Simple Summary
Tumor suppressor genes are critical in the control of many biological functions. They can be classified based on their roles in proliferation, cell-cycle progression, DNA repair/damage, and crucial signaling functions, including apoptosis, autophagy, and necrosis. The absence of functional tumor suppressor genes entails a higher risk of dysfunction of cell growth, differentiation, cell death, and cancer development. Loss of function or mutations of such genes has been identified in many types of cancer, such as breast, bladder, colorectal, head and neck, lung, ovarian, uterine, and pancreatic cancers. Familial cancer syndromes, such as Li–Fraumeni syndrome, are associated with loss of TP53 function. Extensive studies have been carried out to clarify the roles of the products of these genes, as well as their mechanistic link to cancers, to identify novel targets for specific cancer types. Here, we introduce the roles of tumor suppressor gene products in pancreatic cancer development and its therapeutics for tumorigenesis prevention.
Abstract
The high mortality of pancreatic cancer is attributed to the insidious progression of this disease, which results in a delayed diagnosis and advanced disease stage at diagnosis. More than 35% of patients with pancreatic cancer are in stage III, whereas 50% are in stage IV at diagnosis. Thus, understanding the aggressive features of pancreatic cancer will contribute to the resolution of problems, such as its early recurrence, metastasis, and resistance to chemotherapy and radiotherapy. Therefore, new therapeutic strategies targeting tumor suppressor gene products may help prevent the progression of pancreatic cancer. In this review, we discuss several recent clinical trials of pancreatic cancer and recent studies reporting safe and effective treatment modalities for patients with advanced pancreatic cancer.
Keywords:
BRCA1; BRCA2; clinical trial; pancreatic cancer; p53; translational research; tumor suppressor gene 1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a complex and a high-volume cancer that is managed with a multidisciplinary approach. The incidence of PDAC is gradually increasing worldwide, and its 5-year survival rate is about 10% [1,2]. The aggressive and lethal nature of this type of cancer is attributed to delayed diagnosis and lack of effective treatments. Despite the development by surgeons of many novel surgical techniques, such as superior mesenteric artery-first approach and superior mesenteric vein/portal vein resection and reconstruction, the silent nature of PDAC and its presentation leave a small percentage of patients qualifying for surgery (~20%). The most common therapy for PDAC is chemotherapy using modified FOLFIRINOX (mFOLFIRINOX) or gemcitabine-based regimen with the addition of capecitabine or nano-paclitaxel (Abraxane). Based on three randomized clinical trials (n = 2089), Galvano et al., concluded the optional adjuvant regimen for resected pancreatic cancers is mFOLFIRINOX. This robust scientific evidence strongly supports the pre-operative use of mFOLFIRINOX to increase the chance of R0 resection and reduce the incidence of micro metastases [3]. However, none of these regimen target altered genes. After neoadjuvant chemotherapy, surgical resection, and adjuvant chemotherapy with FOLFIRINOX (folinic acid, fluorouracil, irinotecan, and oxaliplatin), a small subgroup of patients can reach a mean survival of 54 months [4]. Pancreatic cancer is very aggressive and a prolonged treatment from diagnosis-to-initiation for PDAC may impact the survival. A study showed the optimal time was “within 6 weeks from diagnosis” when it was associated with an improved survival [5]. Nearly 80% of patients with PDAC cannot receive surgery at the time of diagnosis, and chemotherapy and radiotherapy do not have a significant impact on the overall survival of these patients. Recent advancements in the modality of irreversible electroporation have shown that it is safe and effective; however, it is not frequently used. Thus, new technologies are required that focus on target genes during the development of treatment algorithms for pancreatic cancer. Here we focus on tumor suppressor genes for new genetic trials for pancreatic cancer [6].
