A New Story of the Three Magi: Scaffolding Proteins and lncRNA Suppressors of Cancer
Simple Summary
Abstract
1. Introduction
2. Implication of MAGI Molecular Scaffolds in the Control of Carcinogenesis
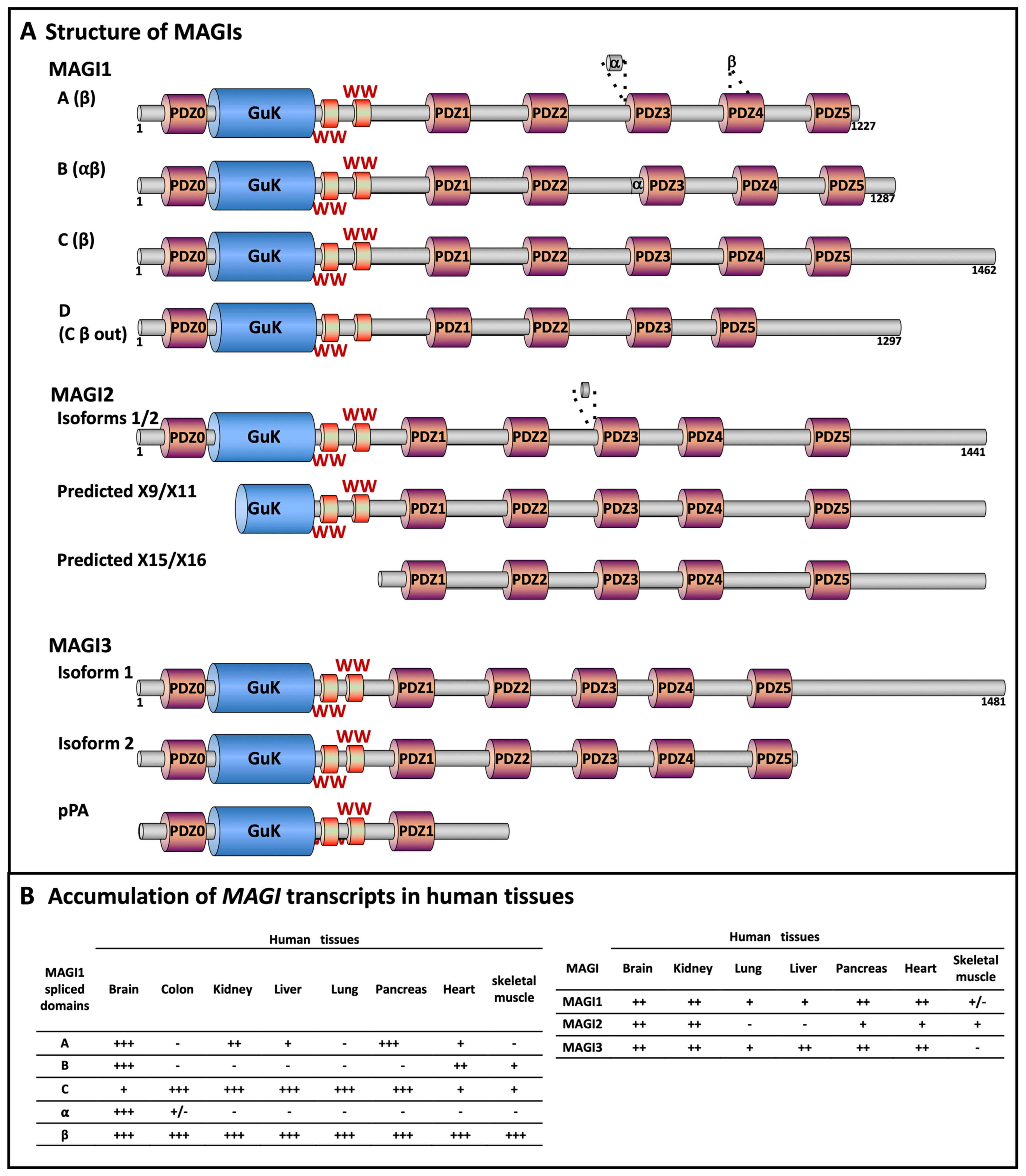
2.1. MAGI Structure
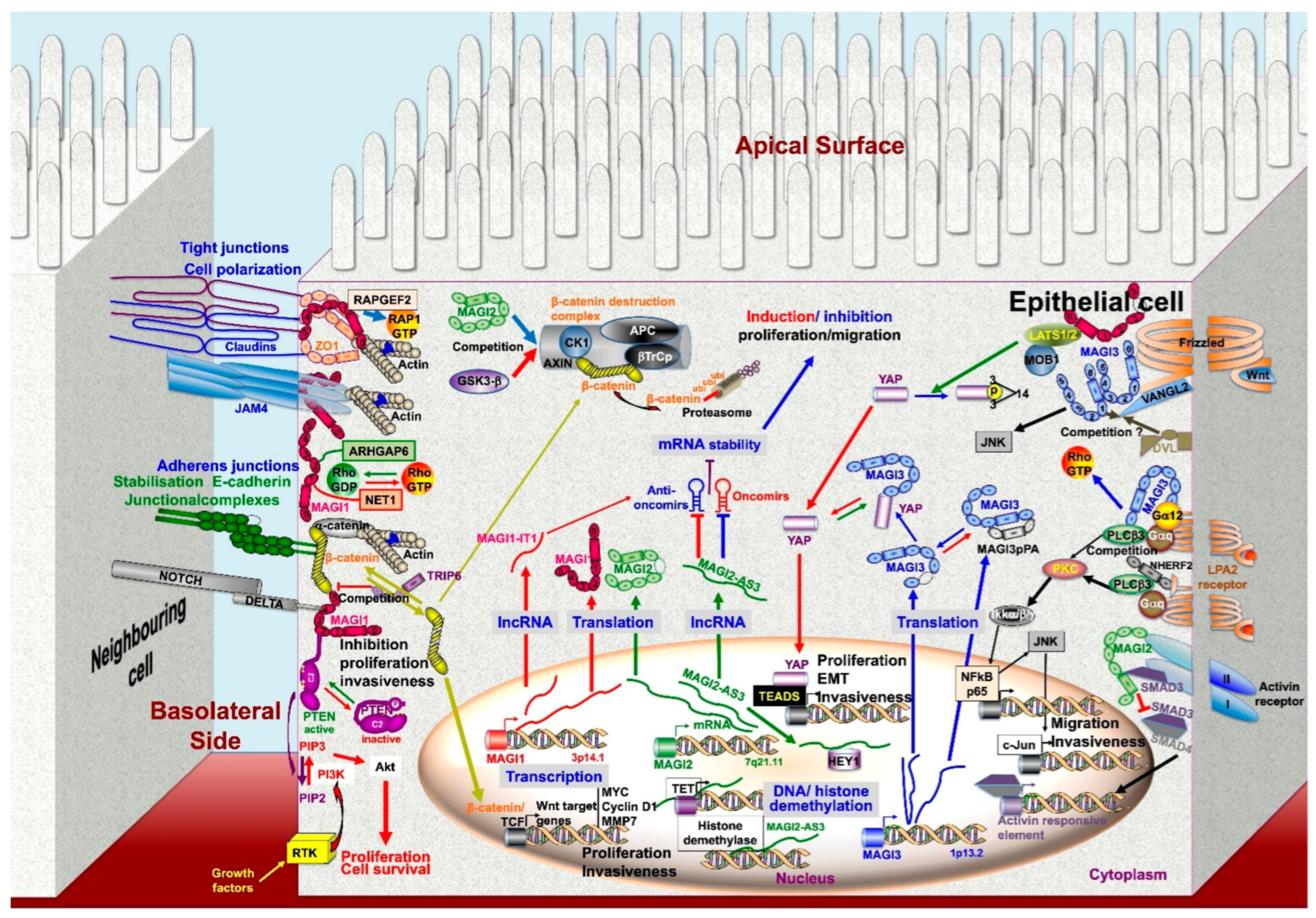
2.2. MAGI Molecular Partners
2.3. MAGIs in Cancer
2.3.1. Genetic Alteration, Silencing
2.3.2. MAGIs Expression and Prognosis
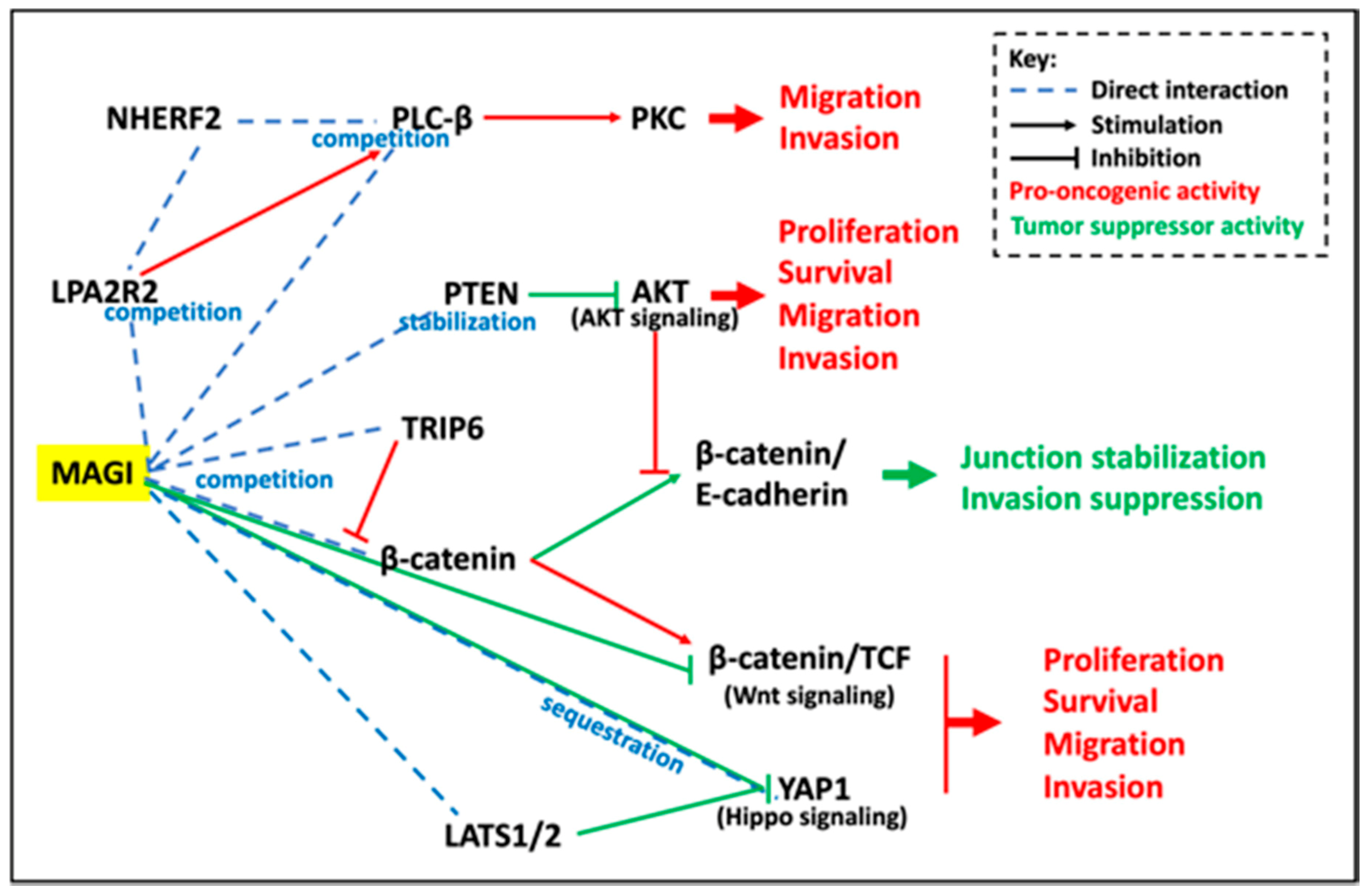
2.3.3. Signaling Pathways and Effector Systems Involved in the Tumor Suppressor Activities of MAGIs
PTEN/PI3K/AKT Signaling Pathway
Beta-Catenin/Wnt Signaling Pathways
HIPPO Signaling Pathway
Junctional Complex Stabilizer, Invasion Suppressor Activity
Competitions between Scaffolding Molecules and Molecular Partners
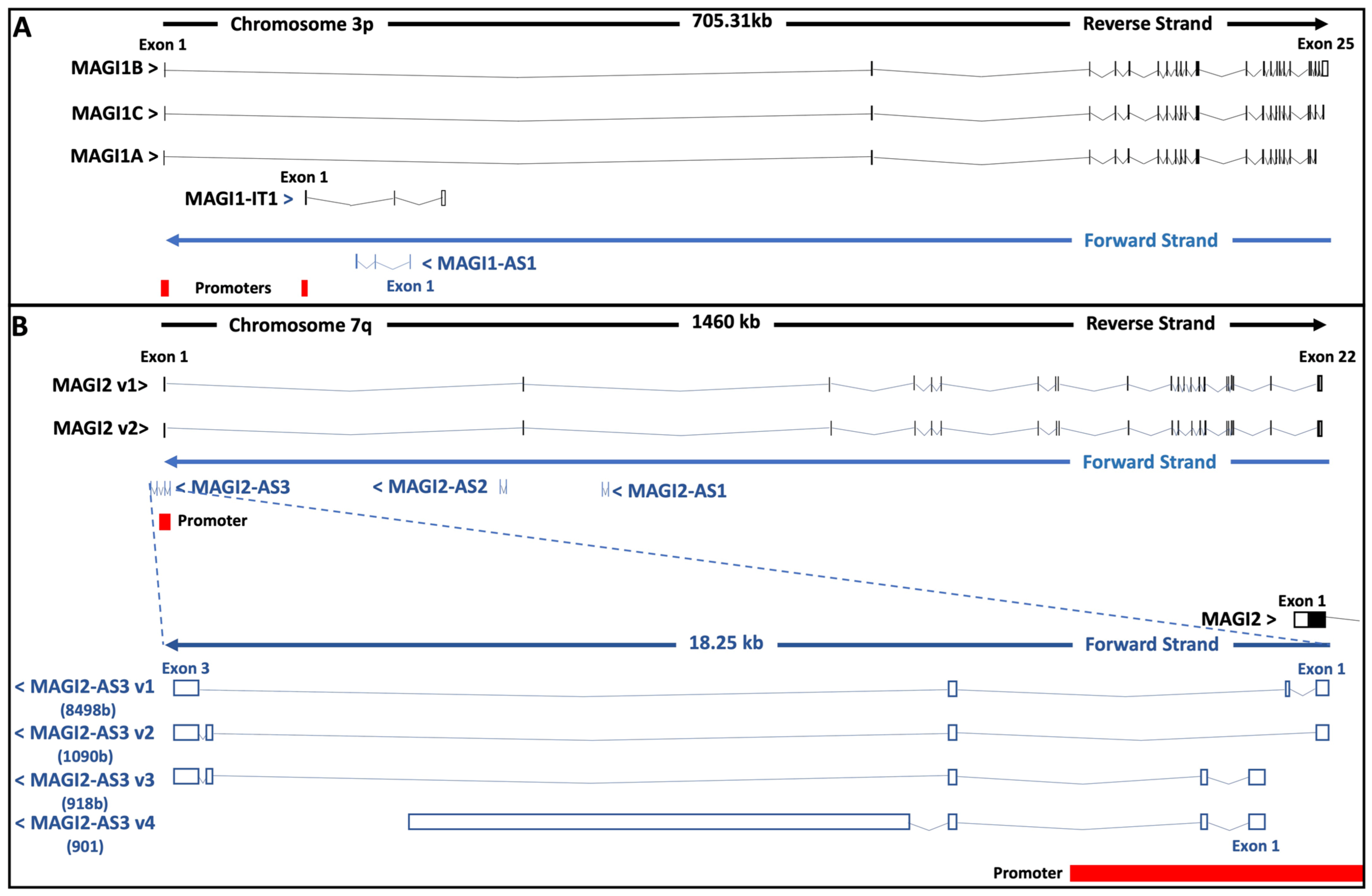
3. MAGI ceRNAs in Carcinogenesis
4. Conclusions, Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kotelevets, L.; Scott, M.G.H.; Chastre, E. Targeted therapy of colorectal cancer subtypes. In Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2019; Volume 1110, pp. 55–73. [Google Scholar]
- Kotelevets, L.; Trifault, B.; Chastre, E.; Scott, M.G.H. Posttranslational Regulation and Conformational Plasticity of PTEN; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2020; Volume 10. [Google Scholar]
- Laporte, S.A.; Scott, M.G.H. β-Arrestins: Multitask scaffolds orchestrating the where and when in cell signalling. Methods Mol. Biol. 2019, 1957, 9–55. [Google Scholar] [CrossRef]
- Kotelevets, L.; Chastre, E. Rac1 signaling: From intestinal homeostasis to colorectal cancer metastasis. Cancers 2020, 12, 665. [Google Scholar] [CrossRef]
- Ivarsson, Y. Plasticity of PDZ domains in ligand recognition and signaling. FEBS Lett. 2012, 586, 2638–2647. [Google Scholar] [CrossRef]
- Manjunath, G.P.; Ramanujam, P.L.; Galande, S. Structure function relations in PDZ-domain-containing proteins: Implications for protein networks in cellular signalling. J. Biosci. 2018, 43, 155–171. [Google Scholar] [CrossRef]
- Christensen, N.R.; Čalyševa, J.; Fernandes, E.F.A.; Lüchow, S.; Clemmensen, L.S.; Haugaard-Kedström, L.M.; Strømgaard, K. PDZ Domains as drug targets. Adv. Ther. 2019, 2, 1800143. [Google Scholar] [CrossRef] [PubMed]
- Amacher, J.F.; Brooks, L.; Hampton, T.H.; Madden, D.R. Specificity in PDZ-peptide interaction networks: Computational analysis and review. J. Struct. Biol. X 2020, 4, 100022. [Google Scholar] [CrossRef]
- Dunn, H.A.; Ferguson, S.S.G. PDZ Protein Regulation of G protein-coupled receptor trafficking and signaling pathways. Mol. Pharmacol. 2015, 88, 624–639. [Google Scholar] [CrossRef]
- Heinemann, U.; Schuetz, A. Structural features of Tight-junction proteins. Int. J. Mol. Sci. 2019, 20, 6020. [Google Scholar] [CrossRef]
- Liu, X.; Fuentes, E.J. Chapter Five—Emerging themes in PDZ domain signaling structure, function, and inhibition. Int. Rev. Cell Mol. Biol. 2019, 343, 129–218. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, C.; Schwietzer, Y.A.; Otani, T.; Furuse, M.; Ebnet, K. Physiological functions of junctional adhesion molecules (JAMs) in tight junctions. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183299. [Google Scholar] [CrossRef] [PubMed]
- Santoni, M.-J.; Kashyap, R.; Camoin, L.; Borg, J.-P. The scribble family in cancer: Twentieth anniversary. Oncogene 2020, 39, 7019–7033. [Google Scholar] [CrossRef]
- Zimmermann, P.; Meerschaert, K.; Reekmans, G.; Leenaerts, I.; Small, J.V.; Vandekerckhove, J.; David, G.; Gettemans, J. PIP2-PDZ domain binding controls the association of syntenin with the plasma membrane. Mol. Cell 2002, 9, 1215–1225. [Google Scholar] [CrossRef]
- Chen, Y.; Sheng, R.; Källberg, M.; Silkov, A.; Tun, M.P.; Bhardwaj, N.; Kurilova, S.; Hall, R.A.; Honig, B.; Lu, H.; et al. Genome-wide functional annotation of dual-specificity protein- and lipid-binding modules that regulate protein interactions. Mol. Cell 2012, 46, 226–237. [Google Scholar] [CrossRef]
- Ponting, C.P.; Phillips, C.; Davies, K.E.; Blake, D.J. PDZ Domains: Targeting signalling molecules to sub-membranous sites. Bioessays 1997, 19, 469–479. [Google Scholar] [CrossRef]
- Songyang, Z.; Fanning, A.S.; Fu, C.; Xu, J.; Marfatia, S.M.; Chishti, A.H.; Crompton, A.; Chan, A.C.; Anderson, J.M.; Cantley, L.C. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 1997, 275, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Von Nandelstadh, P.; Ismail, M.; Gardin, C.; Suila, H.; Zara, I.; Belgrano, A.; Valle, G.; Carpen, O.; Faulkner, G. A Class III PDZ Binding Motif in the Myotilin and FATZ Families Binds Enigma Family Proteins: A Common Link for Z-Disc Myopathies. Mol. Cell Biol 2009, 29, 822–834. [Google Scholar] [CrossRef] [PubMed]
- Tonikian, R.; Zhang, Y.; Sazinsky, S.L.; Currell, B.; Yeh, J.-H.; Reva, B.; Held, H.A.; Appleton, B.A.; Evangelista, M.; Wu, Y.; et al. A Specificity Map for the PDZ Domain Family. PLoS Biol. 2008, 6, e239. [Google Scholar] [CrossRef] [PubMed]
- Gujral, T.S.; Karp, E.S.; Chan, M.; Chang, B.H.; MacBeath, G. Family-Wide investigation of PDZ domain-mediated protein-protein interactions implicates β-catenin in maintaining the integrity of tight junctions. Chem. Biol. 2013, 20, 816–827. [Google Scholar] [CrossRef] [PubMed]
- Dobrosotskaya, I.Y.; James, G.L. MAGI-1 interacts with β-catenin and is associated with cell-cell adhesion structures. Biochem. Biophys. Res. Commun. 2000, 270, 903–909. [Google Scholar] [CrossRef] [PubMed]
- Kotelevets, L.; Hengel, J.; Bruyneel, E.; Mareel, M.; Roy, F.; Chastre, E. Implication of the MAGI-1b/PTEN signalosome in stabilization of adherens junctions and suppression of invasiveness. FASEB J. 2005, 19, 115–117. [Google Scholar] [CrossRef]
- Patrie, K.M.; Drescher, A.J.; Goyal, M.; Wiggins, R.C.; Margolis, B. The membrane-associated guanylate kinase protein MAGI-1 binds megalin and is present in glomerular podocytes. J. Am. Soc. Nephrol. 2001, 12, 667–677. [Google Scholar] [CrossRef]
- Patrie, K.M.; Drescher, A.J.; Welihinda, A.; Mundel, P.; Margolis, B. Interaction of two actin-binding proteins, synaptopodin and α-actinin-4, with the tight junction protein MAGI-1. J. Biol. Chem. 2002, 277, 30183–30190. [Google Scholar] [CrossRef]
- Ohno, H.; Hirabayashi, S.; Kansaku, A.; Yao, I.; Tajima, M.; Nishimura, W.; Ohnishi, H.; Mashima, H.; Fujita, T.; Omata, M.; et al. Carom: A novel membrane-associated guanylate kinase-interacting protein with two SH3 domains. Oncogene 2003, 22, 8422–8431. [Google Scholar] [CrossRef][Green Version]
- Chastre, E.; Abdessamad, M.; Kruglov, A.; Bruyneel, E.; Bracke, M.; Gioia, Y.D.; Beckerle, M.C.; Roy, F.; Kotelevets, L. TRIP6, a novel molecular partner of the MAGI-1 scaffolding molecule, promotes invasiveness. FASEB J. 2009, 23, 916–928. [Google Scholar] [CrossRef]
- Kaufman, L.; Potla, U.; Coleman, S.; Dikiy, S.; Hata, Y.; Kurihara, H.; He, J.C.; D’Agati, V.D.; Klotman, P.E. Up-Regulation of the homophilic adhesion molecule sidekick-1 in podocytes contributes to glomerulosclerosis. J. Biol. Chem. 2010, 285, 25677–25685. [Google Scholar] [CrossRef]
- Ito, H.; Morishita, R.; Iwamoto, I.; Mizuno, M.; Nagata, K. MAGI-1 acts as a scaffolding molecule for NGF receptor-mediated signaling pathway. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 2302–2310. [Google Scholar] [CrossRef]
- Dobrosotskaya, I.; Guy, R.K.; James, G.L. MAGI-1, a membrane-associated guanylate kinase with a unique arrangement of protein-protein interaction domains. J. Biol. Chem. 1997, 272, 31589–31597. [Google Scholar] [CrossRef]
- Hirao, K.; Hata, Y.; Ide, N.; Takeuchi, M.; Irie, M.; Yao, I.; Deguchi, M.; Toyoda, A.; Sudhof, T.C.; Takai, Y. A novel multiple PDZ domain-containing molecule interacting with N-methyl-d-aspartate receptors and neuronal cell adhesion proteins. J. Biol. Chem. 1998, 273, 21105–21110. [Google Scholar] [CrossRef]
- Wu, Y.; Dowbenko, D.; Spencer, S.; Laura, R.; Lee, J.; Gu, Q.; Lasky, L.A. Interaction of the tumor suppressor PTEN/MMAC with a PDZ domain of MAGI3, a novel membrane-associated guanylate kinase. J. Biol. Chem. 2000, 275, 21477–21485. [Google Scholar] [CrossRef]
- Laura, R.P.; Ross, S.; Koeppen, H.; Lasky, L.A. MAGI-1: A widely expressed, alternatively spliced tight junction protein. Exp. Cell Res. 2002, 275, 155–170. [Google Scholar] [CrossRef]
- Franklin, J.L.; Yoshiura, K.; Dempsey, P.J.; Bogatcheval, G.; Jeyakumar, L.; Meise, K.S.; Pearsall, R.S.; Threadgill, D.; Coffey, R.J. Identification of MAGI-3 as a transforming growth factor-α tail binding protein. Exp. Cell Res. 2005, 303, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Yang, Z.; Xie, R.; Ji, Z.; Guan, K.; Zhang, M. Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity. Elife 2019, 8, e49439. [Google Scholar] [CrossRef] [PubMed]
- Kawajiri, A.; Itoh, N.; Fukata, M.; Nakagawa, M.; Yamaga, M.; Iwamatsu, A.; Kaibuchi, K. Identification of a novel β-catenin-interacting protein. Biochem. Biophys. Res. Commun. 2000, 273, 712–717. [Google Scholar] [CrossRef]
- Nishimura, W.; Yao, I.; Iida, J.; Tanaka, N.; Hata, Y. Interaction of synaptic scaffolding molecule and β-catenin. J. Neurosci. 2002, 22, 757–765. [Google Scholar] [CrossRef]
- Subauste, M.C.; Nalbant, P.; Adamson, E.D.; Hahn, K.M. Vinculin Controls PTEN protein level by maintaining the interaction of the adherens junction protein β-catenin with the scaffolding protein MAGI-2. J. Biol. Chem. 2005, 280, 5676–5681. [Google Scholar] [CrossRef]
- Rouaud, F.; Sluysmans, S.; Flinois, A.; Shah, J.; Vasileva, E.; Citi, S. Scaffolding proteins of vertebrate apical junctions: Structure, functions and biophysics. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183399. [Google Scholar] [CrossRef]
- Van Doorslaer, K.; DeSalle, R.; Einstein, M.H.; Burk, R.D. Degradation of human PDZ-proteins by human Alphapapillomaviruses represents an evolutionary adaptation to a novel cellular niche. PLoS Pathog. 2015, 11, e1004980. [Google Scholar] [CrossRef]
- Hirao, K.; Hata, Y.; Yao, I.; Deguchi, M.; Kawabe, H.; Mizoguchi, A.; Takai, Y. Three isoforms of synaptic scaffolding molecule and their characterization multimerization between the isoforms and their interaction with N-methyl-d-aspartate receptors and SAP90/PSD-95-associated protein. J. Biol. Chem. 2000, 275, 2966–2972. [Google Scholar] [CrossRef]
- Rehfeld, A.; Plass, M.; Døssing, K.; Knigge, U.; Kjær, A.; Krogh, A.; Friis-Hansen, L. Alternative polyadenylation of tumor suppressor genes in small intestinal neuroendocrine tumors. Front. Endocrinol. 2014, 5, 46. [Google Scholar] [CrossRef]
- Ni, T.K.; Kuperwasser, C. Premature polyadenylation of MAGI3 produces a dominantly-acting oncogene in human breast cancer. Elife 2016, 5, e14730. [Google Scholar] [CrossRef]
- Shiratsuchi, T.; Futamura, M.; Oda, K.; Nishimori, H.; Nakamura, Y.; Tokino, T. Cloning and characterization of BAI-associated protein 1: A PDZ domain-containing protein that interacts with BAI1. Biochem. Biophys. Res. Commun. 1998, 247, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.D.; Yuan, J.; Margolis, R.L.; Colomer, V.; Duan, K.; Kushi, J.; Kaminsky, Z.; Kleiderlein, J.J.; Sharp, A.H.; Ross, C.A. Atrophin-1, the DRPLA gene product, interacts with two families of WW domain-containing proteins. Mol. Cell. Neurosci. 1998, 11, 149–160. [Google Scholar] [CrossRef]
- Ishikawa, K.; Nagase, T.; Suyama, M.; Miyajima, N.; Tanaka, A.; Kotani, H.; Nomura, N.; Ohara, O. Prediction of the coding sequences of unidentified human genes. X. The complete sequences of 100 New cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1998, 5, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Nagase, T.; Kikuno, R.; Nakayama, M.; Hirosawa, M.; Ohara, O. Prediction of the coding sequences of unidentified human genes. XVIII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 2000, 7, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Balbas, M.D.; Burgess, M.R.; Murali, R.; Wongvipat, J.; Skaggs, B.J.; Mundel, P.; Weins, A.; Sawyers, C.L. MAGI-2 Scaffold protein is critical for kidney barrier function. Proc. Natl. Acad. Sci. USA 2014, 111, 14876–14881. [Google Scholar] [CrossRef]
- Alghamri, M.S.; Sharma, P.; Williamson, T.L.; Readler, J.M.; Yan, R.; Rider, S.D.; Hostetler, H.A.; Cool, D.R.; Kolawole, A.O.; Excoffon, K.J.D.A. MAGI-1 PDZ2 domain blockade averts adenovirus infection via enhanced proteolysis of the apical coxsackievirus and adenovirus receptor. J. Virol. 2021, 95, 13. [Google Scholar] [CrossRef]
- Stephenson, J.R.; Paavola, K.J.; Schaefer, S.A.; Kaur, B.; Meir, E.G.V.; Hall, R.A. Brain-specific angiogenesis inhibitor-1 signaling, regulation, and enrichment in the postsynaptic density. J. Biol. Chem. 2013, 288, 22248–22256. [Google Scholar] [CrossRef]
- Xu, J.; Paquet, M.; Lau, A.G.; Wood, J.D.; Ross, C.A.; Hall, R.A. Β1-Adrenergic receptor association with the synaptic scaffolding protein membrane-associated guanylate kinase inverted-2 (MAGI-2) differential regulation of receptor internalization by MAGI-2 and PSD-95. J. Biol. Chem. 2001, 276, 41310–41317. [Google Scholar] [CrossRef]
- He, J.; Bellini, M.; Inuzuka, H.; Xu, J.; Xiong, Y.; Yang, X.; Castleberry, A.M.; Hall, R.A. Proteomic analysis of Β1-adrenergic receptor interactions with PDZ scaffold proteins. J. Biol. Chem. 2006, 281, 2820–2827. [Google Scholar] [CrossRef]
