Newly-Discovered Neural Features Expand the Pathobiological Knowledge of Blastic Plasmacytoid Dendritic Cell Neoplasm
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Case Collection and Study Design
2.1.1. RNA Sequencing Validation Sets
2.1.2. H3K27me3/ac ChIP Sequencing Independent Validation Set
2.2. MicroRNA Expression Profiling Discovery Set
2.3. MicroRNA Expression Quantification by Quantitative Reverse Transcription PCR (RT-qPCR)
2.4. BPDCN Gene Expression Profiling Analysis Discovery Set
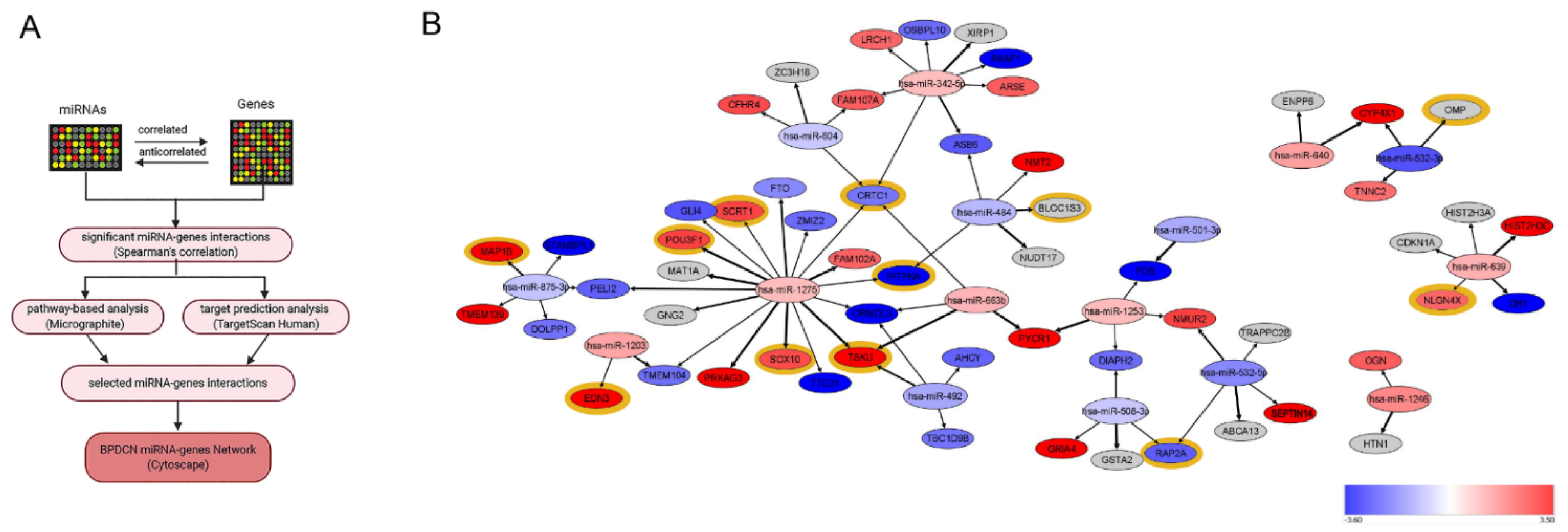
2.5. MicroRNA Network Analysis
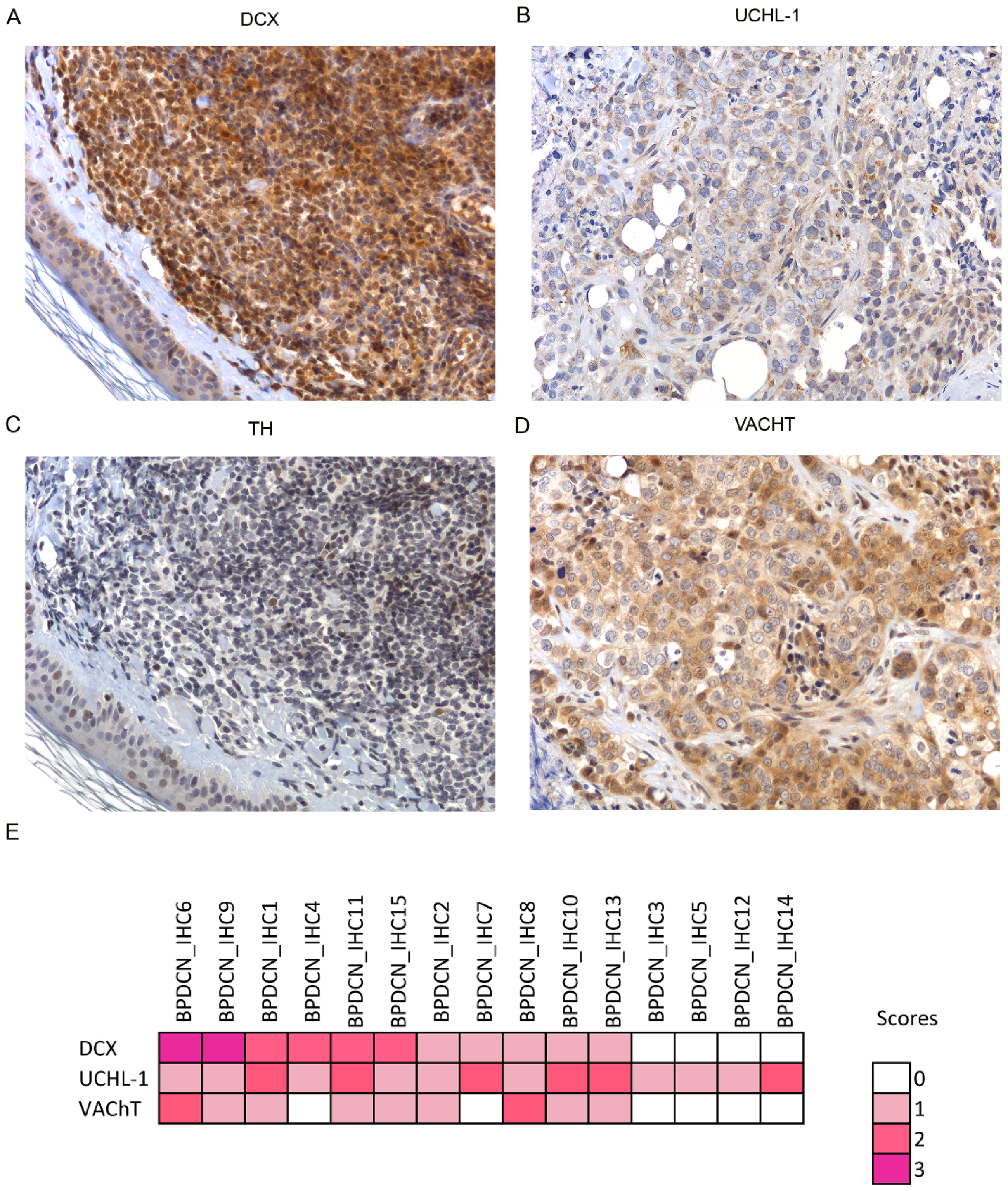
2.6. Immunohistochemistry Validation Set
3. Results
3.1. BPDCN Cases Display a Set of 51 miRNAs Differentially Expressed from Normal pDCs
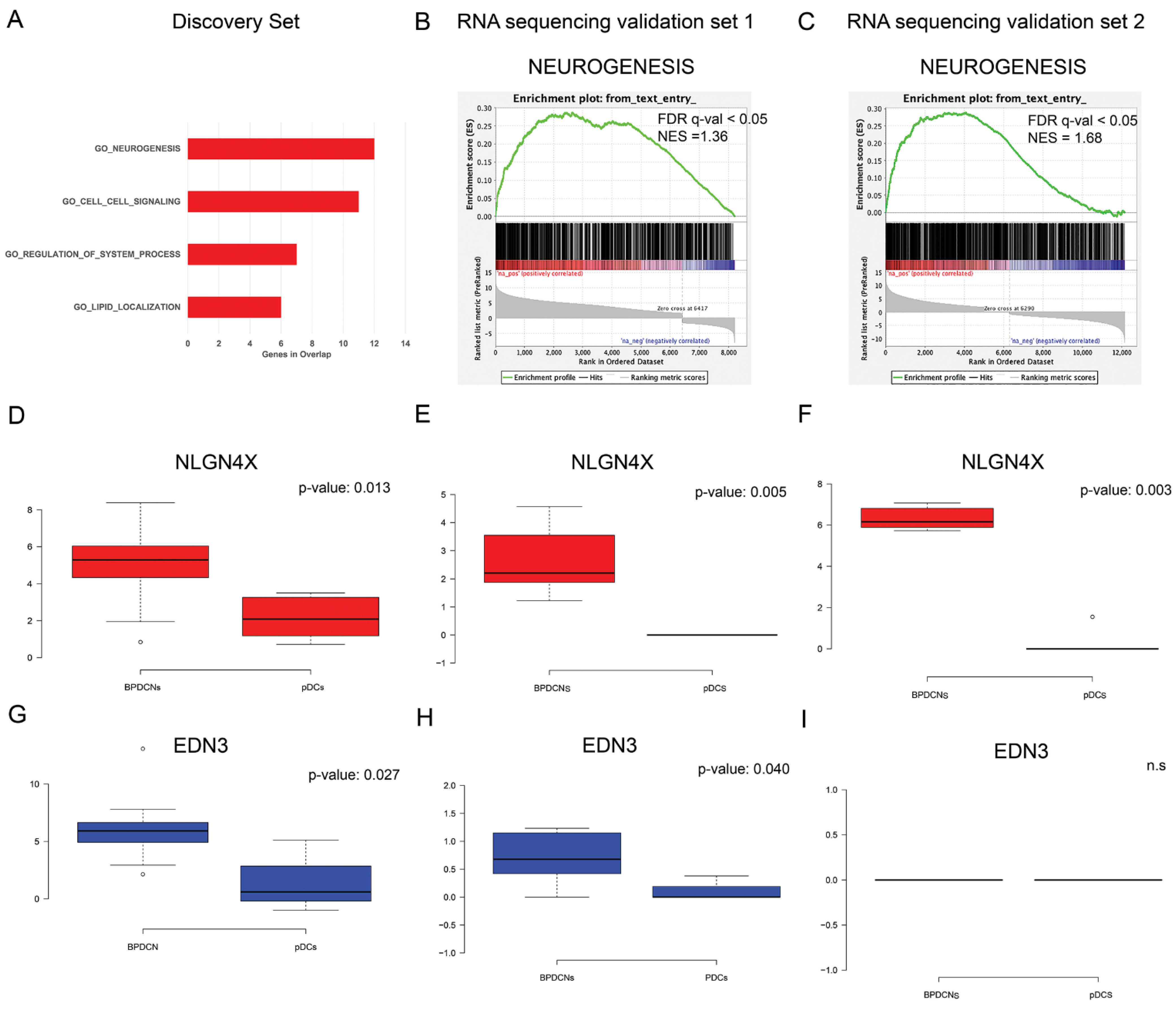
3.2. BPDCN miRNA-mRNA Regulatory Network May Primarily Affect Neurogenesis
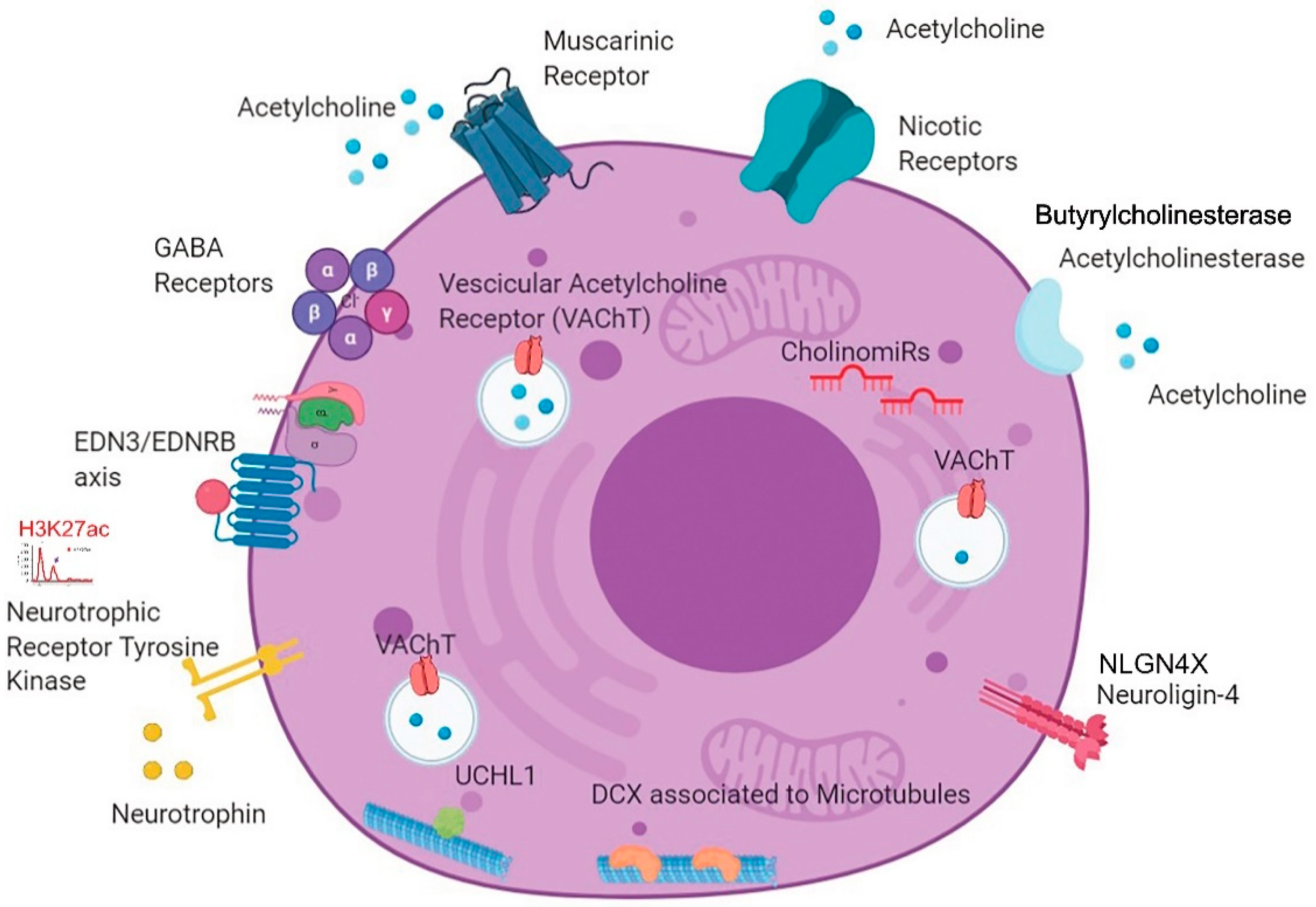
3.3. Unexpected Neuronal Features in BPDCN
3.4. A New BPDCN Cell Model
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J. (Eds.) WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. In World Health Organization Classification of Tumours, 4th ed.; International Agency for Research on Cancer: Lyon, France, 2017; p. 173. ISBN 9789283244943. [Google Scholar]
- Martín-Martín, L.; Almeida, J.; Pomares, H.; González-Barca, E.; Bravo, P.; Giménez, T.; Heras, C.; Queizán, J.-A.; Pérez-Ceballos, E.; Martínez, V.; et al. Blastic Plasmacytoid Dendritic Cell Neoplasm Frequently Shows Occult Central Nervous System Involvement at Diagnosis and Benefits from Intrathecal Therapy. Oncotarget 2016, 7, 10174–10181. [Google Scholar] [CrossRef] [Green Version]
