polymorphisms increase susceptibility to human invasive cervical cancer. Cancer Res. 2007, 67, 11202–11208. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hu, L.; Liu, J.; Chen, X.; Zhang, Y.; Liu, L.; Zhu, J.; Chen, J.; Shen, H.; Qiang, F.; Hu, Z. CTLA-4 gene polymorphism +49 A/G contributes to genetic susceptibility to two infection-related cancers-hepatocellular carcinoma and cervical cancer. Hum. Immunol. 2010, 71, 888–891. [Google Scholar] [CrossRef]
- Yin, J.; Wen, J.; Hang, D.; Han, J.; Jiang, J.; Song, C.; Liu, Y.; Liu, J.; Liu, L.; Zhu, L.; et al. Expression quantitative trait loci for CARD8 contributes to risk of two infection-related cancers—Hepatocellular carcinoma and cervical cancer. PLoS ONE 2015, 10, e0132352. [Google Scholar] [CrossRef][Green Version]
- Jin, Y. Association of Single Nucleotide Polymorphisms in Tumor Necrosis Factor-Alpha with Cervical Cancer Susceptibility. Cell Biochem. Biophys. 2015, 71, 77–84. [Google Scholar] [CrossRef]
- Kohaar, I.; Thakur, N.; Salhan, S.; Batra, S.; Singh, V.; Sharma, A.; Sodhani, P.; Das, B.C.; Sarkar, D.P.; Bharadwaj, M. TNF α –308G/A Polymorphism as a Risk Factor for HPV Associated Cervical Cancer in Indian Population. Anal. Cell. Pathol. 2007, 29, 249–256. [Google Scholar] [CrossRef]
- Barbisan, G.; Pérez, L.O.; Contreras, A.; Golijow, C.D. TNF-α and IL-10 promoter polymorphisms, HPV infection, and cervical cancer risk. Tumor Biol. 2012, 33, 1549–1556. [Google Scholar] [CrossRef]
- Badano, I.; Stietz, S.M.; Schurr, T.G.; Picconi, A.M.; Fekete, D.; Quintero, I.M.; Cabrera, M.D.E.; Campos, R.H.; Liotta, J.D. Analysis of TNFα promoter SNPs and the risk of cervical cancer in urban populations of Posadas (Misiones, Argentina). J. Clin. Virol. 2012, 53, 54–59. [Google Scholar] [CrossRef]
- Liu, H.; Lyu, D.; Zhang, Y.; Sheng, L.; Tang, N. Association Between the IL-6 rs1800795 Polymorphism and the Risk of Cervical Cancer: A Meta-Analysis of 1210 Cases and 1525 Controls. Technol. Cancer Res. Treat. 2017, 16, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Pu, X.; Gu, Z.; Wang, X. Polymorphisms of the interleukin 6 gene and additional gene–gene interaction contribute to cervical cancer susceptibility in Eastern Chinese women. Arch. Gynecol. Obstet. 2016, 294, 1305–1310. [Google Scholar] [CrossRef] [PubMed]
- Han, S.S.; Cho, E.Y.; Lee, T.S.; Kim, J.W.; Park, N.H.; Song, Y.S.; Kim, J.G.; Lee, H.P.; Kang, S.B. Interleukin-12 p40 gene (IL12B) polymorphisms and the risk of cervical caner in Korean women. Eur. J. Obstet. Gynecol. Reprod. Biol. 2008, 140, 71–75. [Google Scholar] [CrossRef]
- Chagas, B.S.; Gurgel, A.P.A.D.; da Cruz, H.L.A.; Amaral, C.M.M.; Cardoso, M.V.; Neto, J.d.S.; Silva, L.A.Ô.F.d.; Albuquerque, E.M.B.d.; Muniz, M.T.C.; Freitas, A.C.d. An interleukin-10 gene polymorphism associated with the development of cervical lesions in women infected with Human Papillomavirus and using oral contraceptives. Infect. Genet. Evol. 2013, 19, 32–37. [Google Scholar] [CrossRef][Green Version]
