A Perspective on the Role of Microbiome for Colorectal Cancer Treatment
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. Epidemiology
1.2. The Microbiome
1.3. Treatment for CRC
2. Effect of Microbiome on Chemotherapy Efficacy and Toxicity
2.1. Effects of Microbiome on Chemotherapy Efficacy
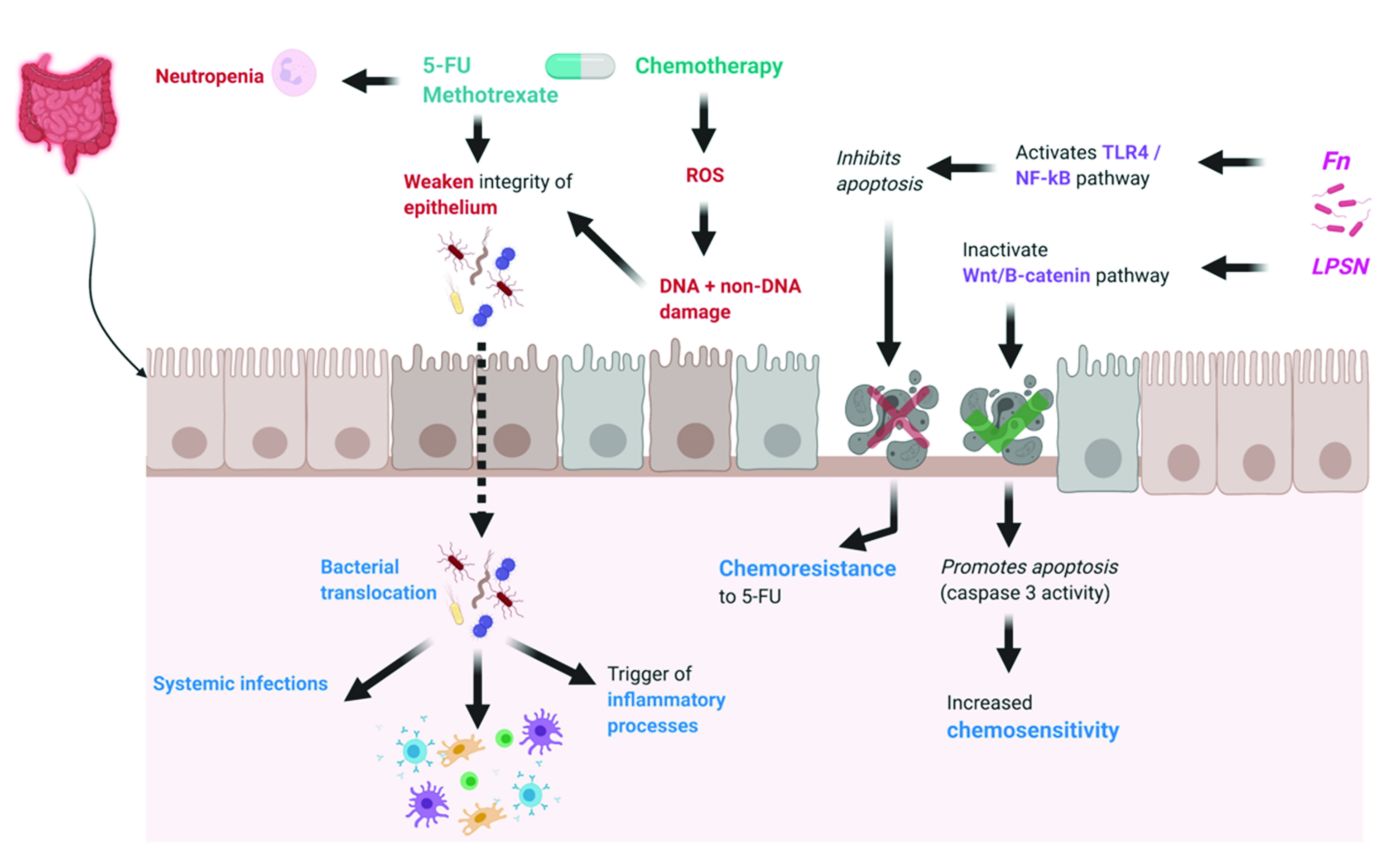
2.2. Effects of Microbiome on Chemotherapy Toxicity
2.3. Effects of Microbiome on Immunotherapy Efficacy
2.4. Effects of Microbiome on Immunotherapy Toxicity
3. Effects of Antineoplastic Treatment on the Microbiome
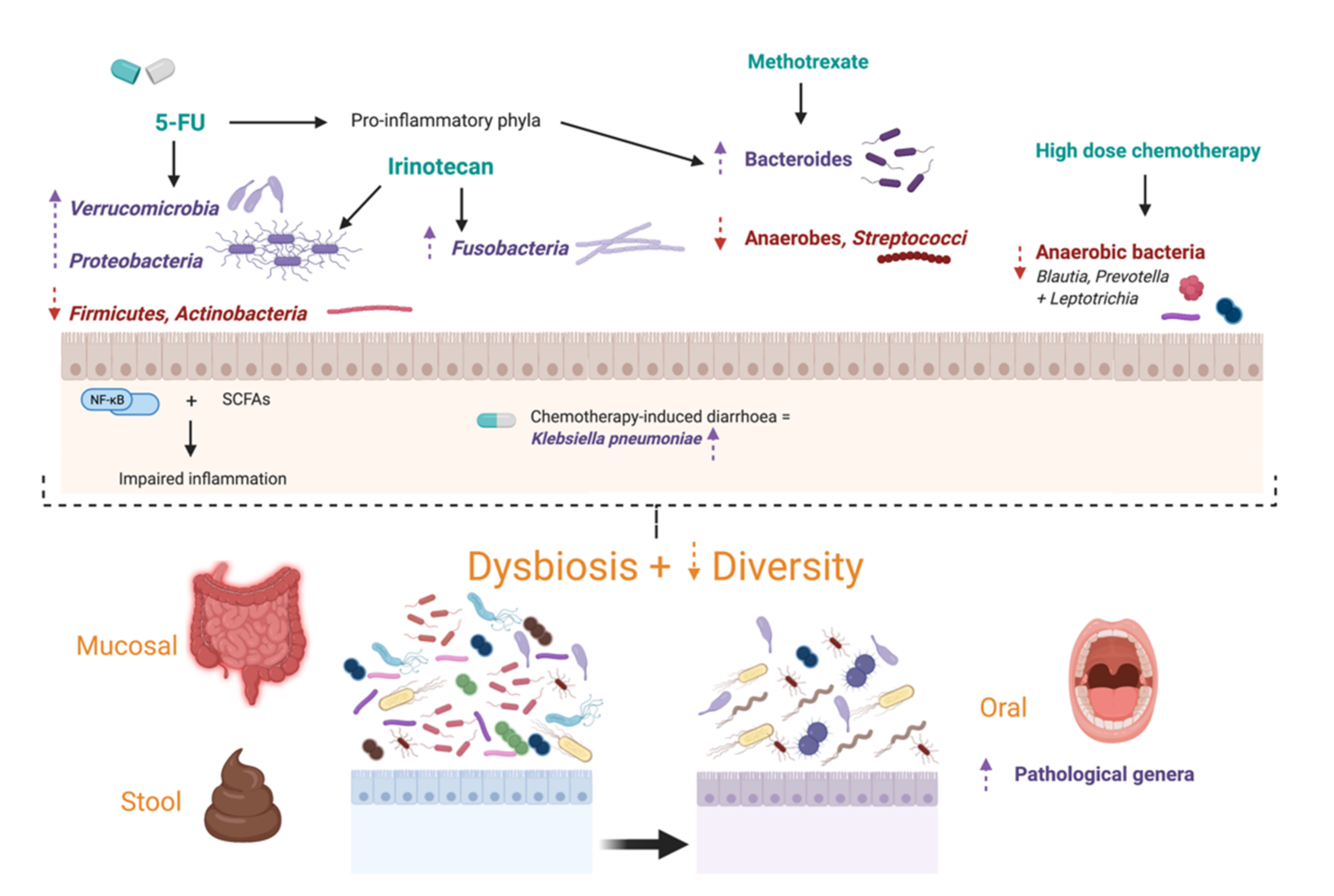
3.1. Specific Chemotherapy Regimens
3.2. Oral Dysbiosis
3.3. Effects of Immunotherapy on the Microbiome
4. Effects of Synbiotics
4.1. Probiotics
4.2. Prebiotics
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Alcoholado, L.; Ramos-Molina, B.; Otero, A.; Laborda-Illanes, A.; Ordóñez, R.; Medina, J.A.; Gómez-Millán, J.; Queipo-Ortuño, M.I. The Role of the Gut Microbiome in Colorectal Cancer Development and Therapy Response. Cancers 2020, 12, 1406. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Keum, N.; Giovannucci, E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef]
- Visnovska, T.; Biggs, P.J.; Schmeier, S.; Frizelle, F.A.; Purcell, R.V. Metagenomics and transcriptomics data from human colorectal cancer. Sci. Data 2019, 6, 116. [Google Scholar] [CrossRef]
- Favoriti, P.; Carbone, G.; Greco, M.; Pirozzi, F.; Pirozzi, R.E.; Corcione, F. Worldwide burden of colorectal cancer: A review. Updates Surg. 2016, 68, 7–11. [Google Scholar] [CrossRef]
- Ternes, D.; Karta, J.; Tsenkova, M.; Wilmes, P.; Haan, S.; Letellier, E. Microbiome in Colorectal Cancer: How to Get from Meta-omics to Mechanism? Trends Microbiol. 2020, 28, 401–423. [Google Scholar] [CrossRef]
- Ahmed, I.; Umar, S. Microbiome and Colorectal Cancer. Curr. Colorectal Cancer Rep. 2018, 14, 217–225. [Google Scholar] [CrossRef]
- Schreuders, E.H.; Ruco, A.; Rabeneck, L.; Schoen, R.E.; Sung, J.J.Y.; Young, G.P.; Kuipers, E.J. Colorectal cancer screening: A global overview of existing programmes. Gut 2015, 64, 1637. [Google Scholar] [CrossRef]
- Ganesh, K.; Stadler, Z.K.; Cercek, A.; Mendelsohn, R.B.; Shia, J.; Segal, N.H.; Diaz, L.A., Jr. Immunotherapy in colorectal cancer: Rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 361–375. [Google Scholar] [CrossRef]
- Takiishi, T.; Fenero, C.I.M.; Câmara, N.O.S. Intestinal barrier and gut microbiota: Shaping our immune responses throughout life. Tissue Barriers 2017, 5, e1373208. [Google Scholar] [CrossRef]
- Gagnière, J.; Raisch, J.; Veziant, J.; Barnich, N.; Bonnet, R.; Buc, E.; Bringer, M.A.; Pezet, D.; Bonnet, M. Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 2016, 22, 501–518. [Google Scholar] [CrossRef]
- Nakatsu, G.; Li, X.; Zhou, H.; Sheng, J.; Wong, S.H.; Wu, W.K.K.; Ng, S.C.; Tsoi, H.; Dong, Y.; Zhang, N.; et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat. Commun. 2015, 6, 8727. [Google Scholar] [CrossRef]
- Brennan, C.A.; Garrett, W.S. Gut Microbiota, Inflammation, and Colorectal Cancer. Ann. Rev. Microbiol. 2016, 70, 395–411. [Google Scholar] [CrossRef] [Green Version]
- Saus, E.; Iraola-Guzmán, S.; Willis, J.R.; Brunet-Vega, A.; Gabaldón, T. Microbiome and colorectal cancer: Roles in carcinogenesis and clinical potential. Mol. Asp. Med. 2019, 69, 93–106. [Google Scholar] [CrossRef]
- Mori, G.; Rampelli, S.; Orena, B.S.; Rengucci, C.; De Maio, G.; Barbieri, G.; Passardi, A.; Casadei Gardini, A.; Frassineti, G.L.; Gaiarsa, S.; et al. Shifts of Faecal Microbiota During Sporadic Colorectal Carcinogenesis. Sci. Rep. 2018, 8, 10329. [Google Scholar] [CrossRef]
- Rezasoltani, S.; Asadzadeh Aghdaei, H.; Dabiri, H.; Akhavan Sepahi, A.; Modarressi, M.H.; Nazemalhosseini Mojarad, E. The association between fecal microbiota and different types of colorectal polyp as precursors of colorectal cancer. Microb. Pathog. 2018, 124, 244–249. [Google Scholar] [CrossRef]
- Tang, Q.; Jin, G.; Wang, G.; Liu, T.; Liu, X.; Wang, B.; Cao, H. Current Sampling Methods for Gut Microbiota: A Call for More Precise Devices. Front. Cell. Infect. Microbiol. 2020, 10, 151. [Google Scholar] [CrossRef]
- Kumar, R.; Eipers, P.; Little, R.B.; Crowley, M.; Crossman, D.K.; Lefkowitz, E.J.; Morrow, C.D. Getting Started with Microbiome Analysis: Sample Acquisition to Bioinformatics. Curr. Protoc. Hum. Genet. 2014, 82, 18.8.1–18.8.29. [Google Scholar] [CrossRef] [Green Version]