- Assaf, C.; Gellrich, S.; Whittaker, S.; Robson, A.; Cerroni, L.; Massone, C.; Kerl, H.; Rose, C.; Chott, A.; Chimenti, S.; et al. CD56-Positive Haematological Neoplasms of the Skin: A Multicentre Study of the Cutaneous Lymphoma Project Group of the European Organisation for Research and Treatment of Cancer. J. Clin. Pathol. 2006, 60, 981–989. [Google Scholar] [CrossRef]
- Campo, E.; Swerdlow, S.H.; Harris, N.L.; Pileri, S.; Stein, H.; Jaffe, E.S. The 2008 WHO Classification of Lymphoid Neoplasms and beyond: Evolving Concepts and Practical Applications. Blood 2011, 117, 5019–5032. [Google Scholar] [CrossRef] [Green Version]
- Pagano, L.; Valentini, C.G.; Grammatico, S.; Pulsoni, A. Blastic Plasmacytoid Dendritic Cell Neoplasm: Diagnostic Criteria and Therapeutical Approaches. Br. J. Haematol. 2016, 174, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Lucioni, M.; Novara, F.; Fiandrino, G.; Riboni, R.; Fanoni, D.; Arra, M.; Venegoni, L.; Nicola, M.; Dallera, E.; Arcaini, L.; et al. Twenty-One Cases of Blastic Plasmacytoid Dendritic Cell Neoplasm: Focus on Biallelic Locus 9p21.3 Deletion. Blood 2011, 118, 4591–4594. [Google Scholar] [CrossRef] [Green Version]
- Sapienza, M.R.; Fuligni, F.; Agostinelli, C.; Tripodo, C.; Righi, S.; Laginestra, M.A.; Pileri, A.; Mancini, M.; Rossi, M.; Ricci, F.; et al. Molecular Profiling of Blastic Plasmacytoid Dendritic Cell Neoplasm Reveals a Unique Pattern and Suggests Selective Sensitivity to NF-KB Pathway Inhibition. Leukemia 2014, 28, 1606–1616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceroi, A.; Masson, D.; Roggy, A.; Roumier, C.; Chagué, C.; Gauthier, T.; Philippe, L.; Lamarthée, B.; Angelot-Delettre, F.; Bonnefoy, F.; et al. LXR Agonist Treatment of Blastic Plasmacytoid Dendritic Cell Neoplasm Restores Cholesterol Efflux and Triggers Apoptosis. Blood 2016, 128, 2694–2707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menezes, J.; Acquadro, F.; Wiseman, M.; Gómez-López, G.; Salgado, R.N.; Talavera-Casañas, J.G.; Buño, I.; Cervera, J.V.; Montes-Moreno, S.; Hernández-Rivas, J.M.; et