Copy Number Changes and Allele Distribution Patterns of Chromosome 21 in B Cell Precursor Acute Lymphoblastic Leukemia
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Cytogenetic Analysis
2.3. Fluorescence In Situ Hybridization (FISH)
2.4. CGH/SNP Array Analysis
2.5. Short Tandem Repeat (STR) Analysis
2.6. RNA Sequencing (RNAseq)
3. Results and Discussion
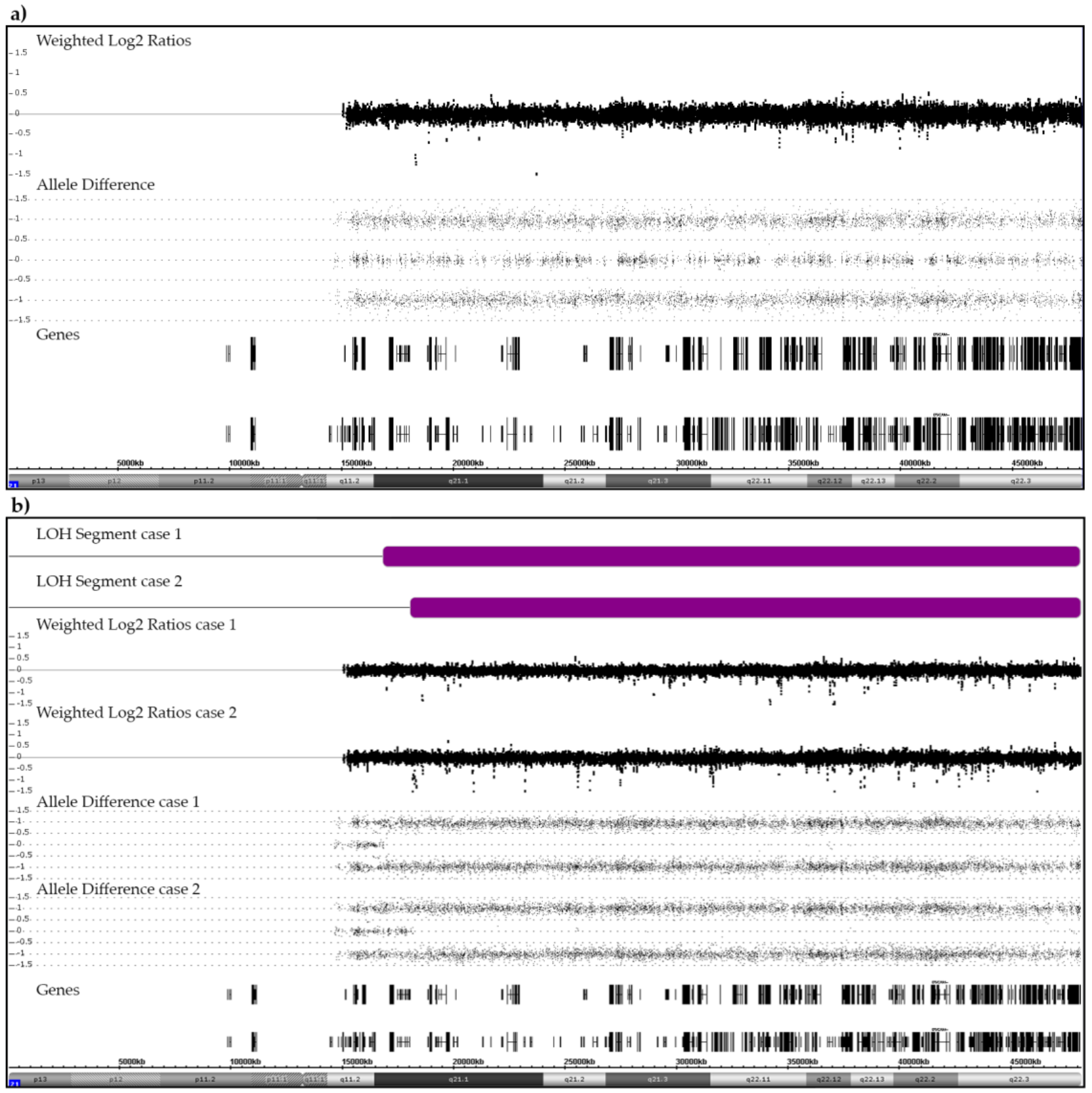
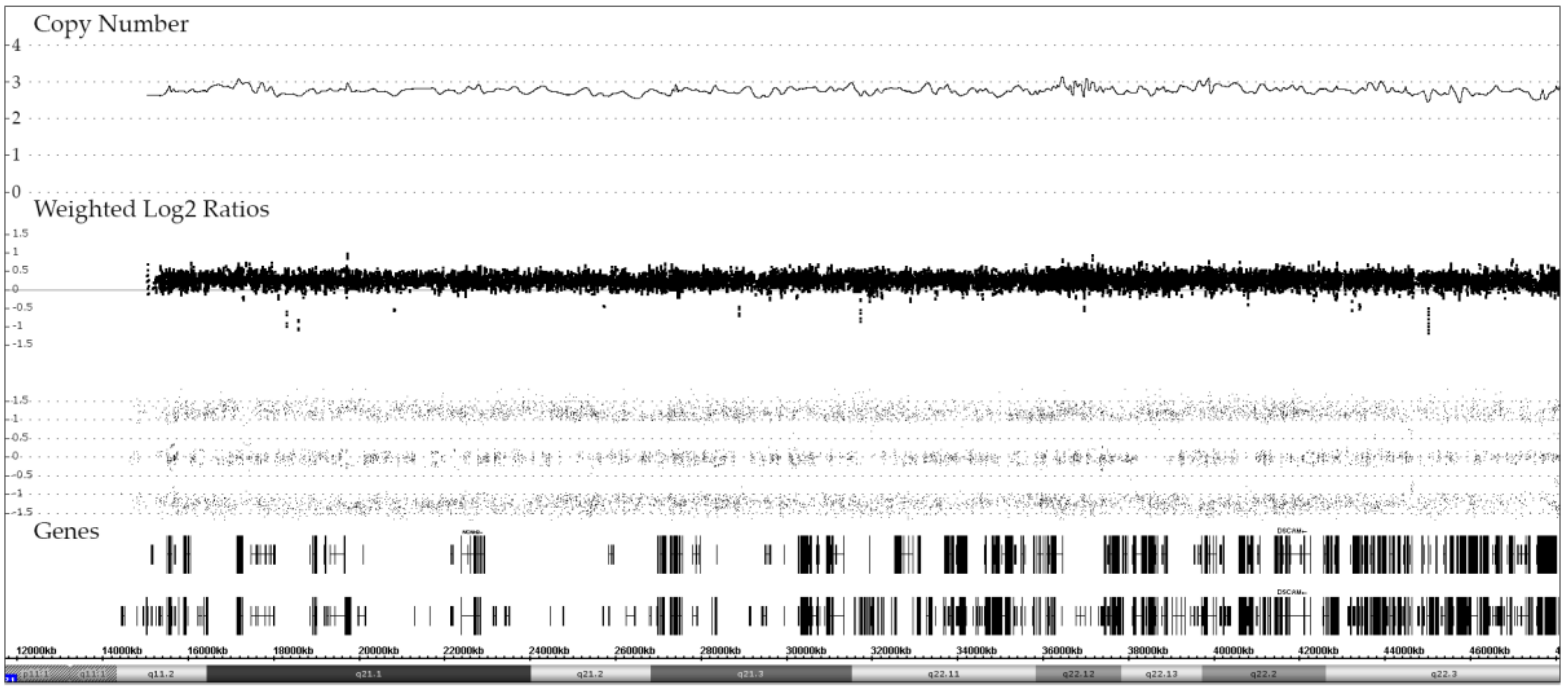
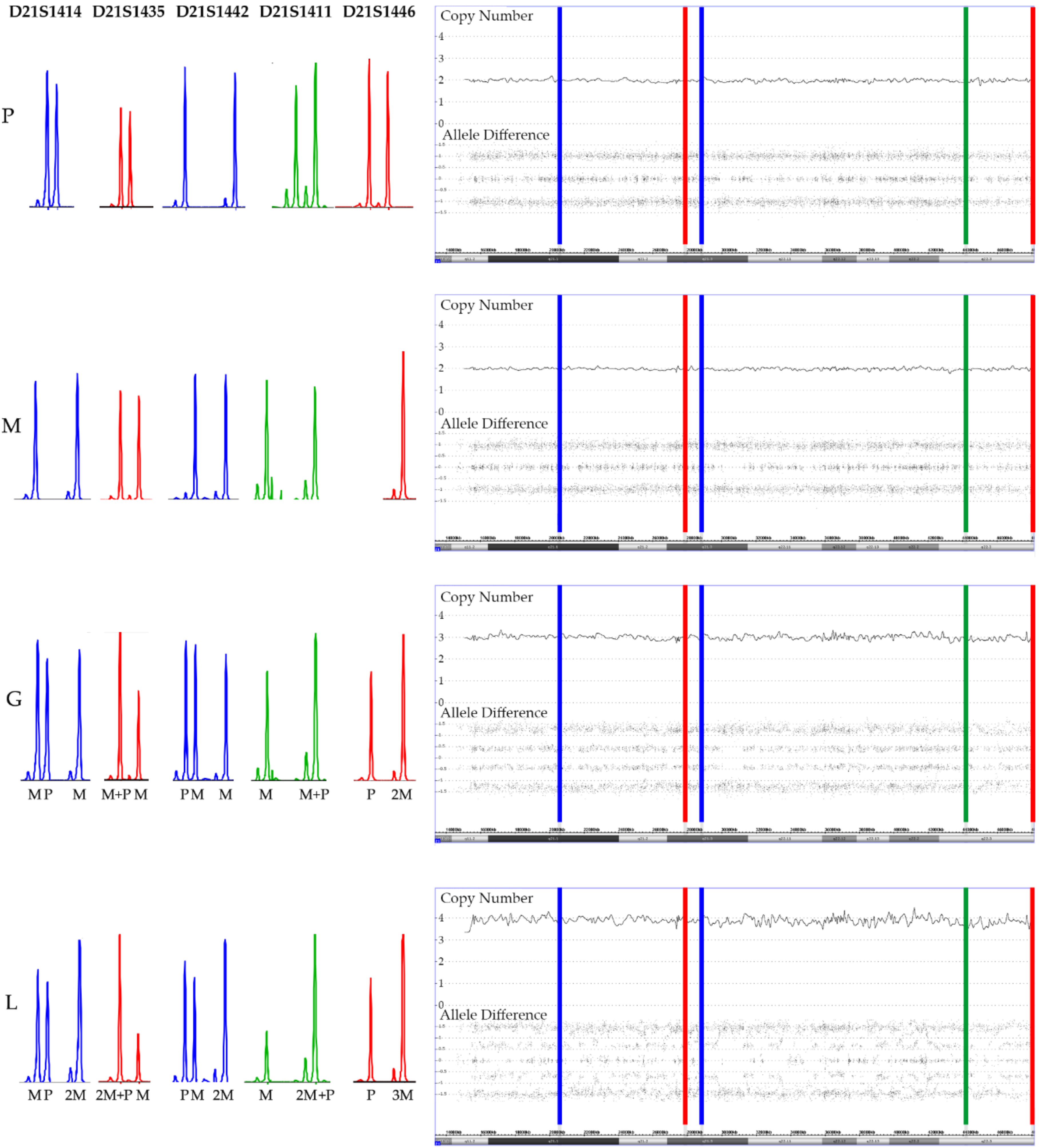
3.1. Disomy 21
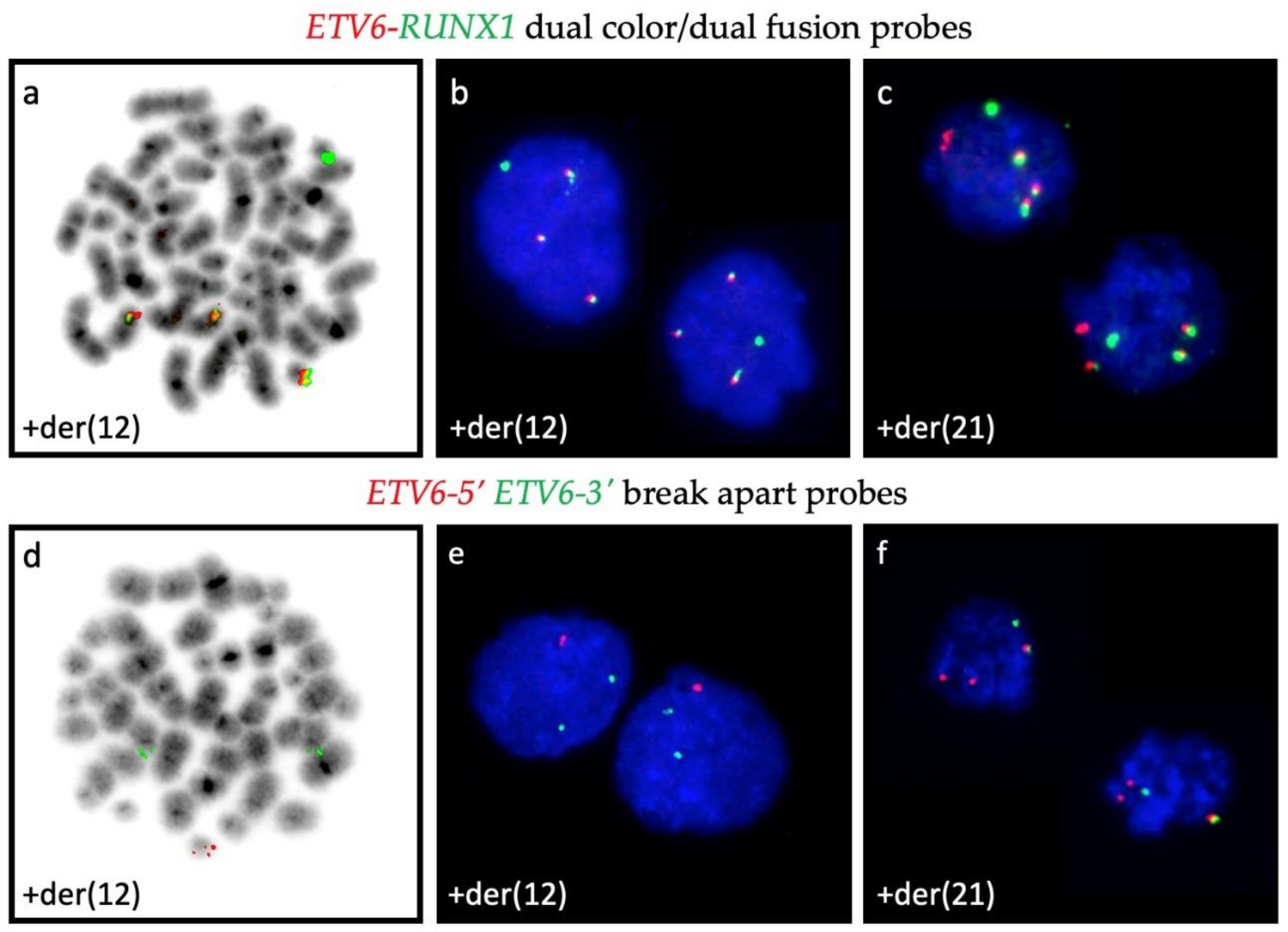
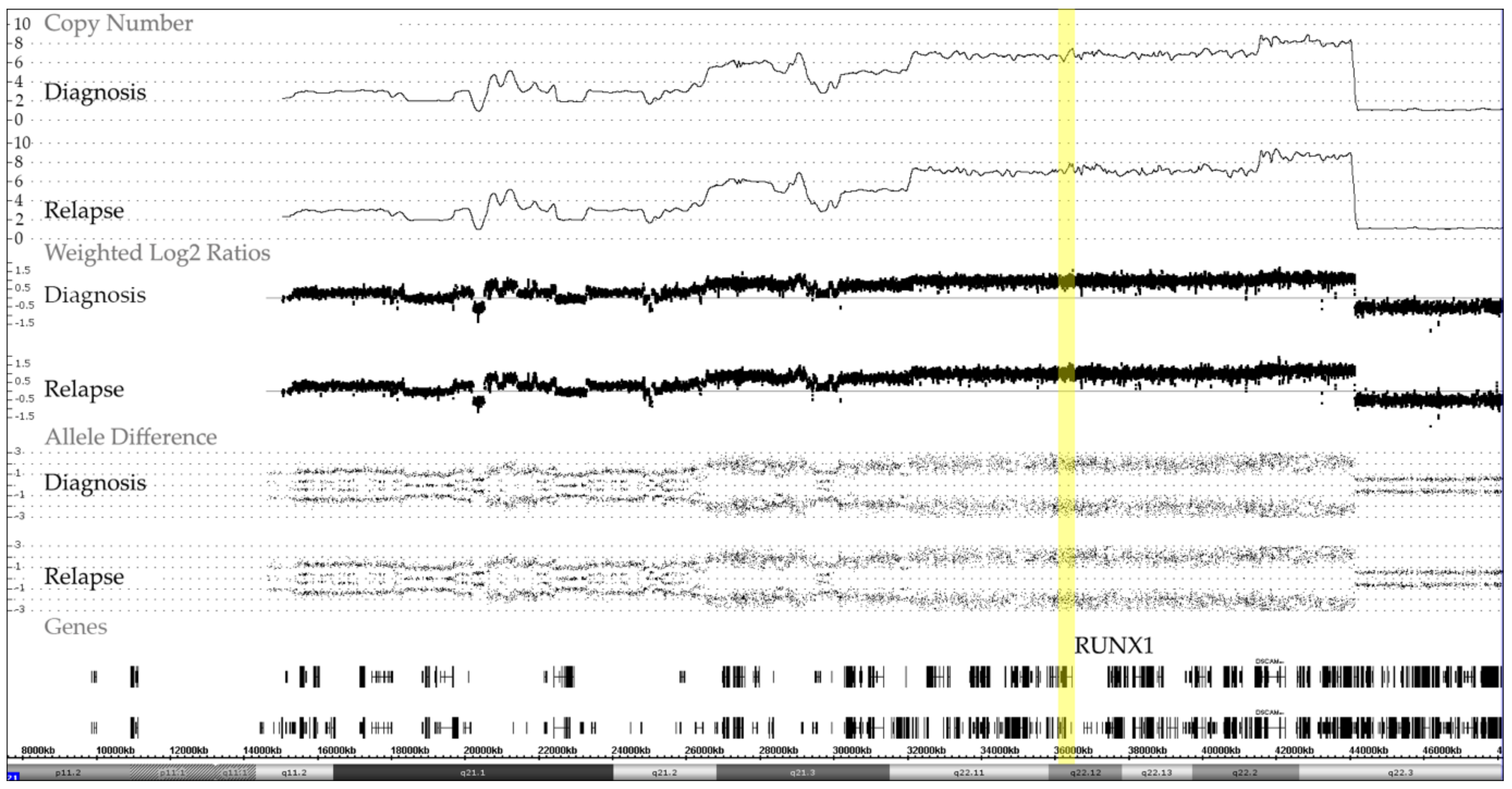
3.2. CNA in ETV6-RUNX1-Positive Cases
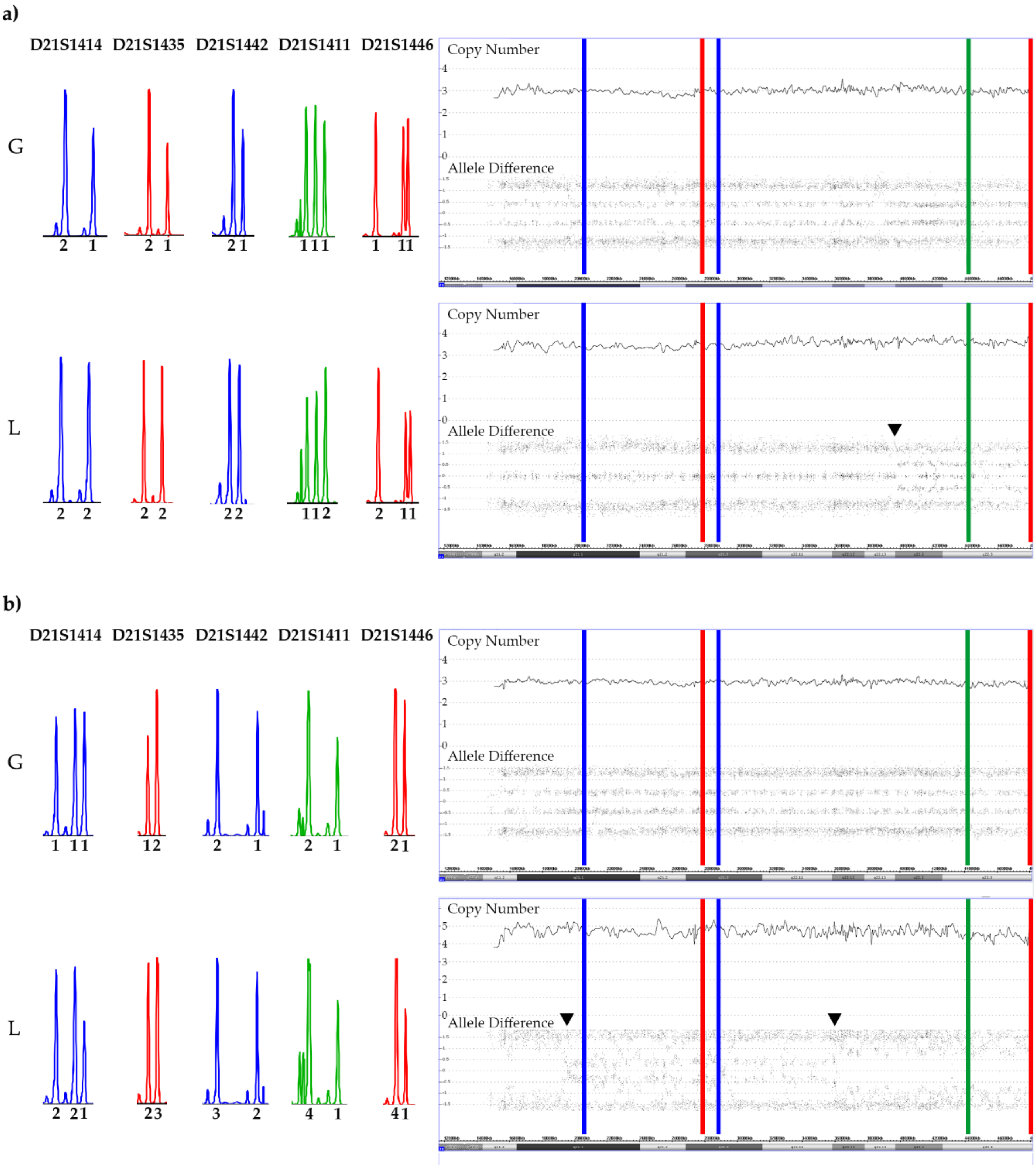
3.3. CNA and Allele Patterns in iAMP21 Cases
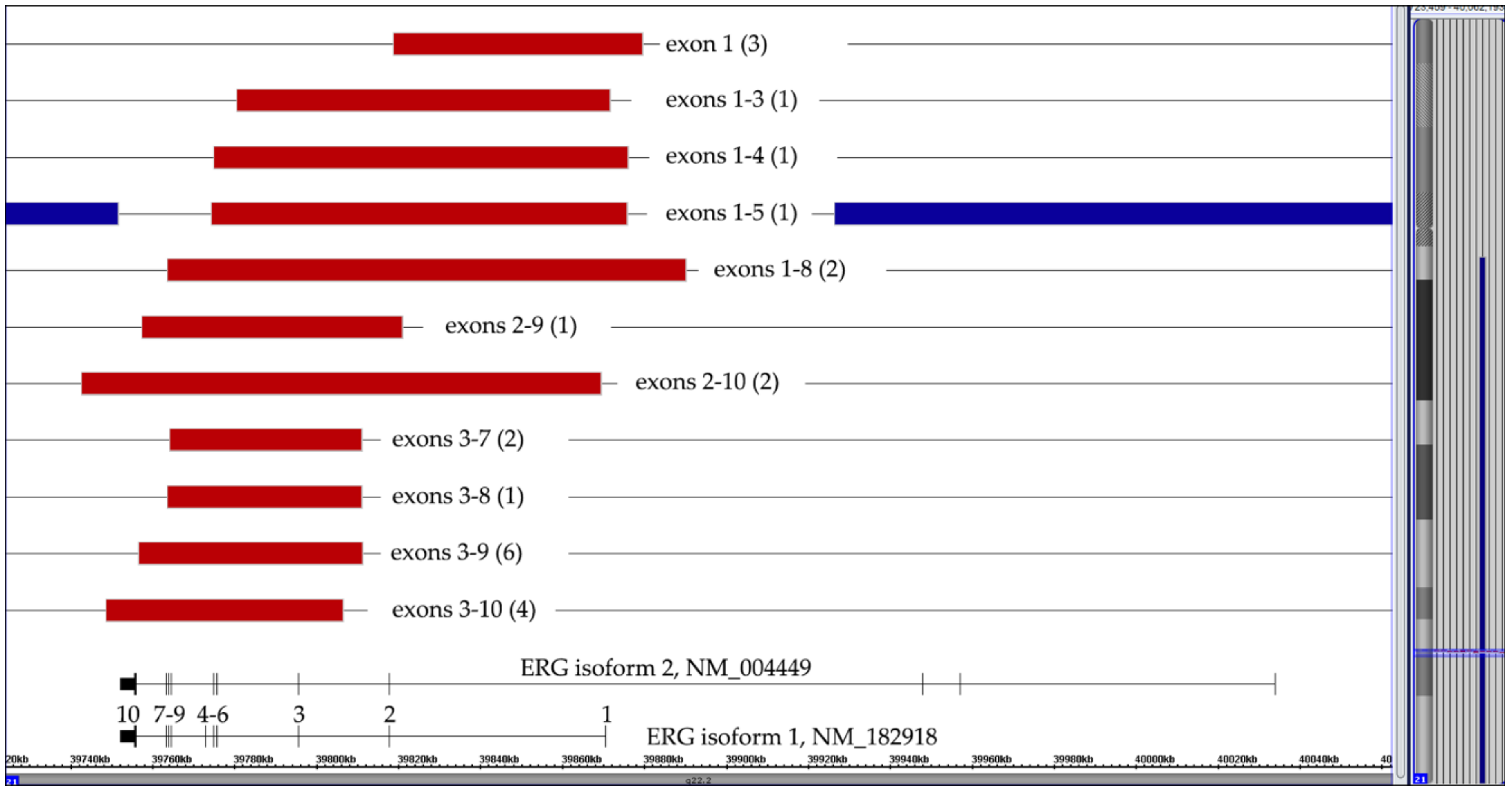
3.4. ERG Deletions
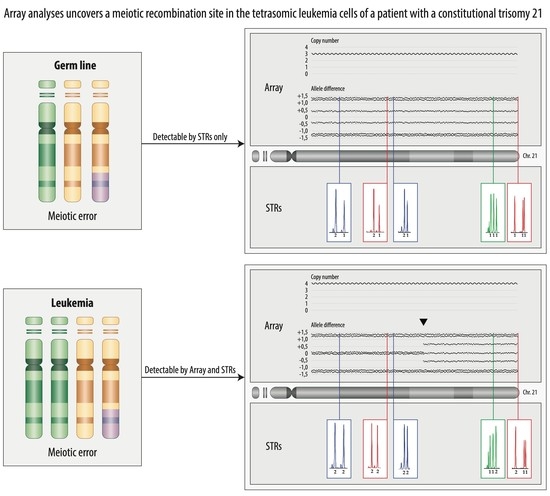
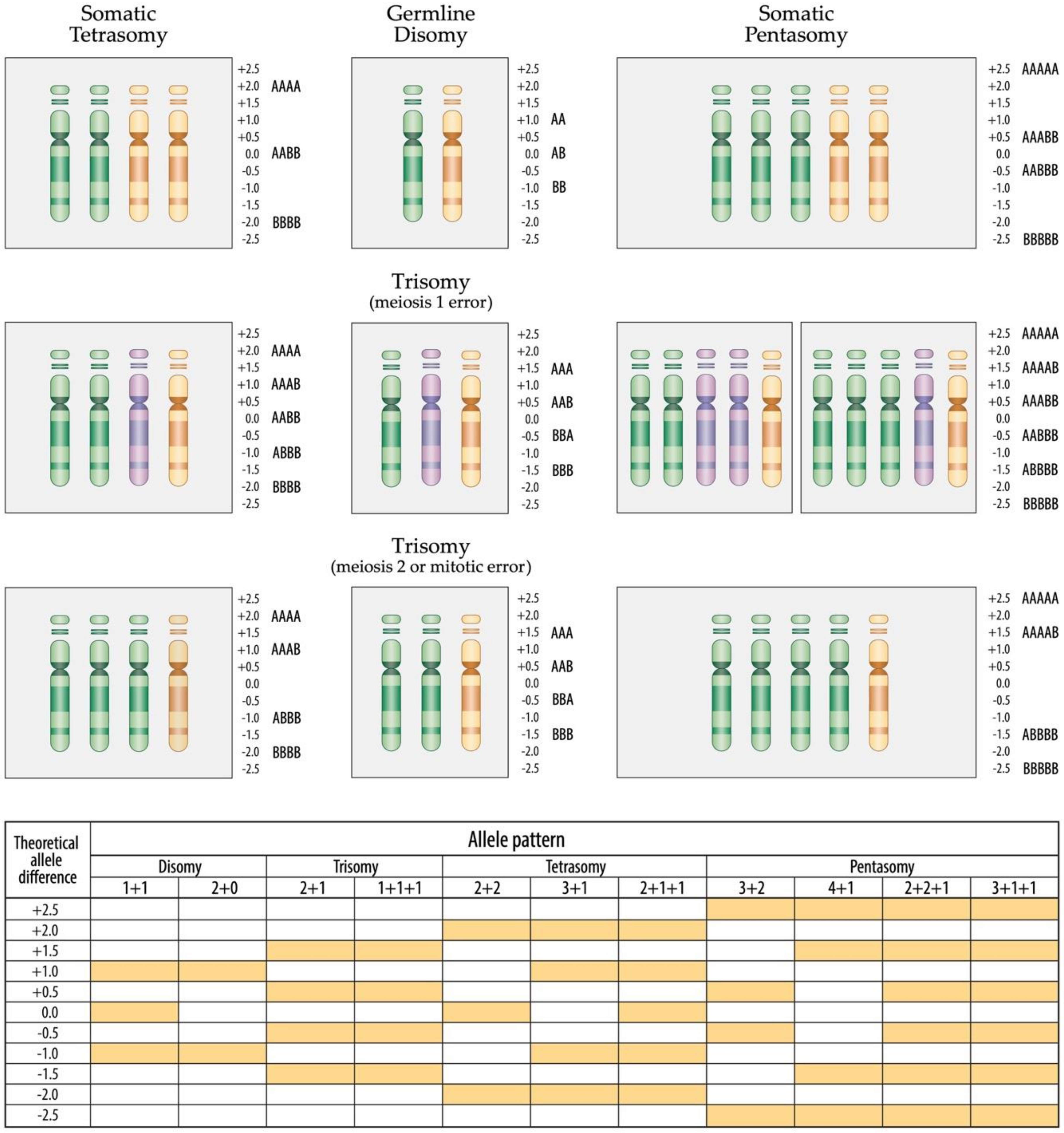
3.5. Constitutional Trisomy 21
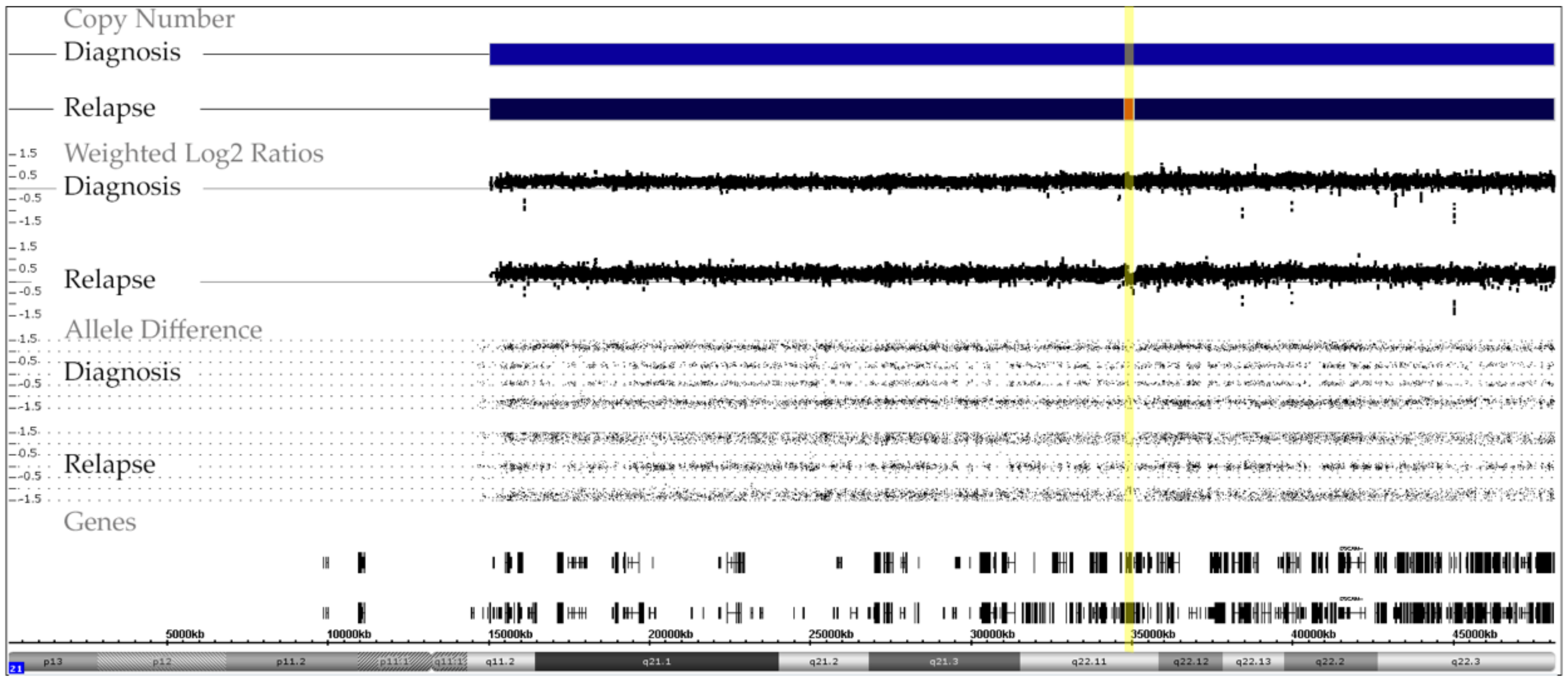
3.6. Acquired Trisomy 21
3.7. Tetrasomy 21
3.8. Pentasomy 21
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Izraeli, S. The acute lymphoblastic leukemia of Down Syndrome—Genetics and pathogenesis. Eur. J. Med. Genet. 2016, 59, 158–161. [Google Scholar] [CrossRef]
- Mateos, M.K.; Barbaric, D.; Byatt, S.A.; Sutton, R.; Marshall, G.M. Down syndrome and leukemia: Insights into leukemogenesis and translational targets. Transl. Pediatr. 2015, 4, 76–92. [Google Scholar] [CrossRef]
- Maloney, K.W.; Taub, J.W.; Ravindranath, Y.; Roberts, I.; Vyas, P. Down syndrome preleukemia and leukemia. Pediatr Clin. N. Am. 2015, 62, 121–137. [Google Scholar] [CrossRef]
- Buitenkamp, T.D.; Izraeli, S.; Zimmermann, M.; Forestier, E.; Heerema, N.A.; van den Heuvel-Eibrink, M.M.; Pieters, R.; Korbijn, C.M.; Silverman, L.B.; Schmiegelow, K.; et al. Acute lymphoblastic leukemia in children with Down syndrome: A retrospective analysis from the Ponte di Legno study group. Blood 2014, 123, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Lundin, C.; Forestier, E.; Klarskov Andersen, M.; Autio, K.; Barbany, G.; Cavelier, L.; Golovleva, I.; Heim, S.; Heinonen, K.; Hovland, R.; et al. Clinical and genetic features of pediatric acute lymphoblastic leukemia in Down syndrome in the Nordic countries. J. Hematol. Oncol. 2014, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Roberts, I.; Izraeli, S. Haematopoietic development and leukaemia in Down syndrome. Br. J. Haematol. 2014, 167, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Hasle, H. Pattern of malignant disorders in individuals with Down’s syndrome. Lancet Oncol. 2001, 2, 429–436. [Google Scholar] [CrossRef]
