Best Practices for Spatial Profiling for Breast Cancer Research with the GeoMx® Digital Spatial Profiler
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. Biology and Prognostic Biomarkers in Breast Cancer
1.2. Opportunity of Spatial Biology in Breast Cancer
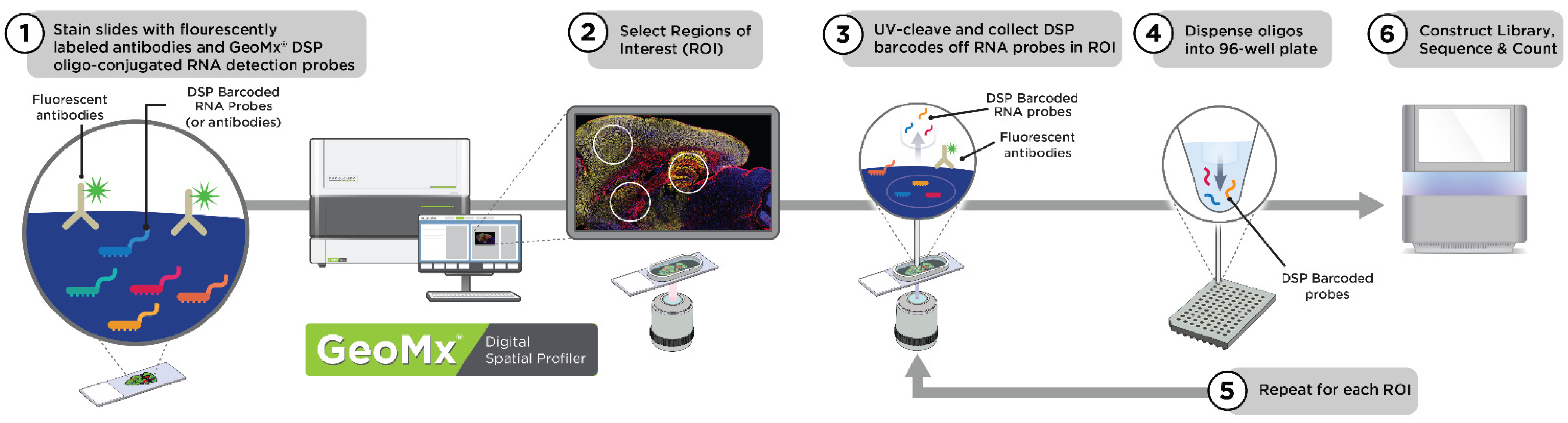
1.3. GeoMx Digital Spatial Profiler (DSP) for Spatially Resolved Biology
2. Defining the Questions in Breast Cancer Biology
2.1. Profiling across Location
2.2. Profiling across Time
2.3. Profiling across Preclinical and Clinical Samples
3. Applications of GeoMx DSP in Breast Cancer
3.1. Characteristics and Spatially-Defined Immune (Micro)Landscapes of Early-Stage PD-L1-Positive Triple-Negative Breast Cancer (Carter JM et al., Clinical Cancer Research 2021)
3.2. Spatial Proteomic Characterization of HER2-Positive Breast Tumors through Neoadjuvant Therapy Predicts Response (McNamara KL et al., Nature Cancer 2021)
3.3. Spatially-Resolved Quantification of Proteins in Triple Negative Breast Cancers Reveals Differences in the Immune Microenvironment Associated with Prognosis (Stewart RL et al., Scientific Reports 2020)
3.4. Multiplexed Digital Spatial Profiling of Invasive Breast Tumors from Black and White Women (Omilian AR et al., Molecular Oncology 2021)
4. Considerations for Robust Experimental Designs
4.1. Sample Considerations
4.1.1. Sample Preparation and Tissue Size
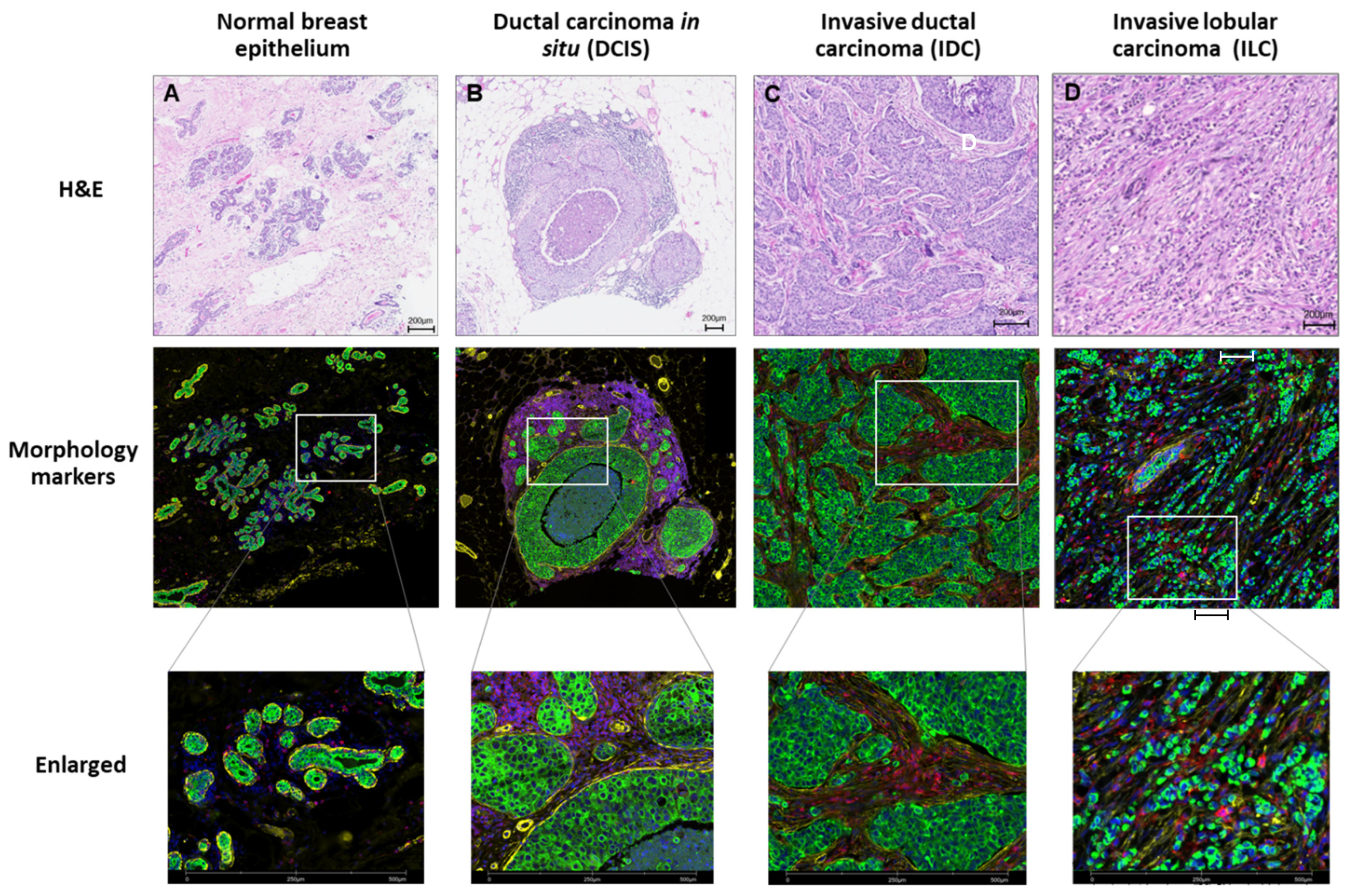
4.1.2. Sample Tissue Features
4.2. ROI Placement and Compartmentalization/Segmentation Strategies
4.2.1. Selecting Visualization Markers
4.2.2. ROI Placement and Tissue Considerations
4.2.3. ROI Size and Quantity
4.2.4. Area of Illumination (AOI) Segmentation Strategies
4.3. Workflow Considerations
5. Tools for Data Analysis and Image Sharing
5.1. Analysis Guidelines for GeoMx Studies
5.1.1. Data Normalization
5.1.2. Analytical Considerations for Running Single- and Multi-Site Studies
5.1.3. Interpreting a Protein’s Counts from an AOI
5.1.4. Comparing Pre- Versus Post-Treatment Data
5.1.5. Reference Sample Inclusion in Ongoing Studies
5.2. Integration with Digital Pathology Toolkits
6. Opportunities for Future Development
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Glossary
| Area of Illumination (AOI) | The area illuminated by ultraviolet (UV) light to photo-release the photocleavable linker that are bound to targets for readout. It must be smaller than a field of view. There is a one-to-one correspondence between AOIs and collected “aspirates”, each to a single well in the AOI collection plate on the GeoMx DSP instrument. If a region of interest (ROI) does not fit into a single field of view, it is broken into multiple AOIs. |
| Digital Spatial Profiling (DSP) | DSP is intended to provide quantitative expression data in a spatial context. Through visual assessment of biological samples using microscopic imaging, researchers can specify ROIs to collect the quantitative expression information of RNA and protein targets. |
| Flow Cell | A general NGS term that refers to the sequencing surface and chamber. |
| GeoMx DSP System | The combination of the GeoMx DSP instruments, assays and software required to generate highly multiplexed DSP data. |
| Mask | Mask is binary ROI image metadata that encodes where the GeoMx must illuminate UV light within an ROI, thus resulting in the area of illumination (AOI). |
| Normalization | A data transformation that flattens differences in counts not arising by biology but by technical or sample variability (i.e., pipetting errors, difference in sample quality, etc.) between lanes, allowing the making of meaningful biological comparisons. |
| Readout (nCounter or NGS) | Digital counting of the hybridization target oligos in collected aspirates on either the nCounter or NGS system. |
| Region of Interest (ROI) | A region of interest as defined by the researcher. It may be arbitrarily large and have any shape the researcher considers appropriate. ROIs are specific to a given scan. |
| Resolution | The minimum UV illumination area that results in signal greater than background plus two times the standard deviation. |
| Scan | An image of a slide. Scans will have one or more imaging channels, may have additional metadata and may have annotated ROIs. A given slide can be scanned multiple times. Scans can originate in the GeoMx DSP system or can include imported images. |