al. Exome Sequencing Reveals Novel and Recurrent Mutations with Clinical Impact in Blastic Plasmacytoid Dendritic Cell Neoplasm. Leukemia 2014, 28, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Sapienza, M.R.; Abate, F.; Melle, F.; Orecchioni, S.; Fuligni, F.; Etebari, M.; Tabanelli, V.; Laginestra, M.A.; Pileri, A.; Motta, G.; et al. Blastic Plasmacytoid Dendritic Cell Neoplasm: Genomics Mark Epigenetic Dysregulation as a Primary Therapeutic Target. Haematologica 2019, 104, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Ceribelli, M.; Hou, Z.E.; Kelly, P.N.; Huang, D.W.; Wright, G.; Ganapathi, K.; Evbuomwan, M.O.; Pittaluga, S.; Shaffer, A.L.; Marcucci, G.; et al. A Druggable TCF4- and BRD4-Dependent Transcriptional Network Sustains Malignancy in Blastic Plasmacytoid Dendritic Cell Neoplasm. Cancer Cell 2016, 30, 764–778. [Google Scholar] [CrossRef] [Green Version]
- Montero, J.; Stephansky, J.; Cai, T.; Griffin, G.K.; Cabal-Hierro, L.; Togami, K.; Hogdal, L.J.; Galinsky, I.; Morgan, E.A.; Aster, J.C.; et al. Blastic Plasmacytoid Dendritic Cell Neoplasm Is Dependent on BCL2 and Sensitive to Venetoclax. Cancer Discov. 2017, 7, 156–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pemmaraju, N.; Lane, A.A.; Sweet, K.L.; Stein, A.S.; Vasu, S.; Blum, W.; Rizzieri, D.A.; Wang, E.S.; Duvic, M.; Sloan, J.M.; et al. Tagraxofusp in Blastic Plasmacytoid Dendritic-Cell Neoplasm. N. Engl. J. Med. 2019, 380, 1628–1637. [Google Scholar] [CrossRef]
- Sapienza, M.R.; Fuligni, F.; Melle, F.; Tabanelli, V.; Indio, V.; Laginestra, M.A.; Motta, G.; Mazzara, S.; Cerroni, L.; Pileri, A.; et al. MicroRNA Profiling of Blastic Plasmacytoid Dendritic Cell Neoplasm and Myeloid Sarcoma. Hematol. Oncol. 2020, 38, 831–833. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.-G.; Alder, H.; et al. MicroRNA Signatures in Human Ovarian Cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef] [Green Version]