- Torres-Poveda, K.; Burguete-García, A.I.; Bahena-Román, M.; Méndez-Martínez, R.; Zurita-Díaz, M.A.; López-Estrada, G.; Delgado-Romero, K.; Peralta-Zaragoza, O.; Bermúdez-Morales, V.H.; Cantú, D.; et al. Risk allelic load in Th2 and Th3 cytokines genes as biomarker of susceptibility to HPV-16 positive cervical cancer: A case control study. BMC Cancer 2016, 16, 330. [Google Scholar] [CrossRef]
- Wang, S.S.; Gonzalez, P.; Yu, K.; Porras, C.; Li, Q.; Safaeian, M.; Rodriguez, A.C.; Sherman, M.E.; Bratti, C.; Schiffman, M.; et al. Common genetic variants and risk for HPV persistence and progression to cervical cancer. PLoS ONE 2010, 5, 1–7. [Google Scholar] [CrossRef]
- Klug, S.J.; Ressing, M.; Koenig, J.; Abba, M.C.; Agorastos, T.; Brenna, S.M.; Ciotti, M.; Das, B.; Del Mistro, A.; Dybikowska, A.; et al. TP53 codon 72 polymorphism and cervical cancer: A pooled analysis of individual data from 49 studies. Lancet Oncol. 2009, 10, 772–784. [Google Scholar] [CrossRef]
- Sud, A.; Kinnersley, B.; Houlston, R.S. Genome-wide association studies of cancer: Current insights and future perspectives. Nat. Rev. Cancer 2017, 17, 692–704. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Mbatchou, J.; Barnard, L.; Backman, J.; Marcketta, A.; Kosmicki, J.A.; Ziyatdinov, A.; Benner, C.; O’Dushlaine, C.; Barber, M.; Boutkov, B.; et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 2021, 53, 1097–1103. [Google Scholar] [CrossRef] [PubMed]
- Cano-Gamez, E.; Trynka, G. From GWAS to Function: Using Functional Genomics to Identify the Mechanisms Underlying Complex Diseases. Front. Genet. 2020, 11, 1–21. [Google Scholar] [CrossRef]
- Freedman, M.L.; Monteiro, A.N.A.; Gayther, S.A.; Coetzee, G.A.; Risch, A.; Plass, C.; Casey, G.; De Biasi, M.; Carlson, C.; Duggan, D.; et al. Principles for the post-GWAS functional characterization of cancer risk loci. Nat. Genet. 2011, 43, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Bush, W.S.; Moore, J.H. Chapter 11: Genome-Wide Association Studies. PLoS Comput. Biol. 2012, 8, e1002822. [Google Scholar] [CrossRef]
- Gallagher, M.D.; Chen-Plotkin, A.S. The Post-GWAS Era: From Association to Function. Am. J. Hum. Genet. 2018, 102, 717–730. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Ward, L.D.; Kellis, M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012, 40, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Dayem Ullah, A.Z.; Oscanoa, J.; Wang, J.; Nagano, A.; Lemoine, N.R.; Chelala, C. SNPnexus: Assessing the functional relevance of genetic variation to facilitate the promise of precision medicine. Nucleic Acids Res. 2018, 46, W109–W113. [Google Scholar] [CrossRef]
- Arnold, M.; Raffler, J.; Pfeufer, A.; Suhre, K.; Kastenmüller, G. SNiPA: An interactive, genetic variant-centered annotation browser. Bioinformatics 2015, 31, 1334–1336. [Google Scholar] [CrossRef]
- Loos, R.J.F. 15 Years of Genome-Wide Association Studies and No Signs of Slowing Down. Nat. Commun. 2020, 11, 10–12. [Google Scholar] [CrossRef]
- Graff, R.E.; Cavazos, T.B.; Thai, K.K.; Kachuri, L.; Rashkin, S.R.; Hoffman, J.D.; Alexeeff, S.E.; Blatchins, M.; Meyers, T.J.; Leong, L.; et al. Cross-cancer evaluation of polygenic risk scores for 16 cancer types in two large cohorts. Nat. Commun. 2021, 12, 970. [Google Scholar] [CrossRef] [PubMed]