- Yang, X.; Zheng, J.; Xiong, Y.; Shen, H.; Sun, L.; Huang, Y.; Sun, C.; Li, Y.; He, J. Beta-2 adrenergic receptor mediated ERK activation is regulated by interaction with MAGI-3. FEBS Lett. 2010, 584, 2207–2212. [Google Scholar] [CrossRef]
- Shoji, H.; Tsuchida, K.; Kishi, H.; Yamakawa, N.; Matsuzaki, T.; Liu, Z.; Nakamura, T.; Sugino, H. Identification and characterization of a PDZ protein that interacts with activin Type II receptors. J. Biol. Chem. 2000, 275, 5485–5492. [Google Scholar] [CrossRef]
- Deng, F.; Price, M.G.; Davis, C.F.; Mori, M.; Burgess, D.L. Stargazin and other transmembrane AMPA receptor regulating proteins interact with synaptic scaffolding protein MAGI-2 in brain. J. Neurosci. 2006, 26, 7875–7884. [Google Scholar] [CrossRef]
- Ma, Q.; Yang, Y.; Feng, D.; Zheng, S.; Meng, R.; Fa, P.; Zhao, C.; Liu, H.; Song, R.; Tao, T.; et al. MAGI3 negatively regulates Wnt/β-catenin Signaling and suppresses malignant phenotypes of glioma cells. Oncotarget 2015, 6, 35851–35865. [Google Scholar] [CrossRef]
- Lu, Y.; Sun, W.; Zhang, L.; Li, J. Silencing of MAGI1 promotes the proliferation and inhibits apoptosis of glioma cells via The Wnt/β-catenin and PTEN/AKT signaling pathways. Oncotargets Ther. 2019, 12, 9639–9650. [Google Scholar] [CrossRef]
- Ide, N.; Hata, Y.; Deguchi, M.; Hirao, K.; Yao, I.; Takai, Y. Interaction of S-SCAM with Neural Plakophilin-related Armadillo-repeat protein/δ-catenin. Biochem. Biophys. Res. Commun. 1999, 256, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Peng, A.W.; Oshima, K.; Heller, S. MAGI-1, A candidate stereociliary scaffolding protein, associates with the tip-link component cadherin 23. J. Neurosci. 2008, 28, 11269–11276. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Oshima, K.; Heller, S. PIST Regulates the intracellular trafficking and plasma membrane expression of Cadherin 23. BMC Cell Biol. 2010, 11, 80. [Google Scholar] [CrossRef]
- Bender, J.; Engeholm, M.; Ederer, M.S.; Breu, J.; Møller, T.C.; Michalakis, S.; Rasko, T.; Wanker, E.E.; Biel, M.; Martinez, K.L.; et al. Corticotropin-releasing hormone receptor Type 1 (CRHR1) clustering with MAGUKs is mediated via its C-terminal PDZ binding motif. PLoS ONE 2015, 10, e0136768. [Google Scholar] [CrossRef] [PubMed]
- Hammad, M.; Dunn, H.; Ferguson, S. MAGI proteins regulate the trafficking and signaling of corticotropin-releasing factor receptor 1 via a compensatory mechanism. J. Mol. Signal. 2016, 11, 5. [Google Scholar] [CrossRef]
- Gupta, S.; Abd-Elrahman, K.S.; Albaker, A.; Dunn, H.A.; Ferguson, S.S.G. Structural determinants governing β-arrestin2 interaction with PDZ proteins and recruitment to CRFR1. Cell Signal. 2019, 63, 109361. [Google Scholar] [CrossRef] [PubMed]
- Pfister, S.; Przemeck, G.K.H.; Gerber, J.-K.; Beckers, J.; Adamski, J.; de Angelis, M.H. Interaction of the MAGUK family member acvrinp1 and the cytoplasmic domain of the notch ligand delta1. J. Mol. Biol. 2003, 333, 229–235. [Google Scholar] [CrossRef]
- Wright, G.J.; Leslie, J.D.; Ariza-McNaughton, L.; Lewis, J. Delta proteins and MAGI proteins: An interaction of notch ligands with intracellular scaffolding molecules and its significance for zebrafish development. Development 2004, 131, 5659–5669. [Google Scholar] [CrossRef]
- Mizuhara, E.; Nakatani, T.; Minaki, Y.; Sakamoto, Y.; Ono, Y.; Takai, Y. MAGI1 recruits Dll1 to Cadherin-based adherens junctions and stabilizes it on the cell surface. J. Biol. Chem. 2005, 280, 26499–26507. [Google Scholar] [CrossRef]
- Buxbaum, J.D.; Georgieva, L.; Young, J.J.; Plescia, C.; Kajiwara, Y.; Jiang, Y.; Moskvina, V.; Norton, N.; Peirce, T.; Williams, H.; et al. Molecular dissection of NRG1-ERBB4 signaling implicates PTPRZ1 as a potential schizophrenia susceptibility gene. Mol. Psychiatry 2008, 13, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Luck, K.; Fournane, S.; Kieffer, B.; Masson, M.; Nominé, Y.; Travé, G. Putting into practice domain-linear motif interaction predictions for exploration of protein networks. PLoS ONE 2011, 6, e25376. [Google Scholar] [CrossRef]
- Yao, R.; Natsume, Y.; Noda, T. MAGI-3 is involved in the regulation of the JNK signaling pathway as a scaffold protein for frizzled and Ltap. Oncogene 2004, 23, 6023–6030. [Google Scholar] [CrossRef]
- Wawrzak, D.; Luyten, A.; Lambaerts, K.; Zimmermann, P. Frizzled-PDZ scaffold interactions in the control of Wnt signaling. Adv. Enzym. Regul. 2009, 49, 98–106. [Google Scholar] [CrossRef]
- Iida, J.; Ishizaki, H.; Okamoto-Tanaka, M.; Kawata, A.; Sumita, K.; Ohgake, S.; Sato, Y.; Yorifuji, H.; Nukina, N.; Ohashi, K.; et al. Synaptic scaffolding molecule α is a scaffold to mediate N-methyl-d-aspartate receptor-dependent RhoA activation in dendrites. Mol. Cell. Biol. 2007, 27, 4388–4405. [Google Scholar] [CrossRef]
- Danielson, E.; Zhang, N.; Metallo, J.; Kaleka, K.; Shin, S.M.; Gerges, N.; Lee, S.H. S-SCAM/MAGI-2 is an essential synaptic scaffolding molecule for the GluA2-containing maintenance pool of AMPA receptors. J. Neurosci. 2012, 32, 6967–6980. [Google Scholar] [CrossRef]
- Hammad, M.M.; Dunn, H.A.; Ferguson, S.S.G. MAGI Proteins can differentially regulate the signaling pathways of 5-HT2AR by enhancing receptor trafficking and PLC recruitment. Cell Signal. 2018, 47, 109–121. [Google Scholar] [CrossRef]
- Hirabayashi, S.; Tajima, M.; Yao, I.; Nishimura, W.; Mori, H.; Hata, Y. JAM4, a junctional cell adhesion molecule interacting with a tight junction protein, MAGI-1. Mol. Cell. Biol. 2003, 23, 4267–4282. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, D.; Sun, H.; Hall, R.A.; Yun, C.C. MAGI-3 Regulates LPA-Induced Activation of Erk and RhoA. Cell Signal. 2007, 19, 261–268. [Google Scholar] [CrossRef]
- Lee, S.; Ritter, S.L.; Zhang, H.; Shim, H.; Hall, R.A.; Yun, C.C. MAGI-3 competes with NHERF-2 to negatively regulate LPA2 receptor signaling in colon cancer cells. Gastroenterology 2011, 140, 924–934. [Google Scholar] [CrossRef] [PubMed]
- Meyer, G.; Varoqueaux, F.; Neeb, A.; Oschlies, M.; Brose, N. The complexity of PDZ domain-mediated interactions at glutamatergic synapses: A case study on neuroligin. Neuropharmacology 2004, 47, 724–733. [Google Scholar] [CrossRef]
- Iida, J.; Hirabayashi, S.; Sato, Y.; Hata, Y. Synaptic scaffolding molecule is involved in the synaptic clustering of neuroligin. Mol. Cell. Neurosci. 2004, 27, 497–508. [Google Scholar] [CrossRef]
- Lehtonen, S.; Ryan, J.J.; Kudlicka, K.; Iino, N.; Zhou, H.; Farquhar, M.G. Cell junction-associated proteins IQGAP1, MAGI-2, CASK, spectrins, and α-actinin are components of the nephrin multiprotein complex. Proc. Natl. Acad. Sci. USA 2005, 102, 9814–9819. [Google Scholar] [CrossRef]
- Hirabayashi, S.; Mori, H.; Kansaku, A.; Kurihara, H.; Sakai, T.; Shimizu, F.; Kawachi, H.; Hata, Y. MAGI-1 Is a component of the glomerular slit diaphragm that is tightly associated with nephrin. Lab. Investig. 2005, 85, 1528–1543. [Google Scholar] [CrossRef]
- Ni, J.; Bao, S.; Johnson, R.I.; Zhu, B.; Li, J.; Vadaparampil, J.; Smith, C.M.; Campbell, K.N.; Grahammer, F.; Huber, T.B.; et al. MAGI-1 Interacts with nephrin to maintain slit diaphragm structure through enhanced Rap1 activation in podocytes. J. Biol. Chem. 2016, 291, 24406–24417. [Google Scholar] [CrossRef]
- Weng, Z.; Shang, Y.; Ji, Z.; Ye, F.; Lin, L.; Zhang, R.; Zhu, J. Structural Basis of highly specific interaction between nephrin and MAGI1 in slit diaphragm assembly and signaling. J. Am. Soc. Nephrol. 2018, 29, 2362–2371. [Google Scholar] [CrossRef] [PubMed]
- Yamada, H.; Shirata, N.; Makino, S.; Miyake, T.; Trejo, J.A.O.; Yamamoto-Nonaka, K.; Kikyo, M.; Empitu, M.A.; Kadariswantiningsih, I.N.; Kimura, M.; et al. MAGI-2 orchestrates the localization of backbone proteins in the slit diaphragm of podocytes. Kidney Int. 2021, 99, 382–395. [Google Scholar] [CrossRef]
- Adamsky, K.; Arnold, K.; Sabanay, H.; Peles, E. Junctional protein MAGI-3 interacts with receptor tyrosine phosphataseβ (RPTPβ) and tyrosine-phosphorylated proteins. J. Cell Sci. 2003, 116, 1279–1289. [Google Scholar] [CrossRef]
- Fukada, M.; Fujikawa, A.; Chow, J.P.H.; Ikematsu, S.; Sakuma, S.; Noda, M. Protein tyrosine phosphatase receptor Type z is inactivated by ligand-induced oligomerization. FEBS Lett. 2006, 580, 4051–4056. [Google Scholar] [CrossRef]
- Yamagata, M.; Sanes, J.R. Synaptic Localization and function of sidekick recognition molecules require MAGI scaffolding proteins. J. Neurosci. 2010, 30, 3579–3588. [Google Scholar] [CrossRef][Green Version]
- Babayeva, S.; Zilber, Y.; Torban, E. Planar cell polarity pathway regulates actin rearrangement, cell shape, motility, and nephrin distribution in podocytes. Am. J. Physiol. Renal Physiol. 2011, 300, F549–F560. [Google Scholar] [CrossRef]
- Gee, H.Y.; Kim, Y.W.; Jo, M.J.; Namkung, W.; Kim, J.Y.; Park, H.W.; Kim, K.S.; Kim, H.; Baba, A.; Yang, J.; et al. Synaptic scaffolding molecule binds to and regulates vasoactive intestinal polypeptide Type-1 receptor in epithelial cells. Gastroenterology 2009, 137, 607–617.e4. [Google Scholar] [CrossRef]
- Kantar, D.; Mur, E.B.; Mancini, M.; Slaninova, V.; Salah, Y.B.; Costa, L.; Forest, E.; Lassus, P.; Géminard, C.; Boissière-Michot, F.; et al. MAGI1 inhibits the AMOTL2/P38 stress pathway and prevents luminal breast tumorigenesis. Sci. Rep. 2021, 11, 5752. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, S.; Nishimura, W.; Iida, J.; Kansaku, A.; Kishida, S.; Kikuchi, A.; Tanaka, N.; Hata, Y. Synaptic scaffolding molecule interacts with axin. J. Neurochem. 2004, 90, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Ni, T.K.; Elman, J.S.; Jin, D.X.; Gupta, P.B.; Kuperwasser, C. Premature polyadenylation of MAGI3 is associated with diminished N6-methyladenosine in its large internal exon. Sci. Rep. 2018, 8, 1415. [Google Scholar] [CrossRef] [PubMed]
- Yao, I.; Hata, Y.; Ide, N.; Hirao, K.; Deguchi, M.; Nishioka, H.; Mizoguchi, A.; Takai, Y. MAGUIN, a novel neuronal membrane-associated guanylate kinase-interacting protein. J. Biol. Chem. 1999, 274, 11889–11896. [Google Scholar] [CrossRef] [PubMed]
- Gou, H.; Liang, J.Q.; Zhang, L.; Chen, H.; Zhang, Y.; Li, R.; Wang, X.; Ji, J.; Tong, J.H.; To, K.-F.; et al. TTPAL promotes colorectal tumorigenesis by stabilizing TRIP6 to activate Wnt/β-catenin signaling. Cancer Res. 2019, 79, 3332–3346. [Google Scholar] [CrossRef] [PubMed]
- Couzens, A.L.; Knight, J.D.R.; Kean, M.J.; Teo, G.; Weiss, A.; Dunham, W.H.; Lin, Z.-Y.; Bagshaw, R.D.; Sicheri, F.; Pawson, T.; et al. Protein interaction network of the mammalian hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci. Signal. 2013, 6, rs15. [Google Scholar] [CrossRef]
- Zmajkovicova, K.; Jesenberger, V.; Catalanotti, F.; Baumgartner, C.; Reyes, G.; Baccarini, M. MEK1 Is Required for PTEN membrane recruitment, AKT regulation, and the maintenance of peripheral tolerance. Mol. Cell 2013, 50, 43–55. [Google Scholar] [CrossRef]
- Dobrosotskaya, I.Y. Identification of MNET1 as a candidate Ligand for the first PDZ domain of MAGI-1. Biochem. Biophys. Res. Commun. 2001, 283, 969–975. [Google Scholar] [CrossRef]
- Wu, X.; Hepner, K.; Castelino-Prabhu, S.; Do, D.; Kaye, M.B.; Yuan, X.-J.; Wood, J.; Ross, C.; Sawyers, C.L.; Whang, Y.E. Evidence for regulation of the PTEN Tumor suppressor by a membrane-localized multi-PDZ domain containing scaffold protein MAGI-2. Proc. Natl. Acad. Sci. USA 2000, 97, 4233–4238. [Google Scholar] [CrossRef] [PubMed]
- Tolkacheva, T.; Boddapati, M.; Sanfiz, A.; Tsuchida, K.; Kimmelman, A.C.; Chan, A.M.-L. Regulation of PTEN Binding to MAGI-2 by two putative phosphorylation sites at threonine 382 and 383. Cancer Res. 2001, 61, 4985–4989. [Google Scholar]
- Valiente, M.; Andrés-Pons, A.; Gomar, B.; Torres, J.; Gil, A.; Tapparel, C.; Antonarakis, S.E.; Pulido, R. Binding of PTEN to specific PDZ domains contributes to PTEN protein stability and phosphorylation by microtubule-associated serine/threonine kinase. J. Biol. Chem. 2005, 280, 28936–28943. [Google Scholar] [CrossRef]
- Ohtsuka, T.; Hata, Y.; Ide, N.; Yasuda, T.; Inoue, E.; Inoue, T.; Mizoguchi, A.; Takai, Y. NRap GEP: A novel neural GDP/GTP exchange Protein for Rap1 small G Protein that interacts with synaptic scaffolding molecule (S-SCAM). Biochem. Biophys. Res. Commun. 1999, 265, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Mino, A.; Ohtsuka, T.; Inoue, E.; Takai, Y. Membrane-associated guanylate kinase with inverted orientation (MAGI)-1/brain angiogenesis inhibitor 1-associated protein (BAP1) as a scaffolding molecule for rap small G protein GDP/GTP exchange protein at tight junctions. Genes Cells 2000, 5, 1009–1016. [Google Scholar] [CrossRef]
- Sakurai, A.; Fukuhara, S.; Yamagishi, A.; Sako, K.; Kamioka, Y.; Masuda, M.; Nakaoka, Y.; Mochizuki, N. MAGI-1 is required for Rap1 activation upon cell-cell contact and for enhancement of vascular endothelial cadherin-mediated cell adhesion. Mol. Biol. Cell 2006, 17, 966–976. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Cao, A.; Li, J.; Young, J.; Wong, J.; Ashraf, S.; Bierzynska, A.; Menon, M.C.; Hou, S.; Sawyers, C.; et al. Disruption of MAGI2-RapGEF2-Rap1 signaling contributes to podocyte dysfunction in congenital nephrotic syndrome caused by mutations in MAGI2. Kidney Int. 2019, 96, 642–655. [Google Scholar] [CrossRef]
- Gregorc, U.; Ivanova, S.; Thomas, M.; Guccione, E.; Glaunsinger, B.; Javier, R.; Turk, V.; Banks, L.; Turk, B. Cleavage of MAGI-1, a tight junction PDZ protein, by caspases is an important step for cell-cell detachment in apoptosis. Apoptosis 2007, 12, 343–354. [Google Scholar] [CrossRef][Green Version]