- Bharti, R.; Grimm, D.G. Current challenges and best-practice protocols for microbiome analysis. Brief. Bioinform. 2021, 22, 178–193. [Google Scholar] [CrossRef] [Green Version]
- Shuwen, H.; Xi, Y.; Yuefen, P.; Jiamin, X.; Quan, Q.; Haihong, L.; Yizhen, J.; Wei, W. Effects of postoperative adjuvant chemotherapy and palliative chemotherapy on the gut microbiome in colorectal cancer. Microb. Pathog. 2020, 149, 104343. [Google Scholar] [CrossRef]
- Gaines, S.; Shao, C.; Hyman, N.; Alverdy, J.C. Gut microbiome influences on anastomotic leak and recurrence rates following colorectal cancer surgery. Br. J. Surg. 2018, 105, e131–e141. [Google Scholar] [CrossRef] [Green Version]
- Sonis, S.T.; Elting, L.S.; Keefe, D.; Peterson, D.E.; Schubert, M.; Hauer-Jensen, M.; Bekele, B.N.; Raber-Durlacher, J.; Donnelly, J.P.; Rubenstein, E.B. Perspectives on cancer therapy-induced mucosal injury: Pathogenesis, measurement, epidemiology, and consequences for patients. Cancer 2004, 100, 31. [Google Scholar]
- Besse, B.; Charrier, M.; Lapierre, V.; Dansin, E.; Lantz, O.; Planchard, D.; Le Chevalier, T.; Livartoski, A.; Barlesi, F.; Laplanche, A.; et al. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology 2016, 5, e1071008. [Google Scholar] [CrossRef] [Green Version]
- Emens, L.A.; Middleton, G. The interplay of immunotherapy and chemotherapy: Harnessing potential synergies. Cancer Immunol. Res. 2015, 3, 436–443. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.; Trinchieri, G. Microbiota: A key orchestrator of cancer therapy. Nat. Rev. Cancer 2017, 17, 271–285. [Google Scholar] [CrossRef]
- Alexander, J.L.; Wilson, I.D.; Teare, J.; Marchesi, J.R.; Nicholson, J.K.; Kinross, J.M. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 356. [Google Scholar] [CrossRef]
- Scott, T.A.; Quintaneiro, L.M.; Norvaisas, P.; Lui, P.P.; Wilson, M.P.; Leung, K.Y.; Herrera-Dominguez, L.; Sudiwala, S.; Pessia, A.; Clayton, P.T.; et al. Host-Microbe Co-metabolism Dictates Cancer Drug Efficacy in C. elegans. Cell 2017, 169, 442–456. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Gonzalez, A.P.; Ritter, A.D.; Shrestha, S.; Andersen, E.C.; Yilmaz, L.S.; Walhout, A.J.M. Bacterial Metabolism Affects the C. elegans Response to Cancer Chemotherapeutics. Cell 2017, 169, 431–441. [Google Scholar] [CrossRef] [Green Version]
- Yuan, L.; Zhang, S.; Li, H.; Yang, F.; Mushtaq, N.; Ullah, S.; Shi, Y.; An, C.; Xu, J. The influence of gut microbiota dysbiosis to the efficacy of 5-Fluorouracil treatment on colorectal cancer. Biomed. Pharmacother. Biomed. Pharmacother. 2018, 108, 184–193. [Google Scholar] [CrossRef]
- Wang, Z.; Hopson, L.M.; Singleton, S.S.; Yang, X.; Jogunoori, W.; Mazumder, R.; Obias, V.; Lin, P.; Nguyen, B.N.; Yao, M.; et al. Mice with Dysfunctional TGF-β Signaling Develop Altered Intestinal Microbiome and Colorectal Cancer Resistant to 5FU. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166179. [Google Scholar] [CrossRef]
- Meng, C.; Bai, C.; Brown, T.D.; Hood, L.E.; Tian, Q. Human Gut Microbiota and Gastrointestinal Cancer. Genom. Proteom. Bioinform. 2018, 16, 33–49. [Google Scholar] [CrossRef]
- An, J.; Ha, E.M. Combination Therapy of Lactobacillus plantarum Supernatant and 5-Fluouracil Increases Chemosensitivity in Colorectal Cancer Cells. J. Microbiol. Biotechnol. 2016, 26, 1490–1503. [Google Scholar] [CrossRef]
- Iida, N.; Dzustev, A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; Dai, R.M.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 4. [Google Scholar]
- Viaud, S.; Saccheri, F.; Mignot, F.; Yamazaki, T.; Daillere, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 6. [Google Scholar]
- Lehouritis, P.; Cummins, J.; Stanton, M.; Murphy, C.T.; McCarthy, F.O.; Reid, G.; Urbaniak, C.; Byrne, W.L.; Tangney, M. Local bacteria affect the efficacy of chemotherapeutic drugs. Sci. Rep. 2015, 5, 14554. [Google Scholar]
- Daillere, R.; Vetizou, M.; Waldschmitt, N.; Yamazaki, T.; Isnard, C.; Poirier-Colame, V.; Duong, C.P.M.; Flament, C.; Lepage, P.; Roberti, M.P.; et al. Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity 2016, 45, 931–943. [Google Scholar]
- Pagès, F.; Mlecnik, B.; Marliot, F.; Bindea, G.; Ou, F.S.; Bifulco, C.; Lugli, A.; Zlobec, I.; Rau, T.T.; Berger, M.D.; et al. International validation of the consensus Immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet 2018, 391, 2128–2139. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Waldner, M.; Obenauf, A.C.; Angell, H.; Fredriksen, T.; Lafontaine, L.; Berger, A.; et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013, 39, 782–795. [Google Scholar] [CrossRef] [Green Version]