| Segment | A uniquely defined sub-region of a given ROI. Segments may not overlap; the sum of all segments in an ROI does not need to be the entire ROI. |
Abbreviations
| AI | artificial intelligence |
| AOIs | areas of illumination |
| CNBs | core needle biopsies |
| CT | tumor center |
| CTA | Cancer Transcriptome Atlas |
| DCIS | ductal carcinoma in situ |
| DFS | disease free survival |
| DSP | Digital Spatial Profiler |
| ECM | extracellular matrix |
| ER | estrogen receptor |
| FFPE | formalin-fixed paraffin-embedded |
| GBCC | GeoMx Breast Cancer Consortium |
| GSEA | gene set enrichment analysis |
| H&E | hematoxylin and eosin |
| HCA | Human Cell Atlas |
| HER2 | epidermal growth factor receptor 2 |
| HK | housekeeping |
| HuBMAP | Human Biomolecular Atlas Program |
| IDC | invasive ductal carcinoma |
| IgG | immunoglobulin G |
| IHC | immunohistochemistry |
| ILC | invasive lobular carcinoma |
| IM | invasive margin |
| LCIS | lobular carcinoma in situ |
| MIAME | Minimum Information About a Microarray Experiment |
| MINSEQE | Minimum Information About a Next-generation Sequencing Experiment |
| NGS | next generation sequencing |
| PanCK | pan-cytokeratin |
| PCA | principal component analysis |
| pCR | pathological complete response |
| PD | pharmacodynamic |
| PR | progesterone receptor |
| ROIs | regions of interest |
| TILs | tumor-infiltrating lymphocytes |
| TMA | tissue microarray |
| TME | tumor microenvironment |
| TMM | trimmed mean of M values |
| TNBC | triple-negative breast cancer |
| tSNE | t-distributed stochastic neighbor embedding |
| UMAP | uniform manifold approximation and projection |
| UV | ultraviolet |
| WTA | Whole Transcriptome Atlas |
References
- Prat, A.; Cheang, M.C.; Galván, P.; Nuciforo, P.; Paré, L.; Adamo, B.; Muñoz, M.; Viladot, M.; Press, M.F.; Gagnon, R.; et al. Prognostic Value of Intrinsic Subtypes in Hormone Re-ceptor-Positive Metastatic Breast Cancer Treated with Letrozole with or without Lapatinib. JAMA Oncol. 2016, 2, 1287–1294. [Google Scholar] [CrossRef] [Green Version]
- Prat, A.; Cheang, M.C.; Martín, M.; Parker, J.S.; Carrasco, E.; Caballero, R.; Tyldesley, S.; Gelmon, K.; Bernard, P.S.; Nielsen, T.O.; et al. Prognostic significance of progesterone recep-tor-positive tumor cells within immunohistochemically defined luminal A breast cancer. J. Clin. Oncol. 2013, 31, 203–209. [Google Scholar] [CrossRef]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. 2017, 4, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walens, A.; Olsson, L.T.; Gao, X.; Hamilton, A.M.; Kirk, E.L.; Cohen, S.M.; Midkiff, B.R.; Xia, Y.; Sherman, M.E.; Nikolaishvili-Feinberg, N.; et al. Protein-based immune profiles of basal-like vs. luminal breast cancers. Lab. Investig. 2021, 101, 785–793. [Google Scholar] [CrossRef]
- Akbar, M.W.; Isbilen, M.; Belder, N.; Canlı, S.D.; Kucukkaraduman, B.; Turk, C.; Özgür, S.; Gure, A.O. A Stemness and EMT Based Gene Expression Signature Identifies Phenotypic Plasticity and is A Predictive but Not Prognostic Biomarker for Breast Cancer. J. Cancer 2020, 11, 949–961. [Google Scholar] [CrossRef] [Green Version]
- Lindström, L.S.; Yau, C.; Czene, K.; Thompson, C.K.; Hoadley, K.; Veer, L.J.V.; Balassanian, R.; Bishop, J.W.; Carpenter, P.M.; Chen, Y.-Y.; et al. Intratumor Heterogeneity of the Estrogen Receptor and the Long-term Risk of Fatal Breast Cancer. J. Natl. Cancer Inst. 2018, 110, 726–733. [Google Scholar] [CrossRef] [Green Version]
- Tekpli, X.; Lien, T.; Røssevold, A.H.; Nebdal, D.; Borgen, E.; Ohnstad, H.O.; Kyte, J.A.; Vallon-Christersson, J.; Fongaard, M.; Due, E.U.; et al. An independent poor-prognosis subtype of breast cancer defined by a distinct tumor immune microenvironment. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Fougner, C.; Bergholtz, H.; Norum, J.H.; Sørlie, T. Re-definition of claudin-low as a breast cancer phenotype. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Aleskandarany, M.A.; Vandenberghe, M.E.; Marchiò, C.; Ellis, I.; Sapino, A.; Rakha, E.A. Tumour Heterogeneity of Breast Cancer: From Morphology to Personalised Medicine. Pathobiology 2018, 85, 23–34. [Google Scholar] [CrossRef]
- Bareche, Y.; Buisseret, L.; Gruosso, T.; Girard, E.; Venet, D.; Dupont, F.; Desmedt, C.; Larsimont, D.; Park, M.; Rothé, F.; et al. Unraveling Triple-Negative Breast Cancer Tumor Microenvironment Heterogeneity: Towards an Optimized Treatment Approach. J. Natl. Cancer Inst. 2020, 112, 708–719. [Google Scholar] [CrossRef] [Green Version]