- Falini, B.; Flenghi, L.; Pileri, S.; Gambacorta, M.; Bigerna, B.; Durkop, H.; Eitelbach, F.; Thiele, J.; Pacini, R.; Cavaliere, A. PG-M1: A New Monoclonal Antibody Directed against a Fixative-Resistant Epitope on the Macrophage-Restricted Form of the CD68 Molecule. Am. J. Pathol. 1993, 142, 1359–1372. [Google Scholar] [PubMed]
- Sales, G.; Calura, E.; Cavalieri, D.; Romualdi, C. Graphite—A Bioconductor Package to Convert Pathway Topology to Gene Network. BMC Bioinform. 2012, 13, 20. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting Effective MicroRNA Target Sites in Mammalian MRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Shannon, P. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. MiRNet 2.0: Network-Based Visual Analytics for MiRNA Functional Analysis and Systems Biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef]
- Xie, B.; Ding, Q.; Han, H.; Wu, D. MiRCancer: A MicroRNA-Cancer Association Database Constructed by Text Mining on Literature. Bioinformatics 2013, 29, 638–644. [Google Scholar] [CrossRef]
- Huang, X.; Schwind, S.; Santhanam, R.; Eisfeld, A.-K.; Chiang, C.; Lankenau, M.; Yu, B.; Hoellerbauer, P.; Jin, Y.; Tarighat, S.S.; et al. Targeting the RAS/MAPK Pathway with MiR-181a in Acute Myeloid Leukemia. Oncotarget 2016, 7, 59273–59286. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Guo, B.; Chen, Z.; Wang, N.; Iacovino, M.; Cheng, J.; Roden, C.; Pan, W.; Khan, S.; Chen, S.; et al. MiR-125b Promotes MLL-AF9–Driven Murine Acute Myeloid Leukemia Involving a VEGFA-Mediated Non–Cell-Intrinsic Mechanism. Blood 2017, 129, 1491–1502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallace, J.A.; O’Connell, R.M. MicroRNAs and Acute Myeloid Leukemia: Therapeutic Implications and Emerging Concepts. Blood 2017, 130, 1290–1301. [Google Scholar] [CrossRef] [Green Version]