A genetically engineered mouse model of PDAC has successfully recapitulated human PDAC cancer biology [7,8,9]. This model was established using a combination of a mutant Kras oncogene with one deleted or mutated tumor suppressor gene (TSG, such as p53, Smad4, p16Ink4a (cdkn2a), or Brca1/2) [10,11,12]. The activated Kras oncogene can change the morphology of normal epithelial cells to the pancreatic intraepithelial neoplasm-1 (PanIN-1) type and initiate tumorigenesis [13,14,15]. In the later stage of cancer development, mutation/deletion of p53 occurs, thus accelerating the disease and leading to its evolution to invasive, advanced PDAC [15].
Kras is a notorious oncogene for PDAC. About 95% of patients with PDAC carry mutant and activated Kras. Kras mutation occurs in the earliest precancerous lesions (such as PanIN-1) [13]. Furukawa et al. [16,17] proposed “the RAS–MAPK pathway with abrogation of dual specificity phosphatase 6 (DUSP6)” as the molecular mechanism underlying PDAC development (Figure 1).
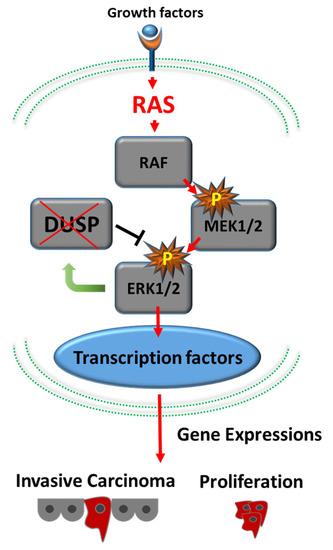
Figure 1.
Development of pancreatic adenocarcinoma via the RAS–MAPK signaling cascade. Active RAS generated by mutated Kras activates downstream cascades, including RAF1–MAP2K1–MAPK1. Loss of expression of DUSP6 results in abrogation of the feedback loop between MAPK1 and DUSP6 and leads to constitutive activation of MAPK1, which eventually results in invasive phenotypes.
Activation of Kras and inactivation of p16Ink4a (CDKN2A) lead to the formation of low-grade PanIN-2 lesions [18]. Moreover, cell proliferation increases after Kras oncogene activation. To maintain the balance between cell division and apoptosis, TSGs are turned on and cell-cycle arrest is triggered. However, if TSGs are inactivated because of deletion or mutation, the increased cell cycles are no longer inhibited. Cell proliferation increases when the Kras oncogene is activated.
Additional loss of p53 and Smad4 functions will accelerate disease progression, and tumors become high-grade PanIN-3 lesions [7,19,20]. Finally, additional inactivation of DUSP6 results in advanced pancreatic ductal adenocarcinoma [16,17]. In this case, TSGs were found to help cellular DNA-repair homeostasis, control cell division, and induce apoptosis. p53, Smad4, p16, Brca1/2, and PTEN are common examples of TSGs that are involved in PDAC carcinogenesis [10,11,12,21,22,23] (Figure 2).
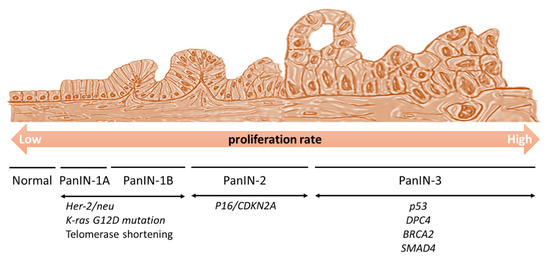
Figure 2.
Generation of pancreatic intraepithelial neoplasia (PanIN). The Kras oncogene is activated and increases cell proliferation. Her-2/neu expression and telomere shortening also occur at the initial stages of the disease. If tumor suppressor genes are inactivated, there is no stopper, and the cell cycle proceeds extensively. p16Ink4a is activated at the PanIN-2 stage, and then p53, DPC4, and BRACA2 are activated at the PanIN-3 stage, to induce a high cellular proliferation. Subsequently, the cells enter the neoplastic stage.