- Sakaue, S.; Hirata, J.; Kanai, M.; Suzuki, K.; Akiyama, M.; Lai Too, C.; Arayssi, T.; Hammoudeh, M.; Al Emadi, S.; Masri, B.K.; et al. Dimensionality reduction reveals fine-scale structure in the Japanese population with consequences for polygenic risk prediction. Nat. Commun. 2020, 11, 1569. [Google Scholar] [CrossRef] [PubMed]
- Reay, W.R.; Cairns, M.J. Advancing the use of genome-wide association studies for drug repurposing. Nat. Rev. Genet. 2021. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Gyllensten, U. Lessons and implications from association studies and post-GWAS analyses of cervical cancer. Trends Genet. 2015, 31, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Juko-Pecirep, I.; Hammer, J.; Ivansson, E.; Enroth, S.; Gustavsson, I.; Feuk, L.; Magnusson, P.K.E.; McKay, J.D.; Wilander, E.; et al. Genome-wide association study of susceptibility loci for cervical cancer. J. Natl. Cancer Inst. 2013, 105, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Li, L.; Hu, Z.; Li, S.; Wang, S.S.; Liu, J.J.; Wu, C.; He, L.; Zhou, J.; Li, Z.Z.Z.; et al. A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nat. Genet. 2013, 45, 918–922. [Google Scholar] [CrossRef]
- Chen, D.; Enroth, S.; Liu, H.; Sun, Y.; Wang, H.; Yu, M.; Deng, L.; Xu, S.; Gyllensten, U. Pooled analysis of genome-wide association studies of cervical intraepithelial neoplasia 3 (CIN3) identifies a new susceptibility locus. Oncotarget 2016, 7, 42216–42224. [Google Scholar] [CrossRef]
- Takeuchi, F.; Kukimoto, I.; Li, Z.; Li, S.; Li, N.; Hu, Z.; Takahashi, A.; Inoue, S.; Yokoi, S.; Chen, J.; et al. Genome-wide association study of cervical cancer suggests a role for ARRDC3 gene in human papillomavirus infection. Hum. Mol. Genet. 2019, 28, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Ishigaki, K.; Akiyama, M.; Kanai, M.; Takahashi, A.; Kawakami, E.; Sugishita, H.; Sakaue, S.; Matoba, N.; Low, S.-K.; Okada, Y.; et al. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat. Genet. 2020, 52, 669–679. [Google Scholar] [CrossRef]
- Bowden, S.J.; Bodinier, B.; Kalliala, I.; Zuber, V.; Vuckovic, D.; Doulgeraki, T.; Whitaker, M.D.; Wielscher, M.; Cartwright, R.; Tsilidis, K.K.; et al. Genetic variation in cervical preinvasive and invasive disease: A genome-wide association study. Lancet Oncol. 2021, 22, 548–557. [Google Scholar] [CrossRef]
- Koel, M.; Võsa, U.; Lepamets, M.; Laivuori, H.; Lemmelä, S.; Daly, M.; Biobank Research Team, E.; Palta, P.; Mägi, R.; Laisk, T. GWAS meta-analysis and gene expression data link reproductive tract development, immune response and cellular proliferation/apoptosis with cervical cancer and clarify overlap with other cervical phenotypes. medRxiv 2021. [Google Scholar] [CrossRef]
- Olafsdottir, T.; Stacey, S.N.; Sveinbjornsson, G.; Thorleifsson, G.; Norland, K.; Sigurgeirsson, B.; Thorisdottir, K.; Kristjansson, A.K.; Tryggvadottir, L.; Sarin, K.Y.; et al. Loss-of-Function Variants in the Tumor-Suppressor Gene PTPN14 Confer Increased Cancer Risk. Cancer Res. 2021, 81, 1954–1964. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Low, S.K.; Akiyama, M.; Hirata, M.; Ueda, Y.; Matsuda, K.; Kimura, T.; Murakami, Y.; Kubo, M.; Kamatani, Y.; et al. GWAS of five gynecologic diseases and cross-trait analysis in Japanese. Eur. J. Hum. Genet. 2019, 95–107. [Google Scholar] [CrossRef]