- Inui, M.; Martello, G.; Piccolo, S. MicroRNA Control of Signal Transduction. Nat. Rev. Mol. Cell Biol. 2010, 11, 252–263. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Henderson, H.J.; Karanam, B.; Samant, R.; Vig, K.; Singh, S.R.; Yates, C.; Bedi, D. Neuroligin 4X Overexpression in Human Breast Cancer Is Associated with Poor Relapse-Free Survival. PLoS ONE 2017, 12, e0189662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhalla, A.; Haque, S.; Taylor, I.; Winslet, M.; Loizidou, M. Endothelin Receptor Antagonism and Cancer. Eur. J. Clin. Investig. 2009, 39, 74–77. [Google Scholar] [CrossRef]
- Liu, Y.; Ye, F.; Yamada, K.; Tso, J.L.; Zhang, Y.; Nguyen, D.H.; Dong, Q.; Soto, H.; Choe, J.; Dembo, A.; et al. Autocrine Endothelin-3/Endothelin Receptor B Signaling Maintains Cellular and Molecular Properties of Glioblastoma Stem Cells. Mol. Cancer Res. 2011, 9, 1668–1685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mauffrey, P.; Tchitchek, N.; Barroca, V.; Bemelmans, A.-P.; Firlej, V.; Allory, Y.; Roméo, P.-H.; Magnon, C. Progenitors from the Central Nervous System Drive Neurogenesis in Cancer. Nature 2019, 569, 672–678. [Google Scholar] [CrossRef]
- Hayakawa, Y.; Sakitani, K.; Konishi, M.; Asfaha, S.; Niikura, R.; Tomita, H.; Renz, B.W.; Tailor, Y.; Macchini, M.; Middelhoff, M.; et al. Nerve Growth Factor Promotes Gastric Tumorigenesis through Aberrant Cholinergic Signaling. Cancer Cell 2017, 31, 21–34. [Google Scholar] [CrossRef] [Green Version]
