Ki-67 as a Prognostic Biomarker in Invasive Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
Biomarkers
2. Ki-67 Proliferation Indices
2.1. Ki-67: Inconsistencies in Detection
2.2. Ki-67 Guidelines and Therapeutic Decision Making
2.2.1. Ki-67 Clinical Interpretation
2.2.2. Ki-67 Guidelines
2.2.3. Ki-67 and Endocrine Therapies
2.2.4. Ki-67 and Triple Negative Breast Cancer
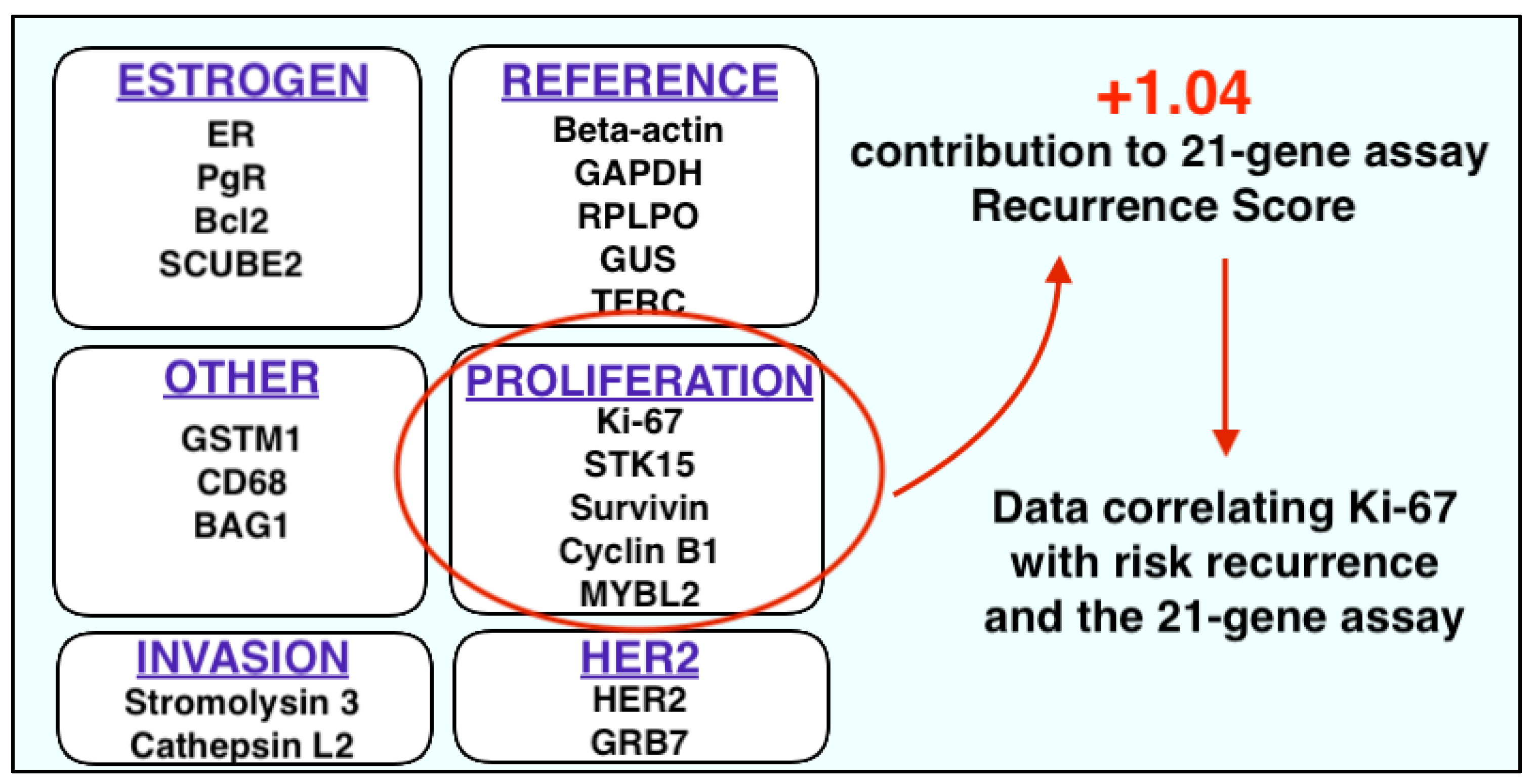
2.3. Ki-67 and Multigene Panels
2.4. Improving Ki-67—Future Considerations
2.4.1. Standardisation
2.4.2. Digital Image Analysis
2.4.3. Ki-67 and miRNA Analysis
2.4.4. Ki-67 and Radiomic Analysis
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Biomarkers Definitions Working Group. Biomarkers and surrogate end points: Preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 2001, 69, 89–95. [Google Scholar] [CrossRef]
- Burke, H.B. Predicting Clinical Outcomes Using Molecular Biomarkers. Biomark. Cancer 2016, 8, 89–99. [Google Scholar] [CrossRef]
- Mayeux, R. Biomarkers: Potential uses and limitations. NeuroRx 2004, 1, 182–188. [Google Scholar] [CrossRef]
- Carlomagno, N.; Incollingo, P.; Tammaro, V.; Peluso, G.; Rupealta, N.; Chiacchio, G.; Sotelo, M.L.S.; Minieri, G.; Pisani, A.; Riccio, E.; et al. Diagnostic, Predictive, Prognostic, and Therapeutic Molecular Biomarkers in Third Millennium: A Breakthrough in Gastric Cancer. BioMed Res. Int. 2017, 2017, 7869802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- National Institutes of Health (US). Understanding Prognostic versus Predictive Biomarkers; National Institutes of Health (US): Bethesda, MD, USA, 2016. Available online: https://www.ncbi.nlm.nih.gov/books/NBK402284/ (accessed on 30 November 2020).