- Casadevall, D.; Li, X.; Powles, R.L.; Wali, V.B.; Buza, N.; Pelekanou, V.; Dhawan, A.; Foldi, J.; Szekely, B.; Lopez-Giraldez, F.; et al. Genomic and Immune Profiling of a Patient with Tri-ple-Negative Breast Cancer That Progressed During Neoadjuvant Chemotherapy Plus PD-L1 Blockade. JCO Precis Oncol. 2019, 3. [Google Scholar] [CrossRef]
- Schroth, W.; Büttner, F.A.; Kandabarau, S.; Hoppe, R.; Fritz, P.; Kumbrink, J.; Kirchner, T.; Brauer, H.A.; Ren, Y.; Henderson, D.A.; et al. Gene expression signatures of BRCAness and tumor inflammation define subgroups of early-stage hormone receptor-positive breast cancer patients. Clin. Cancer Res. 2020, 26, 6523–6534. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.U.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Fumagalli, C.; Ranghiero, A.; Gandini, S.; Corso, F.; Taormina, S.; De Camilli, E.; Rappa, A.; Vacirca, D.; Viale, G.; Guerini-Rocco, E.; et al. Inter-tumor genomic heterogeneity of breast cancers: Comprehensive genomic profile of primary early breast cancers and relapses. Breast Cancer Res. 2020, 22, 1–11. [Google Scholar] [CrossRef]
- Li, X.; Warren, S.; Pelekanou, V.; Wali, V.; Cesano, A.; Liu, M.; Danaher, P.; Elliott, N.; Nahleh, Z.A.; Hayes, D.F.; et al. Immune profiling of pre- and post-treatment breast cancer tissues from the SWOG S0800 neoadjuvant trial. J. Immunother. Cancer 2019, 7, 88. [Google Scholar] [CrossRef] [Green Version]
- Denkert, C.; Von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Heppner, B.I.; Weber, K.E.; Budczies, J.; Huober, J.; Klauschen, F.; Furlanetto, J.; et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: A pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018, 19, 40–50. [Google Scholar] [CrossRef]
- Luen, S.J.; Salgado, R.; Fox, S.; Savas, P.; Eng-Wong, J.; Clark, E.; Kiermaier, A.; Swain, S.; Baselga, J.; Michiels, S.; et al. Tumour-infiltrating lymphocytes in advanced HER2-positive breast cancer treated with pertuzumab or placebo in addition to trastuzumab and docetaxel: A retrospective analysis of the CLEOPATRA study. Lancet Oncol. 2017, 18, 52–62. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Ericsson, P.I.; Stovgaard, E.S.; Sua, L.F.; Reisenbichler, E.; Kos, Z.; Carter, J.M.; Michiels, S.; Quesne, J.L.; Nielsen, T.O.; Lænkholm, A.-V.; et al. The path to a better biomarker: Application of a risk management framework for the implementation of PD-L1 and TILs as immunooncology biomarkers in breast cancer clinical trials and daily practice. J. Pathol. 2020, 250, 667–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amgad, M.; Stovgaard, E.S.; Balslev, E.; Thagaard, J.; Chen, W.; Dudgeon, S.; Sharma, A.; Kerner, J.K.; Denkert, C.; Yuan, Y.; et al. Report on computational assessment of Tumor Infiltrating Lymphocytes from the International Immuno-Oncology Biomarker Working Group. NPJ Breast Cancer 2020, 6, 1–13. [Google Scholar] [CrossRef]
- Watanabe, T.; Hida, A.I.; Inoue, N.; Imamura, M.; Fujimoto, Y.; Akazawa, K.; Hirota, S.; Miyoshi, Y. Abundant tumor infiltrating lymphocytes after primary systemic chemotherapy predicts poor prognosis in estrogen receptor-positive/HER2-negative breast cancers. Breast Cancer Res. Treat. 2017, 168, 135–145. [Google Scholar] [CrossRef]
- Al-Saleh, K.; El-Aziz, N.A.; Ali, A.; Abozeed, W.; El-Warith, A.A.; Ibraheem, A.; Ansari, J.; Al-Rikabi, A.; Husain, S.; Nabholtz, J.-M. Predictive and prognostic significance of CD8+ tumor-infiltrating lymphocytes in patients with luminal B/HER 2 negative breast cancer treated with neoadjuvant chemotherapy. Oncol. Lett. 2017, 14, 337–344. [Google Scholar] [CrossRef] [Green Version]
- Hida, A.I.; Sagara, Y.; Yotsumoto, D.; Kanemitsu, S.; Kawano, J.; Baba, S.; Rai, Y.; Oshiro, Y.; Aogi, K.; Sagara, Y.; et al. Prognostic and predictive impacts of tumor-infiltrating lymphocytes differ between Triple-negative and HER2-positive breast cancers treated with standard systemic therapies. Breast Cancer Res. Treat. 2016, 158, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Crosby, E.J.; Acharya, C.R.; Haddad, A.F.; Rabiola, C.A.; Lei, G.; Wei, J.P.; Yang, X.Y.; Wang, T.; Liu, C.X.; Wagner, K.U.; et al. Stimulation of Oncogene-Specific Tumor-Infiltrating T Cells through Combined Vaccine and αPD-1 Enable Sustained Antitumor Responses against Established HER2 Breast Cancer. Clin. Cancer Res. 2020, 26, 4670–4681. [Google Scholar] [CrossRef]