The next-generation sequencing (NGS) technique can identify a series of somatic mutations, such as fusions (ALK, NRG1, NTRK, and ROS1), mutations (BRAF, BRCA1/2, HER2, KRAS, and PALB2 [24]), and mismatch repair (MMR) genes (MLH1, MSH2, MSH6, and PMS2 [25]) in patients. These are mostly the products of tumor suppressor genes.
In the Ontario Pancreas Cancer Registry study [26], the germline DNA from 290 patients with varying degrees of family history was sequenced using a panel of 13 genes (APC, ATM, BRCA1, BRCA2, CDKN2A, MLH1, MSH2, MSH6, PALB2, PMS2, PRSS1, STK11, and TP53). Chaffee et al. [27,28] reported the results of sequencing using a panel of 25 cancer genes among 303 patients with a family history of pancreatic cancer. They found germline mutations in 10 genes (ATM, BRCA1, BRCA2, CDKN2A, PALB2, PMS2, BARD1, CHEK2, MUTYH, and NBN), which accounted for 11.6% of the prevalence of the overall PDAC cases.
Based on the data mentioned above, we understand that TSGs play important roles in PDAC carcinogenesis. In this review article, we attempt to summarize some of the potential treatments for these TSGs in PDAC.
2. BRCA1/2 Tumor Suppressor Gene—PARP Inhibitors
The loss of the wild-type allele of BRCA, which is considered a classical tumor suppressor gene, increases the risks of breast, ovarian, pancreatic, and prostate cancer, among others [29,30]. The frequency of BRCA1/2 mutation among the whole PDAC population was estimated at around 4–7% [31,32]. Germline mutations mainly in the BRCA2 gene lead to an increased risk of breast cancer, as well as a higher risk of developing PDAC, with a 2–6-fold increase in cancer risk compared with the general population.
The BRCA pathway, including PALB2, FANCC, and FANCG, involves the repair of DNA inter-strand cross-links. Patients with metastatic PDAC were initially frequently treated with platinum-based chemotherapy; currently, the FDA has approved olaparib, a poly (ADP-ribose) polymerase (PARP) inhibitor, as a maintenance regimen for adult patients with PDAC and deleterious germline BRCA mutations. Olaparib can trap the PARP-1 protein at a single-strand break/DNA lesion and disrupt its catalytic cycle, ultimately leading to replication fork progression and consequent double-strand breaks (Figure 3) [33,34]. The efficacy of this regimen was documented in the POLO (NCT 02184195) study [33], which reported a median progression-free survival (PFS) of 7.4 months (95% CI: 4.1, 11.0) among patients who received olaparib compared with 3.8 months (95% CI: 3.5, 4.9) for patients who received the placebo (HR 0.53; 95% CI: 0.35, 0.81; p = 0.0035).
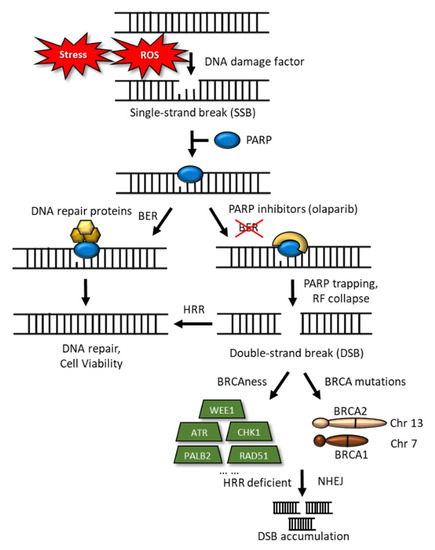
Figure 3.