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Cooper, A. The Principles and Practice of Surgery; Nabu Press: London, UK, 1836. [Google Scholar]
- Bland, C.S. The Halsted mastectomy: Present illness and past history. West. J. Med. 1981, 134, 549–555. [Google Scholar]
- Ellsworth, R.E.; Decewicz, D.J.; Shriver, C.D.; Ellsworth, D.L. Breast cancer in the personal genomics era. Curr. Genom. 2010, 11, 146–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thürlimann, B.; Senn, H.J.; Panel Members. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef] [PubMed]
- Falck, A.-K.; Fernö, M.; Bendahl, P.-O.; Rydén, L. St Gallen molecular subtypes in primary breast cancer and matched lymph node metastases—Aspects on distribution and prognosis for patients with luminal A tumours: Results from a prospective randomised trial. BMC Cancer 2013, 13, 558. [Google Scholar] [CrossRef] [Green Version]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thürlimann, B.; Senn, H.J. Strategies for subtypes—Dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.D.; Wu, J.; Shen, Z.Z.; Shao, Z.M. Hazard of breast cancer-specific mortality among women with estrogen receptor-positive breast cancer after five years from diagnosis: Implication for extended endocrine therapy. J. Clin. Endocrinol. Metab. 2012, 97, E2201–E2209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truong, P.T.; Bernstein, V.; Wai, E.; Chua, B.; Speers, C.; Olivotto, I.A. Age-related variations in the use of axillary dissection: A survival analysis of 8038 women with T1-ST2 breast cancer. Int. J. Radiat. Oncol. Biol. Phys. 2002, 54, 794–803. [Google Scholar] [CrossRef]
- Sopik, V.; Sun, P.; Narod, S.A. The prognostic effect of estrogen receptor status differs for younger versus older breast cancer patients. Breast Cancer Res. Treat. 2017, 165, 391–402. [Google Scholar] [CrossRef]
- Jayasekara, H.; MacInnis, R.J.; Chamberlain, J.A.; Dite, G.S.; Leoce, N.M.; Dowty, J.G.; Bickerstaffe, A.; Win, A.K.; Milne, R.L.; Giles, G.G.; et al. Mortality after breast cancer as a function of time since diagnosis by estrogen receptor status and age at diagnosis. Int. J. Cancer 2019, 145, 3207–3217. [Google Scholar] [CrossRef]
- Zurawska, U.; Baribeau, D.A.; Giilck, S.; Victor, C.; Gandhi, S.; Florescu, A.; Verma, S. Outcomes of her2-positive early-stage breast cancer in the trastuzumab era: A population-based study of Canadian patients. Curr. Oncol. 2013, 20, e539–e545. [Google Scholar] [CrossRef] [Green Version]
- Davey, M.G.; Ryan, É.J.; Folan, P.J.; O’Halloran, N.; Boland, M.R.; Barry, M.K.; Sweeney, K.J.; Malone, C.M.; McLaughlin, R.J.; Kerin, M.J.; et al. The impact of progesterone receptor negativity on oncological outcomes in oestrogen-receptor-positive breast cancer. BJS Open 2021, 5, zrab040. [Google Scholar] [CrossRef]
- Davey, M.G.; Kerin, E.; O’Flaherty, C.; Maher, E.; Richard, V.; McAnena, P.; McLaughlin, R.P.; Sweeney, K.J.; Barry, M.K.; Malone, C.M.; et al. Clinicopathological response to neoadjuvant therapies and pathological complete response as a biomarker of survival in human epidermal growth factor receptor-2 enriched breast cancer—A retrospective cohort study. Breast 2021, 59, 67–75. [Google Scholar] [CrossRef]
- Hammond, M.E.H.; Hayes, D.F.; Dowsett, M.; Allred, D.C.; Hagerty, K.L.; Badve, S.; Fitzgibbons, P.L.; Francis, G.; Goldstein, N.S.; Hayes, M.; et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch. Pathol. Lab. Med. 2010, 134, 907–922. [Google Scholar] [CrossRef] [PubMed]
- Wolff, A.C.; Hammond, M.E.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch. Pathol. Lab. Med. 2014, 138, 241–256. [Google Scholar] [CrossRef] [Green Version]
- Harris, L.N.; Ismaila, N.; McShane, L.M.; Andre, F.; Collyar, D.E.; Gonzalez-Angulo, A.M.; Hammond, E.H.; Kuderer, N.M.; Liu, M.C.; Mennel, R.G.; et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 2016, 34, 1134–1150. [Google Scholar] [CrossRef] [Green Version]
- Taneja, P.; Maglic, D.; Kai, F.; Zhu, S.; Kendig, R.D.; Fry, E.A.; Inoue, K. Classical and Novel Prognostic Markers for Breast Cancer and their Clinical Significance. Clin. Med. Insights Oncol. 2010, 4, 15–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet 2005, 365, 1687–1717. [Google Scholar] [CrossRef]
- Luporsi, E.; André, F.; Spyratos, F.; Martin, P.M.; Jacquemier, J.; Penault-Llorca, F.; Tubiana-Mathieu, N.; Sigal-Zafrani, B.; Arnould, L.; Gompel, A.; et al. Ki-67: Level of evidence and methodological considerations for its role in the clinical management of breast cancer: Analytical and critical review. Breast Cancer Res. Treat. 2012, 132, 895–915. [Google Scholar] [CrossRef] [Green Version]
- De Azambuja, E.; Cardoso, F.; de Castro, G., Jr.; Colozza, M.; Mano, M.S.; Durbecq, V.; Sotiriou, C.; Larsimont, D.; Piccart-Gebhart, M.J.; Paesmans, M. Ki-67 as prognostic marker in early breast cancer: A meta-analysis of published studies involving 12,155 patients. Br. J. Cancer 2007, 96, 1504–1513. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Chen, L.; Huang, B.; Wang, Y.; Ji, L.; Wu, J.; Di, G.; Liu, G.; Yu, K.; Shao, Z.; et al. The prognostic and predictive potential of Ki-67 in triple-negative breast cancer. Sci. Rep. 2020, 10, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schonk, D.M.; Kuijpers, H.J.; van Drunen, E.; van Dalen, C.H.; Geurts van Kessel, A.H.; Verheijen, R.; Ramaekers, F.C. Assignment of the gene(s) involved in the expression of the proliferation-related Ki-67 antigen to human chromosome 10. Hum. Genet. 1989, 83, 297–299. [Google Scholar] [CrossRef] [PubMed]
- Scholzen, T.; Gerdes, J. The Ki-67 protein: From the known and the unknown. J. Cell. Physiol. 2000, 182, 311–322. [Google Scholar] [CrossRef]
- Klöppel, G.; La Rosa, S. Ki67 labeling index: Assessment and prognostic role in gastroenteropancreatic neuroendocrine neoplasms. Virchows Arch. 2018, 472, 341–349. [Google Scholar] [CrossRef]
- Halm, U.; Tannapfel, A.; Breitung, B.; Breidert, M.; Wittekind, C.W.; Mössner, J. Apoptosis and cell proliferation in the metaplasia-dysplasia-carcinoma-sequence of Barrett’s esophagus. Hepatogastroenterology 2000, 47, 962–966. [Google Scholar]
- Rahmanzadeh, R.; Hüttmann, G.; Gerdes, J.; Scholzen, T. Chromophore-assisted light inactivation of pKi-67 leads to inhibition of ribosomal RNA synthesis. Cell Prolif. 2007, 40, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, C.; Sakkab, D.Y.; Scholzen, T.; Dassler, R.; Alison, M.R.; Gerdes, J. Ki-67 expression during rat liver regeneration after partial hepatectomy. Hepatology 1997, 26, 573–578. [Google Scholar] [CrossRef]