- Borst, L.; Wesolowska, A.; Joshi, T.; Borup, R.; Nielsen, F.C.; Andersen, M.K.; Jonsson, O.G.; Wehner, P.S.; Wesenberg, F.; Frost, B.M.; et al. Genome-wide analysis of cytogenetic aberrations in ETV6/RUNX1-positive childhood acute lymphoblastic leukaemia. Br. J. Haematol. 2012, 157, 476–482. [Google Scholar] [CrossRef]
- Lilljebjorn, H.; Soneson, C.; Andersson, A.; Heldrup, J.; Behrendtz, M.; Kawamata, N.; Ogawa, S.; Koeffler, H.P.; Mitelman, F.; Johansson, B.; et al. The correlation pattern of acquired copy number changes in 164 ETV6/RUNX1-positive childhood acute lymphoblastic leukemias. Hum. Mol. Genet. 2010, 19, 3150–3158. [Google Scholar] [CrossRef] [PubMed]
- Forestier, E.; Andersen, M.K.; Autio, K.; Blennow, E.; Borgstrom, G.; Golovleva, I.; Heim, S.; Heinonen, K.; Hovland, R.; Johannsson, J.H.; et al. Cytogenetic patterns in ETV6/RUNX1-positive pediatric B-cell precursor acute lymphoblastic leukemia: A Nordic series of 245 cases and review of the literature. Genes Chromosomes Cancer 2007, 46, 440–450. [Google Scholar] [CrossRef]
- Karrman, K.; Forestier, E.; Andersen, M.K.; Autio, K.; Borgström, G.; Heim, S.; Heinonen, K.; Hovland, R.; Kerndrup, G.; Johansson, B. High incidence of the ETV6/RUNX1 fusion gene in paediatric precursor B-cell acute lymphoblastic leukaemias with trisomy 21 as the sole cytogenetic change: A Nordic series of cases diagnosed 1989?2005. Br. J. Haematol. 2006, 135, 352–354. [Google Scholar] [CrossRef]
- Attarbaschi, A.; Mann, G.; Konig, M.; Dworzak, M.N.; Trebo, M.M.; Muhlegger, N.; Gadner, H.; Haas, O.A.; Austrian Berlin-Frankfurt-Münster (BFM) Study Group. Incidence and relevance of secondary chromosome abnormalities in childhood TEL/AML1+ acute lymphoblastic leukemia: An interphase FISH analysis. Leukemia 2004, 18, 1611–1616. [Google Scholar] [CrossRef]
- Pichler, H.; Moricke, A.; Mann, G.; Teigler-Schlegel, A.; Niggli, F.; Nebral, K.; Konig, M.; Inthal, A.; Krehan, D.; Dworzak, M.N.; et al. Prognostic relevance of dic(9;20)(p11;q13) in childhood B-cell precursor acute lymphoblastic leukaemia treated with Berlin-Frankfurt-Munster (BFM) protocols containing an intensive induction and post-induction consolidation therapy. Br. J. Haematol. 