- Ogino, S.; Nosho, K.; Irahara, N.; Meyerhardt, J.A.; Baba, Y.; Shima, K.; Glickman, J.N.; Ferrone, C.R.; Mino-Kenudson, M.; Tanaka, N.; et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin. Cancer Res. 2009, 15, 6412–6420. [Google Scholar] [CrossRef] [Green Version]
- Roberti, M.P.; Yonekura, S.; Duong, C.P.M.; Picard, M.; Ferrere, G.; Tidjani Alou, M.; Rauber, C.; Iebba, V.; Lehmann, C.H.K.; Amon, L.; et al. Chemotherapy-induced ileal crypt apoptosis and the ileal microbiome shape immunosurveillance and prognosis of proximal colon cancer. Nat. Med. 2020, 26, 919–931. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, Y.; Weng, W.; Guo, B.; Cai, G.; Ma, Y.; Cai, S. Fusobacterium nucleatum promotes chemoresistance to 5-fluorouracil by upregulation of BIRC3 expression in colorectal cancer. J. Exp. Clin. Cancer Res. CR 2019, 38, 14. [Google Scholar] [CrossRef]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017, 170, 548–563. [Google Scholar] [CrossRef] [Green Version]
- Gupta, E.; Lestingi, T.M.; Mick, R.; Ramirez, J.; Vokes, E.E.; Ratain, M.J. Metabolic fate of irinotecan in humans: Correlation of glucuronidation with diarrhea. Cancer Res. 1994, 54, 3. [Google Scholar]
- Nakao, T.; Kurita, N.; Komatsu, M.; Yoshikawa, K.; Iwata, T.; Utusnomiya, T.; Shimada, M. Irinotecan injures tight junction and causes bacterial translocation in rat. J. Surg. Res. 2012, 173, 341–347. [Google Scholar]
- Sparreboom, A.; de Jonge, M.; de Bruijn, P.; Brouwer, E.; Nooter, K.; Loos, W.J.; van Alphen, R.J.; Mathijssen, R.H.; Stoter, G.; Verweij, J. Irinotecan (CPT-11) metabolism and disposition in cancer patients. Clin. Cancer Res. 1998, 4, 2747–2754. [Google Scholar]
- Takasuna, K.; Hagiwara, T.; Hirohashi, M.; Kato, M.; Nomura, M.; Nagai, E.; Yokoi, T.; Kamataki, T. Involvement of beta-glucuronidase in intestinal microflora in the intestinal toxicity of the antitumor camptothecin derivative irinotecan hydrochloride (CPT-11) in rats. Cancer Res. 1996, 56, 3752–3757. [Google Scholar]
- Takasuna, K.; Hagiwara, T.; Hirohashi, M.; Kato, M.; Nomura, M.; Nagai, E.; Yokoi, T.; Kamataki, T. Inhibition of intestinal microflora beta-glucuronidase modifies the distribution of the active metabolite of the antitumor agent, irinotecan hydrochloride (CPT-11) in rats. Cancer Chemother. Pharmacol. 1998, 42, 280–286. [Google Scholar]
- Tanoue, T.; Morita, S.; Plichta, D.R.; Skelly, A.N.; Suda, W.; Sugiura, Y.; Narushima, S.; Vlamakis, H.; Motoo, I.; Sugita, K.; et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 2019, 565, 600–605. [Google Scholar] [CrossRef]
- Wong, S.H.; Yu, J. Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 690–704. [Google Scholar] [CrossRef]
- Nurgali, K.; Jagoe, R.T.; Abalo, R. Editorial: Adverse Effects of Cancer Chemotherapy: Anything New to Improve Tolerance and Reduce Sequelae? Front Pharmacol. 2018, 9, 245. [Google Scholar] [CrossRef]
- Carvalho, R.; Vaz, A.; Pereira, F.L.; Dorella, F.; Aguiar, E.; Chatel, J.-M.; Bermudez, L.; Langella, P.; Fernandes, G.; Figueiredo, H.; et al. Gut microbiome modulation during treatment of mucositis with the dairy bacterium Lactococcus lactis and recombinant strain secreting human antimicrobial PAP. Sci. Rep. 2018, 8, 15072. [Google Scholar] [CrossRef]
- Karin, M.; Jobin, C.; Balkwill, F. Chemotherapy, immunity and microbiota—A new triumvirate? Nat. Med. 2014, 20, 126–127. [Google Scholar] [CrossRef]
- Secombe, K.R.; Coller, J.K.; Gibson, R.J.; Wardill, H.R.; Bowen, J.M. The bidirectional interaction of the gut microbiome and the innate immune system: Implications for chemotherapy-induced gastrointestinal toxicity. Int. J. Cancer 2019, 144, 2365–2376. [Google Scholar] [CrossRef]
- Hong, B.-Y.; Sobue, T.; Choquette, L.; Dupuy, A.K.; Thompson, A.; Burleson, J.A.; Salner, A.L.; Schauer, P.K.; Joshi, P.; Fox, E.; et al. Chemotherapy-induced oral mucositis is associated with detrimental bacterial dysbiosis. Microbiome 2019, 7, 66. [Google Scholar] [CrossRef]
- Thorpe, D.W.; Stringer, A.M.; Gibson, R.J. Chemotherapy-induced mucositis: The role of the gastrointestinal microbiome and toll-like receptors. Exp. Biol. Med. 2013, 238, 1–6. [Google Scholar] [CrossRef]
- Cinausero, M.; Aprile, G.; Ermacora, P.; Basile, D.; Vitale, M.G.; Fanotto, V.; Parisi, G.; Calvetti, L.; Sonis, S.T. New Frontiers in the Pathobiology and Treatment of Cancer Regimen-Related Mucosal Injury. Front. Pharmacol. 2017, 8, 1428. [Google Scholar] [CrossRef] [Green Version]