Schematic representation of DNA damage and DNA-double-strand breaks. DNA-damage inducers, such as stress and ROS, sometimes trigger single-strand breaks (SSBs), to which PARP binds, for their repair. Base excision repair (BER) reverses the DNA damage resulting from oxidation, deamination, and alkylation. In this case, BER DNA glycosylase recognizes and removes the damaged base, leaving an abasic site that is processed further, by short-patch or long-patch repair, which largely uses different proteins to complete BER. PARP inhibitors trap the PARP-1 protein at an SSB/DNA lesion and disrupt its catalytic cycle, this would ultimately lead to replication fork progression and consequent double-strand breaks (DSBs). In the case of BRCA mutations, loss of HRR would result in cell death. HRR deficient and nonhomologous end-joining lead to the accumulation of DSBs.
2.1. Clinical Trials of Inhibitors of PDAC
The recent clinical trials of inhibitors of human pancreatic cancers are summarized as below (Table 1, Table 2, Table 3 and Table 4). We showed the summary of the inhibitors of MDM2, PARP, and gene-based target therapy drugs (Table 3) and immunotherapy drugs (Table 4).

Table 1.
Recent clinical trials of MDM2 inhibitors in human pancreatic cancers.

Table 2.
Recent clinical trials of PARP inhibitors in human pancreatic cancers.

Table 3.
Recent clinical trials based on gene therapy in human pancreatic cancers.

Table 4.
Recent clinical trials based on immunotherapy in human pancreatic cancers.
2.2. Other PARP Inhibitors for the Treatment of Pancreatic Cancer
Similar drugs that function based on the “synthetic lethality” concept are indicated below.
2.2.1. Veliparib
In a phase 1 clinical trial (NCT01908478), dose-escalated veliparib was used in 30 patients with locally advanced or borderline resectable pancreatic cancer, which was combined with weekly gemcitabine treatment and daily radiotherapy. This study confirmed that veliparib is safe and well tolerated in combination therapy with gemcitabine and radiotherapy among patients with PDAC [34].
2.2.2. Talazoparib (MDV3800 or BMN 673)
A novel inhibitor of PARP was developed that is more potent than the previous PARP-1/2 inhibitors. A phase 1 study (NCT01286987) confirmed the antitumor activity and maximal tolerable dose (1.0 mg/day) of talazoparib. Four of the 13 patients with PDAC included in the trial showed clinical benefits (rate, ~31%, ≥16 weeks) [35].
2.2.3. Rucaparib (NCT02042378)
A phase 2 study was carried out to measure the efficacy and safety of rucaparib in patients with BRCA1/2 mutations. Patients with PDAC with measurable locally advanced/metastatic lesions were enrolled in this trial. Nineteen subjects (16 had germline mutations and three had somatic mutations) received oral rucaparib (600 mg twice daily) after the administration of one-to-two prior chemotherapy regimens. Two partial responses and one complete response (CR) were confirmed (objective response rate, 15.8%; 3 out of 19 cases). The disease control rate (CR, partial response, or stable disease for ≥12 weeks) was 31.6% (6 out of 19 cases). This study provided evidence that rucaparib has an acceptable safety profile and is beneficial for patients with advanced PDAC [36].
At present these inhibitors are used for clinical trials for PDACs. Moreover, some of them are also useful for the treatment of breast cancers. Because BRCA1/2 mutations and the PARP activities are closely related to each other, PARP inhibitors are useful for the treatment of PDACs. Trials in combination with other treatments should be explored further.
3. TP53 Tumor Suppressor Gene
There is no doubt that the TP53 tumor suppressor gene is one of the most important genes in many cancers. It is mutated in nearly 50% of human cancers, including PDAC (mutation frequency, 58.7%), esophageal squamous cell carcinoma (93.7%), invasive breast cancer (32.7%), and non-small-cell lung cancer (66.5%) [37,38]. Patients with Li–Fraumeni syndrome who carry TP53 gene mutations in germline cells may have various cancers during their lifetime, with onset of the cancer at a younger age compared with the average patients [39,40]. However, the majority of TP53 gene dysfunctions in sporadic cancers are point or missense mutations triggered by UV light, aflatoxins, smoking, or other environmental factors. Because of the high frequency of TP53 mutation in many types of cancers, therapeutic strategies targeting mutant TP53 have attracted great interest [38].