- Park, I.; Terasaki, P. Origins of the first HLA specificities. Hum. Immunol. 2000, 61, 185–189. [Google Scholar] [CrossRef]
- Horton, R.; Wilming, L.; Rand, V.; Lovering, R.C.; Bruford, E.A.; Khodiyar, V.K.; Lush, M.J.; Povey, S.; Talbot, C.C.; Wright, M.W.; et al. Gene map of the extended human MHC. Nat. Rev. Genet. 2004, 5, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Dendrou, C.A.; Petersen, J.; Rossjohn, J.; Fugger, L. HLA variation and disease. Nat. Rev. Immunol. 2018, 18, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Gyllensten, U. A cis-eQTL of HLA-DRB1 and a frameshift mutation of MICA contribute to the pattern of association of HLA alleles with cervical cancer. Cancer Med. 2014, 3, 445–452. [Google Scholar] [CrossRef]
- Ashiru, O.; Boutet, P.; Fernández-Messina, L.; Agüera-González, S.; Skepper, J.N.; Valés-Gómez, M.; Reyburn, H.T. Natural Killer Cell Cytotoxicity Is Suppressed by Exposure to the Human NKG2D Ligand MICA*008 That Is Shed by Tumor Cells in Exosomes. Cancer Res. 2010, 70, 481–489. [Google Scholar] [CrossRef]
- Chen, D.; Enroth, S.; Ivansson, E.; Gyllensten, U. Pathway analysis of cervical cancer genome-wide association study highlights the MHC region and pathways involved in response to infection. Hum. Mol. Genet. 2014, 23, 6047–6060. [Google Scholar] [CrossRef]
- McKay, J.; Tenet, V.; Franceschi, S.; Chabrier, A.; Gheit, T.; Gaborieau, V.; Chopin, S.; Avogbe, P.H.; Tommasino, M.; Ainouze, M.; et al. Immuno-related polymorphisms and cervical cancer risk: The IARC multicentric case-control study. PLoS ONE 2017, 12, e0177775. [Google Scholar] [CrossRef]
- Ramachandran, D.; Schürmann, P.; Mao, Q.; Wang, Y.; Bretschneider, L.M.; Speith, L.M.; Hülse, F.; Enßen, J.; Bousset, K.; Jentschke, M.; et al. Association of genomic variants at the human leukocyte antigen locus with cervical cancer risk, HPV status and gene expression levels. Int. J. Cancer 2020, 147, 2458–2468. [Google Scholar] [CrossRef]
- Chen, D.; Gyllensten, U. Systematic investigation of contribution of genetic variation in the HLA-DP region to cervical cancer susceptibility. Carcinogenesis 2014, 35, 1765–1769. [Google Scholar] [CrossRef] [PubMed]
- Gregoire, L.; Lawrence, W.D.; Kukuruga, D.; Eisenbrey, A.B.; Lancaster, W.D. Association between HLA-DQB1 alleles and risk for cervical cancer in African-American women. Int. J. Cancer 1994, 57, 504–507. [Google Scholar] [CrossRef]
- Bao, X.; Hanson, A.L.; Madeleine, M.M.; Wang, S.S.; Schwartz, S.M.; Newell, F.; Pettersson-Kymmer, U.; Hemminki, K.; Tiews, S.; Steinberg, W.; et al. HLA and KIR Associations of Cervical Neoplasia. J. Infect. Dis. 2018, 218, 2006–2015. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, G.R.; Olivieri, J.E.; DeBoever, C.; Tanigawa, Y.; Justesen, J.M.; Dilthey, A.; Rivas, M.A. Pervasive additive and non-additive effects within the HLA region contribute to disease risk in the UK Biobank. BioRxiv 2020, 1–19. [Google Scholar] [CrossRef]