- Sloan, E.K.; Priceman, S.J.; Cox, B.F.; Yu, S.; Pimentel, M.A.; Tangkanangnukul, V.; Arevalo, J.M.G.; Morizono, K.; Karanikolas, B.D.W.; Wu, L.; et al. The Sympathetic Nervous System Induces a Metastatic Switch in Primary Breast Cancer. Cancer Res. 2010, 70, 7042–7052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayanlaja, A.A.; Xiong, Y.; Gao, Y.; Ji, G.; Tang, C.; Abdikani Abdullah, Z.; Gao, D. Distinct Features of Doublecortin as a Marker of Neuronal Migration and Its Implications in Cancer Cell Mobility. Front. Mol. Neurosci. 2017, 10, 199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakurai, M.; Ayukawa, K.; Setsuie, R.; Nishikawa, K.; Hara, Y.; Ohashi, H.; Nishimoto, M.; Abe, T.; Kudo, Y.; Sekiguchi, M.; et al. Ubiquitin C-Terminal Hydrolase L1 Regulates the Morphology of Neural Progenitor Cells and Modulates Their Differentiation. J. Cell Sci. 2006, 119, 162–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locher, H.; de Rooij, K.E.; de Groot, J.C.M.J.; van Doorn, R.; Gruis, N.A.; Löwik, C.W.G.M.; Chuva de Sousa Lopes, S.M.; Frijns, J.H.M.; Huisman, M.A. Class III β-Tubulin, a Novel Biomarker in the Human Melanocyte Lineage. Differentiation 2013, 85, 173–181. [Google Scholar] [CrossRef]
- Rao, F.; Zhang, L.; Wessel, J.; Zhang, K.; Wen, G.; Kennedy, B.P.; Rana, B.K.; Das, M.; Rodriguez-Flores, J.L.; Smith, D.W.; et al. Tyrosine Hydroxylase, the Rate-Limiting Enzyme in Catecholamine Biosynthesis: Discovery of Common Human Genetic Variants Governing Transcription, Autonomic Activity, and Blood Pressure In Vivo. Circulation 2007, 116, 993–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Castro, B.M.; De Jaeger, X.; Martins-Silva, C.; Lima, R.D.F.; Amaral, E.; Menezes, C.; Lima, P.; Neves, C.M.L.; Pires, R.G.; Gould, T.W.; et al. The Vesicular Acetylcholine Transporter Is Required for Neuromuscular Development and Function. Mol. Cell. Biol. 2009, 29, 5238–5250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picciotto, M.R.; Higley, M.J.; Mineur, Y.S. Acetylcholine as a Neuromodulator: Cholinergic