3.1. Gain-of-Function Mutant p53 (mutp53)
Mutant p53 proteins, which cannot activate the mouse double minute 2 homolog (MDM2), prolong the half-life of, and become gain-of-function (GOF) molecules in mutp53-harboring cancer cells. These GOF activities have tremendous effects on many important pathways, such as metastasis, proliferation rate, apoptosis/drug resistance, stem cells, chronic inflammation, genomic instability, and metabolism (see Figure 4 and Table 5). These mutp53 proteins function by reversing the effects of each fundamental reaction of cells, to inactivate cell proliferation, differentiation, reprogramming, stem cell function, cell cycle, and apoptosis, as well as metabolic control.
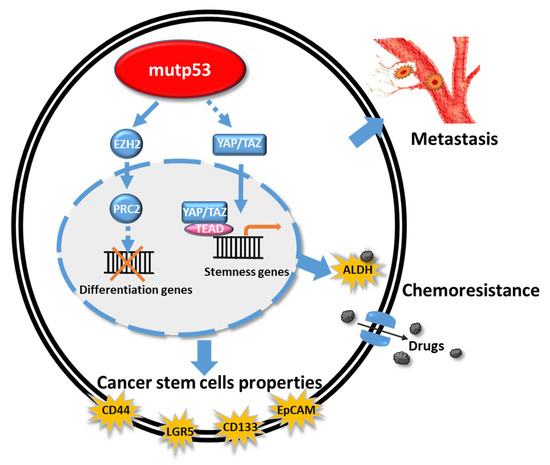
Figure 4.
Schematic representation of the effects of mutp53 on cancer-stem-cell–related genes and chemoresistance function. Mutp53 induces EZH2 to block the function of differentiation genes and triggers YAP/TAZ signaling to upregulate stemness genes, which are induced to chemoresistance-related genes. Mutp53 also induces the cancer stem cell related genes, such as CD44, LGR5, CD133, and EpCAM, which might induce the metastasis and cancer progression.

Table 5.
Models of the multifunctionality of mutant p53. Mut-p53 induces various biological functions, such as chronic inflammation, deregulation of cellular metabolism, high proliferation rate, genomic instability, metastasis, resistance to apoptosis/therapeutic drugs, and stem cells. The figure is adapted and modified to show the various biological functions induced by mutp53 [38].
Donehower et al., reported that GOF mutp53 proteins can activate ~500 downstream genes involving several pathways that mainly regulate cell-cycle arrest, apoptosis, senescence, DNA repair, and genetic stability [41]. GOF mutp53 proteins interact with NF-kB, HIF-1alpha, SP1, Twist1, E2F, or SREBP1 via protein–protein interactions, to inhibit p63 and p73 activities. Di Agostino et al., demonstrated that the mutp53/NF-Y protein complex can aberrantly recruit p300 instead of histone deacetylases, eventually leading to an opposite cell-fate outcome [42]. GOF mutp53 can render cancer cells more aggressive, with aneuploidy, higher proliferation rate, and cancer stem cell phenotypes, such as stem cell markers and resistance to apoptosis and therapeutic drugs [43]. In addition, mutp53 contributes to chronic inflammation and angiogenesis in the cancer cell microenvironment, and eventually enhances distal metastasis. For example, 80% of cases of basal/triple-negative breast cancer have p53 mutations [44]. GOF mutp53 proteins reprogram metabolic and anabolic pathways and enhance the Warburg effect by increasing glucose uptake and lactate formation, despite the presence of functioning mitochondria and oxygen.