- Engelmark, M. Affected sib-pair analysis of the contribution of HLA class I and class II loci to development of cervical cancer. Hum. Mol. Genet. 2004, 13, 1951–1958. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, Y.; Bai, Y. Role of GSDMB in Pyroptosis and Cancer. Cancer Manag. Res. 2020, 12, 3033–3043. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; He, H.; Wang, K.; Shi, X.; Wang, Y.; Su, Y.; Wang, Y.; Li, D.; Liu, W.; Zhang, Y.; et al. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science 2020, 368, eaaz7548. [Google Scholar] [CrossRef]
- He, H.; Yi, L.; Zhang, B.; Yan, B.; Xiao, M.; Ren, J.; Zi, D.; Zhu, L.; Zhong, Z.; Zhao, X.; et al. USP24-GSDMB complex promotes bladder cancer proliferation via activation of the STAT3 pathway. Int. J. Biol. Sci. 2021, 17, 2417–2429. [Google Scholar] [CrossRef]
- Sun, Q.; Yang, J.; Xing, G.; Sun, Q.; Zhang, L.; He, F. Expression of GSDML Associates with Tumor Progression in Uterine Cervix Cancer. Transl. Oncol. 2008, 1, 73-IN1. [Google Scholar] [CrossRef]
- Ramachandran, D.; Wang, Y.; Schürmann, P.; Hülse, F.; Mao, Q.; Jentschke, M.; Böhmer, G.; Strauß, H.G.; Hirchenhain, C.; Schmidmayr, M.; et al. Association of genomic variants at PAX8 and PBX2 with cervical cancer risk. Int. J. Cancer 2021, 149, 893–900. [Google Scholar] [CrossRef]
- Han, J.; Zhou, W.; Jia, M.; Wen, J.; Jiang, J.; Shi, J.; Zhang, K.; Ma, H.; Liu, J.; Ren, J.; et al. Expression quantitative trait loci in long non-coding RNA PAX8-AS1 are associated with decreased risk of cervical cancer. Mol. Genet. Genom. 2016, 291, 1743–1748. [Google Scholar] [CrossRef]
- Kar, S.P.; Adler, E.; Tyrer, J.; Hazelett, D.; Anton-Culver, H.; Bandera, E.V.; Beckmann, M.W.; Berchuck, A.; Bogdanova, N.; Brinton, L.; et al. Enrichment of putative PAX8 target genes at serous epithelial ovarian cancer susceptibility loci. Br. J. Cancer 2017, 116, 524–535. [Google Scholar] [CrossRef]
- Beesley, J.; Pickett, H.A.; Johnatty, S.E.; Dunning, A.M.; Chen, X.; Li, J.; Michailidou, K.; Lu, Y.; Rider, D.N.; Palmieri, R.T.; et al. Functional polymorphisms in the TERT promoter are associated with risk of serous epithelial ovarian and breast cancers. PLoS ONE 2011, 6, e24987. [Google Scholar] [CrossRef]
- O’Mara, T.A.; Glubb, D.M.; Amant, F.; Annibali, D.; Ashton, K.; Attia, J.; Auer, P.L.; Beckmann, M.W.; Black, A.; Bolla, M.K.; et al. Identification of nine new susceptibility loci for endometrial cancer. Nat. Commun. 2018, 9, 3166. [Google Scholar] [CrossRef]
- Haiman, C.A.; Chen, G.K.; Vachon, C.M.; Canzian, F.; Dunning, A.; Millikan, R.C.; Wang, X.; Ademuyiwa, F.; Ahmed, S.; Ambrosone, C.B.; et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor–negative breast cancer. Nat. Genet. 2011, 43, 1210–1214. [Google Scholar] [CrossRef] [PubMed]
- Rafnar, T.; Sulem, P.; Stacey, S.N.; Geller, F.; Gudmundsson, J.; Sigurdsson, A.; Jakobsdottir, M.; Helgadottir, H.; Thorlacius, S.; Aben, K.K.H.; et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 2009, 41, 221–227. [Google Scholar] [CrossRef]