- Torres, J.; Rodriguez, J.; Myers, M.P.; Valiente, M.; Graves, J.D.; Tonks, N.K.; Pulido, R. Phosphorylation-regulated cleavage of the tumor suppressor PTEN by caspase-3 implications for the control of protein stability and PTEN-protein interactions. J. Biol. Chem. 2003, 278, 30652–30660. [Google Scholar] [CrossRef] [PubMed]
- Abe, J.; Ko, K.A.; Kotla, S.; Wang, Y.; Paez-Mayorga, J.; Shin, I.J.; Imanishi, M.; Vu, H.T.; Tao, Y.; Leiva-Juarez, M.M.; et al. MAGI1 as a link between endothelial activation and ER stress drives atherosclerosis. JCI Insight 2019, 4, e125570. [Google Scholar] [CrossRef]
- Abe, R.J.; Savage, H.; Imanishi, M.; Banerjee, P.; Kotla, S.; Paez-Mayorga, J.; Taunton, J.; Fujiwara, K.; Won, J.H.; Yusuf, S.W.; et al. P90RSK-MAGI1 module controls endothelial permeability by post-translational modifications of MAGI1 and hippo pathway. Front. Cardiovasc. Med. 2020, 7, 542485. [Google Scholar] [CrossRef]
- Liu, H.; Golebiewski, L.; Dow, E.C.; Krug, R.M.; Javier, R.T.; Rice, A.P. The ESEV PDZ-binding motif of the avian influenza A virus NS1 protein protects infected cells from apoptosis by directly targeting scribble. J. Virol. 2010, 84, 11164–11174. [Google Scholar] [CrossRef]
- Kumar, M.; Liu, H.; Rice, A.P. Regulation of interferon-β by MAGI-1 and its interaction with influenza A virus NS1 protein with ESEV PBM. PLoS ONE 2012, 7, e41251. [Google Scholar] [CrossRef]
- Fournane, S.; Charbonnier, S.; Chapelle, A.; Kieffer, B.; Orfanoudakis, G.; Travé, G.; Masson, M.; Nominé, Y. Surface plasmon resonance analysis of the binding of high-risk mucosal HPV E6 oncoproteins to the PDZ1 domain of the tight junction protein MAGI-1. J. Mol. Recognit. 2011, 24, 511–523. [Google Scholar] [CrossRef]
- Makokha, G.N.; Takahashi, M.; Higuchi, M.; Saito, S.; Tanaka, Y.; Fujii, M. Human T-cell leukemia virus Type 1 tax protein interacts with and mislocalizes the PDZ domain protein MAGI-1. Cancer Sci. 2013, 104, 313–320. [Google Scholar] [CrossRef]
- Glaunsinger, B.A.; Lee, S.S.; Thomas, M.; Banks, L.; Javier, R. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene 2000, 19, 5270–5280. [Google Scholar] [CrossRef]
- Latorre, I.J.; Roh, M.H.; Frese, K.K.; Weiss, R.S.; Margolis, B.; Javier, R.T. Viral oncoprotein-induced mislocalization of select PDZ proteins disrupts tight junctions and causes polarity defects in epithelial cells. J. Cell Sci. 2005, 118, 4283–4293. [Google Scholar] [CrossRef]
- Chung, S.-H.; Frese, K.K.; Weiss, R.S.; Prasad, B.V.V.; Javier, R.T. A New crucial protein interaction element that targets the adenovirus E4-ORF1 oncoprotein to membrane vesicles. J. Virol. 2007, 81, 4787–4797. [Google Scholar] [CrossRef]
- Chung, S.-H.; Weiss, R.S.; Frese, K.K.; Prasad, B.V.V.; Javier, R.T. Functionally distinct monomers and trimers produced by a viral oncoprotein. Oncogene 2008, 27, 1412–1420. [Google Scholar] [CrossRef]
- Thomas, M.; Glaunsinger, B.; Pim, D.; Javier, R.; Banks, L. HPV E6 and MAGUK protein interactions: Determination of the molecular basis for specific protein recognition and degradation. Oncogene 2001, 20, 5431–5439. [Google Scholar] [CrossRef]
- Thomas, M.; Laura, R.; Hepner, K.; Guccione, E.; Sawyers, C.; Lasky, L.; Banks, L. Oncogenic human papillomavirus E6 proteins target the MAGI-2 and MAGI-3 proteins for degradation. Oncogene 2002, 21, 5088–5096. [Google Scholar] [CrossRef]
- Kranjec, C.; Banks, L. A Systematic analysis of human papillomavirus (HPV) E6 PDZ substrates identifies MAGI-1 as a major target of HPV Type 16 (HPV-16) and HPV-18 whose loss accompanies disruption of tight junctions. J. Virol. 2011, 85, 1757–1764. [Google Scholar] [CrossRef]
- Kranjec, C.; Massimi, P.; Banks, L. Restoration of MAGI-1 Expression in Human Papillomavirus-Positive Tumor Cells Induces Cell Growth Arrest and Apoptosis. J. Virol. 2014, 88, 7155–7169. [Google Scholar] [CrossRef] [PubMed]
- Belyaeva, T.A.; Nicol, C.; Cesur, Ö.; Travé, G.; Blair, G.E.; Stonehouse, N.J. An RNA aptamer targets the PDZ-binding motif of the HPV16 E6 oncoprotein. Cancers 2014, 6, 1553–1569. [Google Scholar] [CrossRef] [PubMed]
- Caillet-Saguy, C.; Durbesson, F.; Rezelj, V.V.; Gogl, G.; Tran, Q.D.; Twizere, J.; Vignuzzi, M.; Vincentelli, R.; Wolff, N. Host PDZ-containing proteins targeted by SARS-CoV-2. FEBS J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Ravi, N.; Yang, M.; Mylona, N.; Wennerberg, J.; Paulsson, K. Global RNA expression and DNA methylation patterns in primary anaplastic thyroid cancer. Cancers 2020, 12, 680. [Google Scholar] [CrossRef] [PubMed]
- Kuang, S.-Q.; Tong, W.-G.; Yang, H.; Lin, W.; Lee, M.K.; Fang, Z.H.; Wei, Y.; Jelinek, J.; Issa, J.-P.; Garcia-Manero, G. Genome-wide identification of aberrantly methylated promoter associated CpG islands in acute lymphocytic leukemia. Leukemia 2008, 22, 1529–1538. [Google Scholar] [CrossRef] [PubMed]
- Kozakai, T.; Takahashi, M.; Higuchi, M.; Hara, T.; Saito, K.; Tanaka, Y.; Masuko, M.; Takizawa, J.; Sone, H.; Fujii, M. MAGI-1 expression is decreased in several types of human t-cell leukemia cell lines, including adult T-Cell leukemia. Int. J. Hematol. 2018, 107, 337–344. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, R.; Huang, Y.; Liao, Y.; Su, P.; Wang, H.; Chang, C.; Lin, Y.; Yu, M.; Chu, T.; et al. Methylomics analysis identifies epigenetically silenced genes and implies an activation of β-catenin signaling in cervical cancer. Int. J. Cancer 2014, 135, 117–127. [Google Scholar] [CrossRef]
- Qu, Y.; Gao, N.; Wu, T. Expression and clinical significance of SYNE1 and MAGI2 gene promoter methylation in gastric cancer. Medicine 2021, 100, e23788. [Google Scholar] [CrossRef]
- Chang, C.-C.; Wang, H.-C.; Liao, Y.-P.; Chen, Y.-C.; Weng, Y.-C.; Yu, M.-H.; Lai, H.-C. The feasibility of detecting endometrial and ovarian cancer using DNA methylation biomarkers in cervical scrapings. J. Gynecol. Oncol. 2018, 29, e17. [Google Scholar] [CrossRef]
- Valle, B.L.; Rodriguez-Torres, S.; Kuhn, E.; Díaz-Montes, T.; Parrilla-Castellar, E.; Lawson, F.P.; Folawiyo, O.; Ili-Gangas, C.; Brebi-Mieville, P.; Eshleman, J.R.; et al. HIST1H2BB and MAGI2 methylation and somatic mutations as precision medicine biomarkers for diagnosis and prognosis of high-grade serous ovarian cancer. Cancer Prev. Res. 2020, 13, 783–794. [Google Scholar] [CrossRef]
- Xu, X.; Yuan, X.; Ni, J.; Guo, J.; Gao, Y.; Yin, W.; Li, F.; Wei, L.; Zhang, J. MAGI2-AS3 Inhibits Breast Cancer by Downregulating DNA Methylation of MAGI2. J. Cell. Physiol. 2021, 236, 1116–1130. [Google Scholar] [CrossRef]
- Salmerón-Bárcenas, E.G.; Illades-Aguiar, B.; Moral-Hernández, O.D.; Ortega-Soto, A.; Hernández-Sotelo, D. HOTAIR knockdown decreased the activity Wnt/β-catenin signaling pathway and increased the mRNA levels of Its negative regulators in Hela cells. Cell Physiol. Biochem. 2019, 53, 948–960. [Google Scholar] [CrossRef]
- Pleasance, E.D.; Cheetham, R.K.; Stephens, P.J.; McBride, D.J.; Humphray, S.J.; Greenman, C.D.; Varela, I.; Lin, M.-L.; Ordóñez, G.R.; Bignell, G.R.; et al. A Comprehensive Catalogue of Somatic Mutations from a Human Cancer Genome. Nature 2010, 463, 191–196. [Google Scholar] [CrossRef]
- Berger, M.F.; Lawrence, M.S.; Demichelis, F.; Drier, Y.; Cibulskis, K.; Sivachenko, A.Y.; Sboner, A.; Esgueva, R.; Pflueger, D.; Sougnez, C.; et al. The genomic complexity of primary human prostate cancer. Nature 2011, 470, 214–220. [Google Scholar] [CrossRef]
- Banerji, S.; Cibulskis, K.; Rangel-Escareno, C.; Brown, K.K.; Carter, S.L.; Frederick, A.M.; Lawrence, M.S.; Sivachenko, A.Y.; Sougnez, C.; Zou, L.; et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 2012, 486, 405–409. [Google Scholar] [CrossRef]
- Mosquera, J.-M.; Varma, S.; Pauli, C.; MacDonald, T.Y.; Yashinskie, J.J.; Varga, Z.; Sboner, A.; Moch, H.; Rubin, M.A.; Shin, S.J. MAGI3-AKT3 fusion in breast cancer amended. Nature 2015, 520, E11–E12. [Google Scholar] [CrossRef][Green Version]
- Pugh, T.J.; Banerji, S.; Meyerson, M. Pugh et al. Reply. Nature 2015, 520, E12–E14. [Google Scholar] [CrossRef]
- Sakuta, K.; Sasaki, Y.; Abe, Y.; Sato, H.; Shoji, M.; Yaoita, T.; Yagi, M.; Mizumoto, N.; Onozato, Y.; Kon, T.; et al. Somatic alterations and mutational burden are potential predictive factors for metachronous development of early gastric cancer. Sci. Rep. 2020, 10, 22071. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, X.; Luo, J.; Liang, B.; Peng, J.; Chen, C.; Guo, H.; Wang, Q.; Xing, X.; Deng, Q.; et al. MiR-486-5p-directed MAGI1/Rap1/RASSF5 Signaling pathway contributes to hydroquinone-induced inhibition of erythroid differentiation in K562 cells. Toxicol. Vitro 2020, 66, 104830. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yang, Y.; Chen, X.; Shao, S.; Hu, S.; Zhang, T. MAGI1 mediates tumor metastasis through C-Myb/MiR-520h/MAGI1 signaling pathway in renal cell carcinoma. Apoptosis 2019, 24, 837–848. [Google Scholar] [CrossRef]
- Wu, C.; Zhao, Y.; Liu, Y.; Yang, X.; Yan, M.; Min, Y.; Pan, Z.; Qiu, S.; Xia, S.; Yu, J.; et al. Identifying miRNA-mRNA regulation network of major depressive disorder in ovarian cancer patients. Oncol. Lett. 2018, 16, 5375–5382. [Google Scholar] [CrossRef]
- Ma, M.; He, M.; Jiang, Q.; Yan, Y.; Guan, S.; Zhang, J.; Yu, Z.; Chen, Q.; Sun, M.; Yao, W.; et al. MiR-487a promotes TGF-Β1-induced EMT, the migration and invasion of breast cancer cells by directly targeting MAGI2. Int J. Biol. Sci. 2016, 12, 397–408. [Google Scholar] [CrossRef]
- Sachdeva, M.; Wu, H.; Ru, P.; Hwang, L.; Trieu, V.; Mo, Y.-Y. MicroRNA-101-Mediated Akt activation and estrogen-independent growth. Oncogene 2011, 30, 822–831. [Google Scholar] [CrossRef]
- Kitamura, K.; Seike, M.; Okano, T.; Matsuda, K.; Miyanaga, A.; Mizutani, H.; Noro, R.; Minegishi, Y.; Kubota, K.; Gemma, A. MiR-134/487b/655 Cluster Regulates TGF-β–induced epithelial-mesenchymal Transition and drug resistance to gefitinib by Targeting MAGI2 in lung adenocarcinoma cells. Mol. Cancer Ther. 2014, 13, 444–453. [Google Scholar] [CrossRef]
- Yao, X.; Tu, Y.; Xu, Y.; Guo, Y.; Yao, F.; Zhang, X. Endoplasmic reticulum stress-induced exosomal MiR-27a-3p promotes immune escape in breast cancer via regulating PD-L1 expression in macrophages. J. Cell. Mol. Med. 2020, 24, 9560–9573. [Google Scholar] [CrossRef]
- Weng, Q.; Chen, M.; Yang, W.; Li, J.; Fan, K.; Xu, M.; Weng, W.; Lv, X.; Fang, S.; Zheng, L.; et al. Integrated analyses identify MiR-34c-3p/MAGI3 axis for the warburg metabolism in hepatocellular carcinoma. FASEB J. 2020, 34, 5420–5434. [Google Scholar] [CrossRef]
- Yu, Q.; Li, X.; Feng, T. GLIDR promotes the progression of glioma by regulating the MiR-4677-3p/MAGI2 axis. Exp. Cell Res. 2021, 406, 112726. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Liu, T.; Wang, Z. Downregulation of MAGI1 associates with poor prognosis of hepatocellular carcinoma. J. Investig. Surg. 2012, 25, 93–99. [Google Scholar] [CrossRef]
- Jia, S.; Lu, J.; Qu, T.; Feng, Y.; Wang, X.; Liu, C.; Ji, J. MAGI1 inhibits migration and invasion via blocking MAPK/ERK signaling pathway in gastric cancer. Chin. J. Cancer Res. 2017, 29, 25–35. [Google Scholar] [CrossRef]
- Alday-Parejo, B.; Richard, F.; Wörthmüller, J.; Rau, T.; Galván, J.A.; Desmedt, C.; Santamaria-Martinez, A.; Rüegg, C. MAGI1, a new potential tumor suppressor gene in estrogen receptor positive breast cancer. Cancers 2020, 12, 223. [Google Scholar] [CrossRef]
- Zaric, J.; Joseph, J.-M.; Tercier, S.; Sengstag, T.; Ponsonnet, L.; Delorenzi, M.; Rüegg, C. Identification of MAGI1 as a tumor-suppressor protein induced by Cyclooxygenase-2 inhibitors in colorectal cancer cells. Oncogene 2012, 31, 48–59. [Google Scholar] [CrossRef]
- Cao, Z.; Ji, J.; Wang, F.-B.; Kong, C.; Xu, H.; Xu, Y.-L.; Chen, X.; Yu, Y.-W.; Sun, Y.-H. MAGI-2 Downregulation: A potential predictor of tumor progression and early recurrence in Han Chinese patients with prostate cancer. Asian J. Androl. 2020, 22, 616. [Google Scholar] [CrossRef]
- Mahdian, R.; Nodouzi, V.; Asgari, M.; Rezaie, M.; Alizadeh, J.; Yousefi, B.; Shahrokh, H.; Abolhasani, M.; Nowroozi, M. expression profile of MAGI2 gene as a novel biomarker in combination with major deregulated genes in prostate cancer. Mol. Biol. Rep. 2014, 41, 6125–6131. [Google Scholar] [CrossRef] [PubMed]
- David, S.N.; Egloff, S.A.A.; Goyal, R.; Clark, P.E.; Phillips, S.; Gellert, L.L.; Hameed, O.; Giannico, G.A. MAGI2 is an independent predictor of biochemical recurrence in prostate cancer. Prostate 2018, 78, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, J.; Borowsky, A.D.; Goyal, R.; Roland, J.T.; Arnold, S.A.; Gellert, L.L.; Clark, P.E.; Hameed, O.; Giannico, G.A. MAGI-2 in prostate cancer: An immunohistochemical study. Hum. Pathol. 2016, 52, 83–91. [Google Scholar] [CrossRef]
- Goldstein, J.; Goyal, R.; Roland, J.T.; Gellert, L.L.; Clark, P.E.; Hameed, O.; Giannico, G.A. MAGI-2 is a sensitive and specific marker of prostatic adenocarcinoma: A comparison with AMACR. Am. J. Clin. Pathol. 2016, 146, 294–302. [Google Scholar] [CrossRef]
- Giannico, G.A.; Arnold, S.A.; Gellert, L.L.; Hameed, O. New and Emerging Diagnostic and Prognostic Immunohistochemical Biomarkers in Prostate Pathology. Adv. Anat. Pathol. 2017, 24, 35–44. [Google Scholar] [CrossRef]