Because p53 mutation is one of the most common genetic alterations in cancers, approaches aimed at restoring “the tumor-suppressive function of wild-type p53 (wtp53)” are urgent and necessary. In turn, this can enhance chemo-radiotherapy sensitivities in mutp53-harboring cancer cells. However, the development of a strategy that effectively targets the GOF mutp53 proteins is challenging and difficult, mainly because a vast number of genes and pathways are altered by these proteins.
3.1.1. Hotspots of mutp53
The six most common mutation hotspots of mutp53 have been identified, including the R175H, G245S, R248Q, R248W, R273H, and R282H residues. These hotspots account for over 28% of the total p53 mutations in various cancers [45]. GOF mutp53 proteins exhibit an altered structural conformation (structural mutations, such as R175H) or reduced capacity of binding to DNA (contact mutations, such as R273H) [46,47]. GOF mutp53 proteins not only lose their original wild-type tumor-suppressive function, but also accumulate to very high levels and exhibit GOF activities in mutp53-harboring cancer cells.
3.1.2. Restoring wtp53 Function
Several small molecules can bind to mutp53 proteins and change their structural conformation to allow binding to DNA in the same manner as does the wtp53 protein (i.e., they can restore wtp53 protein function).
CP-31398
This synthetic small-molecule compound can restore the wtp53 transcription function and induce apoptosis in cancer cells by stabilizing the mutant protein. However, its mechanisms are controversial [48,49].
STIMA-1
Zache et al. [50] found that this low-molecular-weight compound has some structural similarities to CP-31398, stimulates mutant p53 DNA binding in vitro, induces the expression of p53 target proteins, and triggers apoptosis in mutant-p53-harboring tumor cells.
PRIMA-1 and APR-246
PRIMA-1 and its analog PRIMA-1MET, now termed APR-246 [51], can reactivate mutp53 and induce a wtp53 biological response, such as apoptosis, in tumor cells, thus inhibiting tumor growth in mice. Both PRIMA-1 and APR-246 have been tested in clinical trials that included patients with hematological malignancies or hormone-refractory prostate cancer (www.clinicaltrials.gov; NCT03268382, NCT03931291, NCT04214860) [52].
p53R3
p53R3 is a p53 rescue compound that inhibits the proliferation of cancer cells expressing mutp53 by inducing the expression of p53 target genes, including p21Cip1, PUMA, and BAX, to induce cell-cycle arrest and apoptosis in cancer cells [53].
PK083 and PK7088
PK083 and PK7088 were designed to bind the cavity created by mutp53Y220C. The Y220C mutation of p53 is an excellent “druggable” target [54,55]. Both PK083 and PK7088 bind to the Y220C mutant, restore the wtp53 conformation, and induce Y220C-dependent cell-cycle arrest and apoptosis [56,57].
RITA
RITA is another compound that can reactivate several mutant p53 proteins, such as those carrying the R175H, R248W, and R273H mutations [58].
Chetomin
Chetomin is a small molecule that can specifically reactivate the mutp53R175H protein to the wild-type p53 conformation by increasing its binding capacity through HSP40. This drug selectively inhibits the growth of cancer cells harboring mutp53R175H, but not of those carrying mutp53R273H [59]. Furthermore, it can also enhance the radiosensitivity of cancer cells, regardless of p53 status. The underlying mechanism occurs via interference with the hypoxia-inducible factor (HIF) pathway [60,61,62].
Phenethyl Isothiocyanate
Phenethyl isothiocyanate (PEI), which is a natural dietary-related compound that is present in cruciferous vegetables, can restore the wild-type conformation and transcriptional activity of mutp53R175H, sensitize mutp53R175H to proteasomal degradation, and have a growth inhibitory effect on cancer cells expressing mutp53R175H. Dietary supplementation with PEI led to the reactivation of wtp53 activity in vivo and the inhibition of tumor growth in a xenograft mouse model. This represents the first example of mutant p53 reactivation by a dietary compound and may have important implications for cancer prevention and therapy [63].