2010, 149, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Forestier, E.; Gauffin, F.; Andersen, M.K.; Autio, K.; Borgstrom, G.; Golovleva, I.; Gustafsson, B.; Heim, S.; Heinonen, K.; Heyman, M.; et al. Clinical and cytogenetic features of pediatric dic(9;20)(p13.2;q11.2)-positive B-cell precursor acute lymphoblastic leukemias: A Nordic series of 24 cases and review of the literature. Genes Chromosomes Cancer 2008, 47, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Safavi, S.; Paulsson, K. Near-haploid and low-hypodiploid acute lymphoblastic leukemia: Two distinct subtypes with consistently poor prognosis. Blood 2017, 129, 420–423. [Google Scholar] [CrossRef]
- Holmfeldt, L.; Wei, L.; Diaz-Flores, E.; Walsh, M.; Zhang, J.; Ding, L.; Payne-Turner, D.; Churchman, M.; Andersson, A.; Chen, S.C.; et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat. Genet. 2013, 45, 242–252. [Google Scholar] [CrossRef]
- Paulsson, K.; Johansson, B. High hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer 2009, 48, 637–660. [Google Scholar] [CrossRef]
- Harrison, C.J. Blood Spotlight on iAMP21 acute lymphoblastic leukemia (ALL), a high-risk pediatric disease. Blood 2015, 125, 1383–1386. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C.J.; Moorman, A.V.; Schwab, C.; Carroll, A.J.; Raetz, E.A.; Devidas, M.; Strehl, S.; Nebral, K.; Harbott, J.; Teigler-Schlegel, A.; et al. An international study of intrachromosomal amplification of chromosome 21 (iAMP21): Cytogenetic characterization and outcome. Leukemia 2014, 28, 1015–1021. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Schwab, C.; Ryan, S.; Papaemmanuil, E.; Robinson, H.M.; Jacobs, P.; Moorman, A.V.; Dyer, S.; Borrow, J.; Griffiths, M.; et al. Constitutional and somatic rearrangement of chromosome 21 in acute lymphoblastic leukaemia. Nature 2014, 508, 98–102. [Google Scholar] [CrossRef]
- Zaliova, M.; Potuckova, E.; Hovorkova, L.; Musilova, A.; Winkowska, L.; Fiser, K.; Stuchly, J.; Mejstrikova, E.; Starkova, J.; Zuna, J.; et al. ERG deletions in childhood acute lymphoblastic leukemia with DUX4 rearrangements are mostly polyclonal, prognostically relevant and their detection rate strongly depends on screening method sensitivity. Haematologica 2019, 104, 1407–1416. [Google Scholar] [CrossRef] [PubMed]
- Schinnerl, D.; Mejstrikova, E.; Schumich, A.; Zaliova, M.; Fortschegger, K.; Nebral, K.; Attarbaschi, A.; Fiser, K.; Kauer, M.O.; Popitsch, N.; et al. CD371 cell surface expression: A unique feature of DUX4-rearranged acute lymphoblastic leukemia. Haematologica 2019, 104, e352–e355. [Google Scholar] [CrossRef]