- Luen, S.; Virassamy, B.; Savas, P.; Salgado, R.; Loi, S. The genomic landscape of breast cancer and its interaction with host immunity. Breast 2016, 29, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Tse, L.A.; Wang, D.; Koka, H.; Zhang, T.; Abubakar, M.; Lee, P.; Wang, F.; Wu, C.; Tsang, K.H.; et al. Immune gene expression profiling reveals heterogeneity in luminal breast tumors. Breast Cancer Res. 2019, 21, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quintana, Á.; Peg, V.; Prat, A.; Moliné, T.; Villacampa, G.; Paré, L.; Galván, P.; Dientsmann, R.; Schmid, P.; Curigliano, G.; et al. Immune analysis of lymph nodes in relation to the presence or absence of tumor infiltrating lymphocytes in triple-negative breast cancer. Eur. J. Cancer 2021, 148, 134–145. [Google Scholar] [CrossRef]
- Ali, H.R.; Provenzano, E.; Dawson, S.-J.; Blows, F.M.; Liu, B.; Shah, M.; Earl, H.M.; Poole, C.J.; Hiller, L.; Dunn, J.A.; et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann. Oncol. 2014, 25, 1536–1543. [Google Scholar] [CrossRef]
- Hida, A.I.; Watanabe, T.; Sagara, Y.; Kashiwaba, M.; Sagara, Y.; Aogi, K.; Ohi, Y.; Tanimoto, A. Diffuse distribution of tumor-infiltrating lymphocytes is a marker for better prognosis and chemotherapeutic effect in triple-negative breast cancer. Breast Cancer Res. Treat. 2019, 178, 283–294. [Google Scholar] [CrossRef]
- Heindl, A.; Sestak, I.; Naidoo, K.; Cuzick, J.; Dowsett, M.; Yuan, Y. Relevance of Spatial Heterogeneity of Immune Infiltration for Predicting Risk of Recurrence After Endocrine Therapy of ER+ Breast Cancer. J. Natl. Cancer Inst. 2018, 110, 166–175. [Google Scholar] [CrossRef]
- Mani, N.L.; Schalper, K.A.; Hatzis, C.; Saglam, O.; Tavassoli, F.; Butler, M.; Chagpar, A.B.; Pusztai, L.; Rimm, D.L. Quantitative assessment of the spatial heterogeneity of tumor-infiltrating lymphocytes in breast cancer. Breast Cancer Res. 2016, 18, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Prat, A.; Perou, C.M. Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 2011, 5, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Fitzal, F.; Filipits, M.; Fesl, C.; Rudas, M.; Greil, R.; Balic, M.; Moinfar, F.; Herz, W.; Dubsky, P.; Bartsch, R.; et al. PAM-50 predicts local recurrence after breast cancer surgery in postmenopausal patients with ER+/HER2− disease: Results from 1204 patients in the randomized ABCSG-8 trial. BJS 2021, 108, 308–314. [Google Scholar] [CrossRef]
- Buus, R.; Szijgyarto, Z.; Schuster, E.F.; Xiao, H.; Haynes, B.P.; Sestak, I.; Cuzick, J.; Paré, L.; Seguí, E.; Chic, N.; et al. Development and validation for research assessment of Oncotype DX® Breast Recurrence Score, EndoPredict® and Prosigna®. NPJ Breast Cancer 2021, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.B.; Sørlie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- McNamara, K.L.; Caswell-Jin, J.L.; Joshi, R.; Ma, Z.; Kotler, E.; Bean, G.R.; Kriner, M.; Zhou, Z.; Hoang, M.; Beechem, J.; et al. Spatial proteomic characterization of HER2-positive breast tumors through neoadjuvant therapy predicts response. Nat. Rev. Cancer 2021, 2, 400–413. [Google Scholar] [CrossRef]
- Gruosso, T.; Gigoux, M.; Manem, V.S.K.; Bertos, N.; Zuo, D.; Perlitch, I.; Saleh, S.M.I.; Zhao, H.; Souleimanova, M.; Johnson, R.M.; et al. Spatially distinct tumor immune microenvironments stratify triple-negative breast cancers. J. Clin. Investig. 2019, 129, 1785–1800. [Google Scholar] [CrossRef] [Green Version]
- Aleskandarany, M.A.; Green, A.R.; Ashankyty, I.; Elmouna, A.; Diez-Rodriguez, M.; Nolan, C.C.; Ellis, I.O.; Rakha, E.A. Impact of intratumoural heterogeneity on the assessment of Ki67 expression in breast cancer. Breast Cancer Res. Treat. 2016, 158, 287–295. [Google Scholar] [CrossRef] [Green Version]
- von Wahlde, M.K.; Timms, K.M.; Chagpar, A.; Wali, V.B.; Jiang, T.; Bossuyt, V.; Saglam, O.; Reid, J.; Gutin, A.; Neff, C.; et al. Intratumor Heterogeneity of Homologous Recombination Deficiency in Primary Breast Cancer. Clin. Cancer Res. 2017, 23, 1193–1199. [Google Scholar] [CrossRef] [Green Version]
- Rueda, O.M.; Sammut, S.-J.; Seoane, J.A.; Chin, S.-F.; Caswell-Jin, J.L.; Callari, M.; Batra, R.; Pereira, B.; Bruna, A.; Ali, H.R.; et al. Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature 2019, 567, 399–404. [Google Scholar] [CrossRef]