- Hou, J.; Zheng, H.; Li, P.; Liu, H.; Zhou, H.; Yang, X. Distinct shifts in the oral microbiota are associated with the progression and aggravation of mucositis during radiotherapy. Radiother. Oncol. 2018, 129, 44–51. [Google Scholar] [CrossRef]
- Vasconcelos, R.M.; Sanfilippo, N.; Paster, B.J.; Kerr, A.R.; Li, Y.; Ramalho, L.; Queiroz, E.L.; Smith, B.; Sonis, S.T.; Corby, P.M. Host-Microbiome Cross-talk in Oral Mucositis. J. Dent. Res. 2016, 95, 725–733. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Gao, W.; Shi, X.; Ding, J.; Liu, W.; He, H.; Wang, K.; Shao, F. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a Gasdermin. Nature 2017, 547, 99–103. [Google Scholar] [CrossRef]
- Kanarek, N.; Grivennikov, S.I.; Leshets, M.; Lasry, A.; Alkalay, I.; Horwitz, E.; Shaul, Y.D.; Stachler, M.; Voronov, E.; Apte, R.N.; et al. Critical role for IL-1β in DNA damage-induced mucositis. Proc. Natl. Acad. Sci. USA 2014, 111, E702–E711. [Google Scholar] [CrossRef] [Green Version]
- Papanicolas, L.E.; Gordon, D.L.; Wesselingh, S.L.; Rogers, G.B. Not Just Antibiotics: Is Cancer Chemotherapy Driving Antimicrobial Resistance? Trends Microbiol. 2018, 26, 393–400. [Google Scholar] [CrossRef]
- Pedroso, S.H.S.P.; Vieira, A.T.; Bastos, R.W.; Oliveira, J.S.; Cartelle, C.T.; Arantes, R.M.E.; Soares, P.M.G.; Generoso, S.V.; Cardoso, V.N.; Teixeira, M.M.; et al. Evaluation of mucositis induced by irinotecan after microbial colonization in germ-free mice. Microbiology 2015, 161, 1950–1960. [Google Scholar] [CrossRef]
- Brandi, G.; Dabard, J.; Raibaud, P.; Di Battista, M.; Bridonneau, C.; Pisi, A.M.; Morselli Labate, A.M.; Pantaleo, M.A.; De Vivo, A.; Biasco, G. Intestinal microflora and digestive toxicity of irinotecan in mice. Clin. Cancer Res. 2006, 12, 1299. [Google Scholar] [CrossRef] [Green Version]
- Stringer, A.M.; Al-Dasooqi, N. Biomarkers of chemotherapy-induced diarrhoea: A clinical study of intestinal microbiome alterations, inflammation and circulating matrix metalloproteinases. Supportive Care Cancer 2013, 21, 1843–1852. [Google Scholar]
- Rooks, M.G.; Veiga, P.; Wardwell-Scott, L.H.; Tickle, T.; Segata, N.; Michaud, M.; Gallini, C.A.; Beal, C.; van Hylckama-Vlieg, J.E.; Ballal, S.A.; et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014, 8, 1403–1417. [Google Scholar] [CrossRef]
- Thi, T.D.; Lopez, E.; Rodriguez-Rojas, A.; Rodriguez-Beltran, J.; Couce, A.; Guelfo, J.R.; Castaneda-Garcia, A.; Blazquez, J. Effect of recA inactivation on mutagenesis of Escherichia coli exposed to sublethal concentrations of antimicrobials. J. Antimicrob. Chemother. 2011, 66, 531–538. [Google Scholar] [CrossRef] [Green Version]
- Fessler, J.; Matson, V.; Gajewski, T.F. Exploring the emerging role of the microbiome in cancer immunotherapy. J. Immunother. Cancer 2019, 7, 108. [Google Scholar] [CrossRef]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Tian, X.; Ji, L.; Zhang, X.; Cao, Y.; Shen, C.; Hu, Y.; Wong, J.W.H.; Fang, J.-Y.; Hong, J.; et al. A tumor microenvironment-specific gene expression signature predicts chemotherapy resistance in colorectal cancer patients. NPJ Precis. Oncol. 2021, 5, 7. [Google Scholar] [CrossRef]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Cowey, C.L.; Lao, C.D.; Schadendorf, D.; Dummer, R.; Smylie, M.; Rutkowski, P.; et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N. Engl. J. Med. 2015, 373, 23–34. [Google Scholar] [CrossRef] [Green Version]
- Saâda-Bouzid, E.; Defaucheux, C.; Karabajakian, A.; Coloma, V.P.; Servois, V.; Paoletti, X.; Even, C.; Fayette, J.; Guigay, J.; Loirat, D.; et al. Hyperprogression during anti-PD-1/PD-L1 therapy in patients with recurrent and/or metastatic head and neck squamous cell carcinoma. Ann. Oncol. 2017, 28, 1605–1611. [Google Scholar] [CrossRef]
- Chaput, N.; Lepage, P.; Coutzac, C.; Soularue, E.; Le Roux, K.; Monot, C.; Boselli, L.; Routier, E.; Cassard, L.; Collins, M.; et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017, 28, 1368–1379. [Google Scholar] [CrossRef]
- Sun, J.-Y.; Yin, T.-L.; Zhou, J.; Xu, J.; Lu, X.-J. Gut microbiome and cancer immunotherapy. J. Cell. Physiol. 2020, 235, 4082–4088. [Google Scholar] [CrossRef]
- Dubin, K.; Callahan, M.K.; Ren, B.; Khanin, R.; Viale, A.; Ling, L.; No, D.; Gobourne, A.; Littmann, E.; Huttenhower, C.; et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun. 2016, 7, 10391. [Google Scholar] [CrossRef] [Green Version]