- Zhang, J.; McCastlain, K.; Yoshihara, H.; Xu, B.; Chang, Y.; Churchman, M.L.; Wu, G.; Li, Y.; Wei, L.; Iacobucci, I.; et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat. Genet. 2016, 48, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Stanulla, M.; Dagdan, E.; Zaliova, M.; Moricke, A.; Palmi, C.; Cazzaniga, G.; Eckert, C.; Te Kronnie, G.; Bourquin, J.P.; Bornhauser, B.; et al. IKZF1(plus) Defines a New Minimal Residual Disease-Dependent Very-Poor Prognostic Profile in Pediatric B-Cell Precursor Acute Lymphoblastic Leukemia. J. Clin. Oncol. 2018, 36, 1240–1249. [Google Scholar] [CrossRef]
- Schwab, C.; Harrison, C.J. Advances in B-cell Precursor Acute Lymphoblastic Leukemia Genomics. Hemasphere 2018, 2, e53. [Google Scholar] [CrossRef]
- Olsson, L.; Lundin-Strom, K.B.; Castor, A.; Behrendtz, M.; Biloglav, A.; Noren-Nystrom, U.; Paulsson, K.; Johansson, B. Improved cytogenetic characterization and risk stratification of pediatric acute lymphoblastic leukemia using single nucleotide polymorphism array analysis: A single center experience of 296 cases. Genes Chromosomes Cancer 2018, 57, 604–607. [Google Scholar] [CrossRef]
- Baughn, L.B.; Biegel, J.A.; South, S.T.; Smolarek, T.A.; Volkert, S.; Carroll, A.J.; Heerema, N.A.; Rabin, K.R.; Zweidler-McKay, P.A.; Loh, M.; et al. Integration of cytogenomic data for furthering the characterization of pediatric B-cell acute lymphoblastic leukemia: A multi-institution, multi-platform microarray study. Cancer Genet. 2015, 208, 1–18. [Google Scholar] [CrossRef][Green Version]
- Conlin, L.K.; Thiel, B.D.; Bonnemann, C.G.; Medne, L.; Ernst, L.M.; Zackai, E.H.; Deardorff, M.A.; Krantz, I.D.; Hakonarson, H.; Spinner, N.B. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum. Mol. Genet. 2010, 19, 1263–1275. [Google Scholar] [CrossRef]
- Schieck, M.; Lentes, J.; Thomay, K.; Hofmann, W.; Behrens, Y.L.; Hagedorn, M.; Ebersold, J.; Davenport, C.F.; Fazio, G.; Moricke, A.; et al. Implementation of RNA sequencing and array CGH in the diagnostic workflow of the AIEOP-BFM ALL 2017 trial on acute lymphoblastic leukemia. Ann. Hematol. 2020, 99, 809–818. [Google Scholar] [CrossRef]
- Makishima, H.; Maciejewski, J.P. Pathogenesis and consequences of uniparental disomy in cancer. Clin. Cancer Res. 2011, 17, 3913–3923. [Google Scholar] [CrossRef] [PubMed]
- O’Keefe, C.; McDevitt, M.A.; Maciejewski, J.P. Copy neutral loss of heterozygosity: A novel chromosomal lesion in myeloid malignancies. Blood 2010, 115, 2731–2739. [Google Scholar] [CrossRef]
- Al-Shehhi, H.; Konn, Z.J.; Schwab, C.J.; Erhorn, A.; Barber, K.E.; Wright, S.L.; Gabriel, A.S.; Harrison, C.J.; Moorman, A.V. Abnormalities of the der(12)t(12;21) in ETV6-RUNX1 acute lymphoblastic leukemia. Genes Chromosomes Cancer 2013, 52, 202–213. [Google Scholar] [CrossRef]