- Kim, C.; Gao, R.; Sei, E.; Brandt, R.; Hartman, J.; Hatschek, T.; Crosetto, N.; Foukakis, T.; Navin, N.E. Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing. Cell 2018, 173, 879–893. [Google Scholar] [CrossRef] [Green Version]
- Beechem, J.M. High-Plex Spatially Resolved RNA and Protein Detection Using Digital Spatial Profiling: A Technology Designed for Immuno-Oncology Biomarker Discovery and Translational Research. Methods Mol. Biol. 2020, 2055, 563–583. [Google Scholar]
- Merritt, C.R.; Ong, G.T.; Church, S.E.; Barker, K.; Danaher, P.; Geiss, G.; Hoang, M.; Jung, J.; Liang, Y.; McKay-Fleisch, J.; et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 2020, 38, 586–599. [Google Scholar] [CrossRef] [PubMed]
- Zollinger, D.R.; Lingle, S.E.; Sorg, K.; Beechem, J.M.; Merritt, C.R. GeoMx™ RNA Assay: High Multiplex, Digital, Spatial Analysis of RNA in FFPE Tissue. Methods Mol. Biol. 2020, 2148, 331–345. [Google Scholar] [CrossRef] [PubMed]
- GeoMx DSP Sample Prep Guidelines; MAN-10091-08 for v2.2; NanoString Technologies Inc.: Seattle, WA, USA, 2021; Available online: https://blog.nanostring.com/geomx-online-user-manual/Content/GuidanceDocs/DSPSamplePrepGuidance.htm (accessed on 11 June 2021).
- Omilian, A.R.; Sheng, H.; Hong, C.C.; Bandera, E.V.; Khoury, T.; Ambrosone, C.B.; Yao, S. Multiplexed digital spatial profiling of invasive breast tumors from Black and White women. Mol. Oncol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.L.; Matynia, A.P.; Factor, R.E.; Varley, K.E. Spatially-resolved quantification of proteins in triple negative breast cancers reveals differences in the immune microenvironment associated with prognosis. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCart Reed, A.E.; Bennett, J.; Kutasovic, J.R.; Kalaw, E.; Ferguson, K.; Yeong, J. Digital spatial profiling application in breast cancer: A user’s perspective. Virchows Arch. 2020, 477, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Decalf, J.; Albert, M.L.; Ziai, J. New tools for pathology: A user’s review of a highly multiplexed method for in situ analysis of protein and RNA expression in tissue. J. Pathol. 2019, 247, 650–661. [Google Scholar] [CrossRef] [Green Version]
- Toki, M.I.; Merritt, C.R.; Wong, P.F.; Smithy, J.W.; Kluger, H.M.; Syrigos, K.N.; Ong, G.T.; Warren, S.E.; Beechem, J.M.; Rimm, D.L. High-Plex Predictive Marker Discovery for Melanoma Immunotherapy-Treated Patients Using Digital Spatial Profiling. Clin. Cancer Res. 2019, 25, 5503–5512. [Google Scholar] [CrossRef] [PubMed]
- Zugazagoitia, J.; Gupta, S.; Liu, Y.; Fuhrman, K.; Gettinger, S.; Herbst, R.S.; Schalper, K.A.; Rimm, D.L. Biomarkers Associated with Beneficial PD-1 Checkpoint Blockade in Non–Small Cell Lung Cancer (NSCLC) Identified Using High-Plex Digital Spatial Profiling. Clin. Cancer Res. 2020, 26, 4360–4368. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Seow, J.J.W.; Dutertre, C.-A.; Pai, R.; Blériot, C.; Mishra, A.; Wong, R.M.M.; Singh, G.S.N.; Sudhagar, S.; Khalilnezhad, S.; et al. Onco-fetal Reprogramming of Endothelial Cells Drives Immunosuppressive Macrophages in Hepatocellular Carcinoma. Cell 2020, 183, 377–394. [Google Scholar] [CrossRef] [PubMed]
- Monkman, J.; Taheri, T.; Ebrahimi Warkiani, M.; O’Leary, C.; Ladwa, R.; Richard, D.; O’Byrne, K.; Kulasinghe, A. High-Plex and High-Throughput Digital Spatial Profiling of Non-Small-Cell Lung Cancer (NSCLC). Cancers 2020, 12, 3551. [Google Scholar] [CrossRef]
- Muñoz, N.M.; Dupuis, C.; Williams, M.; Dixon, K.; McWatters, A.; Avritscher, R.; Bouchard, R.; Kaseb, A.; Schachtschneider, K.M.; Rao, A.; et al. Molecularly targeted photothermal ablation improves tumor specificity and immune modulation in a rat model of hepatocellular carcinoma. Commun. Biol. 2020, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tavernari, D.; Battistello, E.; Dheilly, E.; Petruzzella, A.S.; Mina, M.; Sordet-Dessimoz, J.; Peters, S.; Krueger, T.; Gfeller, D.; Riggi, N.; et al. Nongenetic Evolution Drives Lung Adenocarcinoma Spatial Heterogeneity and Progression. Cancer Discov. 2021, 11, 1490–1507. [Google Scholar] [CrossRef]
- Amaria, R.N.; Reddy, S.M.; Tawbi, H.A.; Davies, M.A.; Ross, M.I.; Glitza, I.C.; Wargo, J.A. Neoadjuvant immune checkpoint blockade in high-risk resectable melanoma. Nat. Med. 2018, 24, 1649–1654. [Google Scholar] [CrossRef] [PubMed]