- Frankel, A.E.; Coughlin, L.A.; Kim, J.; Froehlich, T.W.; Xie, Y.; Frenkel, E.P.; Koh, A.Y. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia 2017, 19, 848–855. [Google Scholar] [CrossRef]
- Montassier, E.; Gastinne, T.; Vangay, P.; Al-Ghalith, G.A.; Bruley des Varannes, S.; Massart, S. Chemotherapy-driven dysbiosis in the intestinal microbiome. Aliment. Pharmacol. Ther. 2015, 42. [Google Scholar] [CrossRef]
- Fijlstra, M.; Ferdous, M.; Koning, A.M.; Rings, E.H.H.M.; Harmsen, H.J.M.; Tissing, W.J.E. Substantial decreases in the number and diversity of microbiota during chemotherapy-induced gastrointestinal mucositis in a rat model. Supportive Care Cancer 2015, 23, 10. [Google Scholar]
- Von Bultzingslowen, I.; Adlerberth, I.; Wold, A.E.; Dahlen, G.; Jontell, M. Oral and intestinal microflora in 5-fluorouracil treated rats, translocation to cervical and mesenteric lymph nodes and effects of probiotic bacteria. Oral Microbiol. Immunol. 2003, 18, 278–284. [Google Scholar]
- van Vliet, M.J.; Harmsen, H.J.; de Bont, E.S.; Tissing, W.J. The role of intestinal microbiota in the development and severity of chemotherapy-induced mucositis. PLoS Pathog. 2010, 6, e1000879. [Google Scholar] [CrossRef] [Green Version]
- Galloway-Peña, J.R.; Smith, D.P.; Sahasrabhojane, P.; Ajami, N.J.; Wadsworth, W.D.; Daver, N.G.; Chemaly, R.F.; Marsh, L.; Ghantoji, S.S.; Pemmaraju, N.; et al. The role of the gastrointestinal microbiome in infectious complications during induction chemotherapy for acute myeloid leukemia. Cancer 2016, 122, 2186–2196. [Google Scholar] [CrossRef] [Green Version]
- Zwielehner, J.; Lassl, C.; Hippe, B.; Pointner, A.; Switzeny, O.J.; Remely, M.; Kitzweger, E.; Ruckser, R.; Haslberger, A.G. Changes in human fecal microbiota due to chemotherapy analyzed by TaqMan-PCR, 454 sequencing and PCR-DGGE fingerprinting. PLoS ONE 2011, 6, e28654. [Google Scholar] [CrossRef] [Green Version]
- Vanlancker, E.; Vanhoecke, B.; Stringer, A.; Van de Wiele, T. 5-Fluorouracil and irinotecan (SN-38) have limited impact on colon microbial functionality and composition in vitro. PeerJ 2017, 5, e4017. [Google Scholar] [CrossRef]
- Fei, Z.; Lijuan, Y.; Xi, Y.; Wei, W.; Jing, Z.; Miao, D.; Shuwen, H. Gut microbiome associated with chemotherapy-induced diarrhea from the CapeOX regimen as adjuvant chemotherapy in resected stage III colorectal cancer. Gut Pathog. 2019, 11, 18. [Google Scholar] [CrossRef]
- Stojanovska, V.; McQuade, R.M.; Fraser, S.; Prakash, M.; Gondalia, S.; Stavely, R.; Palombo, E.; Apostolopoulos, V.; Sakkal, S.; Nurgali, K. Oxaliplatin-induced changes in microbiota, TLR4+ cells and enhanced HMGB1 expression in the murine colon. PLoS ONE 2018, 13, e0198359. [Google Scholar] [CrossRef]
- Yuan, W.; Xiao, X.; Yu, X.; Xie, F.; Feng, P.; Malik, K.; Wu, J.; Ye, Z.; Zhang, P.; Li, X. Probiotic Therapy (BIO-THREE) Mitigates Intestinal Microbial Imbalance and Intestinal Damage Caused by Oxaliplatin. Probiotics Antimicrob. Proteins 2021. [Google Scholar] [CrossRef]
- Forsgard, R.A.; Marrachelli, V.G.; Korpela, K.; Frias, R.; Collado, M.C.; Korpela, R.; Monleon, D.; Spillmann, T.; Osterlund, P. Chemotherapy-induced gastrointestinal toxicity is associated with changes in serum and urine metabolome and fecal microbiota in male Sprague-Dawley rats. Cancer Chemother. Pharmacol. 2017, 80, 317–332. [Google Scholar] [CrossRef] [Green Version]
- Stringer, A.M.; Gibson, R.J.; Logan, R.; Bowen, J.; Yeoh, A.; Keefe, D.M. Faecal microflora and beta-glucuronidase expression are altered in an irinotecan-induced diarrhea model in rats. Cancer Biol. Ther. 2008, 7, 1919–1925. [Google Scholar]
- Lin, X.B.; Dieleman, L.A.; Ketabi, A.; Bibova, I.; Sawyer, M.B.; Xue, H.; Field, C.J.; Baracos, V.E.; Gänzle, M.G. Irinotecan (CPT-11) Chemotherapy Alters Intestinal Microbiota in Tumour Bearing Rats. PLoS ONE 2012, 7, e39764. [Google Scholar] [CrossRef]
- Napeñas, J.J.; Brennan, M.T.; Bahrani-Mougeot, F.K.; Fox, P.C.; Lockhart, P.B. Relationship between mucositis and changes in oral microflora during cancer chemotherapy. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2007, 103, 48–59. [Google Scholar] [CrossRef]
- de Mendonca, R.M.; de Araujo, M.; Levy, C.E.; Morari, J.; Silva, R.A.; Yunes, J.A.; Brandalise, S.R. Prospective evaluation of HSV, Candida spp., and oral bacteria on the severity of oral mucositis in pediatric acute lymphoblastic leukemia. Supportive Care Cancer Off. J. Multinatl. Assoc. Supportive Care Cancer 2012, 20, 1101–1107. [Google Scholar] [CrossRef]