- Dawoud, M.M.; Aiad, A.-S.S.; Bahbah, A.M.N.H.; Shaban, M.I. Comparative Study of Immunohistochemical Expression of ERG and MAGI2 in Prostatic Carcinoma. Ann. Diagn. Pathol. 2021, 52, 151727. [Google Scholar] [CrossRef]
- Jia, Y.; Zhou, J.; Tan, T.K.; Chung, T.-H.; Wong, R.W.J.; Chooi, J.-Y.; Lim, J.S.L.; Sanda, T.; Ooi, M.; Mel, S.D.; et al. Myeloma-Specific Superenhancers Affect Genes of Biological and Clinical Relevance in Myeloma. Blood Cancer J. 2021, 11, 32. [Google Scholar] [CrossRef]
- Zhang, Y.; Ye, Q.; He, J.; Chen, P.; Wan, J.; Li, J.; Yang, Y.; Li, X. Recurrence-Associated Multi-RNA Signature to Predict Disease-Free Survival for Ovarian Cancer Patients. Biomed. Res. Int. 2020, 2020, 1–19. [Google Scholar] [CrossRef]
- Zhang, Y.; Dasgupta, J.; Ma, R.Z.; Banks, L.; Thomas, M.; Chen, X.S. Structures of a Human Papillomavirus (HPV) E6 Polypeptide Bound to MAGUK Proteins: Mechanisms of Targeting Tumor Suppressors by a High-Risk HPV Oncoprotein. J. Virol. 2007, 81, 3618–3626. [Google Scholar] [CrossRef]
- Yoshimatsu, Y.; Nakahara, T.; Tanaka, K.; Inagawa, Y.; Narisawa-Saito, M.; Yugawa, T.; Ohno, S.; Fujita, M.; Nakagama, H.; Kiyono, T. Roles of the PDZ-binding motif of HPV 16 E6 protein in oncogenic transformation of human cervical keratinocytes. Cancer Sci. 2017, 108, 1303–1309. [Google Scholar] [CrossRef]
- Boon, S.S.; Tomaić, V.; Thomas, M.; Roberts, S.; Banks, L. Cancer-causing human papillomavirus E6 proteins display major differences in the phospho-regulation of their PDZ interactions. J. Virol. 2015, 89, 1579–1586. [Google Scholar] [CrossRef]
- Yokota, M.; Kojima, M.; Higuchi, Y.; Nishizawa, Y.; Kobayashi, A.; Ito, M.; Saito, N.; Ochiai, A. Gene expression profile in the activation of subperitoneal fibroblasts reflects prognosis of patients with colon cancer. Int. J. Cancer 2016, 138, 1422–1431. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Gomez-Rueda, H.; Palacios-Corona, R.; Gutiérrez-Hermosillo, H.; Trevino, V. A robust biomarker of differential correlations improves the diagnosis of cytologically indeterminate thyroid cancers. Int. J. Mol. Med. 2016, 37, 1355–1362. [Google Scholar] [CrossRef]
- Feng, X.; Jia, S.; Martin, T.A.; Jiang, W.G. Regulation and Involvement in cancer and pathological Conditions of MAGI1, A tight JUNCTION protein. Anticancer Res. 2014, 34, 3251–3256. [Google Scholar] [PubMed]
- Nagashima, S.; Kodaka, M.; Iwasa, H.; Hata, Y. MAGI2/S-SCAM outside brain. J. Biochem. 2015, 157, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Wörthmüller, J.; Rüegg, C. MAGI1, a scaffold protein with tumor suppressive and vascular functions. Cells 2021, 10, 1494. [Google Scholar] [CrossRef]
- Vazquez, F.; Grossman, S.R.; Takahashi, Y.; Rokas, M.V.; Nakamura, N.; Sellers, W.R. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J. Biol. Chem. 2001, 276, 48627–48630. [Google Scholar] [CrossRef]
- Hu, Y.; Li, Z.; Guo, L.; Wang, L.; Zhang, L.; Cai, X.; Zhao, H.; Zha, X. MAGI-2 Inhibits cell migration and proliferation via PTEN in human hepatocarcinoma cells. Arch. Biochem. Biophys. 2007, 467, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Z.; Li, N.; Qi, J.; Fan, K.; Yin, P.; Zhao, C.; Liu, Y.; Yao, W.; Cai, X.; et al. MAGI2 enhances the sensitivity of BEL-7404 human hepatocellular carcinoma cells to staurosporine-induced apoptosis by increasing PTEN stability. Int. J. Mol. Med. 2013, 32, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Kotelevets, L.; van Hengel, J.; Bruyneel, E.; Mareel, M.; van Roy, F.; Chastre, E. The lipid phosphatase activity of PTEN Is critical for stabilizing intercellular junctions and reverting invasiveness. J. Cell Biol. 2001, 155, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Wang, Z. MAGI1 inhibits cancer cell migration and invasion of hepatocellular carcinoma via regulating PTEN. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2011, 36, 381–385. [Google Scholar] [CrossRef]
- Ma, Q.; Zhang, Y.; Meng, R.; Xie, K.M.; Xiong, Y.; Lin, S.; He, Z.L.K.; Tao, T.; Yang, Y.; Zhao, J.Z.; et al. MAGI3 suppresses glioma cell proliferation via upregulation of PTEN expression. Biomed. Environ. Sci. 2015, 28, 502–509. [Google Scholar] [CrossRef]
- Alimonti, A.; Carracedo, A.; Clohessy, J.G.; Trotman, L.C.; Nardella, C.; Egia, A.; Salmena, L.; Sampieri, K.; Haveman, W.J.; Brogi, E.; et al. Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet. 2010, 42, 454–458. [Google Scholar] [CrossRef]
- Zhang, G.; Gan, Y.-H. Synergistic antitumor effects of the combined treatment with an HDAC6 inhibitor and a cox-2 inhibitor through Activation of PTEN. Oncol. Rep. 2017, 38, 2657–2666. [Google Scholar] [CrossRef]
- Mohammad, M.A.; Greenman, J.; Maraveyas, A.; Ettelaie, C. Activation of PAR2 by tissue factor induces the release of the PTEN from MAGI proteins and regulates PTEN and Akt activities. Sci. Rep. 2020, 10, 20908. [Google Scholar] [CrossRef]
- Nusse, R.; Clevers, H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef]
- Fang, D.; Hawke, D.; Zheng, Y.; Xia, Y.; Meisenhelder, J.; Nika, H.; Mills, G.B.; Kobayashi, R.; Hunter, T.; Lu, Z. Phosphorylation of β-catenin by AKT promotes β-catenin transcriptional Activity. J. Biol. Chem. 2007, 282, 11221–11229. [Google Scholar] [CrossRef]
- Yuan, H.; Mao, J.; Li, L.; Wu, D. Suppression of glycogen synthase kinase activity is not sufficient for leukemia enhancer factor-1 activation. J. Biol. Chem. 1999, 274, 30419–30423. [Google Scholar] [CrossRef][Green Version]
- Hermida, M.A.; Kumar, J.D.; Leslie, N.R. GSK3 and its interactions with the PI3K/AKT/MTOR signalling network. Adv. Biol. Regul. 2017, 65, 5–15. [Google Scholar] [CrossRef]
- Veeman, M.T.; Axelrod, J.D.; Moon, R.T. A second canon functions and mechanisms of β-catenin-independent Wnt signaling. Dev. Cell 2003, 5, 367–377. [Google Scholar] [CrossRef]
- Heng, B.C.; Zhang, X.; Aubel, D.; Bai, Y.; Li, X.; Wei, Y.; Fussenegger, M.; Deng, X. An overview of signaling pathways regulating YAP/TAZ activity. Cell. Mol. Life Sci. 2021, 78, 497–512. [Google Scholar] [CrossRef]
- Thompson, B.J. YAP/TAZ: Drivers of tumor growth, metastasis, and resistance to therapy. Bioessays 2020, 42, 1900162. [Google Scholar] [CrossRef]
- Emami, S.S.; Zhang, D.; Yang, X. Interaction of the Hippo pathway and phosphatases in tumorigenesis. Cancers 2020, 12, 2438. [Google Scholar] [CrossRef]
- Mohseni, M.; Sun, J.; Lau, A.; Curtis, S.; Goldsmith, J.; Fox, V.L.; Wei, C.; Frazier, M.; Samson, O.; Wong, K.-K.; et al. A genetic screen identifies an LKB1–MARK signalling Axis controlling the Hippo-Yap pathway. Nat. Cell Biol. 2014, 16, 108–117. [Google Scholar] [CrossRef]
- Xu, W.; Yang, Z.; Xie, C.; Zhu, Y.; Shu, X.; Zhang, Z.; Li, N.; Chai, N.; Zhang, S.; Wu, K.; et al. PTEN lipid phosphatase inactivation links the Hippo and PI3K/Akt pathways to induce gastric tumorigenesis. J. Exp. Clin. Cancer Res. 2018, 37, 198. [Google Scholar] [CrossRef]
- Huang, W.; Lv, X.; Liu, C.; Zha, Z.; Zhang, H.; Jiang, Y.; Xiong, Y.; Lei, Q.-Y.; Guan, K.-L. The N-terminal phosphodegron targets TAZ/WWTR1 Protein for SCFβ-TrCP-dependent degradation in response to phosphatidylinositol 3-kinase inhibition. J. Biol. Chem. 2012, 287, 26245–26253. [Google Scholar] [CrossRef]
- Wapenaar, M.C.; Monsuur, A.J.; van Bodegraven, A.A.; Weersma, R.K.; Bevova, M.R.; Linskens, R.K.; Howdle, P.; Holmes, G.; Mulder, C.J.; Dijkstra, G.; et al. Associations with tight junction genes PARD3 and MAGI2 in Dutch patients point to a common barrier defect for coeliac disease and ulcerative colitis: An unusual case of ascites. Gut 2008, 57, 463. [Google Scholar] [CrossRef]
- McGovern, D.P.B.; Taylor, K.D.; Landers, C.; Derkowski, C.; Dutridge, D.; Dubinsky, M.; Ippoliti, A.; Vasiliauskas, E.; Mei, L.; Mengesha, E.; et al. MAGI2 genetic variation and inflammatory bowel disease. Inflamm. Bowel Dis. 2009, 15, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Domènech, E.; Julià, A.; Panés, J.; García-Sánchez, V.; Mateu, P.N.; Gutiérrez, A.; Gomollón, F.; Mendoza, J.L.; Garcia-Planella, E.; et al. Identification of risk loci for Crohn’s Disease phenotypes using a genome-wide association study. Gastroenterology 2015, 148, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Norén, E.; Almer, S.; Söderman, J. Genetic variation and expression levels of tight junction genes identifies association between MAGI3 and inflammatory bowel disease. BMC Gastroenterol. 2017, 17, 68. [Google Scholar] [CrossRef] [PubMed]
- Norén, E.; Mellander, M.-R.; Almer, S.; Söderman, J. Genetic variation and gene expression levels of tight junction genes indicates relationships between PTEN as well as MAGI1 and microscopic colitis. Digest. Dis. Sci. 2018, 63, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Kotelevets, L.; Chastre, E.; Desmaële, D.; Couvreur, P. Nanotechnologies for the treatment of colon cancer: From old drugs to new hope. Int. J. Pharmaceut. 2016, 514, 24–40. [Google Scholar] [CrossRef] [PubMed]
- Terzić, J.; Grivennikov, S.; Karin, E.; Karin, M. Inflammation and colon cancer. Gastroenterology 2010, 138, 2101–2114.e5. [Google Scholar] [CrossRef]
- Fantini, M.C.; Guadagni, I. From inflammation to colitis-associated colorectal cancer in inflammatory bowel disease: Pathogenesis and Impact of current therapies. Digest. Liver Dis. 2021, 53, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-Y.; Li, X.-H.; Tian, G.-W.; Zhang, D.-Y.; Gao, H.; Wang, Z.-Y. MAGI1 inhibits the proliferation, migration and invasion of glioma cells. Oncotargets Ther. 2019, 12, 11281–11290. [Google Scholar] [CrossRef] [PubMed]
- Alday-Parejo, B.; Ghimire, K.; Coquoz, O.; Albisetti, G.W.; Tamò, L.; Zaric, J.; Stalin, J.; Rüegg, C. MAGI1 localizes to mature focal adhesion and modulates endothelial cell adhesion, migration and angiogenesis. Cell Adhes. Migr. 2021, 15, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Xu, X.; Tang, Y.; Zhu, X. TRIP6 functions as a potential oncogene and facilitated proliferation and metastasis of gastric cancer. Biol. Targets Ther. 2019, 13, 101–110. [Google Scholar] [CrossRef]
- Zhao, W.; Dai, Y.; Dai, T.; Xie, T.; Su, X.; Li, J.; Zhou, X.; Meng, K.; Zhao, X. TRIP6 promotes cell proliferation in hepatocellular carcinoma via suppression of FOXO3a. Biochem. Biophys. Res. Commun. 2017, 494, 594–601. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, B.; Xu, X.; Zhu, L.; Zhu, X. TRIP6 promotes tumorigenic capability through regulating FOXC1 in hepatocellular carcinoma. Pathol. Res. Pract. 2020, 216, 152850. [Google Scholar] [CrossRef]
- Zhao, X.; Jiang, C.; Xu, R.; Liu, Q.; Liu, G.; Zhang, Y. TRIP6 Enhances stemness property of breast cancer cells through activation of Wnt/β-catenin. Cancer Cell Int. 2020, 20, 51. [Google Scholar] [CrossRef]
- Dutta, S.; Mana-Capelli, S.; Paramasivam, M.; Dasgupta, I.; Cirka, H.; Billiar, K.; McCollum, D. TRIP6 inhibits hippo signaling in response to tension at adherens junctions. EMBO Rep. 2018, 19, 337–350. [Google Scholar] [CrossRef]
- Yang, F.; Li, L.; Zhang, J.; Zhang, J.; Yang, L. TRIP6 accelerates the proliferation and invasion of cervical cancer by upregulating oncogenic YAP signaling. Exp. Cell Res. 2020, 396, 112248. [Google Scholar] [CrossRef]
- Song, C.; Zhang, J.; Liu, Y.; Pan, H.; Qi, H.; Cao, Y.; Zhao, J.; Li, S.; Guo, J.; Sun, H.; et al. Construction and analysis of cardiac hypertrophy-associated LncRNA-mRNA Network based on competitive endogenous RNA reveal functional LncRNAs in cardiac hypertrophy. Oncotarget 2016, 7, 10827–10840. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, F.; Wang, F.; Wu, N. Long noncoding RNA MAGI1-IT1 regulates cardiac hypertrophy by modulating MiR-302e/DKK1/Wnt/Beta-catenin signaling pathway. J. Cell. Physiol. 2020, 235, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chen, H.-X.; Yang, S.-N.; Zhao, J. MAGI1-IT1 Stimulates proliferation in non-small cell lung cancer by upregulating AKT1 as a CeRNA. Eur. Rev. Med. Pharmacol. 2020, 24, 691–698. [Google Scholar] [CrossRef]
- Gao, H.; Li, X.; Zhan, G.; Zhu, Y.; Yu, J.; Wang, J.; Li, L.; Wu, W.; Liu, N.; Guo, X. Long noncoding RNA MAGI1-IT1 promoted invasion and metastasis of epithelial ovarian cancer via the MiR-200a/ZEB axis. Cell Cycle 2019, 18, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gu, M.; Zhuang, Y.; Chen, J. The long noncoding RNA MAGI1-IT1 regulates the MiR-302d-3p/IGF1 axis to control gastric cancer cell proliferation. Cancer Manag. Res. 2021, 13, 2959–2967. [Google Scholar] [CrossRef]
- Wang, F.; Zu, Y.; Zhu, S.; Yang, Y.; Huang, W.; Xie, H.; Li, G. Long noncoding RNA MAGI2-AS3 regulates CCDC19 expression by sponging MiR-15b-5p and suppresses bladder cancer progression. Biochem. Biophys. Res. Commun. 2018, 507, 231–235. [Google Scholar] [CrossRef]
- Gokulnath, P.; de Cristofaro, T.; Manipur, I.; Palma, T.D.; Soriano, A.A.; Guarracino, M.R.; Zannini, M. Long non-coding RNA MAGI2-AS3 Is a new player with a tumor suppressive role in high grade serous ovarian carcinoma. Cancers 2019, 11, 2008. [Google Scholar] [CrossRef]
- Hao, X.-Z.; Yang, K. LncRNA MAGI2-AS3 Suppresses the proliferation and invasion of non-small cell lung carcinoma through MiRNA-23a-3p/PTEN axis. Eur. Rev. Med. Pharmacol. 2019, 23, 7399–7407. [Google Scholar] [CrossRef]
- He, J.; Zhou, X.; Li, L.; Han, Z. Long noncoding MAGI2-AS3 suppresses several cellular processes of lung squamous cell carcinoma cells by regulating MiR-374a/b-5p/CADM2 axis. Cancer Manag. Res. 2020, 12, 289–302. [Google Scholar] [CrossRef]