- Shirendeb, U.; Hishikawa, Y.; Moriyama, S.; Win, N.; Thu, M.M.M.; Mar, K.S.; Khatanbaatar, G.; Masuzaki, H.; Koji, T. Human papillomavirus infection and its possible correlation with p63 expression in cervical cancer in Japan, Mongolia, and Myanmar. Acta Histochem. Cytochem. 2009, 42, 181–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hooghe, B.; Hulpiau, P.; van Roy, F.; De Bleser, P. ConTra: A promoter alignment analysis tool for identification of transcription factor binding sites across species. Nucleic Acids Res. 2008, 36, W128–W132. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Diederichs, S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012, 9, 703–719. [Google Scholar] [CrossRef] [Green Version]
- Cuylen, S.; Blaukopf, C.; Politi, A.Z.; Müller-Reichert, T.; Neumann, B.; Poser, I.; Ellenberg, J.; Hyman, A.A.; Gerlich, D.W. Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature 2016, 535, 308–312. [Google Scholar] [CrossRef]
- Modlin, I.M.; Moss, S.F.; Chung, D.C.; Jensen, R.T.; Snyderwine, E. Priorities for improving the management of gastroenteropancreatic neuroendocrine tumors. J. Natl. Cancer Inst. 2008, 100, 1282–1289. [Google Scholar] [CrossRef]
- Miller, I.; Min, M.; Yang, C.; Tian, C.; Gookin, S.; Carter, D.; Spencer, S.L. Ki67 is a Graded Rather than a Binary Marker of Proliferation versus Quiescence. Cell Rep. 2018, 24, 1105–1112.e5. [Google Scholar] [CrossRef] [Green Version]
- Ishihara, M.; Mukai, H.; Nagai, S.; Onozawa, M.; Nihei, K.; Shimada, T.; Wada, N. Retrospective analysis of risk factors for central nervous system metastases in operable breast cancer: Effects of biologic subtype and Ki67 overexpression on survival. Oncology 2013, 84, 135–140. [Google Scholar] [CrossRef]
- Sorbye, S.W.; Kilvaer, T.K.; Valkov, A.; Donnem, T.; Smeland, E.; Al-Shibli, K.; Bremnes, R.M.; Busund, L.-T. Prognostic impact of Jab1, p16, p21, p62, Ki67 and Skp2 in soft tissue sarcomas. PLoS ONE 2012, 7, e47068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciancio, N.; Galasso, M.G.; Campisi, R.; Bivona, L.; Migliore, M.; Di Maria, G.U. Prognostic value of p53 and Ki67 expression in fiberoptic bronchial biopsies of patients with non small cell lung cancer. Multidiscip. Respir. Med. 2012, 7, 29. [Google Scholar] [CrossRef]
- Josefsson, A.; Wikström, P.; Egevad, L.; Granfors, T.; Karlberg, L.; Stattin, P.; Bergh, A. Low endoglin vascular density and Ki67 index in Gleason score 6 tumours may identify prostate cancer patients suitable for surveillance. Scand. J. Urol. Nephrol. 2012, 46, 247–257. [Google Scholar] [CrossRef]
- Zhao, W.-Y.; Xu, J.; Wang, M.; Zhang, Z.-Z.; Tu, L.; Wang, C.-J.; Lin, T.-L.; Shen, Y.-Y.; Liu, Q.; Cao, H. Prognostic value of Ki67 index in gastrointestinal stromal tumors. Int. J. Clin. Exp. Pathol. 2014, 7, 2298–2304. [Google Scholar]
- Nadler, A.; Cukier, M.; Rowsell, C.; Kamali, S.; Feinberg, Y.; Singh, S.; Law, C.H. Ki-67 is a reliable pathological grading marker for neuroendocrine tumors. Virchows Arch. 2013, 462, 501–505. [Google Scholar] [CrossRef]
- Kim, K.I.; Lee, K.H.; Kim, T.R.; Chun, Y.S.; Lee, T.H.; Park, H.K. Ki-67 as a predictor of response to neoadjuvant chemotherapy in breast cancer patients. J. Breast Cancer 2014, 17, 40–46. [Google Scholar] [CrossRef] [Green Version]
- Fasching, P.A.; Heusinger, K.; Haeberle, L.; Niklos, M.; Hein, A.; Bayer, C.M.; Rauh, C.; Schulz-Wendtland, R.; Bani, M.R.; Schrauder, M.; et al. Ki67, chemotherapy response, and prognosis in breast cancer patients receiving neoadjuvant treatment. BMC Cancer 2011, 11, 486. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, R.; Osako, T.; Nishiyama, Y.; Tashima, R.; Nakano, M.; Fujisue, M.; Toyozumi, Y.; Arima, N. Prognostic significance of Ki-67 index value at the primary breast tumor in recurrent breast cancer. Mol. Clin. Oncol. 2014, 2, 1062–1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abubakar, M.; Orr, N.; Daley, F.; Coulson, P.; Ali, H.R.; Blows, F.; Benitez, J.; Milne, R.; Brenner, H.; Stegmaier, C.; et al. Prognostic value of automated KI67 scoring in breast cancer: A centralised evaluation of 8088 patients from 10 study groups. Breast Cancer Res. 2016, 18, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietiläinen, T.; Lipponen, P.; Aaltomaa, S.; Eskelinen, M.; Kosma, V.M.; Syrjänen, K. The important prognostic value of Ki-67 expression as determined by image analysis in breast cancer. J. Cancer Res. Clin. Oncol. 1996, 122, 687–692. [Google Scholar] [CrossRef]
- Denkert, C.; Budczies, J.; von Minckwitz, G.; Wienert, S.; Loibl, S.; Klauschen, F. Strategies for developing Ki67 as a useful biomarker in breast cancer. Breast 2015, 24 (Suppl. 2), S67–S72. [Google Scholar] [CrossRef] [PubMed]
- Dowsett, M.; Nielsen, T.O.; A’Hern, R.; Bartlett, J.; Coombes, R.C.; Cuzick, J.; Ellis, M.; Henry, N.L.; Hugh, J.C.; Lively, T.; et al. Assessment of Ki67 in breast cancer: Recommendations from the International Ki67 in Breast Cancer working group. J. Natl. Cancer Inst. 2011, 103, 1656–1664. [Google Scholar] [CrossRef] [Green Version]
- Arima, N.; Nishimura, R.; Osako, T.; Nishiyama, Y.; Fujisue, M.; Okumura, Y.; Nakano, M.; Tashima, R.; Toyozumi, Y. The importance of tissue handling of surgically removed breast cancer for an accurate assessment of the Ki-67 index. J. Clin. Pathol. 2016, 69, 255–259. [Google Scholar] [CrossRef] [Green Version]
- Hammond, M.E.H.; Hayes, D.F.; Wolff, A.C.; Mangu, P.B.; Temin, S. American society of clinical oncology/college of american pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Oncol. Pract. 2010, 6, 195–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mengel, M.; von Wasielewski, R.; Wiese, B.; Rüdiger, T.; Müller-Hermelink, H.K.; Kreipe, H. Inter-laboratory and inter-observer reproducibility of immunohistochemical assessment of the Ki-67 labelling index in a large multi-centre trial. J. Pathol. 2002, 198, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Benini, E.; Rao, S.; Daidone, M.G.; Pilotti, S.; Silvestrini, R. Immunoreactivity to MIB-1 in breast cancer: Methodological assessment and comparison with other proliferation indices. Cell Prolif. 1997, 30, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Faratian, D.; Munro, A.; Twelves, C.; Bartlett, J.M. Membranous and cytoplasmic staining of Ki67 is associated with HER2 and ER status in invasive breast carcinoma. Histopathology 2009, 54, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Urruticoechea, A.; Smith, I.E.; Dowsett, M. Proliferation marker Ki-67 in early breast cancer. J. Clin. Oncol. 2005, 23, 7212–7220. [Google Scholar] [CrossRef]