- Helmink, B.A.; Reddy, S.M.; Gao, J.; Zhang, S.; Basar, R.; Thakur, R.; Wargo, J.A. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 2020, 577, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Cabrita, R.; Lauss, M.; Sanna, A.; Donia, M.; Skaarup Larsen, M.; Mitra, S.; Jönsson, G. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 2020, 577, 561–565. [Google Scholar] [CrossRef]
- Brady, L.; Kriner, M.; Coleman, I.; Morrissey, C.; Roudier, M.; True, L.D.; Gulati, R.; Plymate, S.R.; Zhou, Z.; Birditt, B.; et al. Inter- and intra-tumor heterogeneity of metastatic prostate cancer determined by digital spatial gene expression profiling. Nat. Commun. 2021, 12, 1–16. [Google Scholar] [CrossRef]
- Ihle, C.; Provera, M.D.; Straign, D.M.; Smith, E.E.; Edgerton, S.M.; Van Bokhoven, A.; Lucia, M.S.; Owens, P. Distinct tumor microenvironments of lytic and blastic bone metastases in prostate cancer patients. J. Immunother. Cancer 2019, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Keam, S.P.; Halse, H.; Nguyen, T.; Wang, M.; Van Kooten Losio, N.; Mitchell, C.; Caramia, F.; Byrne, D.J.; Haupt, S.; Ryland, G.; et al. High dose-rate brachytherapy of localized prostate cancer converts tumors from cold to hot. J. Immunother. Cancer 2020, 8, e000792. [Google Scholar] [CrossRef]
- Fittall, M.W.; Van Loo, P. Translating insights into tumor evolution to clinical practice: Promises and challenges. Genome Med. 2019, 11, 20. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Biological and Therapeutic Impact of Intratumor Heterogeneity in Cancer Evolution. Cancer Cell 2015, 27, 15–26. [Google Scholar] [CrossRef] [Green Version]
- Jamal-Hanjani, M.; Quezada, S.; Larkin, J.; Swanton, C. Translational Implications of Tumor Heterogeneity. Clin. Cancer Res. 2015, 21, 1258–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, P.; Rugo, H.S.; Adams, S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Henschel, V.; Molinero, L.; Chui, S.Y.; et al. Atezolizumab plus nabpaclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): Updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2020, 21, 44–59. [Google Scholar] [CrossRef]
- Cortes, J.; Cescon, D.W.; Rugo, H.S.; Nowecki, Z.; Im, S.A.; Yusof, M.M.; de las Heras Begona, B. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEY-NOTE-355): A randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet 2020, 396, 1817–1828. [Google Scholar] [CrossRef]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 2015, 3, 83–92. [Google Scholar] [CrossRef] [Green Version]
- Goddard, E.T.; Bassale, S.; Schedin, T.; Jindal, S.; Johnston, J.; Cabral, E.; Borges, V.F. Association Between Postpartum Breast Cancer Diag-nosis and Metastasis and the Clinical Features Underlying Risk. JAMA Netw. Open 2019, 2, e186997. [Google Scholar] [CrossRef] [Green Version]
- Borges, V.F.; Lyons, T.R.; Germain, D.; Schedin, P. Postpartum Involution and Cancer: An Opportunity for Targeted Breast Can-cer Prevention and Treatments? Cancer Res. 2020, 80, 1790–1798. [Google Scholar] [CrossRef] [Green Version]
- Holokai, L.; Chakrabarti, J.; Lundy, J.; Croagh, D.; Adhikary, P.; Richards, S.S.; Woodson, C.; Steele, N.; Kuester, R.; Scott, A.; et al. Murine- and Human-Derived Autologous Organoid/Immune Cell Co-Cultures as Pre-Clinical Models of Pancreatic Ductal Adenocarcinoma. Cancers 2020, 12, 3816. [Google Scholar] [CrossRef]
- Freed-Pastor, W.A.; Lambert, L.J.; Ely, Z.A.; Pattada, N.B.; Bhutkar, A.; Eng, G.; Mercer, K.L.; Garcia, A.P.; Lin, L.; Rideout, W.M., 3rd; et al. The CD155/TIGIT axis promotes and maintains immune evasion in neoantigen-expressing pancreatic cancer. Cancer Cell 2021, S1535-6108, 00384-6. [Google Scholar] [CrossRef]
- Carter, J.M.; Polley, M.C.; Leon-Ferre, R.A.; Sinnwell, J.; Thompson, K.J.; Wang, X.; Ma, Y.; Zahrieh, D.; Kachergus, J.M.; Solanki, M.; et al. Characteristics and spatially-defined im-mune (micro)landscapes of early-stage PD-L1-positive triple-negative breast cancer. Clin. Cancer Res. 2021. [Google Scholar] [CrossRef]
- GeoMx-nCounter/NGS User Manuals; MAN-10091-07 for v2.1; NanoString Technologies Inc.: Seattle, WA, USA, 2021; Available online: https://blog.nanostring.com/geomx-online-user-manual-v2.1/Content/PDF_Quickstarts_and_Manuals.htm (accessed on 11 June 2021).