- Sui, Y.; Chi, W.; Feng, L.; Jiang, J. LncRNA MAGI2-AS3 Is Downregulated in non-small cell lung cancer and may be a sponge of MiR-25. BMC Pulm. Med. 2020, 20, 59. [Google Scholar] [CrossRef]
- Tang, C.; Cai, Y.; Jiang, H.; Lv, Z.; Yang, C.; Xu, H.; Li, Z.; Li, Y. LncRNA MAGI2-AS3 inhibits bladder cancer progression by targeting the MiR-31-5p/TNS1 axis. Aging 2020, 12, 25547–25563. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Hu, Q.; Pang, Z.; Xu, X. LncRNA MAGI2-AS3 upregulates cytokine signaling 1 by sponging MiR-155 in non-small cell lung cancer. Cancer Biother. Radiopharm. 2020, 35, 72–76. [Google Scholar] [CrossRef]
- Hou, A.; Zhang, Y.; Fan, Y.; Zheng, Y.; Zhou, X.; Liu, H. LncRNA MAGI2-AS3 affects cell invasion and migration of cervical squamous cell carcinoma (CSCC) via sponging MiRNA-233/EPB41L3 axis. Cancer Manag. Res. 2020, 12, 4209–4216. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Hu, W.; Zhao, Y.; Zhou, H.; Wen, W.; Xu, M.; Zhao, P.; Liu, K. Long non-coding RNA MAGI2-AS3 Inhibits breast cancer cell migration and invasion via sponging MicroRNA-374a. Cancer Biomark. 2019, 24, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Ma, T.; Yan, J.; Shi, N.; Zhang, C.; Lu, X.; Hou, B.; Jian, Z. LncRNA MAGI2-AS3 inhibits hepatocellular carcinoma cell proliferation and migration by targeting the MiR-374b-5p/SMG1 signaling pathway. J. Cell. Physiol. 2019, 234, 18825–18836. [Google Scholar] [CrossRef]
- Chang, H.; Zhang, X.; Li, B.; Meng, X. MAGI2-AS3 suppresses MYC signaling to inhibit cell proliferation and migration in ovarian cancer through targeting MiR-525-5p/MXD1 axis. Cancer Med. 2020, 9, 6377–6386. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Shi, X.; Lin, M.; Yao, Q.; Ma, J.; Li, J. LncRNA MAGI2-AS3 overexpression Sensitizes esophageal cancer cells to irradiation through down-regulation of HOXB7 via EZH2. Front. Cell Dev. Biol. 2020, 8, 552822. [Google Scholar] [CrossRef]
- Wang, G.; Li, H.; Hou, Y. LncRNA MAGI2-AS3 inhibits tumor progression and angiogenesis by regulating acy1 via interacting with transcription factor HEY1 in clear cell renal cell Carcinoma. Cancer Gene Ther. 2021. [Google Scholar] [CrossRef]
- Pu, J.; Wang, J.; Wei, H.; Lu, T.; Wu, X.; Wu, Y.; Shao, Z.; Luo, C.; Lu, Y. LncRNA MAGI2-AS3 prevents the development of HCC via Recruiting KDM1A and PROMOTING H3K4me2 demethylation of the RACGAP1 promoter. Mol. Ther. Nucleic Acids 2019, 18, 351–362. [Google Scholar] [CrossRef]
- Chen, L.; Fan, X.; Zhu, J.; Chen, X.; Liu, Y.; Zhou, H. LncRNA MAGI2-AS3 inhibits the self-renewal of leukaemic stem cells by promoting TET2-dependent DNA demethylation of the LRIG1 promoter in acute myeloid leukaemia. RNA Biol. 2020, 17, 784–793. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, H.; Xu, M.; Zhang, H.; Sun, M.; Mu, P.; Dong, T.; Du, S.; Liu, K. Long non-coding RNA (LncRNA) MAGI2-AS3 inhibits breast cancer cell growth by targeting the Fas/FasL signalling pathway. Hum. Cell 2018, 31, 232–241. [Google Scholar] [CrossRef]
- Fang, G.; Wang, J.; Sun, X.; Xu, R.; Zhao, X.; Shao, L.; Sun, C.; Wang, Y. LncRNA MAGI2-AS3 Is downregulated in the distant recurrence of hepatocellular carcinoma after surgical resection and affects migration and invasion via ROCK2. Ann. Hepatol. 2020, 19, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, J.; Zhang, M.; Hu, X.; She, J.; Qiu, X.; Zhang, X.; Xu, L.; Liu, Y.; Qin, S. LncRNA MAGI2-AS3 is regulated by BRD4 and promotes gastric cancer progression via maintaining ZEB1 overexpression by sponging MiR-141/200a. Mol. Ther. Nucleic Acids 2020, 19, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Hou, J.; Wu, Y.; Liu, J.; Li, G.; Zhao, W.; Ma, G.; Chen, B.; Song, Y. Integrated analysis of a competing endogenous rna network reveals key LncRNAs as potential prognostic biomarkers for human bladder cancer. Medicine 2018, 97, e11887. [Google Scholar] [CrossRef]
- Adam, L.; Zhong, M.; Choi, W.; Qi, W.; Nicoloso, M.; Arora, A.; Calin, G.; Wang, H.; Siefker-Radtke, A.; McConkey, D.; et al. MiR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin. Cancer Res. 2009, 15, 5060–5072. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Zhou, S.; Hu, J. Long noncoding RNA MAGI2-AS3/MiR-218-5p/GDPD5/SEC61A1 axis drives cellular proliferation and migration and confers cisplatin resistance in nasopharyngeal carcinoma. Int. Forum Allergy Rhinol. 2020, 10, 1012–1023. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Li, Z.; Tang, Z.; Li, J.; Lang, X. Long noncoding MAGI2-AS3 promotes colorectal cancer progression through regulating MiR-3163/TMEM106B axis. J. Cell. Physiol. 2020, 235, 4824–4833. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.-L.; Xu, Z.-G.; Chen, H.; Ji, J.; Wang, Y.-H.; Hu, W.; Wang, K.; Zhang, W.-W.; Yuan, C.-H.; Wang, F.-B. LncRNAs and EGFRvIII Sequestered in TEPs enable blood-based NSCLC diagnosis. Cancer Manag. Res. 2018, 10, 1449–1459. [Google Scholar] [CrossRef]
- Zhang, X.; Zhuang, J.; Liu, L.; He, Z.; Liu, C.; Ma, X.; Li, J.; Ding, X.; Sun, C. Integrative transcriptome data mining for identification of core LncRNAs in breast cancer. PeerJ 2019, 7, e7821. [Google Scholar] [CrossRef]
- Tian, T.; Gong, Z.; Wang, M.; Hao, R.; Lin, S.; Liu, K.; Guan, F.; Xu, P.; Deng, Y.; Song, D.; et al. Identification of Long non-coding RNA Signatures in triple-negative breast cancer. Cancer Cell Int. 2018, 18, 103. [Google Scholar] [CrossRef]
- Rastogi, M.; Singh, S.K. Modulation of Type-I interferon response by Hsa-MiR-374b-5p during Japanese encephalitis virus infection in human microglial cells. Front. Cell. Infect. Microbiol. 2019, 9, 291. [Google Scholar] [CrossRef]
- Chen, X.-D.; Zhu, M.-X.; Wang, S.-J. Expression of long non-coding RNA MAGI2-AS3 in human gliomas and its prognostic significance. Eur. Rev. Med. Pharmacol. 2019, 23, 3455–3460. [Google Scholar] [CrossRef]
- Shen, D.; Xu, J.; Cao, X.; Cao, X.; Tan, H.; Deng, H. Long noncoding RNA MAGI2-AS3 inhibits bladder cancer progression through MAGI2/PTEN/epithelial-mesenchymal transition (EMT) axis. Cancer Biomark. 2021, 30, 155–165. [Google Scholar] [CrossRef]
- Yang, X.; Wu, S.; Li, X.; Yin, Y.; Chen, R. MAGI2-AS3 Rs7783388 Polymorphism contributes to colorectal cancer risk through altering the binding affinity of the transcription factor GR to the MAGI2-AS3 promoter. J. Clin. Lab. Anal. 2020, 34, e23431. [Google Scholar] [CrossRef]
- Poursheikhani, A.; Abbaszadegan, M.R.; Nokhandani, N.; Kerachian, M.A. Integration Analysis of Long Non-Coding RNA (LncRNA) Role in Tumorigenesis of Colon Adenocarcinoma. BMC Med. Genom. 2020, 13, 108. [Google Scholar] [CrossRef]
- Cai, J.; Chen, Z.; Chen, X.; Huang, H.; Lin, X.; Miao, B. Coexpression network analysis identifies a novel nine-RNA signature to improve prognostic prediction for prostate cancer patients. Biomed. Res. Int. 2020, 2020, 1–17. [Google Scholar] [CrossRef]
- Xue, C.; Li, G.; Lu, J.; Luo, J.; Jia, J. Novel insights for LncRNA MAGI2-AS3 in solid tumors. Biomed. Pharmacother. 2021, 137, 111429. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, Y.; Xie, Y.; Leng, X.; Song, F. Comprehensive analysis of LncRNAs associated with the pathogenesis and prognosis of gastric cancer. DNA Cell Biol. 2020, 39, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, Y.; Du, P.; Zhang, X.; Li, X.; Cheng, G. Hypomethylation of the LncRNA SOX21-AS1 Has clinical prognostic value in cervical cancer. Life Sci. 2019, 233, 116708. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, S.; Wang, X.; Zhang, J.; Liu, K. LncRNA MAGI2-AS3 Is Involved in Cervical Squamous Cell Carcinoma Development through CDK6 up-Regulation. Infect. Agents Cancer 2019, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Erlendsson, S.; Thorsen, T.S.; Vauquelin, G.; Ammendrup-Johnsen, I.; Wirth, V.; Martinez, K.L.; Teilum, K.; Gether, U.; Madsen, K.L. Mechanisms of PDZ domain scaffold assembly illuminated by use of supported cell membrane sheets. Elife 2019, 8, e39180. [Google Scholar] [CrossRef]
- McCann, J.J.; Choi, U.B.; Bowen, M.E. Reconstitution of multivalent PDZ domain binding to the scaffold protein PSD-95 reveals ternary-complex specificity of combinatorial inhibition. Structure 2014, 22, 1458–1466. [Google Scholar] [CrossRef][Green Version]
- Gannon, F. Understanding life—Now for the hard part! EMBO Rep. 2002, 3, 287. [Google Scholar] [CrossRef]
- Nicholson, D.J. Is the cell really a machine? J. Theor. Biol. 2019, 477, 108–126. [Google Scholar] [CrossRef]
- Wu, L.; Wu, D.; Ning, J.; Liu, W.; Zhang, D. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer 2019, 19, 326. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zou, X.; Chen, Y.; Cho, W.C.; Zhou, X. Effect of N6-methyladenosine regulators on progression and prognosis of triple-negative breast cancer. Front. Genet. 2021, 11, 580036. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Samanta, D.; Lu, H.; Bullen, J.W.; Zhang, H.; Chen, I.; He, X.; Semenza, G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated M6A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. USA 2016, 113, E2047–E2056. [Google Scholar] [CrossRef]
- Lan, Q.; Liu, P.Y.; Haase, J.; Bell, J.L.; Hüttelmaier, S.; Liu, T. The critical role of RNA M6A methylation in cancer. Cancer Res. 2019, 79, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Selberg, S.; Seli, N.; Kankuri, E.; Karelson, M. Rational design of novel anticancer small-molecule RNA m6A demethylase ALKBH5 inhibitors. ACS Omega 2021, 6, 13310–13320. [Google Scholar] [CrossRef]
- Zheng, F.; Jewell, H.; Fitzpatrick, J.; Zhang, J.; Mierke, D.F.; Grigoryan, G. Computational design of selective peptides to discriminate between similar PDZ domains in an oncogenic pathway. J. Mol. Biol. 2015, 427, 491–510. [Google Scholar] [CrossRef] [PubMed]

| Molecular Partners | Full Name | Function | MAGI1 | Interaction MAGI2 | MAGI3 | MAGI Binding Domain * | Biological Impact | References |
|---|---|---|---|---|---|---|---|---|
| Receptor Ligands, Membrane Receptors, Cell Adhesion Molecules | ||||||||
| ADAM17; TACE | Disintegrin and metalloproteinase domain-containing protein 17; ADAM17, | Membrane-anchored metalloproteinase | + | ND | ND | PDZ2 | Involved (with ɣ-secretase) in coxsackie virus and adenovirus receptor (CAR Ex8) | [48] |
| ADGRB1; BAI1 | Adhesion G protein-coupled receptor B1; Brain-specific angiogenesis inhibitor-1 | G-protein-coupled receptor involved in inflammation, tumorigenesis, and phagocytosis. Mobilizes Rho signaling pathway through Gα12/13 and Rac1 pathways via ELMO/DOCK GEF. | + | + | + | PDZ4 | BAI1 PDZ-binding motif required for Rho signaling, association with β-Arrestin2, and receptor ubiquitination. MAGI3 potentiates ERK signaling by a constitutively active BAI1. MAGI1 was originally designed BAI1-associated protein 1 | [43,49] |
| ADRB1 | Beta-1 adrenergic receptor (β1AR); Adrenoceptor beta 1; | G protein-coupled receptor (GPCR), a receptor for epinephrine and norepinephrine | + | + | + | PDZ1 (MAGI1, 3); PDZ1 and PDZ2 (MAGI2) | Interaction MAGI2/ β1AR enhanced by agonist stimulation. MAGI2 increases agonist-induced β1AR internalization. Ternary complex involves MAGI2/ β1AR/ β-catenin. MAGI3 impairs β1AR-mediated ERK1/2 activation but has no effect on cAMP generation or β1AR internalization | [50,51] |
| ADRB2 | Beta-2 adrenergic receptor (β2AR) | G protein-coupled receptor (GPCR) | ND | ND | + | PDZ5 | Agonist stimulation of β2AR promotes MAGI3/ β2AR interaction (dependent on β2AR phosphorylation by GRK). MAGI3 inhibits β2AR-mediated ERK activation. | [52] |
| ActRIIA | Activin A receptor type 2A | Ser/Thr kinase, a component of the heterodimeric receptor for activin, a member of the TGFβ family | ND | + | ND | PDZ5 | MAGI2 interacts with activin type 2 receptor via its PDZ5 and with SMAD3 via its WW domains. MAGI2 negatively regulates activin/ SMAD3 -induced gene transcription | [53] |
| CACNG2; Stargazin | Voltage-dependent calcium channel gamma-2 subunit; Stargazin | Type I transmembrane AMPA receptor regulatory protein. Promotes targeting of AMPA-selective glutamate receptors; Modulates their gating properties. | ND | + | + | PDZ1, PDZ3, and PDZ5 | Stargazin recruits MAGI2 to cell membranes and cell–cell contact. MAGI2 might act as a physical link between AMPAR/ AMPAR regulators complexes and synaptic adherens junctions | [54] |
| CTNNB1 | Catenin beta 1; Beta-catenin | Component of the protein complex that constitutes adherens junctions, involved in cadherin-mediated cell–cell contact and tissue integrity. Effector system of the canonical Wnt signaling pathway: coactivator for the TCF/LEF family of transcription factors and Wnt target genes (Cyclin D1, c-Myc, MMP7). | + | + | + | PDZ5 | MAGIs are recruited to cadherin junctional complexes by PDZ5 binding to β-catenin. The interaction of PTEN with MAGI PDZ2 allows the tuning of PtdIns(3,4,5)P3 and downstream effectors with PH domain (e.g., AKT, RhoGEF) stabilizing junctional complexes and inhibiting invasiveness. MAGIs also restrain β-catenin nuclear translocation and Wnt target genes transactivation | [20,21,22,35,36,37,55,56] |
| CTNND2; NPRAP | Catenin delta 2; Delta-catenin | Member of the armadillo protein family, involved in cell–cell contacts and signal transduction | ND | + | ND | PDZ5 | Junctional organization | [57] |