3.1.3. Zinc-Based Therapy
Zinc is a known regulator of p53 that is essential for its correct binding to target genes. Adding zinc to the mutp53 protein can reduce the effect of substitutions (G245C/G245D) on conformational changes [64]. The combination of adriamycin with zinc inhibited tumor growth in the transgenic MMTI-neu murine breast cancer model [65].
NSC319726/ZMC1
NSC319726/ZMC1 activates the allele-specific mutp53R175 and restores its wild-type structure and function [66]. The p53R175 mutation is the third most frequent missense mutation of this protein. The potential number of patients who may benefit from this compound is estimated at 32,000 per year in the United States [67]. NSC319726, as a p53R175 mutant reactivator, does not bind to mutp53; rather, it increases the intracellular zinc concentration and enhances the folding of R175H mutants.
COTI-2
COTI-2, which is a third-generation thiosemicarbazone compound, can chelate zinc ions and reactivate mutant p53 to its wild-type form. COTI-2 was shown to restore the normal DNA-binding properties of the p53 mutant protein through p53-dependent and -independent mechanisms [68].
It is evident that p53 mutation is one of the fundamental genetic mutations that trigger cancers. Even hemi-allelic mutation initiates the cancer program together with other critical mutations of cancer genes. Thus, recovering the mutation phenotypes to the normal wtp53 presentation is the main target in the prevention of p53-dependent cancers. Further trials targeting GOF mutp53 proteins are required.
4. GOF mutTP53 Proteins and Cancer Stem Cell Phenotypes
GOF of mutp53 proteins have pinpointed the critical steps that are involved in cancer-stem-cell-related genes and chemoresistance.
GOF mutp53 proteins involve a broad spectrum of mechanisms of chemoradio resistance, including resistance to apoptosis, autophagy inhibition, metabolic reprogramming, and increased expression of drug efflux pumps [66]. GOF mutp53 proteins favor self-renewal pathways by increasing EZH2 and YAP/TAZ activity and enhancing the surface expression of cancer stem cell markers, such as CD44, CD133, LGR5, and ALDH [69]. All these factors are important for cancer stem cells (CSCs), which are believed to be the origin of many cancers. Thus, GOF mutp53 plays an important role in CSC formation. Hassen et al., showed that Kras and the mutated GOF p53 are the main drivers of PDAC aggressiveness [70]. Mutant p53 and CREB1 upregulate the FOXA1 transcription factor and promote Wnt–β-catenin signaling to drive tumor metastasis [71]. Capaci et al., reported that the mutp53/HIF1α/miR-30d axis can potentiate the release of the soluble extracellular secretome to remodel the extracellular matrix, thus favoring cancer cell growth and metastatic colonization [72].
Thus, the interrelationship between the signaling of GOF mutp53 and chromatin modifiers, such as PRC2 and EZH2, and the molecular mechanisms underlying the generation of the cancer stem cell markers CD44, CD133, LGR5, and ALDH, should be clarified, and other signaling pathways, such as Wnt–β-catenin and FOXA1 should be elucidated. These molecular signaling pathways of CSCs are also the targets of future clinical research.
5. Targeting Mutant p53 Protein Stability
Mutant p53 genes are pro-oncogenic drivers [73,74,75], and cancer cells rely on these oncogenes for survival or growth [76]. Reducing mutp53 protein stability leads to their degradation, which in turn leads to cancer cell death. Heat-shock proteins, such as HSP70 and HSP90, can bind to mutp53 proteins to form a chaperone complex. This complex can inactivate endogenous MDM2 and the carboxy terminus of the HSP70-interacting protein (CHIP), resulting in mutp53 protein accumulation in human cancer cells. The pharmacological inhibitor 17-allylamino-17-demethoxygeldanamycin (17AAG) can destroy this chaperone complex, liberate mutp53 proteins, and reactivate endogenous MDM2 and CHIP, to degrade mutp53 proteins [77] (Figure 5).