- Cattoretti, G.; Becker, M.H.; Key, G.; Duchrow, M.; Schlüter, C.; Galle, J.; Gerdes, J. Monoclonal antibodies against recombinant parts of the Ki-67 antigen (MIB 1 and MIB 3) detect proliferating cells in microwave-processed formalin-fixed paraffin sections. J. Pathol. 1992, 168, 357–363. [Google Scholar] [CrossRef]
- Inwald, E.C.; Klinkhammer-Schalke, M.; Hofstädter, F.; Zeman, F.; Koller, M.; Gerstenhauer, M.; Ortmann, O. Ki-67 is a prognostic parameter in breast cancer patients: Results of a large population-based cohort of a cancer registry. Breast Cancer Res. Treat. 2013, 139, 539–552. [Google Scholar] [CrossRef] [Green Version]
- Ekholm, M.; Beglerbegovic, S.; Grabau, D.; Lövgren, K.; Malmström, P.; Hartman, L.; Fernö, M. Immunohistochemical assessment of Ki67 with antibodies SP6 and MIB1 in primary breast cancer: A comparison of prognostic value and reproducibility. Histopathology 2014, 65, 252–260. [Google Scholar] [CrossRef]
- Muftah, A.A.; Aleskandarany, M.A.; Al-Kaabi, M.M.; Sonbul, S.N.; Diez-Rodriguez, M.; Nolan, C.C.; Caldas, C.; Ellis, I.O.; Rakha, E.A.; Green, A.R. Ki67 expression in invasive breast cancer: The use of tissue microarrays compared with whole tissue sections. Breast Cancer Res. Treat. 2017, 164, 341–348. [Google Scholar] [CrossRef] [Green Version]
- Viale, G.; Hanlon Newell, A.E.; Walker, E.; Harlow, G.; Bai, I.; Russo, L.; Dell’Orto, P.; Maisonneuve, P. Ki-67 (30-9) scoring and differentiation of Luminal A- and Luminal B-like breast cancer subtypes. Breast Cancer Res. Treat. 2019, 178, 451–458. [Google Scholar] [CrossRef] [Green Version]
- Owens, R.; Gilmore, E.; Bingham, V.; Cardwell, C.; McBride, H.; McQuaid, S.; Humphries, M.; Kelly, P. Comparison of different anti-Ki67 antibody clones and hot-spot sizes for assessing proliferative index and grading in pancreatic neuroendocrine tumours using manual and image analysis. Histopathology 2020, 77, 646–658. [Google Scholar] [CrossRef]
- Wong, S.C.C.; Chan, J.K.C.; Lo, E.S.F.; Chan, A.K.C.; Wong, M.C.K.; Chan, C.M.L.; Lam, M.Y.Y.; Chan, A.T.C. The Contribution of Bifunctional SkipDewax Pretreatment Solution, Rabbit Monoclonal Antibodies, and Polymer Detection Systems in Immunohistochemistry. Arch. Pathol. Lab. Med. 2007, 131, 1047–1055. [Google Scholar] [CrossRef] [PubMed]
- Zabaglo, L.; Salter, J.; Anderson, H.; Quinn, E.; Hills, M.; Detre, S.; A’Hern, R.; Dowsett, M. Comparative validation of the SP6 antibody to Ki67 in breast cancer. J. Clin. Pathol. 2010, 63, 800–804. [Google Scholar] [CrossRef]
- Cheang, M.C.U.; Chia, S.K.; Voduc, D.; Gao, D.; Leung, S.; Snider, J.; Watson, M.; Davies, S.; Bernard, P.S.; Parker, J.S.; et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J. Natl. Cancer Inst. 2009, 101, 736–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, G.; Sun, X.; Cao, Q.; Courter, D.; Koong, A.; Le, Q.T.; Gambhir, S.S.; Chen, X. Cetuximab-based immunotherapy and radioimmunotherapy of head and neck squamous cell carcinoma. Clin. Cancer Res. 2010, 16, 2095–2105. [Google Scholar] [CrossRef] [Green Version]
- Coates, A.S.; Winer, E.P.; Goldhirsch, A.; Gelber, R.D.; Gnant, M.; Piccart-Gebhart, M.; Thürlimann, B.; Senn, H.J. Tailoring therapies—Improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann. Oncol. 2015, 26, 1533–1546. [Google Scholar] [CrossRef]
- Prat, A.; Cheang, M.C.; Martín, M.; Parker, J.S.; Carrasco, E.; Caballero, R.; Tyldesley, S.; Gelmon, K.; Bernard, P.S.; Nielsen, T.O.; et al. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal A breast cancer. J. Clin. Oncol. 2013, 31, 203–209. [Google Scholar] [CrossRef]
- Enrico, D.H.; Hannois, A.R.; Bravo, I. Evaluation of the best cut-off point for Ki-67 and progesterone receptor as a prognostic factor in hormone receptor-positive (HR+) breast cancer. J. Clin. Oncol. 2018, 36, e12549. [Google Scholar] [CrossRef]
- Petrelli, F.; Viale, G.; Cabiddu, M.; Barni, S. Prognostic value of different cut-off levels of Ki-67 in breast cancer: A systematic review and meta-analysis of 64,196 patients. Breast Cancer Res. Treat. 2015, 153, 477–491. [Google Scholar] [CrossRef]
- Tian, C.; Fu, L.; Wei, J.; Yin, P.; Zhang, H. Ki-67 versus MammaPrint/BluePrint for assessing luminal type breast cancer. J. Clin. Oncol. 2020, 38, e13673. [Google Scholar] [CrossRef]
- Aleskandarany, M.A.; Green, A.R.; Benhasouna, A.A.; Barros, F.F.; Neal, K.; Reis-Filho, J.S.; Ellis, I.O.; Rakha, E.A. Prognostic value of proliferation assay in the luminal, HER2-positive, and triple-negative biologic classes of breast cancer. Breast Cancer Res. BCR 2012, 14, R3. [Google Scholar] [CrossRef] [Green Version]
- Munzone, E.; Botteri, E.; Sciandivasci, A.; Curigliano, G.; Nolè, F.; Mastropasqua, M.; Rotmensz, N.; Colleoni, M.; Esposito, A.; Adamoli, L.; et al. Prognostic value of Ki-67 labeling index in patients with node-negative, triple-negative breast cancer. Breast Cancer Res. Treat. 2012, 134, 277–282. [Google Scholar] [CrossRef]
- Wu, Q.; Ma, G.; Deng, Y.; Luo, W.; Zhao, Y.; Li, W.; Zhou, Q. Prognostic Value of Ki-67 in Patients With Resected Triple-Negative Breast Cancer: A Meta-Analysis. Front. Oncol. 2019, 9, 1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, T.O.; Leung, S.C.Y.; Rimm, D.L.; Dodson, A.; Acs, B.; Badve, S.; Denkert, C.; Ellis, M.J.; Fineberg, S.; Flowers, M.; et al. Assessment of Ki67 in Breast Cancer: Updated Recommendations from the International Ki67 in Breast Cancer Working Group. J. Natl. Cancer Inst. 2020, 113, 808–819. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, M.; Ishida, T.; Ishida, K.; Tamaki, K.; Amari, M.; Watanabe, M.; Ohuchi, N.; Sasano, H. Histopathological subclassification of triple negative breast cancer using prognostic scoring system: Five variables as candidates. Virchows Arch. 2011, 458, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Smith, I.; Robertson, J.; Kilburn, L.; Wilcox, M.; Evans, A.; Holcombe, C.; Horgan, K.; Kirwan, C.; Mallon, E.; Sibbering, M.; et al. Long-term outcome and prognostic value of Ki67 after perioperative endocrine therapy in postmenopausal women with hormone-sensitive early breast cancer (POETIC): An open-label, multicentre, parallel-group, randomised, phase 3 trial. Lancet Oncol. 2020, 21, 1443–1454. [Google Scholar] [CrossRef]