Signaling Shapes Nervous System Function and Behavior. Neuron 2012, 76, 116–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadorp, B.; Soreq, H. Predicted Overlapping MicroRNA Regulators of Acetylcholine Packaging and Degradation in Neuroinflammation-Related Disorders. Front. Mol. Neurosci. 2014, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dijkman, R.; van Doorn, R.; Szuhai, K.; Willemze, R.; Vermeer, M.H.; Tensen, C.P. Gene-Expression Profiling and Array-Based CGH Classify CD4+CD56+ Hematodermic Neoplasm and Cutaneous Myelomonocytic Leukemia as Distinct Disease Entities. Blood 2007, 109, 1720–1727. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Kim, Y.M.; Lim, S.; Nam, Y.K.; Jeong, J.; Kim, H.-J.; Lee, K.-J. Ubiquitin C-Terminal Hydrolase-L1 Is a Key Regulator of Tumor Cell Invasion and Metastasis. Oncogene 2009, 28, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Kawashima, K.; Fujii, T.; Moriwaki, Y.; Misawa, H. Critical Roles of Acetylcholine and the Muscarinic and Nicotinic Acetylcholine Receptors in the Regulation of Immune Function. Life Sci. 2012, 91, 1027–1032. [Google Scholar] [CrossRef]
- Greig, N.H.; Reale, M.; Tata, A.M. New Pharmacological Approaches to the Cholinergic System: An Overview on Muscarinic Receptor Ligands and Cholinesterase Inhibitors. Recent Pat. CNS Drug Discov. 2013, 8, 123–141. [Google Scholar] [CrossRef]
- Song, P.; Sekhon, H.S.; Jia, Y.; Keller, J.A.; Blusztajn, J.K.; Mark, G.P.; Spindel, E.R. Acetylcholine Is Synthesized by and Acts as an Autocrine Growth Factor for Small Cell Lung Carcinoma. Cancer Res. 2003, 63, 214–221. [Google Scholar]
- Cox, M.A.; Duncan, G.S.; Lin, G.H.Y.; Steinberg, B.E.; Yu, L.X.; Brenner, D.; Buckler, L.N.; Elia, A.J.; Wakeham, A.C.; Nieman, B.; et al. Choline Acetyltransferase–Expressing T Cells Are Required to Control Chronic Viral Infection. Science 2019, 363, 639–644. [Google Scholar] [CrossRef]