- Network NCC. Breast Cancer (Version 4.2021). Available online: https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1419 (accessed on 11 June 2021).
- Chao, X.; Liu, L.; Sun, P.; Yang, X.; Li, M.; Luo, R.; Huang, Y.; He, J.; Yun, J. Immune parameters associated with survival in metaplastic breast cancer. Breast Cancer Res. 2020, 22, 1–11. [Google Scholar] [CrossRef]
- Malhotra, G.K.; Zhao, X.; Band, H.; Band, V. Histological, molecular and functional subtypes of breast cancers. Cancer Biol. Ther. 2010, 10, 955–960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snyder, M.P.; Lin, S.; Posgai, A.; Atkinson, M.; Regev, A.; Rood, J. The human body at cellular resolution: The NIH Human Biomolecular Atlas Program. Nature 2019, 574, 187–192. [Google Scholar]
- Rood, J.E.; Stuart, T.; Ghazanfar, S.; Biancalani, T.; Fisher, E.; Butler, A.; Hupalowska, A.; Gaffney, L.; Mauck, W.; Eraslan, G.; et al. Toward a Common Coordinate Framework for the Human Body. Cell 2019, 179, 1455–1467. [Google Scholar] [CrossRef]
- Zhu, G.; Falahat, R.; Wang, K.; Mailloux, A.; Artzi, N.; Mulé, J.J. Tumor-Associated Tertiary Lymphoid Structures: Gene-Expression Profiling and Their Bioengineering. Front. Immunol. 2017, 8, 767. [Google Scholar] [CrossRef] [Green Version]
- Gatti-Mays, M.E.; Balko, J.M.; Gameiro, S.R.; Bear, H.D.; Prabhakaran, S.; Fukui, J.; Disis, M.L.; Nanda, R.; Gulley, J.L.; Kalinsky, K.; et al. If we build it they will come: Targeting the immune response to breast cancer. NPJ Breast Cancer 2019, 5, 37. [Google Scholar] [CrossRef] [Green Version]
- Anandappa, A.J.; Wu, C.J.; Ott, P.A. Directing Traffic: How to Effectively Drive T Cells into Tumors. Cancer Discov. 2020, 10, 185–197. [Google Scholar] [CrossRef]
- Danaher, P.; Kim, Y.; Nelson, B.; Griswold, M.; Yang, Z.; Piazza, E.; Beechem, J.M. Advances in mixed cell deconvolution enable quantifica-tion of cell types in spatially-resolved gene expression data. bioRxiv 2020. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. arXiv 2015, arXiv:1406.5823. [Google Scholar]
- Ortogero, N.; Yang, Z.; Vitancol, R.; Griswold, M.; Henderson, D. GeomxTools: NanoString GeoMx Tools. R Package Version 1.0.0; NanoString Technologies Inc.: Seattle, WA, USA, 2021; Available online: http://bioconductor.org/packages/release/bioc/html/GeomxTools.html (accessed on 11 June 2021).
- Bolstad, B.; Irizarry, R.; Astrand, M.; Speed, T. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [Green Version]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [Green Version]
- Hafemeister, C.; Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019, 20, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.H.; Dudoit, S.; Luu, P.; Lin, D.M.; Peng, V.; Ngai, J.; Speed, T.P. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002, 30, e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudoit, S.; Yang, Y.H.; Callow, M.J.; Speed, T.P. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat. Sin. 2002, 12, 111–139. [Google Scholar]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Wang, R.; Zhang, X.; Li, X.; Liang, Y.; Ding, Z. Spatially-resolved proteomics and transcriptomics: An emerging digital spatial profiling approach for tumor microenvironment. Vis. Cancer Med. 2021, 2, 1. [Google Scholar] [CrossRef]
- Kleshchevnikov, V.; Shmatko, A.; Dann, E.; Aivazidis, A.; King, H.W.; Li, T.; Lomakin, A.; Kedlian, V.; Jain, M.S.; Park, J.S.; et al. Comprehensive mapping of tissue cell architec-ture via integrated single cell and spatial transcriptomics. bioRxiv 2020. [Google Scholar] [CrossRef]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Silva, V.W.K.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef]
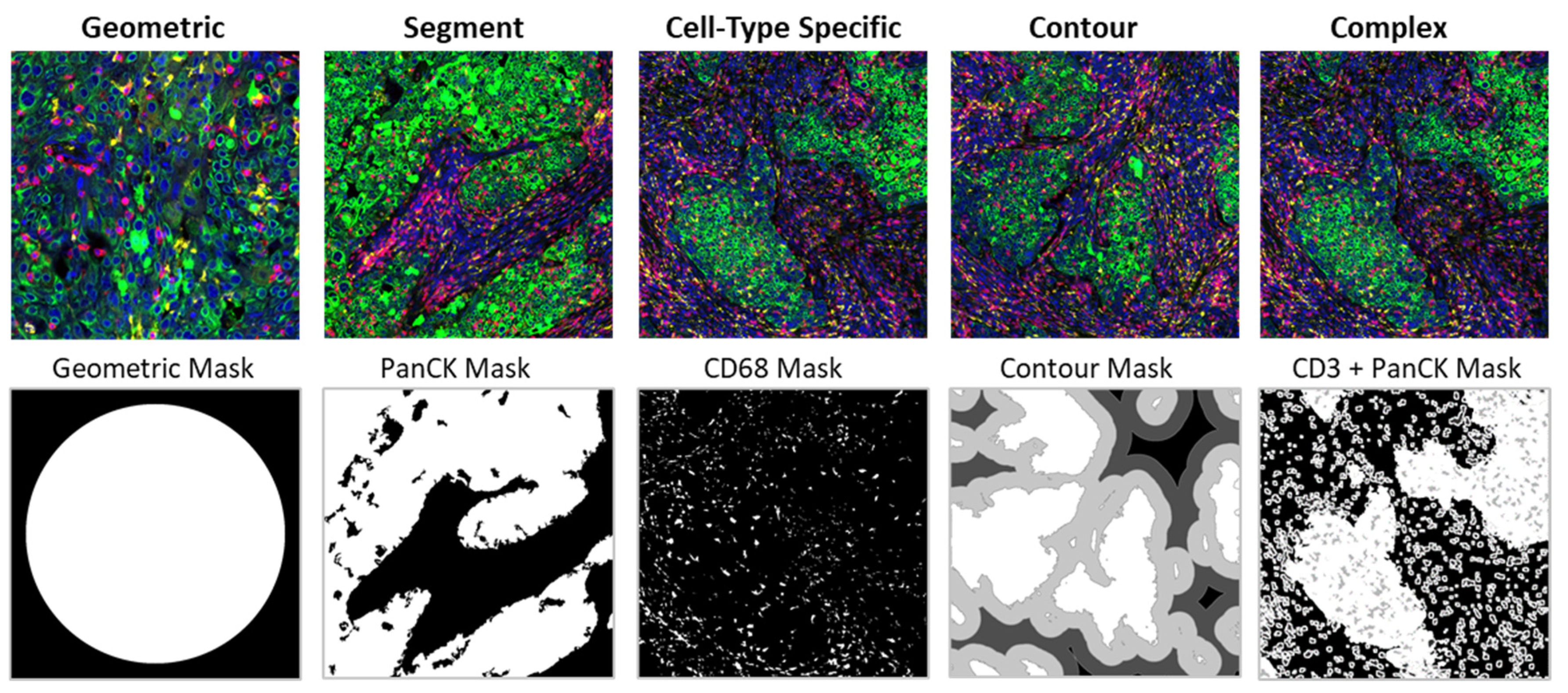
| Profiling Type | Features | Variable AOI Area | GeoMx ROI Selection | Meta-Analysis Integration |
|---|---|---|---|---|
| Geometric Profiling | Assess tissue heterogeneity by profiling geometric shapes and polygons | Sometimes | Native software | Simple |
| Segment Profiling | Use morphology markers to identify and profile distinct biological compartments | Yes | Native software | Simple |
| Cell-Type Specific Profiling | Profile distinct cell populations with cell type-specific morphology markers | Yes | Native software | Difficult |