- Perales-Puchalt, A.; Perez-Sanz, J.; Payne, K.K.; Svoronos, N.; Allegrezza, M.J.; Chaurio, R.A.; Anadon, C.; Calmette, J.; Biswas, S.; Mine, J.A.; et al. Frontline Science: Microbiota reconstitution restores intestinal integrity after cisplatin therapy. J. Leukoc. Biol. 2018, 103, 799–805. [Google Scholar] [CrossRef]
- Botticelli, A.; Vernocchi, P.; Marini, F.; Quagliariello, A.; Cerbelli, B.; Reddel, S.; Del Chierico, F.; Di Pietro, F.; Giusti, R.; Tomassini, A.; et al. Gut metabolomics profiling of non-small cell lung cancer (NSCLC) patients under immunotherapy treatment. J. Transl. Med. 2020, 18, 49. [Google Scholar] [CrossRef]
- Vernocchi, P.; Gili, T.; Conte, F.; Del Chierico, F.; Conta, G.; Miccheli, A.; Botticelli, A.; Paci, P.; Caldarelli, G.; Nuti, M.; et al. Network Analysis of Gut Microbiome and Metabolome to Discover Microbiota-Linked Biomarkers in Patients Affected by Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2020, 21, 8730. [Google Scholar] [CrossRef]
- FAO; WHO. Evaluation of health and nutritional properties of powder milk and live lactic acid bacteria. Prevention 2001, 5, 1–10. [Google Scholar]
- Fuller, R. Probiotics in human medicine. Gut 1991, 32, 439–442. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Liu, C.; Yue, J. Radiotherapy and the gut microbiome: Facts and fiction. Radiat. Oncol. 2021, 16, 9. [Google Scholar] [CrossRef]
- Tsai, Y.L.; Lin, T.L.; Chang, C.J.; Wu, T.R.; Lai, W.F.; Lu, C.C.; Lai, H.C. Probiotics, prebiotics and amelioration of diseases. J. Biomed. Sci. 2019, 26, 3. [Google Scholar] [CrossRef]
- Dikeocha, I.J.; Al-Kabsi, A.M.; Eid, E.E.M.; Hussin, S.; Alshawsh, M.A. Probiotics supplementation in patients with colorectal cancer: A systematic review of randomized controlled trials. Nutr. Rev. 2021. [Google Scholar] [CrossRef]
- Whitford, E.J.; Cummins, A.; Butler, R.N.; Prisciandaro, L.D.; Fauser, J.K.; Yazbeck, R.; Lawrence, A.; Cheah, K.Y.; Wright, T.H.; Lymn, K.A.; et al. Effects of Streptococcus thermophilus TH-4 on intestinal mucositis induced by the chemotherapeutic agent, 5-Fluorouracil (5-FU). Cancer Biol. Ther. 2009, 8, 505–511. [Google Scholar]
- Tooley, K.; Howarth, G.S.; Lymn, K.; Lawrence, A.; Butler, R. Oral ingestion of Streptococcus thermophilus diminishes severity of small intestinal mucositis in methotrexate treated rats. Cancer Biol. Ther. 2006, 5, 593–600. [Google Scholar]
- Tooley, K.; Howarth, G.; Lymn, K.; Lawrence, A.; Butler, R. Oral ingestion of Streptococcus thermophilus does not affect mucositis severity or tumor progression in the tumor-bearing rat. Cancer Biol. Ther. 2011, 12, 131–138. [Google Scholar]
- Justino, P.F.; Melo, L.; Nogueira, A.F.; Morais, C.M.; Mendes, W.O.; Franco, A.X.; Souza, E.P.; Ribeiro, R.A.; Souza, M.H.L.P.; Soares, P.M.G. Regulatory role of Lactobacillus acidophilus on inflammation and gastric dysmotility in intestinal mucositis induced by 5-fluorouracil in mice. Cancer Chemother. Pharmacol. 2015, 75, 559–567. [Google Scholar]
- Prisciandaro, L.D.; Geier, M.S.; Butler, R.N.; Cummins, A.G.; Howarth, G.S. Probiotic factors partially improve parameters of 5-fluorouracil-induced intestinal mucositis in rats. Cancer Biol. Ther. 2011, 11, 671–677. [Google Scholar]
- Chang, C.W.; Liu, C.Y.; Lee, H.C.; Huang, Y.H.; Li, L.H.; Chiau, J.C.; Wang, T.E.; Chu, C.H.; Shih, S.C.; Tsai, T.H.; et al. Lactobacillus casei Variety rhamnosus Probiotic Preventively Attenuates 5-Fluorouracil/Oxaliplatin-Induced Intestinal Injury in a Syngeneic Colorectal Cancer Model. Front Microbiol. 2018, 9, 983. [Google Scholar] [CrossRef]
- Mi, H.; Dong, Y.; Zhang, B.; Wang, H.; Peter, C.C.K.; Gao, P.; Fu, H.; Gao, Y. Bifidobacterium Infantis Ameliorates Chemotherapy-Induced Intestinal Mucositis Via Regulating T Cell Immunity in Colorectal Cancer Rats. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 42, 2330–2341. [Google Scholar] [CrossRef] [Green Version]
- Owens, J.A.; Saeedi, B.J.; Naudin, C.R.; Hunter-Chang, S.; Barbian, M.E.; Eboka, R.U.; Askew, L.; Darby, T.M.; Robinson, B.S.; Jones, R.M. Lactobacillus rhamnosus GG Orchestrates an Anti-tumor Immune Response. Cell. Mol. Gastroenterol. Hepatol. 2021, 21, 115–116. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [Green Version]