- Ellis, M.J.; Tao, Y.; Luo, J.; A’Hern, R.; Evans, D.B.; Bhatnagar, A.S.; Chaudri Ross, H.A.; von Kameke, A.; Miller, W.R.; Smith, I.; et al. Outcome prediction for estrogen receptor-positive breast cancer based on postneoadjuvant endocrine therapy tumor characteristics. J. Natl. Cancer Inst. 2008, 100, 1380–1388. [Google Scholar] [CrossRef]
- Brown, J.R.; DiGiovanna, M.P.; Killelea, B.; Lannin, D.R.; Rimm, D.L. Quantitative assessment Ki-67 score for prediction of response to neoadjuvant chemotherapy in breast cancer. Lab. Investig. 2014, 94, 98–106. [Google Scholar] [CrossRef] [Green Version]
- Niikura, N.; Masuda, S.; Kumaki, N.; Xiaoyan, T.; Terada, M.; Terao, M.; Iwamoto, T.; Oshitanai, R.; Morioka, T.; Tuda, B.; et al. Prognostic significance of the Ki67 scoring categories in breast cancer subgroups. Clin. Breast Cancer 2014, 14, 323–329. [Google Scholar] [CrossRef]
- Polley, M.Y.; Leung, S.C.; McShane, L.M.; Gao, D.; Hugh, J.C.; Mastropasqua, M.G.; Viale, G.; Zabaglo, L.A.; Penault-Llorca, F.; Bartlett, J.M.; et al. An international Ki67 reproducibility study. J. Natl. Cancer Inst. 2013, 105, 1897–1906. [Google Scholar] [CrossRef]
- Polley, M.Y.; Leung, S.C.; Gao, D.; Mastropasqua, M.G.; Zabaglo, L.A.; Bartlett, J.M.; McShane, L.M.; Enos, R.A.; Badve, S.S.; Bane, A.L.; et al. An international study to increase concordance in Ki67 scoring. Mod. Pathol. 2015, 28, 778–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, S.C.Y.; Nielsen, T.O.; Zabaglo, L.; Arun, I.; Badve, S.S.; Bane, A.L.; Bartlett, J.M.S.; Borgquist, S.; Chang, M.C.; Dodson, A.; et al. Analytical validation of a standardized scoring protocol for Ki67: Phase 3 of an international multicenter collaboration. NPJ Breast Cancer 2016, 2, 16014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klintman, M.; Dowsett, M. Early Surrogate Markers of Treatment Activity: Where Are We Now? JNCI Monogr. 2015, 2015, 24–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowsett, M.; Smith, I.E.; Ebbs, S.R.; Dixon, J.M.; Skene, A.; Griffith, C.; Boeddinghaus, I.; Salter, J.; Detre, S.; Hills, M.; et al. Short-term changes in Ki-67 during neoadjuvant treatment of primary breast cancer with anastrozole or tamoxifen alone or combined correlate with recurrence-free survival. Clin. Cancer Res. 2005, 11 Pt 2, 951s–958s. [Google Scholar]
- Baum, M.; Buzdar, A.; Cuzick, J.; Forbes, J.; Houghton, J.; Howell, A.; Sahmoud, T. Anastrozole alone or in combination with tamoxifen versus tamoxifen alone for adjuvant treatment of postmenopausal women with early-stage breast cancer: Results of the ATAC (Arimidex, Tamoxifen Alone or in Combination) trial efficacy and safety update analyses. Cancer 2003, 98, 1802–1810. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.J.; Coop, A.; Singh, B.; Tao, Y.; Llombart-Cussac, A.; Jänicke, F.; Mauriac, L.; Quebe-Fehling, E.; Chaudri-Ross, H.A.; Evans, D.B.; et al. Letrozole inhibits tumor proliferation more effectively than tamoxifen independent of HER1/2 expression status. Cancer Res. 2003, 63, 6523–6531. [Google Scholar]
- Thürlimann, B.; Keshaviah, A.; Coates, A.S.; Mouridsen, H.; Mauriac, L.; Forbes, J.F.; Paridaens, R.; Castiglione-Gertsch, M.; Gelber, R.D.; Rabaglio, M.; et al. A Comparison of Letrozole and Tamoxifen in Postmenopausal Women with Early Breast Cancer. N. Engl. J. Med. 2005, 353, 2747–2757. [Google Scholar] [CrossRef] [Green Version]
- Symmans, W.F.; Peintinger, F.; Hatzis, C.; Rajan, R.; Kuerer, H.; Valero, V.; Assad, L.; Poniecka, A.; Hennessy, B.; Green, M.; et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J. Clin. Oncol. 2007, 25, 4414–4422. [Google Scholar] [CrossRef]
- Jones, R.L.; Salter, J.; A’Hern, R.; Nerurkar, A.; Parton, M.; Reis-Filho, J.S.; Smith, I.E.; Dowsett, M. The prognostic significance of Ki67 before and after neoadjuvant chemotherapy in breast cancer. Breast Cancer Res. Treat. 2009, 116, 53–68. [Google Scholar] [CrossRef] [PubMed]
- von Minckwitz, G.; Schmitt, W.D.; Loibl, S.; Müller, B.M.; Blohmer, J.U.; Sinn, B.V.; Eidtmann, H.; Eiermann, W.; Gerber, B.; Tesch, H.; et al. Ki67 measured after neoadjuvant chemotherapy for primary breast cancer. Clin. Cancer Res. 2013, 19, 4521–4531. [Google Scholar] [CrossRef] [Green Version]
- Sheri, A.; Smith, I.E.; Johnston, S.R.; A’Hern, R.; Nerurkar, A.; Jones, R.L.; Hills, M.; Detre, S.; Pinder, S.E.; Symmans, W.F.; et al. Residual proliferative cancer burden to predict long-term outcome following neoadjuvant chemotherapy. Ann. Oncol. 2015, 26, 75–80. [Google Scholar] [CrossRef]
- Ma, J.; Chi, D.; Wang, Y.; Yan, Y.; Zhao, S.; Liu, H.; Jing, J.; Pu, H.; Zhang, M. Prognostic value of PD-L1 expression in resected lung adenocarcinoma and potential molecular mechanisms. J. Cancer 2018, 9, 3489–3499. [Google Scholar] [CrossRef]
- Li, Y.; He, M.; Zhou, Y.; Yang, C.; Wei, S.; Bian, X.; Christopher, O.; Xie, L. The Prognostic and Clinicopathological Roles of PD-L1 Expression in Colorectal Cancer: A Systematic Review and Meta-Analysis. Front. Pharmacol. 2019, 10, 139. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Sun, J.; Wang, L.; Li, Z.; Wang, L.; Li, Z. Prognostic and Clinicopathological Significance of PD-L1 in Patients With Bladder Cancer: A Meta-Analysis. Front. Pharmacol. 2019, 10, 962. [Google Scholar] [CrossRef] [Green Version]
- Yun, S.; Park, Y.; Moon, S.; Ahn, S.; Lee, K.; Park, H.J.; Lee, H.S.; Choe, G.; Lee, K.S. Clinicopathological and prognostic significance of programmed death ligand 1 expression in Korean melanoma patients. J. Cancer 2019, 10, 3070–3078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Hegg, R.; Im, S.-A.; Shaw Wright, G.; et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef]
- Schmid, P.; Cortes, J.; Pusztai, L.; McArthur, H.; Kümmel, S.; Bergh, J.; Denkert, C.; Park, Y.H.; Hui, R.; Harbeck, N.; et al. Pembrolizumab for Early Triple-Negative Breast Cancer. N. Engl. J. Med. 2020, 382, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Wolchok, J.D.; Chen, L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci. Transl. Med. 2016, 8, 328rv324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schütz, F.; Stefanovic, S.; Mayer, L.; von Au, A.; Domschke, C.; Sohn, C. PD-1/PD-L1 Pathway in Breast Cancer. Oncol. Res. Treat. 2017, 40, 294–297. [Google Scholar] [CrossRef] [PubMed]