- Tsuzuki, S.; Karnan, S.; Horibe, K.; Matsumoto, K.; Kato, K.; Inukai, T.; Goi, K.; Sugita, K.; Nakazawa, S.; Kasugai, Y.; et al. Genetic abnormalities involved in t(12;21) TEL-AML1 acute lymphoblastic leukemia: Analysis by means of array-based comparative genomic hybridization. Cancer Sci. 2007, 98, 698–706. [Google Scholar] [CrossRef]
- Stams, W.A.; Beverloo, H.B.; den Boer, M.L.; de Menezes, R.X.; Stigter, R.L.; van Drunen, E.; Ramakers-van-Woerden, N.L.; Loonen, A.H.; van Wering, E.R.; Janka-Schaub, G.E.; et al. Incidence of additional genetic changes in the TEL and AML1 genes in DCOG and COALL-treated t(12;21)-positive pediatric ALL, and their relation with drug sensitivity and clinical outcome. Leukemia 2006, 20, 410–416. [Google Scholar] [CrossRef]
- Alvarez, Y.; Coll, M.D.; Ortega, J.J.; Bastida, P.; Dastugue, N.; Robert, A.; Cervera, J.; Verdeguer, A.; Tasso, M.; Aventin, A.; et al. Genetic abnormalities associated with the t(12;21) and their impact in the outcome of 56 patients with B-precursor acute lymphoblastic leukemia. Cancer Genet. Cytogenet. 2005, 162, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.A.; Chapuy, B.; Lin, C.Y.; Tivey, T.; Li, H.; Townsend, E.C.; van Bodegom, D.; Day, T.A.; Wu, S.C.; Liu, H.; et al. Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation. Nat. Genet. 2014, 46, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Mowery, C.T.; Reyes, J.M.; Cabal-Hierro, L.; Higby, K.J.; Karlin, K.L.; Wang, J.H.; Kimmerling, R.J.; Cejas, P.; Lim, K.; Li, H.; et al. Trisomy of a Down Syndrome Critical Region Globally Amplifies Transcription via HMGN1 Overexpression. Cell Rep. 2018, 25, 1898.e5–1911.e5. [Google Scholar] [CrossRef] [PubMed]
- Zaliova, M.; Zimmermannova, O.; Dorge, P.; Eckert, C.; Moricke, A.; Zimmermann, M.; Stuchly, J.; Teigler-Schlegel, A.; Meissner, B.; Koehler, R.; et al. ERG deletion is associated with CD2 and attenuates the negative impact of IKZF1 deletion in childhood acute lymphoblastic leukemia. Leukemia 2014, 28, 182–185. [Google Scholar] [CrossRef]
- Clappier, E.; Auclerc, M.F.; Rapion, J.; Bakkus, M.; Caye, A.; Khemiri, A.; Giroux, C.; Hernandez, L.; Kabongo, E.; Savola, S.; et al. An intragenic ERG deletion is a marker of an oncogenic subtype of B-cell precursor acute lymphoblastic leukemia with a favorable outcome despite frequent IKZF1 deletions. Leukemia 2014, 28, 70–77. [Google Scholar] [CrossRef]