- Renosi, F.; Roggy, A.; Giguelay, A.; Soret, L.; Viailly, P.-J.; Cheok, M.; Biichle, S.; Angelot-Delettre, F.; Asnafi, V.; Macintyre, E.; et al. Transcriptomic and Genomic Heterogeneity in Blastic Plasmacytoid Dendritic Cell Neoplasms: From Ontogeny to Oncogenesis. Blood Adv. 2021, 5, 1540–1551. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.K.; Davare, M.A.; Druker, B.J.; Tognon, C.E. Revisiting NTRKs as an Emerging Oncogene in Hematological Malignancies. Leukemia 2019, 33, 2563–2574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chavan, S.S.; Pavlov, V.A.; Tracey, K.J. Mechanisms and Therapeutic Relevance of Neuro-Immune Communication. Immunity 2017, 46, 927–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sapienza, M.R.; Benvenuto, G.; Ferracin, M.; Mazzara, S.; Fuligni, F.; Tripodo, C.; Belmonte, B.; Fanoni, D.; Melle, F.; Motta, G.; et al. Newly-Discovered Neural Features Expand the Pathobiological Knowledge of Blastic Plasmacytoid Dendritic Cell Neoplasm. Cancers 2021, 13, 4680. https://doi.org/10.3390/cancers13184680
Sapienza MR, Benvenuto G, Ferracin M, Mazzara S, Fuligni F, Tripodo C, Belmonte B, Fanoni D, Melle F, Motta G, et al. Newly-Discovered Neural Features Expand the Pathobiological Knowledge of Blastic Plasmacytoid Dendritic Cell Neoplasm. Cancers. 2021; 13(18):4680. https://doi.org/10.3390/cancers13184680
Chicago/Turabian StyleSapienza, Maria Rosaria, Giuseppe Benvenuto, Manuela Ferracin, Saveria Mazzara, Fabio Fuligni, Claudio Tripodo, Beatrice Belmonte, Daniele Fanoni, Federica Melle, Giovanna Motta, and et al. 2021. "Newly-Discovered Neural Features Expand the Pathobiological Knowledge of Blastic Plasmacytoid Dendritic Cell Neoplasm" Cancers 13, no. 18: 4680. https://doi.org/10.3390/cancers13184680