- Bayraktar, S.; Batoo, S.; Okuno, S.; Glück, S. Immunotherapy in breast cancer. J. Carcinog. 2019, 18, 2. [Google Scholar] [CrossRef]
- Ghebeh, H.; Barhoush, E.; Tulbah, A.; Elkum, N.; Al-Tweigeri, T.; Dermime, S. FOXP3+ Tregs and B7-H1+/PD-1+ T lymphocytes co-infiltrate the tumor tissues of high-risk breast cancer patients: Implication for immunotherapy. BMC Cancer 2008, 8, 57. [Google Scholar] [CrossRef] [Green Version]
- Ghebeh, H.; Mohammed, S.; Al-Omair, A.; Qattan, A.; Lehe, C.; Al-Qudaihi, G.; Elkum, N.; Alshabanah, M.; Bin Amer, S.; Tulbah, A.; et al. The B7-H1 (PD-L1) T lymphocyte-inhibitory molecule is expressed in breast cancer patients with infiltrating ductal carcinoma: Correlation with important high-risk prognostic factors. Neoplasia 2006, 8, 190–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghebeh, H.; Tulbah, A.; Mohammed, S.; Elkum, N.; Bin Amer, S.M.; Al-Tweigeri, T.; Dermime, S. Expression of B7-H1 in breast cancer patients is strongly associated with high proliferative Ki-67-expressing tumor cells. Int. J. Cancer 2007, 121, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.G.; Ryan, É.J.; Davey, M.S.; Lowery, A.J.; Miller, N.; Kerin, M.J. Clinicopathological and prognostic significance of programmed cell death ligand 1 expression in patients diagnosed with breast cancer: Meta-analysis. Br. J. Surg. 2021, 108, 622–631. [Google Scholar] [CrossRef] [PubMed]
- Muenst, S.; Schaerli, A.R.; Gao, F.; Däster, S.; Trella, E.; Droeser, R.A.; Muraro, M.G.; Zajac, P.; Zanetti, R.; Gillanders, W.E.; et al. Expression of programmed death ligand 1 (PD-L1) is associated with poor prognosis in human breast cancer. Breast Cancer Res. Treat. 2014, 146, 15–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, S.B.; Cho, H.D.; Oh, M.H.; Lee, J.H.; Jang, S.H.; Hong, S.A.; Cho, J.; Kim, S.Y.; Han, S.W.; Lee, J.E.; et al. Expression of Programmed Death Receptor Ligand 1 with High Tumor-Infiltrating Lymphocytes Is Associated with Better Prognosis in Breast Cancer. J. Breast Cancer 2016, 19, 242–251. [Google Scholar] [CrossRef] [Green Version]
- Asano, Y.; Kashiwagi, S.; Goto, W.; Takada, K.; Takahashi, K.; Morisaki, T.; Fujita, H.; Takashima, T.; Tomita, S.; Ohsawa, M.; et al. Prediction of treatment responses to neoadjuvant chemotherapy in triple-negative breast cancer by analysis of immune checkpoint protein expression. J. Transl. Med. 2018, 16, 87. [Google Scholar] [CrossRef]
- Wang, R.-X.; Chen, S.; Jin, X.; Shao, Z.-M. Value of Ki-67 expression in triple-negative breast cancer before and after neoadjuvant chemotherapy with weekly paclitaxel plus carboplatin. Sci. Rep. 2016, 6, 30091. [Google Scholar] [CrossRef] [Green Version]
- Mukai, H.; Yamaguchi, T.; Takahashi, M.; Hozumi, Y.; Fujisawa, T.; Ohsumi, S.; Akabane, H.; Nishimura, R.; Takashima, T.; Park, Y.; et al. Ki-67 response-guided preoperative chemotherapy for HER2-positive breast cancer: Results of a randomised Phase 2 study. Br. J. Cancer 2020, 122, 1747–1753. [Google Scholar] [CrossRef]
- Pabla, S.; Conroy, J.M.; Nesline, M.K.; Glenn, S.T.; Papanicolau-Sengos, A.; Burgher, B.; Hagen, J.; Giamo, V.; Andreas, J.; Lenzo, F.L.; et al. Proliferative potential and resistance to immune checkpoint blockade in lung cancer patients. J. ImmunoTher. Cancer 2019, 7, 27. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, K.G.; Parra, E.R.; Nelson, D.B.; Zhang, J.; Wistuba, I.I.; Fujimoto, J.; Roth, J.A.; Antonoff, M.B.; Corsini, E.M.; Vaporciyan, A.A.; et al. Tumor cellular proliferation is associated with enhanced immune checkpoint expression in stage I non-small cell lung cancer. J. Thorac. Cardiovasc. Surg. 2019, 158, 911–919.e6. [Google Scholar] [CrossRef] [PubMed]
- Offit, K. The future of clinical cancer genomics. Semin. Oncol. 2016, 43, 615–622. [Google Scholar] [CrossRef] [Green Version]
- Brittain, H.K.; Scott, R.; Thomas, E. The rise of the genome and personalised medicine. Clin. Med. 2017, 17, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.S.; Li, H.; Chan, A.M.Y.; Tudor, R.; Bigras, G.; Morris, D.; Enwere, E.K.; Yang, H. The use of automated Ki67 analysis to predict Oncotype DX risk-of-recurrence categories in early-stage breast cancer. PLoS ONE 2018, 13, e0188983. [Google Scholar] [CrossRef] [Green Version]
- National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines)—Breast Cancer; National Comprehensive Cancer Network: Plymouth Meeting, PA, USA, 2017. [Google Scholar]
- Senkus, E.; Kyriakides, S.; Ohno, S.; Penault-Llorca, F.; Poortmans, P.; Rutgers, E.; Zackrisson, S.; Cardoso, F. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26 (Suppl. 5), v8–v30. [Google Scholar] [CrossRef] [PubMed]
- Gnant, M.; Harbeck, N.; Thomssen, C. St. Gallen/Vienna 2017: A Brief Summary of the Consensus Discussion about Escalation and De-Escalation of Primary Breast Cancer Treatment. Breast Care 2017, 12, 102–107. [Google Scholar] [CrossRef] [Green Version]
- NICE. Gene Expression Profiling and Expanded Immunohistochemistry Tests for Guiding Adjuvant Chemotherapy Decisions in Early Breast Cancer Management: MammaPrint, Oncotype DX, IHC4 and Mammostrat; NICE: London, UK, 2013. [Google Scholar]
- Siow, Z.R.; De Boer, R.H.; Lindeman, G.J.; Mann, G.B. Spotlight on the utility of the Oncotype DX(®) breast cancer assay. Int. J. Womens Health 2018, 10, 89–100. [Google Scholar] [CrossRef] [Green Version]
- McVeigh, T.P.; Kerin, M.J. Clinical use of the Oncotype DX genomic test to guide treatment decisions for patients with invasive breast cancer. Breast Cancer 2017, 9, 393–400. [Google Scholar] [CrossRef] [Green Version]