- Panzer-Grumayer, R.; Kohrer, S.; Haas, O.A. The enigmatic role(s) of P2RY8-CRLF2. Oncotarget 2017, 8, 96466–96467. [Google Scholar] [CrossRef]
- Nussinov, R.; Tsai, C.J. ‘Latent drivers‘ expand the cancer mutational landscape. Curr. Opin. Struct. Biol. 2015, 32, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Castro-Giner, F.; Ratcliffe, P.; Tomlinson, I. The mini-driver model of polygenic cancer evolution. Nat. Rev. Cancer 2015, 15, 680–685. [Google Scholar] [CrossRef]
- Morak, M.; Attarbaschi, A.; Fischer, S.; Nassimbeni, C.; Grausenburger, R.; Bastelberger, S.; Krentz, S.; Cario, G.; Kasper, D.; Schmitt, K.; et al. Small sizes and indolent evolutionary dynamics challenge the potential role of P2RY8-CRLF2-harboring clones as main relapse-driving force in childhood ALL. Blood 2012, 120, 5134–5142. [Google Scholar] [CrossRef] [PubMed]
- Hassold, T.; Sherman, S. Down syndrome: Genetic recombination and the origin of the extra chromosome 21. Clin. Genet. 2000, 57, 95–100. [Google Scholar] [CrossRef]
- Jain, S.; Agarwal, S.; Panigrahi, I.; Tamhankar, P.; Phadke, S. Diagnosis of Down syndrome and detection of origin of nondisjunction by short tandem repeat analysis. Genet Test Mol Biomarkers 2010, 14, 489–491. [Google Scholar] [CrossRef]
- Oliver, T.R.; Feingold, E.; Yu, K.; Cheung, V.; Tinker, S.; Yadav-Shah, M.; Masse, N.; Sherman, S.L. New insights into human nondisjunction of chromosome 21 in oocytes. PLoS Genet. 2008, 4, e1000033. [Google Scholar] [CrossRef]
- Nagaoka, S.I.; Hassold, T.J.; Hunt, P.A. Human aneuploidy: Mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 2012, 13, 493–504. [Google Scholar] [CrossRef]
- Paulsson, K.; Lindgren, D.; Johansson, B. SNP array analysis of leukemic relapse samples after allogeneic hematopoietic stem cell transplantation with a sibling donor identifies meiotic recombination spots and reveals possible correlation with the breakpoints of acquired genetic aberrations. Leukemia 2011, 25, 1358–1361. [Google Scholar] [CrossRef]
- Brown, A.; Niggli, F.; Hengartner, H.; Caflisch, U.; Nobile, L.; Kuhne, T.; Angst, R.; Bourquin, J.P.; Betts, D. Characterization of high-hyperdiploidy in childhood acute lymphoblastic leukemia with gain of a single chromosome 21. Leuk Lymphoma 2007, 48, 2457–2460. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Churchman, M.L.; Roberts, K.G.; Moore, I.; Zhou, X.; Nakitandwe, J.; Hagiwara, K.; Pelletier, S.; Gingras, S.; Berns, H.; et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat. Genet. 