- Miura, K.; Mishima, H.; Kinoshita, A.; Hayashida, C.; Abe, S.; Tokunaga, K.; Masuzaki, H.; Yoshiur, K. Genome-Wide Association Study of HPV-Associated Cervical Cancer in Japanese Women. J. Med. Virol. 2014, 86, 1153–1158. [Google Scholar] [CrossRef] [PubMed]
- Batista, T.M.; Dagdeviren, S.; Carroll, S.H.; Cai, W.; Melnik, V.Y.; Noh, H.L.; Saengnipanthkul, S.; Kim, J.K.; Kahn, C.R.; Lee, R.T. Arrestin domain-containing 3 (Arrdc3) modulates insulin action and glucose metabolism in liver. Proc. Natl. Acad. Sci. USA 2020, 117, 6733–6740. [Google Scholar] [CrossRef]
- Arakaki, A.K.S.; Pan, W.-A.; Wedegaertner, H.; Roca-Mercado, I.; Chinn, L.; Gujral, T.S.; Trejo, J. α-Arrestin ARRDC3 tumor suppressor function is linked to GPCR-induced TAZ activation and breast cancer metastasis. J. Cell Sci. 2021, 134, jcs254888. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Ito, H.; Hirata, J.; Sakaue, S.; Ueda, Y.; Kimura, T.; Takeuchi, F.; Murakami, Y.; Matsuda, K.; Matsuo, K.; et al. Fine Mapping of the Major Histocompatibility Complex Region and Association of the HLA-B*52:01 Allele with Cervical Cancer in Japanese Women. JAMA Netw. Open 2020, 3, 1–9. [Google Scholar] [CrossRef]
- Riancho, J.A.; Vázquez, L.; García-Pérez, M.A.; Sainz, J.; Olmos, J.M.; Hernández, J.L.; Pérez-López, J.; Amado, J.A.; Zarrabeitia, M.T.; Cano, A.; et al. Association of ACACB polymorphisms with obesity and diabetes. Mol. Genet. Metab. 2011, 104, 670–676. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, K.; Zhang, Q.; Zhou, J.; Sun, H.; Tang, F.; Zhou, H.; Hu, T.; Wang, S.; Jia, Y.; et al. Genome-wide association study identifies four SNPs associated with response to platinum-based neoadjuvant chemotherapy for cervical cancer. Sci. Rep. 2017, 7, 41103. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, M.; Pozzoli, U.; Cagliani, R.; Comi, G.P.; Bresolin, N.; Clerici, M.; Sironi, M. Genome-Wide Identification of Susceptibility Alleles for Viral Infections through a Population Genetics Approach. PLoS Genet. 2010, 6, e1000849. [Google Scholar] [CrossRef]
- Chen, D.; McKay, J.D.; Clifford, G.; Gaborieau, V.; Chabrier, A.; Waterboer, T.; Zaridze, D.; Lissowska, J.; Rudnai, P.; Fabianova, E.; et al. Genome-wide association study of HPV seropositivity. Hum. Mol. Genet. 2011, 20, 4714–4723. [Google Scholar] [CrossRef]
- Adebamowo, S.N.; Adeyemo, A.A.; Rotimi, C.N.; Olaniyan, O.; Offiong, R.; Adebamowo, C.A.; Odutola, M.; Dareng, E.O.; Famooto, A.O.; Adebiyi, R. Genome-wide association study of prevalent and persistent cervical high-risk human papillomavirus (HPV) infection. BMC Med. Genet. 2020, 21, 1–10. [Google Scholar] [CrossRef]
- Liu, J.; Tang, W.; Budhu, A.; Forgues, M.; Hernandez, M.O.; Candia, J.; Kim, Y.; Bowman, E.D.; Ambs, S.; Zhao, Y.; et al. A Viral Exposure Signature Defines Early Onset of Hepatocellular Carcinoma. Cell 2020, 182, 317–328. [Google Scholar] [CrossRef]
- Chen, H.; Wang, T.; Huang, S.; Zeng, P. New novel non-MHC genes were identified for cervical cancer with an integrative analysis approach of transcriptome-wide association study. J. Cancer 2021, 12, 840–848. [Google Scholar] [CrossRef]