- Wada, M.; Nagata, S.; Saito, M.; Shimizu, T.; Yamashiro, Y.; Matsuki, T.; Asahara, T.; Nomoto, K. Effects of the enteral administration of Bifidobacterium breve on patients undergoing chemotherapy for pediatric malignancies. Supportive Care Cancer 2010, 18, 751–759. [Google Scholar]
- Ishikawa, H.; Akedo, I.; Otani, T.; Suzuki, T.; Nakamura, T.; Takeyama, I.; Ishiguro, S.; Miyaoka, E.; Sobue, T.; Kakizoe, T. Randomized trial of dietary fiber andLactobacillus casei administration for prevention of colorectal tumors. Int. J. Cancer 2005, 116, 762–767. [Google Scholar] [CrossRef]
- Osterlund, P.; Ruotsalainen, T.; Korpela, R.; Saxelin, M.; Ollus, A.; Valta, P.; Kouri, M.; Elomaa, I.; Joensuu, H. Lactobacillus supplementation for diarrhoea related to chemotherapy of colorectal cancer: A randomised study. Br. J. Cancer 2007, 97, 1028–1034. [Google Scholar]
- Benito, I.; Encío, I.J.; Milagro, F.I.; Alfaro, M.; Martínez-Peñuela, A.; Barajas, M.; Marzo, F. Microencapsulated Bifidobacterium bifidum and Lactobacillus gasseri in Combination with Quercetin Inhibit Colorectal Cancer Development in Apc(Min/+) Mice. Int. J. Mol. Sci. 2021, 22, 4906. [Google Scholar] [CrossRef]
- Yoon, Y.; Kim, G.; Jeon, B.-N.; Fang, S.; Park, H. Bifidobacterium Strain-Specific Enhances the Efficacy of Cancer Therapeutics in Tumor-Bearing Mice. Cancers 2021, 13, 957. [Google Scholar] [CrossRef]
- Liong, M.-T. Safety of probiotics: Translocation and infection. Nutr. Rev. 2008, 66, 192–202. [Google Scholar]
- Zmora, N.; Zilberman-Schapira, G.; Suez, J.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Kotler, E.; Zur, M.; Regev-Lehavi, D.; Brik, R.B.-Z.; et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell 2018, 174, 1388–1405. [Google Scholar] [CrossRef] [Green Version]
- Lara-Villoslada, F.; de Haro, O.; Camuesco, D.; Comalada, M.; Velasco, J.; Zarzuelo, A.; Xaus, J.; Galvez, J. Short-chain fructooligosaccharides, in spite of being fermented in the upper part of the large intestine, have anti-inflammatory activity in the TNBS model of colitis. Eur. J. Nutr. 2006, 45, 418–425. [Google Scholar]
- Ten Bruggencate, S.J.; Bovee-Oudenhoven, I.M.; Lettink-Wissink, M.L.; Katan, M.B.; van der Meer, R. Dietary fructooligosaccharides affect intestinal barrier function in healthy men. J. Nutri. 2006, 136, 70–74. [Google Scholar]
- Scharlau, D.; Borowicki, A.; Habermann, N.; Hofmann, T.; Klenow, S.; Miene, C.; Munjal, U.; Stein, K.; Glei, M. Mechanisms of primary cancer prevention by butyrate and other products formed during gut flora-mediated fermentation of dietary fibre. Mutat. Res. 2009, 628, 39–53. [Google Scholar]
- Looijer-van Langen, M.; Dieleman, L. Prebiotics in chronic intestinal inflammation. Inflamm. Bowel Dis. 2009, 15, 454–462. [Google Scholar]
- Sauer, J.; Richter, K.K.; Pool-Zobel, B.L. Products formed during fermentation of the prebiotic inulin with human gut flora enhance expression of biotransformation genes in human primary colon cells. Br. J. Nutri. 2007, 97, 928–937. [Google Scholar]
- Manhart, N.; Vierlinger, K.; Spittler, A.; Bergmeister, H.; Sautner, T.; Roth, E. Oral feeding with glutamine prevents lymphocyte and glutathione depletion of Peyer’s patches in endotoxemic mice. Ann. Surg. 2001, 234, 92–97. [Google Scholar]
- Decker-Baumann, C.; Buhl, K.; Frohmuller, S.; von Herbay, A.; Dueck, M.; Schlag, P.M. Reduction of chemotherapy-induced side effects by parenteral glutamine supplementation in patients with metastatic colorectal cancer. Eur. J. Cancer 1999, 35, 202–207. [Google Scholar]
- Guo, C.; Guo, D.; Fang, L.; Sang, T.; Wu, J.; Guo, C.; Wang, Y.; Wang, Y.; Chen, C.; Chen, J.; et al. Ganoderma lucidum polysaccharide modulates gut microbiota and immune cell function to inhibit inflammation and tumorigenesis in colon. Carbohydr. Polym. 2021, 267, 118231. [Google Scholar] [CrossRef]
- Smith, C.; Geier, M.S.; Yazbeck, R.; Torres, D.; Butler, R.; Howarth, G.S. Lactobacillus fermentum BR11 and fructo-oligosaccharide partially reduce jejunal inflammation in a model of intestinal mucositis in rats. Nutr. Cancer 2008, 60, 757–767. [Google Scholar]
- So, D.; Whelan, K.; Rossi, M.; Morrison, M.; Holtmann, G.; Kelly, J.T.; Shanahan, E.R.; Staudacher, H.M.; Campbell, K.L. Dietary fiber intervention on gut microbiota composition in healthy adults: A systematic review and meta-analysis. Am. J. Clin. Nutr. 2018, 107, 965–983. [Google Scholar] [CrossRef] [Green Version]