| CDH23 | Cadherin-23 | Member of the cadherin superfamily; Involved in the organization of the stereocilia bundle of hair cells in the cochlea | + | − | − | PDZ4 | MAGI1, PIST, and harmonin collaborate in intracellular trafficking and plasma membrane targeting of cadherin 23 | [58,59] |
| CRHR1; CRFR1 | Corticotropin-releasing hormone receptor 1; | G-protein (Gα) coupled receptor, binds neuropeptides of the corticotropin-releasing hormone family | + | + | + | PDZ1 | Receptor clustering; MAGIs regulate CRFR1 internalization by mediating β-arrestin recruitment. MAGI modulate CRFR1 signaling via the MAPK pathway but not cAMP formation | [60,61,62] |
| CXADR; CAR | Coxsackievirus and adenovirus receptor | Component of epithelial apical junction complexes, a membrane receptor for group B coxsackieviruses, and subgroup C adenoviruses. Isoforms originate from alternative splicing, e.g., CAR Ex8 | + | ND | ND | PDZ1 and PDZ3 | CAR Ex8 localizes to the apical surface of epithelial cells. Interaction with MAGI1 PDZ1 protects CAR Ex8 from degradation, whereas MAGI PDZ3 interaction triggers CAR Ex8 cleavage by ADAM17. | [48] |
| DLL1 | Delta-like protein; Delta1 | Involved in cell–cell communication. Delta proteins activate Notch through their extracellular domains; the intracellular domains also exert biological functions. | + | + | + | PDZ4 | MAGI1 recruits Delta1 to adherens junctions through its interaction with β-catenin. In Zebrafish, MAGI1, 2, and 3 interact with Delta1 and control neuron migration | [63,64,65] |
| ERBB4; HER4 | Receptor tyrosine-protein kinase erbB-4 | Member of EGF receptor family tyrosine kinases. Binds to and is activated by neuregulins. Mutated in various cancer types | + | + | ND | PDZ1 | MAGI1, 2 bind ERBB4 and R-PTP-zeta, and form a ternary phosphotyrosine kinase/ phosphotyrosine phosphatase complex. Bring ERBB4 and R-PTP-zeta to specific cellular domains. ERBB4 phosphorylation of MAGI enhanced by ERB4 ligand. | [66] |
| FZD4 | Frizzled 4, Fz-4 | Receptor for Wnt proteins; Activates disheveled proteins leading to nuclear accumulation of β-catenin and transactivation of Wnt target genes | + | ND | + | PDZ1 | MAGI3 forms a ternary complex with Fz-4 and VANGL2 (involved in planar cell polarity) at cell–cell contact. Both Fz-4 and Vangl2 interact with MAGI3 PDZ1suggesting MAGI3 oligomerization. This ternary complex activates JNK via Rac1 | [67,68,69] |
| GRIN2A; NMDAR2A; NR2A | Glutamate receptor ionotropic, NMDA 2A | Component of NMDA receptor complexes, ligand-gated ion channels with high calcium permeability, and voltage-dependent sensitivity to magnesium. | ND | + | ND | PDZ5 | Clustering of NMDA receptors? Required for NMDA receptor-induced RhoA. Hampered NMDA 2 internalization. | [30,70,71] |
| HTR2A | 5-Hydroxytryptamine receptor 2A | One of the receptors for serotonin (this also binds mescaline and LSD psychoactive substances); Gq/G11-protein-coupled, increases cellular levels of IP3 and DAG via PLC and modulates PKC activity and Ca2+ release from intracellular stores | + | + | + | PDZ nd | MAGIs differentially regulate 5-HT2AR activity: MAGI1, 3 favor receptor internalization; MAGI2, 3 enhance receptor trafficking. MAGIs increase PLC-β3 recruitment to the receptor and IP3 formation. MAGI knockdown increases ERK1/2 activation by 5-HT-2A, independently of the PDZ binding motif | [72] |
| IGSF5; JAM4 | Immunoglobulin superfamily member 5; Junctional adhesion molecule 4 | Cell adhesion molecule at tight junctions of kidney glomerulus and small intestinal epithelial cells. Mediates calcium-independent homophilic adhesion. | + | ND | ND | PDZ1 and PDZ4 | MAGI1 induces clustering of IgSF5 and strengthens cell adhesion. May regulate the permeability of kidney glomerulus and small intestinal epithelium | [73] |
| LRP2; Megalin | Low-density lipoprotein receptor-related protein 2; Megalin | Endocytic receptor expressed in absorptive epithelial tissues. Involved in reuptake of sterols, lipoproteins, vitamin-binding proteins, and hormones | + | ND | ND | PDZ5 | [23] | |
| LPAR2; LPA2 receptor | Lysophosphatidic acid receptor 2 | G protein (Gi/Go, G12/G13, & Gq)-coupled receptor. Involved in phospholipase C-beta (PLC-β) signaling pathway | − | + | + | PDZ5 | MAGI3 inhibits NHERF2 –induced migration and invasion of colon cancer cells by competing with NHERF2 for binding to LPAR2 and its downstream effector PLC-β3, leading to decreased NFkB and JNK activities | [74,75] |
| NGFR; TNFRSF16; P75NTR | Tumor necrosis factor receptor superfamily member 16; NGF receptor | Low-affinity receptor for neurotrophins (NGF, BDNF, NTF3, and NTF4). Mediates RhoA-dependent inhibition of growth of regenerating axons. | + | − | − | PDZ0 | MAGI1 interacts with NGF-R and SHC, positive regulation of NGF-stimulated SHC-ERK pathway | [28] |
| NLGN1 | Neuroligin-1 | Neuronal cell surface protein. Transsynaptic adhesion molecules. Role in synapse function and synaptic signal transmission. | + | + | + | PDZ1 | S-SCAM (MAGI2) is tethered by β-catenin and triggers the accumulation of neuroligin, which subsequently recruits PSD-95 to synapses | [30,76,77] |
| NPHS1; Nephrin | Nephrin; Renal glomerulus-specific cell adhesion receptor | Transmembrane protein, member of the immunoglobulin family of cell adhesion molecules. Involved in glomerular filtration barrier in the kidney (slit diaphragm of glomerular podocytes) | + | + | - | PDZ3 (MAGI1); Indirect or PDZ3-5 (MAGI2) | In podocytes, MAGI1 connects Nephrin and JAM4 to the actin cytoskeleton, critical for stable assembly of the slit diaphragm. Nephrin/ MAGI1 interaction enhances Rap1 activation, essential for maintaining slit diaphragm function. | [78,79,80,81,82] |
| PTPRZ1; RPTPB; RPTPbeta | Receptor-type tyrosine-protein phosphatase zeta; R-PTP-zeta | Role in cell adhesion, neurite growth, and neuronal differentiation, blood vessel remodeling, and angiogenesis | + | + | + | PDZ2 | Favors interactions of R-PTP-zeta with its substrates at the plasma membrane. MAGI1, 2 form a ternary phosphotyrosine kinase/ phosphotyrosine phosphatase complex with ERBB4 and R-PTP-zeta. R-PTP-zeta dephosphorylates Tyr-373 and Tyr-858 of MAGI1 | [66,83,84] |
| SDK1; Sidekick-1 | Protein sidekick-1 | Transmembrane protein, immunoglobulin family of cell adhesion molecules; promotes synaptic connectivity via homophilic interactions | + | + | ND | WW and PDZ4 and/or PDZ5; PDZ2 and/or PDZ3 | Localizes Sidekick1, 2 in distinct subsets of retinal synapses. Upregulation of Sidekick during focal segmental glomerulosclerosis impairs MAGI1 function, promotes actin cytoskeleton disruption, leading to podocyte dysfunction | [27,85] |
| TGFA; TGF-alpha | Protransforming growth factor alpha | Growth factor; ligand for EGFR; acts as transmembrane-bound or a soluble ligand | − | + | + | PDZ1 | Role in the trafficking of TGF-alpha to the cell surface in polarized epithelial cells | [33] |
| VANGL2, LTAP | Vang-like protein 2 (Vangl2); Loop-tail-associated protein (LTAP) | Membrane protein; Involved in regulation of planar cell polarity (non-canonical Wnt signaling pathway) | ND | + | + | PDZ1 | MAGI3 forms a ternary complex with Fz-4 and Vangl2 at cell–cell contact and activates JNK via Rac1. Both Fz-4 and Vangl2 interact with MAGI3 PDZ1 suggesting MAGI3 oligomerization. In podocytes, MAGI2 forms a ternary complex with nephrin and Vangl2 | [68,86] |
| VIPR1; VPAC1 | Vasoactive intestinal polypeptide receptor 1; VIP-R-1 | G protein-coupled receptor; receptor for vasoactive intestinal peptide, involved in smooth muscle relaxation, exocrine and endocrine secretion, and hydro-electrolytic flux | ND | + | ND | PDZ1 and/or PDZ2 | MAGI2 reduces VIP-R-1 activation, leading to decreased receptor internalization (GRK dependent). Recruitment of VIP-R-1 close to CFTR enables localized cAMP generation and hydroelectrolytic secretion with a minimal cellular response. | [87] |
| Adaptors, Scaffolding Molecules | ||||||||
| AMOTL2 | Angiomotin-like protein 2 | Member of the angiomotin membrane-associated scaffold proteins. Involved in cadherin linkage to the cytoskeleton and in YAP1 signaling | + | ND | ND | 2nd WW | MAGI1 downregulation is associated with enhanced accumulation of AMOTL2 and E-cadherin in MCF7 cells. Junctional dysfunction and cytoskeletal tension might activate the p38 stress pathway and tumorigenesis | [88] |
| ARRB2; BAAR2 | Arrestin beta 2; Beta-arrestin-2 | Member of arrestin protein family; scaffolding molecule; involved in agonist-mediated desensitization of GPCR, and in signaling regulation, e.g., TGFβ, MAPK, PTEN, AKT, NFkB, TP53 | + | ND | ND | PDZ nd | Role in regulating β-arrestin-2 recruitment to CRFR1 and receptor signaling. Involved in receptor/β-arrestin complex stability and trafficking, and in signaling functions of receptors. | [61,62] |
| Axin | Axin | Tumor suppressor involved in the control of Wnt pathway. Cytoplasmic protein interacting with APC, β-catenin, GSK 3β, PP 2A, and itself. Component of the complexities involved in β-catenin destruction. | ND | + | ND | GuK | MAGI2 forms a complex with β-catenin and Axin and competes with GSK3β for Axin-binding. MAGI2 may protect β-catenin from degradation by inhibiting its phosphorylation by GSK3β. | [89] |
| DLGAP4; SAPAP4 | Disks large-associated protein 4 | Membrane-associated guanylate kinase found at the postsynaptic density in neuronal cells | ND | + | ND | GuK | Interaction of DLGAP with MAGI2 (S-SCAM) | [30] |
| DLG4; SAP-90; PSD95 | Disks large homolog 4; Synapse-associated protein 90; Post-synaptic density protein 95 | MAGUK family, contains 1 domain SH3, 1 Guk domain, and 3 PDZ domains. Postsynaptic scaffolding protein involved in synaptogenesis and synaptic plasticity | ND | + | ND | PDZ4 + PDZ5 | The Guk domain of PSD95/ SAP90 interacts with PDZ4+PDZ5 of MAGI2. Role in clustering NMDA receptor? | [40] |
| FCHSD2 | F-BAR and double SH3 domains protein 2; Carom | Adapter protein. Promotes endocytosis of EGFR and down-regulation of EGFR signaling. Contributes to internalization of integrin β1 and transferrin receptor. Binds to membranes enriched in PtdIns(3,4)-P2 or PtdIns(3,4,5)P3. Promotes actin polymerization. | + | ND | ND | PDZ5 | MAGI1 colocalizes with Carom at immature cell contacts of epithelial cells. MAGI1 competes with CASK (calcium/calmodulin-dependent Ser protein kinase) for Carom binding. | [25] |
| MAGI2; SSCAM; ACVRIP1; AIP1; ARIP1 | Membrane-associated guanylate kinase, WW, and PDZ domain-containing protein 2 | Scaffolding molecule containing 1 Guk domain, 2 WW domains, and 6 PDZ domains | ND | + | ND | PDZ4 + PDZ5 | MAGI2 dimerization might be involved in scaffolding, clustering, signaling | [30,40,44] |
| MAGI3 | Membrane-associated guanylate kinase, WW, and PDZ domain-containing protein 3 | Scaffolding molecule containing 1 Guk domain, 2 WW domains, and 6 PDZ domains | ND | ND | + | ND | Frizzled-4 and Vangl2 form a ternary complex with MAGI3 via binding to PDZ1, suggesting MAGI3 oligomerization. Assumption supported by the interaction of wild-type MAGI3 with MAGI3pPA | [68] |
| MAGI3pPA | MAGI3 with premature polyadenylation | A truncated form of MAGI3 (depletion PDZ2-5) resulting from the use of a cryptic intronic polyadenylation signal | ND | ND | + | ND | Prooncogenic; competition with full-length MAGI3 for controlling YAP1 signaling; increases nuclear YAP1 localization. Expressed especially in breast cancer | [42,90] |
| CNKSR2; KSR2 | Connector enhancer of kinase suppressor of Ras 2 | Scaffolding protein with SAM, PDZ, and PH domains. Involved in MAPK pathways | ND | + | ND | PDZ4 and/or PDZ5 | Role in linking cell surface receptors to MAPK pathways? | [91] |
| SHC | SHC-transforming protein | Couple activated receptor tyrosine kinases to the RAS pathway by recruitment of GRB2/SOS complex | + | ND | ND | PDZ4 and/or PDZ5 | MAGI1 interacts directly with NGF-R and SHC, positive regulation of NGF-stimulated SHC-ERK pathway | [28] |
| TRIP6; ZRP1 | Thyroid receptor-interacting protein 6 | Member of the zyxin family contains 3 LIM domains. Localizes to focal adhesion and actin stress fibers. Regulates lysophosphatidic acid-induced cell migration. Modulates gene expression by nuclear translocation and interaction with some transcriptional factors, e.g., NFkB | + | ND | ND | PDZ5 | TRIP6 competes with β-catenin for binding MAGI1 PDZ5. Hence, TRIP6 impairs PTEN recruitment to E-cadherin/ β-catenin complexes and adherens junctions stabilization. In conjunction with AKT and NFkB activation by its core protein, TRIP6 promotes invasiveness. Displacement from MAGI1 allows β-catenin nuclear translocation and Wnt oncogenic signaling | [26,92] |
| Intracellular Signaling Proteins | ||||||||
| ARHGAP6 | Rho GTPase-activating protein 6 | GTPase activator for the Rho-type GTPases; Involved in the regulation of actin polymerization at the plasma membrane | + | ND | ND | PDZ2 | The antagonistic activities of NET1 and ARHGAP6 and their recruitment in the same molecular scaffold via two adjacent MAGI1 PDZ domains allow the fine-tuning of Rho signaling. The four last residues of NET1 and ARHGAP6 are identical, thus their selective binding to PDZ1 and PDZ2 of MAGI1 must be defined by residues upstream. | [67] |