- Campbell, J.D.; Yau, C.; Bowlby, R.; Liu, Y.; Brennan, K.; Fan, H.; Taylor, A.M.; Wang, C.; Walter, V.; Akbani, R.; et al. Genomic, Pathway Network, and Immunologic Features Distinguishing Squamous Carcinomas. Cell Rep. 2018, 23, 194–212. [Google Scholar] [CrossRef] [PubMed]
- Kachuri, L.; Graff, R.E.; Smith-Byrne, K.; Meyers, T.J.; Rashkin, S.R.; Ziv, E.; Witte, J.S.; Johansson, M. Pan-cancer analysis demonstrates that integrating polygenic risk scores with modifiable risk factors improves risk prediction. Nat. Commun. 2020, 11, 6084. [Google Scholar] [CrossRef] [PubMed]
- Mavaddat, N.; Michailidou, K.; Dennis, J.; Lush, M.; Fachal, L.; Lee, A.; Tyrer, J.P.; Chen, T.H.; Wang, Q.; Bolla, M.K.; et al. Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. Am. J. Hum. Genet. 2019, 104, 21–34. [Google Scholar] [CrossRef]
- Franceschi, S. Genomic characterisation of cervical cancer and human papillomavirus: New opportunities for precision medicine. Lancet Oncol. 2021, 22, 419–420. [Google Scholar] [CrossRef]
- Masuda, T.; Ogawa, K.; Kamatani, Y.; Murakami, Y.; Kimura, T.; Okada, Y. A Mendelian randomization study identified obesity as a causal risk factor of uterine endometrial cancer in Japanese. Cancer Sci. 2020, 111, 4646–4651. [Google Scholar] [CrossRef] [PubMed]
- Seddighi, S.; Houck, A.L.; Rowe, J.B.; Pharoah, P.D.P. Evidence of a Causal Association Between Cancer and Alzheimer’s Disease: A Mendelian Randomization Analysis. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Kar, S.; Carter, P.; Vithayathil, M.; Mason, A.M.; Burgess, S.; Larsson, S.C. Is type 2 diabetes causally associated with cancer risk? Evidence from a two-sample mendelian randomization study. Diabetes 2020, 69, 1588–1596. [Google Scholar] [CrossRef]
- Thorsby, E. Invited anniversary review: HLA associated diseases. Hum. Immunol. 1997, 53, 1–11. [Google Scholar] [CrossRef]
- Trowsdale, J.; Knight, J.C. Major Histocompatibility Complex Genomics and Human Disease. Annu. Rev. Genom. Hum. Genet. 2013, 14, 301–323. [Google Scholar] [CrossRef]
- Liu, Z.; Derkach, A.; Yu, K.J.; Yeager, M.; Chang, Y.-S.; Chen, C.-J.; Gyllensten, U.; Lan, Q.; Lee, M.-H.; McKay, J.D.; et al. Patterns of Human Leukocyte Antigen Class I and Class II Associations and Cancer. Cancer Res. 2021, 81, 1148–1152. [Google Scholar] [CrossRef]
- Chen, D.; Hammer, J.; Lindquist, D.; Idahl, A.; Gyllensten, U. A variant upstream of HLA-DRB1 and multiple variants in MICA influence susceptibility to cervical cancer in a Swedish population. Cancer Med. 2014, 3, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Pereyra, F.; Jia, X.; McLaren, P.J.; Telenti, A.; de Bakker, P.I.W.; Walker, B.D.; Ripke, S.; Brumme, C.J.; Pulit, S.L.; Carrington, M.; et al. The Major Genetic Determinants of HIV-1 Control Affect HLA Class I Peptide Presentation. Science 2010, 330, 1551–1557. [Google Scholar] [CrossRef]
- McLaren, P.J.; Fellay, J. HIV-1 and human genetic variation. Nat. Rev. Genet. 2021, 22, 645–657. [Google Scholar] [CrossRef]
- Monteiro, A.N.A.; Freedman, M.L. Lessons from postgenome-wide association studies: Functional analysis of cancer predisposition loci. J. Intern. Med. 2013, 274, 414–424. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).