- Alberts, D.S.; Martínez, M.E.; Roe, D.J.; Guillén-Rodríguez, J.M.; Marshall, J.R.; Van Leeuwen, J.B.; Reid, M.E.; Ritenbaugh, C.; Vargas, P.A.; Bhattacharyya, A.B.; et al. Lack of Effect of a High-Fiber Cereal Supplement on the Recurrence of Colorectal Adenomas. N. Engl. J. Med. 2000, 342, 1156–1162. [Google Scholar] [CrossRef]
- Schatzkin, A.; Lanza, E.; Corle, D.; Lance, P.; Iber, F.; Caan, B.; Shike, M.; Weissfeld, J.; Burt, R.; Cooper, M.R.; et al. Lack of Effect of a Low-Fat, High-Fiber Diet on the Recurrence of Colorectal Adenomas. N. Engl. J. Med. 2000, 342, 1149–1155. [Google Scholar] [CrossRef]
- Lanza, E.; Yu, B.; Murphy, G.; Albert, P.S.; Caan, B.; Marshall, J.R.; Lance, P.; Paskett, E.D.; Weissfeld, J.; Slattery, M.; et al. The Polyp Prevention Trial Continued Follow-up Study: No Effect of a Low-Fat, High-Fiber, High-Fruit, and -Vegetable Diet on Adenoma Recurrence Eight Years after Randomization. Cancer Epidemiol. Biomark. Prev. 2007, 16, 1745–1752. [Google Scholar] [CrossRef] [Green Version]
| Author | Study Subjects (n) | Specimen Types | Method | Microbiota Changes |
|---|---|---|---|---|
| Montassier, Gastinne, Vangay, Al-Ghalith, Bruley des Varannes and Massart [76] | Non-Hodgkin’s Lymphoma (28) | Stool | 16S rRNA | ↓ Firmicutes Actinobacteria Proteobacteria |
| Galloway-Peña, Smith, Sahasrabhojane, Ajami, Wadsworth, Daver, Chemaly, Marsh, Ghantoji, Pemmaraju, Garcia-Manero, Rezvani, Alousi, Wargo, Shpall, Futreal, Guindani, Petrosino, Kontoyiannis and Shelburne [80] | Acute myelogenous leukaemia patients (34) | Buccal specimens Stool | 16S rRNA (V4) | Oral and stool -diversity (with carbapenem) Lactobacillus (oral and buccal) Anaerobes e.g., Blautia, Prevotella, Leptotrichia |
| Zwielehner, Lassl, Hippe, Pointner, Switzeny, Remely, Kitzweger, Ruckser and Haslberger [81] | Chemotherapy patients (17) | Stool | 16S rRNA | Diversity of Clostridium clusters IV and XIVa Bacteroides Bifidobacterium |
| Fei, Lijuan, Xi, Wei, Jing, Miao and Shuwen [83] | Resected stage III CRC patients undergoing CapeOX regimen (17) | Stool | 16S rRNA | Microbial richness and diversity Klebsiella pneumoniae |
| Vanlancker, Vanhoecke, Stringer and Van de Wiele [82] | In vitro mucosal stimulator treating stool and mucosa with 5-FU and SN-38 (active metabolite of irinotecan) (6 human donors) | Stool Mucosal | 16S rRNA (V3–4) | Proteobacteria (Escherichia/Shigella) Bacteroidetes (Bacteroides) Firmicutes (Clostridium cluster XIVa, Veillonella) Bacteroides Escherichia/Shigella |
| Author | Study Subjects (n) | Specimen Types | Method | Microbiota Changes |
|---|---|---|---|---|
| Fijlstra et al. [77] | Rats treated with methotrexate | Stool | FISH | Anaerobes Streptococci Bacteroides |
| Forsgard, Marrachelli, Korpela, Frias, Collado, Korpela, Monleon, Spillmann and Osterlund [86] | Sprague-Dawley rats injected with 5-FU, oxaliplatin or irinotecan (48) | Stool | 16S rRNA | Irinotecan: Fusobacteria Proteobacteria 5-FU and Oxaliplatin caused minor shifts |
| Stringer et al. [87] | Irinotecan-treated rats (81) | Stool | DNA extraction + PCR | E. coli Staphylococcus spp. Clostridium spp. Lactobacillus spp. Bifidobacterium spp. Bacteroides spp. |
| Lin et al. [88] | Tumour-bearing rats receiving irinotecan +/− oral glutamine bolus (6) Rats receiving two cycles of irinotecan followed by 5-FU (6) | Stool | DNA extraction + PCR | Clostridium cluster XI Enterobacteriaceae Glutamate caused: Clostridium cluster VI Bacteroides |
| Carvalho, Vaz, Pereira, Dorella, Aguiar, Chatel, Bermudez, Langella, Fernandes, Figueiredo, Goes-Neto and Azevedo [52] | 5-FU treated mice (72) | Stool | 16s rRNA (V4) | Bacteroidetes Firmicutes Proteobacteria |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalasabail, S.; Engelman, J.; Zhang, L.Y.; El-Omar, E.; Yim, H.C.H. A Perspective on the Role of Microbiome for Colorectal Cancer Treatment. Cancers 2021, 13, 4623. https://doi.org/10.3390/cancers13184623
Kalasabail S, Engelman J, Zhang LY, El-Omar E, Yim HCH. A Perspective on the Role of Microbiome for Colorectal Cancer Treatment. Cancers. 2021; 13(18):4623. https://doi.org/10.3390/cancers13184623
Chicago/Turabian StyleKalasabail, Sanjna, Jared Engelman, Linda Yun Zhang, Emad El-Omar, and Howard Chi Ho Yim. 2021. "A Perspective on the Role of Microbiome for Colorectal Cancer Treatment" Cancers 13, no. 18: 4623. https://doi.org/10.3390/cancers13184623
APA StyleKalasabail, S., Engelman, J., Zhang, L. Y., El-Omar, E., & Yim, H. C. H. (2021). A Perspective on the Role of Microbiome for Colorectal Cancer Treatment. Cancers, 13(18), 4623. https://doi.org/10.3390/cancers13184623