| Contour Profiling | Evaluate the proximity around a central structure using radiating ROIs | Sometimes | Custom | Moderate |
| Complex Profiling | Combine the above approaches to profile multiple cell types and/or complex regions of tissue | Yes | Native software and custom | Difficult |
| Parameter | Features |
|---|---|
| Segment Definition | Segment definition determines the cellular composition of each AOI within an ROI based on the fluorescent morphology markers to profile distinct cell populations (e.g., CD3 for T cells, CD68 for macrophages) (Figure 4). |
| Erosion | Erosion enables the uniform removal of the UV-light mask boundaries that define an AOI. This parameter accounts for the subcellular spread in the focused UV light and is commonly set between 1 and 3 µm. For tumor AOIs that present with larger nuclei and more cell aggregation, erosion can be higher. For immune cell-specific AOIs that are more punctate in appearance, erosion is set lower to maximize signal and minimize removal of cells from the mask. To ensure the purity of the signal from the particular cell population, it can be helpful to erode the signal away from the cell membrane boundary by 2–5 μm to minimize contamination from surrounding cells. |
| N-Dilation | N-dilation uniformly expands the UV-light mask in an AOI, whose segment definition requires a positive signal for the nuclear marker (e.g., Syto13). This setting is only applicable for nuclear-tagged segments. |
| Hole Size | Hole size fills gaps in the AOI masks that are smaller than the value (in µm2) the researcher sets. This technique is particularly useful for segmenting tumor cells that have large nuclei that are negative for the tumor marker but are still intended to be part of the AOI of the tumor marker. |
| Particle Size | Any small segment areas (particles) less than this value (in µm2) are removed from the AOI mask. When segmenting immune cells, lowering the particle size value (to ~5 µm2) is important because a high value would subtract out immune cells from the mask. |
| Collection Order | Collection order determines the order in which UV light illuminates each AOI within an ROI. Since an AOI collection can be accompanied by minor signal loss near the AOI boundaries, it is valuable to collect AOIs from low to high abundance to maximize signal. |
| Threshold | Thresholds are adjustable per ROI and enable the researcher to tune the fluorescence-based masking that defines an AOI. This approach ensures specific illumination of target cell types or regions without bias or contamination. With the exception of thresholding, it is important to keep all other parameters above consistent across all the ROIs and slides in a given study. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bergholtz, H.; Carter, J.M.; Cesano, A.; Cheang, M.C.U.; Church, S.E.; Divakar, P.; Fuhrman, C.A.; Goel, S.; Gong, J.; Guerriero, J.L.; et al. Best Practices for Spatial Profiling for Breast Cancer Research with the GeoMx® Digital Spatial Profiler. Cancers 2021, 13, 4456. https://doi.org/10.3390/cancers13174456
Bergholtz H, Carter JM, Cesano A, Cheang MCU, Church SE, Divakar P, Fuhrman CA, Goel S, Gong J, Guerriero JL, et al. Best Practices for Spatial Profiling for Breast Cancer Research with the GeoMx® Digital Spatial Profiler. Cancers. 2021; 13(17):4456. https://doi.org/10.3390/cancers13174456
Chicago/Turabian StyleBergholtz, Helga, Jodi M. Carter, Alessandra Cesano, Maggie Chon U Cheang, Sarah E. Church, Prajan Divakar, Christopher A. Fuhrman, Shom Goel, Jingjing Gong, Jennifer L. Guerriero, and et al. 2021. "Best Practices for Spatial Profiling for Breast Cancer Research with the GeoMx® Digital Spatial Profiler" Cancers 13, no. 17: 4456. https://doi.org/10.3390/cancers13174456
APA StyleBergholtz, H., Carter, J. M., Cesano, A., Cheang, M. C. U., Church, S. E., Divakar, P., Fuhrman, C. A., Goel, S., Gong, J., Guerriero, J. L., Hoang, M. L., Hwang, E. S., Kuasne, H., Lee, J., Liang, Y., Mittendorf, E. A., Perez, J., Prat, A., Pusztai, L., ... on behalf of the GeoMx Breast Cancer Consortium. (2021). Best Practices for Spatial Profiling for Breast Cancer Research with the GeoMx® Digital Spatial Profiler. Cancers, 13(17), 4456. https://doi.org/10.3390/cancers13174456