2019, 51, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Haas, O.A. Somatic Sex: On the Origin of Neoplasms With Chromosome Counts in Uneven Ploidy Ranges. Front. Cell Dev. Biol. 2021, 9, 631946. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.F.; Ketterling, R.P.; Huang, L.; Finn, L.E.; Shi, M.; Hoppman, N.L.; Greipp, P.T.; Baughn, L.B. A near-haploid clone harboring a BCR/ABL1 gene fusion in an adult patient with newly diagnosed B-lymphoblastic leukemia. Genes Chromosomes Cancer 2019, 58, 665–668. [Google Scholar] [CrossRef] [PubMed]
- Sunil, S.K.; Prakash, P.N.; Hariharan, S.; Vinod, G.; Preethi, R.T.; Geetha, N.; Ankathil, R. Adult acute lymphoblastic leukemia with near haploidy, hyperdiploidy and Ph positive lines: A rare entity with poor prognosis. Leuk Lymphoma 2006, 47, 561–563. [Google Scholar] [CrossRef] [PubMed]



| Genetic Subtype | Patients | Samples | Male | Female | Diagnosis | Relapse | Matched | Down | Copy Number & Allele Distribution | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | ||||||||||||
| 1 + 1 | 1 * | 2 + 1 | 2 + 2 | 3 + 1 | 2 + 1+1 | 3 + 2 | 3 + 3 | |||||||||
| ETV6-RUNX1 fusion | 66 | 69 | 34 | 32 | 66 | 3 | 2 | 1 | 54 | 0 | 12 | 0 | 0 | 0 | 0 | 0 |
| KMT2A fusions | 17 | 21 | 6 | 11 | 17 | 4 | 3 | 0 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| BCR-ABL1 fusion | 8 | 10 | 7 | 1 | 6 | 4 | 2 | 0 | 7 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| TCF3-PBX1 fusion | 16 | 16 | 8 | 8 | 15 | 1 | 0 | 0 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Aneuploid | 200 | 212 | 115 | 85 | 197 | 15 | 11 | 8 | 3 | 0 | 21 | 156 | 8 | 1 | 11 | 0 |
| Hyperdiploid, classical | 186 | 198 | 106 | 80 | 184 | 14 | 11 | 8 | 0 | 0 | 21 | 145 | 8 | 1 | 11 | 0 |
| Hyperdiploid, GW CN-LOH ** | 5 | 5 | 2 | 3 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 |
| Hyperhaploid/hyperdiploid | 2 | 2 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| Hypodiploid only | 3 | 3 | 3 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hypodiploid/hyperdiploid | 4 | 4 | 3 | 1 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 |
| B-other | 271 | 294 | 157 | 114 | 264 | 30 | 22 | 17 | 196 | 2 | 48 | 5 | 0 | 0 | 1 | 0 |
| iAMP21 | 19 | 24 | 7 | 12 | 17 | 7 | 5 | 0 | ||||||||
| P2RY8-CRLF2 fusion | 26 | 30 | 18 | 8 | 26 | 4 | 3 | 9 | 10 | 0 | 15 | 1 | 0 | 0 | 0 | 0 |
| IGH-CRLF2 fusion | 12 | 12 | 5 | 7 | 11 | 1 | 0 | 3 | 8 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| dic(9;20) *** | 20 | 24 | 9 | 11 | 20 | 4 | 4 | 0 | 10 | 0 | 10 | 0 | 0 | 0 | 0 | 0 |