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E., Jr.; Dees, E.C.; Perez, E.A.; Olson, J.A., Jr.; et al. Prospective Validation of a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2015, 373, 2005–2014. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, L.J.; Gray, R.; Badve, S.; Childs, B.H.; Yoshizawa, C.; Rowley, S.; Shak, S.; Baehner, F.L.; Ravdin, P.M.; Davidson, N.E.; et al. Prognostic utility of the 21-gene assay in hormone receptor-positive operable breast cancer compared with classical clinicopathologic features. J. Clin. Oncol. 2008, 26, 4063–4071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowsett, M.; Cuzick, J.; Wale, C.; Forbes, J.; Mallon, E.A.; Salter, J.; Quinn, E.; Dunbier, A.; Baum, M.; Buzdar, A.; et al. Prediction of risk of distant recurrence using the 21-gene recurrence score in node-negative and node-positive postmenopausal patients with breast cancer treated with anastrozole or tamoxifen: A TransATAC study. J. Clin. Oncol. 2010, 28, 1829–1834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Schutter, H.; Van Damme, N.; Colpaert, C.; Galant, C.; Lambein, K.; Cornelis, A.; Neven, P.; Van Eycken, E. Quality of pathology reporting is crucial for cancer care and registration: A baseline assessment for breast cancers diagnosed in Belgium in 2008. Breast 2015, 24, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.G.; Ryan, É.J.; Abd Elwahab, S.; Elliott, J.A.; McAnena, P.F.; Sweeney, K.J.; Malone, C.M.; McLaughlin, R.; Barry, M.K.; Keane, M.M.; et al. Clinicopathological correlates, oncological impact, and validation of Oncotype DX™ in a European Tertiary Referral Centre. Breast J. 2021, 27, 521–528. [Google Scholar] [CrossRef]
- Xin, L.; Liu, Y.-H.; Martin, T.A.; Jiang, W.G. The Era of Multigene Panels Comes? The Clinical Utility of Oncotype DX and MammaPrint. World J. Oncol. 2017, 8, 34–40. [Google Scholar] [CrossRef] [Green Version]
- Sahebjam, S.; Aloyz, R.; Pilavdzic, D.; Brisson, M.L.; Ferrario, C.; Bouganim, N.; Cohen, V.; Miller, W.H., Jr.; Panasci, L.C. Ki 67 is a major, but not the sole determinant of Oncotype Dx recurrence score. Br. J. Cancer 2011, 105, 1342–1345. [Google Scholar] [CrossRef] [Green Version]
- Tan, A.C.; Li, B.T.; Nahar, K.; Danieletto, S.; Fong, E.S.; Currer, T.; Parasyn, A.; Middleton, P.; Wong, H.; Smart, D.; et al. Correlating Ki67 and other prognostic markers with Oncotype DX recurrence score in early estrogen receptor-positive breast cancer. Asia Pac. J. Clin. Oncol. 2018, 14, e161–e166. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Roepman, P.; Van’t Veer, L.J.; Bernards, R.; de Snoo, F.; Glas, A.M. Biological functions of the genes in the mammaprint breast cancer profile reflect the hallmarks of cancer. Biomark. Insights 2010, 5, 129–138. [Google Scholar] [CrossRef] [Green Version]
- van de Vijver, M.J.; He, Y.D.; van’t Veer, L.J.; Dai, H.; Hart, A.A.; Voskuil, D.W.; Schreiber, G.J.; Peterse, J.L.; Roberts, C.; Marton, M.J.; et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002, 347, 1999–2009. [Google Scholar] [CrossRef] [Green Version]
- Van ’t Veer, L.J.; Dai, H.; van de Vijver, M.J.; He, Y.D.; Hart, A.A.M.; Mao, M.; Peterse, H.L.; van der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef] [Green Version]
- Pronzato, P.; Mustacchi, G.; Generali, D.G.; Bottini, A. Complementary role of Ki67 index and 70-gene signature (MammaPrint) high-risk patients in the St Gallen risk group with uncertain chemotherapy suggestion. J. Clin. Oncol. 2012, 30, 579. [Google Scholar] [CrossRef]
- Davey, M.G.; Ryan, É.J.; McAnena, P.F.; Boland, M.R.; Barry, M.K.; Sweeney, K.J.; Malone, C.M.; McLaughlin, R.J.; Lowery, A.J.; Kerin, M.J. Disease recurrence and oncological outcome of patients treated surgically with curative intent for estrogen receptor positive, lymph node negative breast cancer. Surg. Oncol. 2021, 37, 101531. [Google Scholar] [CrossRef]
- Cardoso, F.; van ’t Veer, L.; Poncet, C.; Lopes Cardozo, J.; Delaloge, S.; Pierga, J.-Y.; Vuylsteke, P.; Brain, E.; Viale, G.; Kuemmel, S.; et al. MINDACT: Long-term results of the large prospective trial testing the 70-gene signature MammaPrint as guidance for adjuvant chemotherapy in breast cancer patients. J. Clin. Oncol. 2020, 38, 506. [Google Scholar] [CrossRef]
- Stemmer, S.M.; Steiner, M.; Rizel, S.; Geffen, D.B.; Nisenbaum, B.; Peretz, T.; Soussan-Gutman, L.; Bareket-Samish, A.; Isaacs, K.; Rosengarten, O.; et al. Clinical outcomes in ER+ HER2 -node-positive breast cancer patients who were treated according to the Recurrence Score results: Evidence from a large prospectively designed registry. NPJ Breast Cancer 2017, 3, 32. [Google Scholar] [CrossRef] [PubMed]
- Aung, T.N.; Acs, B.; Warrell, J.; Bai, Y.; Gaule, P.; Martinez-Morilla, S.; Vathiotis, I.; Shafi, S.; Moutafi, M.; Gerstein, M.; et al. A new tool for technical standardization of the Ki67 immunohistochemical assay. Mod. Pathol. 2021, 34, 1261–1270. [Google Scholar] [CrossRef] [PubMed]
- Riber-Hansen, R.; Vainer, B.E.N.; Steiniche, T. Digital image analysis: A review of reproducibility, stability and basic requirements for optimal results. APMIS 2012, 120, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Gudlaugsson, E.; Skaland, I.; Janssen, E.A.; Smaaland, R.; Shao, Z.; Malpica, A.; Voorhorst, F.; Baak, J.P. Comparison of the effect of different techniques for measurement of Ki67 proliferation on reproducibility and prognosis prediction accuracy in breast cancer. Histopathology 2012, 61, 1134–1144. [Google Scholar] [CrossRef]
- Tadrous, P.J. On the concept of objectivity in digital image analysis in pathology. Pathology 2010, 42, 207–211. [Google Scholar] [CrossRef]
- Klauschen, F.; Wienert, S.; Schmitt, W.D.; Loibl, S.; Gerber, B.; Blohmer, J.U.; Huober, J.; Rüdiger, T.; Erbstößer, E.; Mehta, K.; et al. Standardized Ki67 Diagnostics Using Automated Scoring—Clinical Validation in the GeparTrio Breast Cancer Study. Clin. Cancer Res. 2015, 21, 3651–3657. [Google Scholar] [CrossRef] [Green Version]
- Zhong, F.; Bi, R.; Yu, B.; Yang, F.; Yang, W.; Shui, R. A Comparison of Visual Assessment and Automated Digital Image Analysis of Ki67 Labeling Index in Breast Cancer. PLoS ONE 2016, 11, e0150505. [Google Scholar] [CrossRef]
- Røge, R.; Riber-Hansen, R.; Nielsen, S.; Vyberg, M. Proliferation assessment in breast carcinomas using digital image analysis based on virtual Ki67/cytokeratin double staining. Breast Cancer Res. Treat. 2016, 158, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Stålhammar, G.; Fuentes Martinez, N.; Lippert, M.; Tobin, N.P.; Mølholm, I.; Kis, L.; Rosin, G.; Rantalainen, M.; Pedersen, L.; Bergh, J.; et al. Digital image analysis outperforms manual biomarker assessment in breast cancer. Mod. Pathol. 2016, 29, 318–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kårsnäs, A.; Strand, R.; Doré, J.; Ebstrup, T.; Lippert, M.; Bjerrum, K. A histopathological tool for quantification of biomarkers with sub-cellular resolution. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2015, 3, 25–46. [Google Scholar] [CrossRef]