| ATN1 | Atrophin | Transcriptional corepressor. Associated with dentatorubral pallidoluysian atrophy, a rare neurodegenerative disorder | + | + | ND | WW | The first designation of MAGI2 was atrophin-1-interacting protein 1 (AIP1) as a novel partner of atrophin | [44] |
| KRAS; Ki-Ras | GTPase KRas; K-Ras 2 | Proto-oncogene member of the small GTPase superfamily; upstream effector of PI3K and MAPK signaling pathways. | + | ND | ND | PDZ1 | Role in K-Ras 2 targeting to plasma membrane? Interaction not validated in mammalian cells | [29] |
| LATS1/2 | Serine/threonine-protein kinase LATS1/2; Large tumor suppressor homolog 1/ 2 | Ser, Thr kinases. Negative regulators of YAP1 (Hippo signaling pathway). Involved in the control of cell cycle, a negative regulator of CDK1/cyclin A | + | + | + | WW | The PY motifs of LATS1/2 interact with the WW domain tandem of MAGIs. This tandem enables high affinity and specificity towards their ligands. | [34,93] |
| MAP2K1, MEK1 | Dual specificity mitogen-activated protein kinase kinase 1; MEK1 MKK1 | Dual specificity protein kinase. Phosphorylates a Thr and a Tyr residue in a Thr-Glu-Tyr sequence of MAPK3/ERK1 and MAPK1/ERK2, leading to their activation and transduction of the signal within the MAPK/ERK cascade | + | + | + | WW; GuK | MEK1 is necessary for PTEN membrane recruitment as part of a ternary complex involving MAGI1. Complex formation requires MEK1 phosphorylation by ERK and leads to the negative regulation of the AKT pathway | [94] |
| NET1; ARHGEF8 | Neuroepithelial cell-transforming gene 1 protein | Rho guanine nucleotide exchange factor | + | ND | ND | PDZ1 | The antagonistic activities of NET1 and ARHGAP6 and their recruitment in the same scaffold via two adjacent MAGI1 PDZ domains allow the fine-tuning of Rho signaling. The four last residues of NET1 and ARHGAP6 are identical, thus their selective binding to PDZ1 and PDZ2 of MAGI1 must be defined by residues upstream. | [67,95] |
| PLCB3 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3; PLC-beta-3 | Member of the phospholipase C-β family generates DAG and InsP3 from phosphatidylinositol in response to G-protein-linked receptor stimulation. | + | + | + | PDZ nd | MAGI3 competes with NHERF2 for binding to both LPAR2 and PLC-β3, decreases the activities of PLC-β3 and its downstream effector NFκB by facilitating LPAR2 coupling to different G proteins | [72,75] |
| PTEN | Phosphatase and Tensin homolog deleted on chromosome TEN | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase; Tumor suppressor | + | + | + | PDZ2 | PTEN stabilization, PTEN recruitment to high molecular weight molecular complexes, including E-cadherin junctional complexes. Stabilization of cell–cell junctions, decreased cell migration | [22,37,96,97,98] |
| PTPN14; PTPD2; PEZ | Protein tyrosine phosphatase non-receptor type 14 | Tyr phosphatase with a FERM domain (cytoskeletal-associated proteins). Putative tumor suppressor. Dephosphorylates β-catenin at junctional complexes and stabilizes cell–cell contacts. Negatively regulates YAP1 (phosphatase dependent and independent manner) | +/− | + | + | WW | The PY motifs of PTPN14 interact with the WW domain tandem of MAGIs. This tandem enables high affinity and specificity towards their ligands. The inter-domain between the WW tandem might modulate the affinity of WW domains | [34] |
| PTPN21; PTPD1 | Protein tyrosine phosphatase non-receptor Type 21 | Protein tyrosine phosphatase. Contains a FERM found in cytoskeletal-associated proteins. | ND | + | + | WW | The PY motifs of PTPN21 interact with the WW domain tandem of MAGIs. This tandem enables high affinity and specificity towards their ligands. | [34] |
| RAPGEF2; NRap GEP; PDZ-GEF1 | Rap guanine nucleotide exchange factor 2; RA-GEF-1 | Guanine nucleotide exchange factor (GEF); activates the Rap and Ras family of small GTPases by exchanging bound GDP for free GTP in a cAMP-dependent manner | + | + | ND | PDZ0 (MAGI1; PDZ1 (MAGI2) | MAGI1 recruits RA-GEF-1 at tight junctions in epithelial cells. In endothelial cells, MAGI1 is required for Rap1 activation and adherens junction formation. Translocation of MAGI1 to cell–cell contacts is ascribed to its interaction with β-catenin. MAGI2 co-expressed with RA-GEF-1 enhances activation of Rap1 in podocytes, signaling essential for normal podocyte function | [99,100,101,102] |
| SMAD3 | Mothers against decapentaplegic homolog 3; SMAD family member 3 | Effector of TGFβ and activin type 1 receptor kinases. Transmits signals from the cell surface to the nucleus. Forms heterodimer with SMAD4 and activates transcription. | ND | + | ND | WW | MAGI2 interacts with activin type 2 receptor via its PDZ5 and with SMAD3 via its WW domains. MAGI2 negatively regulates activin/ SMAD3 -induced gene transcription | [53] |
| YAP1 | Yes-associated protein 1 | Regulator for TEAD transcription factors; coactivator and corepressor of gene expression downstream Hippo signaling pathway; Involved in cell growth, anchorage-independent growth, and EMT | ND | ND | + | PDZ5 | MAGI3 interacts with and sequesters YAP1 oncoprotein in the cytoplasm, inhibiting YAP1-induced malignant transformation of human mammary epithelial cells. | [42,90] |
| Actin Associated Proteins | ||||||||
| ACTN4 | Alpha-actinin-4 | Actin-binding protein | + | ND | ND | PDZ5 | Role in actin cytoskeleton dynamics? | [24] |
| SYNPO | Synaptopodin | Actin-associated protein, involved in actin-based cell shape and motility | + | ND | ND | 2nd WW | Role in actin cytoskeleton dynamics? | [24] |
| Posttranslational Modifications | ||||||||
| CASP3 | Caspase-3; CASP-3 | Cysteine-aspartic acid protease. Effector caspase is involved in the execution phase of apoptosis. Activated by proteolytic cleavage. CASP-3 shares several substrates with CASP-7 but exerts distinct functions during apoptosis. | + | + | ND | Asp761 (MAGI1) | MAGI1 is cleaved by CASP-3 at Asp761, between PDZ2 and PDZ3. The N-term fragment translocates in the cytoplasm and the C-term accumulates in the nucleus. MAGI1 cleavages induce disruption of cell–cell contacts and apoptosis. MAGI2 cleavage site was not determined. | [103,104] |
| CASP7 | Caspase7; CASP-7 | Cysteine-aspartic acid protease. Effector caspase is involved in the execution phase of apoptosis. Activated by proteolytic cleavage. CASP-7 shares several substrates with CASP-3 but exerts distinct functions during apoptosis. | + | ND | ND | Asp761 | MAGI1 is cleaved by CASP-7 at Asp761, generating 97 kDa and 68 kDa fragments. The N-term fragment translocates in the cytoplasm and the C-term accumulates in the nucleus. MAGI1 cleavages induce disruption of cell–cell contacts and apoptosis. | [103] |
| RPS6KA1, P90RSK | Ribosomal protein S6 kinase alpha-1; S6K-alpha-1 | Ser/Thr kinase. Acts downstream ERK1/2. Mediates mitogenic and stress-induced activation via CREB1. Regulates translation through RPS6 and EIF4B. Mediates cellular proliferation, survival, and differentiation by modulating mTOR signaling and repressing pro-apoptotic function of BAD and DAPK1. | + | ND | ND | Ser741 | Thrombin stimulates P90RSK that directly phosphorylates MAGI1 at Ser741 and induces MAGI1 deSUMOylation at Lys 931, leading to endothelial cells activation. This process involves Rap1 activation by phosphorylated MAGI1. SUMOylation impairs MAGI1 nuclear translocation and stabilizes endothelial barrier function. | [105,106] |
| SENP2 | Sentrin/SUMO-Specific Protease SENP2 | Cysteine protease targeting members of small ubiquitin-like modifier (SUMO) protein family. Deconjugates sumoylated proteins | + | ND | ND | Lys 931 | MAGI1-K931 deSUMOylation induces both nuclear translocation of p90RSK-MAGI1 and ATF-6-MAGI1 complexes, which enhances endothelial cells activation and apoptosis, respectively. | [105] |
| Viral (onco/) Proteins | ||||||||
| Molecular Partners | Virus | Viral Protein Function | MAGI1 | Interaction MAGI2 | MAGI3 | MAGI Binding Domain | Biological Impact | References |
| NS1 | Influenza A virus. RNA virus | Non-structural (NS) protein; non-essential virulence factor. Involved in inhibition of host immune responses (IFN production/ response) | + | + | + | PDZ nd | Sequesters MAGI1 from the plasma membrane. Does not seem to confer benefit to viral replication | [107,108] |
| Tax | Human T-cell leukemia virus type 1 (HTLV-1). Retrovirus | Viral oncoprotein, immortalization of human T-cells. Promotes viral gene transcription and regulates expression of human genes by modulating CREB/ATF, NFkB, AP-1, and SRF pathways | + | MAGI2 mRNA not detectable in T-cell lines | + | PDZ1 | Downregulation of MAGI1 transcripts. Mis-localization of MAGI1, 3 to perinuclear region (from detergent-soluble to detergent-insoluble fraction). MAGI3 increases Tax1 accumulation | [109,110] |
| E4-ORF1 | Human adenovirus type 9. DNA virus | Viral oncoprotein; PDZ binding motif required for E4-ORF1 -induced PI3K activation and cell transformation | + | ND | ND | PDZ1 and PDZ3 | Ad9 E4-ORF1 protein sequesters MAGI1 in the cytoplasm (from detergent-soluble to detergent-insoluble fraction). Altered tight junction assembly and cell polarity | [111,112,113,114] |
| E6 | Human papillomavirus (HPV); high-risk HPV (HPV 16 and 18) causes cervical cancer. DNA virus | Viral oncoprotein interacts with and promotes degradation of TP53 as well as proteins with PDZ domains, including MAGIs | + | + | + | PDZ1 | HPV E6 proteins targets MAGI1 for proteasome degradation, leading to tight junction disruption. Restoration of MAGI1 expression in HPV positive cervix cancer HeLa cell line induces cell growth arrest and apoptosis. RNA aptamers to E6 inhibiting interaction with PDZ domain trigger cancer cell apoptosis | [111,115,116,117,118,119] |
| E protein | Coronavirus (MERS-CoV). RNA virus | The envelope protein forms a transmembrane ion channel. PDZ binding motifs of E proteins from MERS and SARS-CoV/SARS-COV-2 differs and interact with distinct and shared cellular proteins with the PDZ domain | + | + | ND | PDZ5 | [120] | |
| Molecular Alterations | MAGI | Types of Cancer | Clinical Significance | References | ||
|---|---|---|---|---|---|---|
| Promoter Methylation | ||||||
| MAGI1 | Anaplastic thyroid cancer | Malignant tumor associated with poor survival | [121] | |||
| Lymphocytic leukemia | Decreased overall survival | [122] | ||||
| MAGI2 | Breast cancer | Decreased overall survival | [128] | |||
| Cervical cancer | [124] | |||||
| Colon cancer | 32.8% exhibit MAGI2 hypermethylation | https://cancer.sanger.ac.uk/cosmic/ accessed on 22 July 2021 | ||||
| Endometrium cancer | 34.9% exhibit MAGI2 hypermethylation | [126]; https://cancer.sanger.ac.uk/cosmic/ accessed on 22 July 2021 | ||||
| Gastric cancer | Decreased progression-free survival | [125] | ||||
| Ovarian cancer | Decreased overall survival | [126,127] | ||||
| Rearrangement | ||||||
| MAGI2 | Melanoma cell lines | [130] | ||||
| Prostate cancer | [131] | |||||
| MAGI3/AKT3 | Breast cancer | Enriched in triple-negative breast cancers | [132] | |||
| miRNA Targeting | ||||||
| miR-486-5p | MAGI1 | Erythroid leukemia K562 cell line | Decreased erythroid differentiation | [136] | ||
| miR-520h | Renal cell carcinoma | Decreased overall survival | [137] | |||
| miR-629-5p | MAGI2 | Ovarian cancer | miR-629-5p is downregulated in ovarian cancer | [138] | ||
| miR-27a-3p | Breast cancer/ macrophages | Exosomal miR-27a-3p from breast cancer exhausts MAGI2 in macrophages, leading to downregulation of PTEN, increased PDL1 expression, and cancer cells’ immune escape. | [142] | |||
| miR-487a | Breast cancer | miR-487a expression correlates with lymph node metastases | [139] | |||
| miR-101 | Breast cell line (MCF7 cells) | [140] | ||||
| miR-134/487b/655 cluster | Lung cancer cell lines | Epithelial–mesenchymal transition. Resistance to the EGFR inhibitor gefitinib, | [141] | |||
| miR-4677-3p | Glioma | The GLIDR lncRNA sponges miR-4677-3p that exhausts MAGI2. MAGI2 overexpression promotes glioma cell lines proliferation. | [144] | |||
| miR-34c-3p | MAGI3 | Hepatocarcinoma | Decreased overall survival | [143] | ||
| Premature Polyadenylation | ||||||
| MAGI3 | Breast cancer | Competition MAGI3pPa / wild-type MAGI3; activation YAP1 signaling | [42] | |||
| MAGI1 Point Mutations | MAGI2 Point Mutations | MAGI3 Point Mutations | ||||
| Tissue | % Mutated * | Tissue | % Mutated * | Tissue | % Mutated * | |
| Liver | 16.5 | Prostate | 26.4 | Liver | 9.8 | |
| Skin | 13.6 | Pancreas | 23.5 | Nervous system | 9.8 | |
| Prostate | 13.4 | Liver | 21.2 | Skin | 8.2 | |
| Pancreas | 12.3 | Breast | 17.8 | Prostate | 7.7 | |
| Breast | 10.8 | Stomach | 14.4 | Biliary tract | 6.7 | |
| Endometrium | 10.1 | Skin | 14.4 | Ovary | 6.2 | |
| Nervous system | 9.8 | Esophagus | 14.3 | Breast | 6.1 | |
| Ovary | 9.5 | Nervous system | 13.3 | Pancreas | 5.9 | |
| Stomach | 9.2 | Ovary | 13.2 | Endometrium | 5,8 | |
| Biliary tract | 8.9 | Endometrium | 10.2 | Esophagus | 5.6 | |
| MAGI | Types of Cancer | Transcript Accumulation * | Protein Expression * | Clinical Significance | References |
|---|---|---|---|---|---|
| MAGI1 | Breast cancer luminal | down | down | Decreased disease-free survival | [88] |
| Breast cancer ER+/HER2− | down | down | High expression good prognosis | [147] | |
| Colon cancer | down | nd | Decreased disease-free survival | [161] | |
| Gastric cancer | down | down | Distant metastases | [146] | |
| Gliomas | down | down | Distant metastases | [56] | |