| i(9q) | 13 | 13 | 11 | 2 | 13 | 0 | 0 | 2 | 11 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ERG deletions | 24 | 25 | 15 | 10 | 24 | 1 | 0 | 0 | 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Not further specified | 157 | 166 | 92 | 64 | 153 | 13 | 10 | 3 | 133 | 2 | 17 | 4 | 0 | 0 | 1 | 0 |
| Total | 578 | 622 | 327 | 251 | 565 | 57 | 40 | 26 | 293 | 2 | 82 | 161 | 8 | 1 | 12 | 0 |
| Patient | IKZF1 N159Y | RNA seq Profile | Array Pattern |
|---|---|---|---|
| 1 | no | no | 47, XY, del(9) (p13.2)/PAX5,+ 21 |
| 2 | yes | yes | 47, XY, del(2)(p12p11.2),del(16)(p11.2),+ 21 |
| 3 | Not analyzed | 47, XX, del(3)(q25.31q25.32),del(3)(q25.32q26.1),del(3)(q26.1),del(3)(q26.32), del(3)(q26.32q26.33),chromothripsis(4)(q22.1q34.1),del(5)(q21.3q22.2),del(5)(q35.1),del(5)(q35.1),del(5)(q35.1q35.2),del(5)(q35.3),+ 21 | |
| 4 | yes | yes | 47, XX, dup(X)(p22.33p11.22),dup(7)(q11.21q36.3),del(17)(q25.3),+21 |
| 5 | yes | yes | 47, XY,+21 |
| 6 | Not analyzed | 50, X,i(X)(p22.33p11.1)x3,der(X)(q),del(9)(p13.2)/PAX5,+ 21 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbasi, M.R.; Nebral, K.; Haslinger, S.; Inthal, A.; Zeitlhofer, P.; König, M.; Schinnerl, D.; Köhrer, S.; Strehl, S.; Panzer-Grümayer, R.; et al. Copy Number Changes and Allele Distribution Patterns of Chromosome 21 in B Cell Precursor Acute Lymphoblastic Leukemia. Cancers 2021, 13, 4597. https://doi.org/10.3390/cancers13184597
Abbasi MR, Nebral K, Haslinger S, Inthal A, Zeitlhofer P, König M, Schinnerl D, Köhrer S, Strehl S, Panzer-Grümayer R, et al. Copy Number Changes and Allele Distribution Patterns of Chromosome 21 in B Cell Precursor Acute Lymphoblastic Leukemia. Cancers. 2021; 13(18):4597. https://doi.org/10.3390/cancers13184597
Chicago/Turabian StyleAbbasi, M. Reza, Karin Nebral, Sabrina Haslinger, Andrea Inthal, Petra Zeitlhofer, Margit König, Dagmar Schinnerl, Stefan Köhrer, Sabine Strehl, Renate Panzer-Grümayer, and et al. 2021. "Copy Number Changes and Allele Distribution Patterns of Chromosome 21 in B Cell Precursor Acute Lymphoblastic Leukemia" Cancers 13, no. 18: 4597. https://doi.org/10.3390/cancers13184597
APA StyleAbbasi, M. R., Nebral, K., Haslinger, S., Inthal, A., Zeitlhofer, P., König, M., Schinnerl, D., Köhrer, S., Strehl, S., Panzer-Grümayer, R., Mann, G., Attarbaschi, A., & Haas, O. A. (2021). Copy Number Changes and Allele Distribution Patterns of Chromosome 21 in B Cell Precursor Acute Lymphoblastic Leukemia. Cancers, 13(18), 4597. https://doi.org/10.3390/cancers13184597