- Davey, M.G.; Davies, M.; Lowery, A.J.; Miller, N.; Kerin, M.J. The Role of MicroRNA as Clinical Biomarkers for Breast Cancer Surgery and Treatment. Int. J. Mol. Sci. 2021, 22, 8290. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Hamam, R.; Hamam, D.; Alsaleh, K.A.; Kassem, M.; Zaher, W.; Alfayez, M.; Aldahmash, A.; Alajez, N.M. Circulating microRNAs in breast cancer: Novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017, 8, e3045. [Google Scholar] [CrossRef] [Green Version]
- Sakurai, M.; Masuda, M.; Miki, Y.; Hirakawa, H.; Suzuki, T.; Sasano, H. Correlation of miRNA Expression Profiling in Surgical Pathology Materials, with Ki-67, HER2, ER and PR in Breast Cancer Patients. Int. J. Biol. Markers 2015, 30, 190–199. [Google Scholar] [CrossRef]
- Wang, X.; Cao, L.; Wang, Y.; Wang, X.; Liu, N.; You, Y. Regulation of let-7 and its target oncogenes (Review). Oncol. Lett. 2012, 3, 955–960. [Google Scholar] [CrossRef]
- Amorim, M.; Lobo, J.; Fontes-Sousa, M.; Estevão-Pereira, H.; Salta, S.; Lopes, P.; Coimbra, N.; Antunes, L.; Palma de Sousa, S.; Henrique, R.; et al. Predictive and Prognostic Value of Selected MicroRNAs in Luminal Breast Cancer. Front. Genet. 2019, 10, 815. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Tang, K.; Yan, W.; Wang, Y.; You, G.; Kang, C.; Jiang, T.; Zhang, W. Identifying Ki-67 specific miRNA–mRNA interactions in malignant astrocytomas. Neurosci. Lett. 2013, 546, 36–41. [Google Scholar] [CrossRef]
- Trang, P.; Medina, P.P.; Wiggins, J.F.; Ruffino, L.; Kelnar, K.; Omotola, M.; Homer, R.; Brown, D.; Bader, A.G.; Weidhaas, J.B.; et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene 2010, 29, 1580–1587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Bellomi, M. Radiomics: The facts and the challenges of image analysis. Eur. Radiol. Exp. 2018, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Sun, J.; Qu, N.; Zhang, G.; Yu, T.; Piao, H. Application of Radiomics for Personalized Treatment of Cancer Patients. Cancer Manag. Res. 2019, 11, 10851–10858. [Google Scholar] [CrossRef] [Green Version]
- Juan, M.-W.; Yu, J.; Peng, G.-X.; Jun, L.-J.; Feng, S.-P.; Fang, L.-P. Correlation between DCE-MRI radiomics features and Ki-67 expression in invasive breast cancer. Oncol. Lett. 2018, 16, 5084–5090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tagliafico, A.S.; Bignotti, B.; Rossi, F.; Matos, J.; Calabrese, M.; Valdora, F.; Houssami, N. Breast cancer Ki-67 expression prediction by digital breast tomosynthesis radiomics features. Eur. Radiol. Exp. 2019, 3, 36. [Google Scholar] [CrossRef] [Green Version]


| Author | Year | N | Patients | Findings |
|---|---|---|---|---|
| Ellis [75] | 2008 | 228 | ER+ stage II/III | Per 2.7% increase in Ki-67 expression levels, there is an increased risk of RFS in patients treated with NET (HR: 1.3, 95% CI: 1.05–1.50) |
| Fasching [47] | 2011 | 552 | Early breast cancer | Using greater than 13% as a cut-off for Ki-67, Ki-67 predicted pCR ro NAC (OR: 3.5, 95% CI: 1.4–10.1) and OS (HR: 8.1, 95% CI: 3.3–20.4) and DDFS (HR: 3.2 95% CI: 1.8–5.9) |
| Brown [76] | 2013 | 105 | Received NAC | Ki-67 expression correlated directly to pCR |
| Niikura [77] | 2014 | 971 | ER+/HER2- | Patients with low Ki-67 expression indices had significantly better RFS and OS than those with intermediate- and high- Ki-67 expression (all p < 0.001) |
| Petrelli [69] | 2015 | 64,196 | All subtypes | In this meta-analysis, Ki-67 expression levels greater than or equal to 25% predicted OS in 64,196 breast cancer patients (HR: 2.05, 95% CI: 1.66–2.53) |
| Enrico [68] | 2018 | 506 | Stage I-III | Illustrated the 20% Ki-67 expression cut off as clinically relevant for recurrence and survival (HR: 7.14, 95% CI: 3.87–13.16) |
| Wu [74] | 2019 | 7,716 | Resected TNBC | In this meta-analysis, Ki-67 expression levels greater than 40% predicted DFS (HR: 2.30, 95% CI: 1.54–3.44) and OS (HR: 2.95, 95% CI: 1.67–5.19) |
| Zhu [27] | 2020 | 1800 | Early stage TNBC | Using a 30%, high Ki-67 indices independently predicted worse OS (HR: 1.947, 95% CI: 1.108–3.421) |
| Tian [70] | 2020 | 1008 | ER+/HER- | Ki-67 expression profiles correlated with the 70-gene assay; for patients with Ki-67 less than 15%, 81.4% were GLR |
| Author and Year | Country | Tissue | N | Technique | MicroRNA and Ki-67 Status |
|---|---|---|---|---|---|
| Sakurai 2018 [144] | Japan | Breast tumour | 21 | qRT-PCR | miR-let-7a, miR-let-7b, miR-let-7e, miR-29a, miR-143, miR-181a, miR-214, and miR-218 were all overexpressed in control breast cancer group, defined as possessing Ki-67 indices of 0–14% miR-7, miR-15b, miR-16, miR-18b, miR-20b, miR-21, miR-25, miR-27a, miR-27b, miR-34a, miR-92a, miR-96, miR-125a-5p, miR-125b, miR-132, miR-133b, miR-146a, miR-148b, miR-149, miR-150, miR-183, miR-184, miR-191, miR-199a-3p, miR-200c, miR-203, miR-301a, miR-355, and miR-363 were all upregulated in breast cancers with high Ki-67 expression (greater than 25%) |
| Amorim 2019 [146] | Portugal | Breast tumour | 139 | qRT-PCR | miR-30c-5p, miR-182-5p, and miR-200-3p expression profiles independently predict endocrine resistance-free survival once adjusted for Ki-67 status miR-30c-5p, miR-200b-3p, and miR-182-5p levels independently predict endocrine resistance-free survival once adjusted for Ki-67 status Predictive of disease-free recurrence, once adjusted for Ki-67 status |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davey, M.G.; Hynes, S.O.; Kerin, M.J.; Miller, N.; Lowery, A.J. Ki-67 as a Prognostic Biomarker in Invasive Breast Cancer. Cancers 2021, 13, 4455. https://doi.org/10.3390/cancers13174455
Davey MG, Hynes SO, Kerin MJ, Miller N, Lowery AJ. Ki-67 as a Prognostic Biomarker in Invasive Breast Cancer. Cancers. 2021; 13(17):4455. https://doi.org/10.3390/cancers13174455
Chicago/Turabian StyleDavey, Matthew G., Sean O. Hynes, Michael J. Kerin, Nicola Miller, and Aoife J. Lowery. 2021. "Ki-67 as a Prognostic Biomarker in Invasive Breast Cancer" Cancers 13, no. 17: 4455. https://doi.org/10.3390/cancers13174455
APA StyleDavey, M. G., Hynes, S. O., Kerin, M. J., Miller, N., & Lowery, A. J. (2021). Ki-67 as a Prognostic Biomarker in Invasive Breast Cancer. Cancers, 13(17), 4455. https://doi.org/10.3390/cancers13174455