| Hepatocellular carcinoma | down | down | Poor prognosis | [145] | |
| Kidney | High RNA level, increased overall survival | [162] http://gepia2.cancer-pku.cn, accessed on 26 July 2021 | |||
| MAGI2 | Kidney | High RNA level, increased overall survival | [162] http://gepia2.cancer-pku.cn, accessed on 26 July 2021 | ||
| Multiple myeloma | up | nd | Decreased overall survival | [156] | |
| Ovarian cancer | down | nd | Decreased overall survival | [127] | |
| Prostate | down | unaffected/down | High mRNA levels better prognosis | [149,150,151] | |
| nd | up | [152,153,155] | |||
| MAGI3 | Gliomas | down | down | Decreased overall survival | [55] |
| Kidney | High RNA level, increased overall survival | [162] http://gepia2.cancer-pku.cn accessed on 26 July 2021 | |||
| Ovarian cancer | down | nd | Decreased disease-free survival | [157] | |
| Thyroid cancer | down | nd | Decreased RNA level in malignant vs benign tumors | [163] |
| MAGI ceRNA | Primary Target | Secondary Target * | Activity, Function of Secondary Target | Physiological Impact | References | ||
|---|---|---|---|---|---|---|---|
| MAGI1-IT1 | |||||||
| MAGI1-IT1 |  | miR-200a |  | ZEB1, ZEB2 (zinc finger E-Box binding homeobox 1/ 2) | Transcriptional repressor. Promotes epithelial to mesenchymal transition | Epithelial–mesenchymal transition, invasiveness. Ovarian cancer cell lines | [206] |
 | miR-302d-3p |  | IGF1 (insulin-like growth factor 1) | Structurally and functionally related to insulin. Growth-promoting activity through binding to IGF1R tyrosine kinase, leading to activation of the PI3K-AKT/PKB and the Ras-MAPK pathways. | Overexpression MAGI1-IT1 in gastric cancer is associated with poor overall survival. Knockdown in gastric cancer cell lines decreases proliferation in vitro and in vivo | [207] | |
 | miR-512p-3p |  | AKT (protein kinase B, PKB) | Proto-oncogene. Ser/Thr kinase downstream of phosphatidylinositol 3-kinase (PI3K). Involved in cell proliferation, survival, and migration. | Increased AKT activity. Increased lung cancer cell lines proliferation | [205] | |
| MAGI2-AS3 Tumor Suppressor Activity | |||||||
| MAGI2-AS3 |  | miR-15b-5p |  | CCDC19/CFAP45 (cilia and flagella associated protein 45) | Tumor suppressor. Inhibits cell proliferation | Inhibition of proliferation, migration, and invasion. Bladder cancer cell lines | [208] |
 | RECK * (reversion inducing cysteine-rich protein with kazal motifs; ST15, suppressor of tumorigenicity 15 protein) | Extracellular protein anchored to the plasma membrane. Decreases diffusible gelatinase/collagenase activity | Downregulation of miR15b-5p and miR-374a/b-5p by MAGI2-AS3 decrease the viability and migration of the epithelial ovarian cancer PEA1, KURAMOCHI, and SKOV3 cell lines. Downregulation of MAGI2-AS3 due to promoter hypermethylation | [209] | |||
 | MTSS1 (metastasis suppressor protein 1) | Tumor suppressor controlling migration and invasion. Involved in Hedgehog and EGF pathways. Interacts with cytoskeleton | Downregulation of miR-15b-5p and miR-374a/b-5p by MAGI2-AS3 decrease the viability and migration of the epithelial ovarian cancer PEA1, KURAMOCHI, and SKOV3 cell lines. Downregulation of MAGI2-AS3 due to promoter hypermethylation | [209] | |||
 | miR-23a-3p |  | PTEN * (phosphatase and tensin homolog deleted on chromosome 10) | Lipid and protein phosphatase. Dephosphorylates PtdIns(3,4,5)P3 produced by PI3K. Inhibits AKT pathway. Dephosphorylates proteins, e.g., FAK, IRS1, Dvl2, PTEN. Exerts activity independent of its phosphatase activity, e.g., DNA damage repair. Induces apoptosis, decreases cell proliferation and migration | Decreased proliferation, migration, and invasion. Increased apoptosis. Lung squamous carcinoma cell lines | [210,211] | |
 | miR-25 |  | RECK * (reversion inducing cysteine-rich protein with kazal motifs; ST15, suppressor of tumorigenicity 15 protein) | Extracellular protein anchored to the plasma membrane. Decreases diffusible gelatinase/ collagenase activity | Decreased cell migration and invasiveness. Non-small cell lung carcinoma H1993 cell line | [212] | |
 | miR-31-5p |  | TNS1 (tensin 1) | Focal adhesion molecule, crosslinks actin filaments. Involved in cell migration | Decreased proliferation, migration, and invasion of bladder cancer cell lines | [213] | |
 | miR-155 |  | SOCS1 (suppressor of cytokine signaling 1) | Negative regulation of cytokines signaling through the JAK/STAT3 pathway | Decreased cell proliferation. Non-small cell lung carcinoma H1993 cell line | [214] | |
 | miR-223-3p (according to the sequence in Ref. [215]) |  | EBP41L3 (erythrocyte membrane protein band 4.1 Like 3) | Tumor suppressor. Member of the band 4.1 family of cytoskeletal proteins. The linker between membrane proteins and cytoskeleton. Involved in cell adhesion and motility | Inhibition of the human cervix cancer SiHa and HeLa cell lines invasion and migration | [215] | |
 | miR374a/b-5p |  | CADM2 (cell adhesion molecule 2) | Transmembrane protein involved in cell aggregation | Decreased proliferation and migration in vitro, enhanced apoptosis, decreased tumor growth, and experimental metastases in nude mice. Lung squamous carcinoma xxxSW900, SK-MES_1, and A549 and cell line. | [211] | |
 | PTEN * (phosphatase and tensin homolog deleted on chromosome 10) | Lipid and protein phosphatase. Dephosphorylates PtdIns(3,4,5)P3 produced by PI3K. Inhibits AKT pathway. Dephosphorylates proteins, e.g., FAK, IRS1, Dvl2, PTEN. Exerts activity independent of its phosphatase activity, e.g., DNA damage repair. Induces apoptosis, decreases cell proliferation and migration | Downregulation of miR15b-5p and miR-374a/b-5p by MAGI2-AS3 decrease the viability and migration of the epithelial ovarian cancer PEA1, KURAMOCHI, and SKOV3 cell lines. Downregulation of MAGI2-AS3 due to promoter hypermethylation. Decreased migration and invasion of breast cancer cell lines | [209,216] | |||
 | miR374b-5p |  | SMG1 (genitalia family member 1) | Ser/Thr protein kinase involved in nonsense-mediated decay of mRNAs containing premature Stop codons. Can phosphorylate and trigger TP53 activation after cell exposure to genotoxic stress | Decreased proliferation and migration. Hepatocarcinoma cell lines | [217] | |
 | RECK * (reversion inducing cysteine-rich protein with kazal motifs; ST15, suppressor of tumorigenicity 15 protein) | Extracellular protein anchored to the plasma membrane. Decreases diffusible gelatinase/collagenase activity | Downregulation of miR15b-5p and miR-374a/b-5p by MAGI2-AS3 decrease the viability and migration of the epithelial ovarian cancer PEA1, KURAMOCHI, and SKOV3 cell lines. Downregulation of MAGI2-AS3 due to promoter hypermethylation | [209] | |||
 | HOXA5 (homeobox A5) | Transcription factor upregulates the tumor suppressor p53 | Downregulation of miR15b-5p and miR-374a/b-5p by MAGI2-AS3 decrease the viability and migration of the epithelial ovarian cancer PEA1, KURAMOCHI, and SKOV3 cell lines. Downregulation of MAGI2-AS3 due to promoter hypermethylation | [209] | |||
 | miR-525-5p |  | MDX1 (MAX dimerization protein) | Competes with MAX (Myc associated factor X) for interaction with the c-Myc proto-oncogene. Inhibits transactivation by this transcriptional activator complex | Decreases proliferation and migration of ovarian cancer cell lines | [218] | |
 | EZH2 (Enhancer of zeste homolog 2; histone-lysine N-methyltransferase) |  | HOXB7 (homeobox B7) | Member of the homeobox family of transcription factors. Involved in cell proliferation and differentiation. Master regulatory gene orchestrating several oncogenic pathways. Overexpressed in various types of cancers including breast, esophageal, gastric, ovarian, and melanoma | MAGI2-AS3 binds to the HOXB7 promoter and recruits EZH2. Methylation of lysine 27 on histone H3 in the promoter region negatively regulates the transcription of HOXB7 and inhibits proliferation and radio-resistance of esophageal cancer cells. | [219] | |
 | HEY1 (Hes related Family BHLH transcription factor with YRPW motif 1) |  | ACY1 (aminoacylase 1) | Involved in the hydrolysis of various N-acetylated amino acids. Putative tumor suppressor in clear cell renal cell carcinoma | MAGI2-AS3 forms a nucleoprotein complex with HEY1 in the nucleus and impairs its transcriptional repressor activity on the ACY1 promoter. Decreased cell viability, EMT, and migration in vitro; reduced tumor growth and angiogenesis in vivo. Clear cell renal cell carcinoma | [220] | |
 | KDM1A (lysine demethylase 1A |  | RacGAP (Rac GTPase activating protein 1) | Negative regulation of Rho GTPase signaling through stimulation of GTP hydrolysis | Demethylation of Lys4 of histone H3 (H3K4Me2) at RACGAP1 promoter reduces transcription. Decreased proliferation and migration, enhanced apoptosis of HEPG2 hepatocarcinoma cells in vitro, and decreased tumor growth in nude mice | [221] | |
 | TET1 (Tet Methylcytosine Dioxygenase 1) |  | MAGI2 (membrane-associated guanylate kinase inverted 2) | Scaffolding molecule with 1 Guanylate Kinase domain, 2 WW domains, and 6 PDZ Domains. Involved in junctional complexes stability and cell signaling | Demethylation of MAGI2 promoter, increased MAGI2 expression, inhibition of AKT and Wnt/ β-catenin pathways; decreased cell proliferation of MCF-7 breast cancer cell line | [128] | |
 | TET2 (Tet Methylcytosine Dioxygenase 2) |  | LRIG1 (leucine-rich repeats and immunoglobulin-like domains 1) | Transmembrane protein that interacts and negatively regulates receptor tyrosine kinases of the EGFR family, MET and RET by enhancing receptor ubiquitination and degradation | Impaired leukemia stem cells self-renewal | [222] | |
| ? | ? |  | FAS (Fas cell surface death receptor); FASL (FAS ligand) | Members of the TNF/ TNF receptor family. Promotes activation of caspase-8 and triggers the apoptotic cascade | Decreased proliferation, enhanced apoptosis of MDA-MB-231 and MCF-7 breast cancer cell lines. The underlying mechanism has not been characterized but might involve miR-374a -a target of MAGI2-AS3- that exhausts FAS transcripts | [223] | |
| ? | ? |  | ROCK2 (Rho-associated coiled-coil containing protein kinase 2) | Ser/Thr kinase, a downstream effector of the Rho GTPase. Involved in the regulation of actin cytoskeleton organization, stress fiber and focal adhesion formation, cell adhesion, and motility | MAGI2-AS3 induces the downregulation of ROCK2 at both RNA and protein levels in the Hep3B and MHCC97-H hepatocarcinoma cell lines decrease cell migration and invasiveness and induce apoptosis. | [224] | |
| MAGI2-AS3 Tumor Promoter Activity | |||||||
| MAGI2-AS3 |  | miR-141 |  | ZEB1 * (zinc finger E-box binding homeobox 1) | Transcriptional repressor. Promotes epithelial to mesenchymal transition | Increased migration and invasion of gastric cancer cell lines | [225] |
 | miR-143 |  | AKT, COX2, ERK5, HMBG1, IGF1-R, PAI-1 ? | MiR-143 functions as a tumor suppressor in human bladder cancers. Known targets of miR-143 in bladder cancers cells are AKT, Cox2, ERK5, HMBG1, IGF1-R, PAI-1 | [226] | ||
 | miR-200a |  | ZEB1 *, ZEB2 (zinc finger E-box binding homeobox 1/ 2) | Transcriptional repressor. Promotes epithelial to mesenchymal transition | Increased migration and invasion of gastric cancer cell lines. The bromodomain-containing 4 (BRD4) protein, an acetylated histone binding protein is a transcriptional activator of MAGI2-AS3 in gastric cancer. MiR-200a increases E-cadherin levels, inhibits migration, and enhances the sensitivity of bladder cancer cell lines to anti-EGFR therapies. | [225,226,227] | |
 | miR-218-5p |  | GDPD5 (glycerophosphodiester phosphodiesterase domain containing 5) | Involved in glycerol metabolism. Mediates the cleavage of glycosylphosphatidylinositol -anchor of RECK, leading to RECK release from the plasma membrane | Increased proliferation, migration, epithelial–mesenchymal transition, and cisplatin resistance of nasopharyngeal epithelial cell lines | [228] | |
 | miR-3163 |  | TMEM106B (transmembrane protein 106B) | Involved in the regulation of lysosomal trafficking | Decreased apoptosis, increased proliferation, and migration of the HCT116 and RKO colon cancer cell lines | [229] | |
 : Inhibition of the target;
: Inhibition of the target;  : Activation of the target; * Secondary targets regulated by different miRNAs exhausted by MAGI2-AS3 are in bold.
: Activation of the target; * Secondary targets regulated by different miRNAs exhausted by MAGI2-AS3 are in bold.Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kotelevets, L.; Chastre, E. A New Story of the Three Magi: Scaffolding Proteins and lncRNA Suppressors of Cancer. Cancers 2021, 13, 4264. https://doi.org/10.3390/cancers13174264
Kotelevets L, Chastre E. A New Story of the Three Magi: Scaffolding Proteins and lncRNA Suppressors of Cancer. Cancers. 2021; 13(17):4264. https://doi.org/10.3390/cancers13174264
Chicago/Turabian StyleKotelevets, Larissa, and Eric Chastre. 2021. "A New Story of the Three Magi: Scaffolding Proteins and lncRNA Suppressors of Cancer" Cancers 13, no. 17: 4264. https://doi.org/10.3390/cancers13174264
APA StyleKotelevets, L., & Chastre, E. (2021). A New Story of the Three Magi: Scaffolding Proteins and lncRNA Suppressors of Cancer. Cancers, 13(17), 4264. https://doi.org/10.3390/cancers13174264