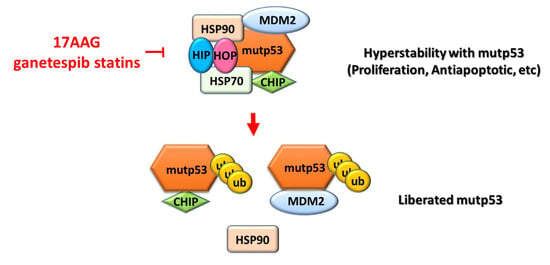
Figure 5.
Schematic modeling of the modulation of mutp53 stability by the HSP90 multichaperone machinery in cancer cells. Heat-shock proteins (such as HSP70 and HSP90) bind to mutp53 proteins to form a chaperone complex [77]. This complex can be inactivated by endogenous MDM2 and the carboxy terminus of the HSP70-interacting protein (CHIP), resulting in the accumulation of mutp53 proteins in human cancer cells. The pharmacological inhibitor 17-allylamino-17-demethoxygeldanamycin (17AAG) can destroy this chaperone complex, liberate mutp53 proteins, and reactivate endogenous MDM2 and CHIP, to degrade mutp53 proteins [77]. CHIP, carboxy terminus of the Hsc70-interacting protein; HIP, heat-shock protein (HSP) 70 co-chaperone; HOP, HSP70/90 organizing protein; Ub, ubiquitinated proteins.
5.1. Geldanamycin
Geldanamycin, which is a highly specific HSP90 inhibitor, decreases the intracellular levels of mutp53 proteins via MDM2 degradation [78]. Because HSPs also regulate wtp53, HSP inhibitors have not received FDA approval for entering clinical trials.
5.2. Ganetespib
Alexandrova et al. [79] reported that ganetespib, which is a potent HSP90 inhibitor, can significantly extend the lifespan of homozygous mutp53R172H mice.
5.3. Alvespimycin Plus SAHA
Similarly, a combination of alvespimycin (17DMAG) and SAHA (histone deacetylase inhibitor) effectively induces mutp53 protein degradation and tumor necrosis and prolongs the lifespan of mutp53R172H mice [79].
The stability of mutp53 proteins is crucial for the progression of many cancers including PDACs. HSPs, MDM2/4, and the CHIP chaperone are among these regulators and might be possible targets for anti-cancer therapeutics.
6. Synthetic Lethality of p53 Loss
Mutp53-harboring cancer cells usually lack G1/S checkpoints and become more dependent on G2/M checkpoints to survive after DNA damage. Therefore, inhibition of G2/M checkpoint regulators, such as CHK1/2 and WEE1, has been reported to induce a mitotic catastrophe and synthetic lethality in mutp53-harboring cancer cells. ARK/CHK1 inhibitors, such as UCN-01, PF477736, and AZD-7762, may potentiate the cytotoxicity of chemoradio-genotoxic effects in mutp53-harboring human cancers. More than 70 clinical trials (various chemoradiotherapy regimens combined with different ARK/CHK1 inhibitors) have been carried out and reviewed in this context [80]. A combination of gemcitabine or olaparib (PARP1 inhibitor) with a potent WEE1 inhibitor (AZD1775) yielded synergistic lethal effects on ovarian and endometrial mutp53-harboring cancer cells [81]. A phase II clinical trial has proven that AZD1775 enhances carboplatin efficacy in patients with mutp53 ovarian cancer who were refractory to first-line platinum-based therapy [82]. Recently, Hartman et al., reported the in vivo and in vitro anti-tumor effects of the combination of AZD1775 with irinotecan or capecitabine/5-FU in patients with PDAC using a patient-derived xenograft model [83]. Xiao et al., combined a chemotherapeutic agent with a PARP inhibitor and showed their synergistic cytotoxicity in GOF mutp53 (particularly mutp53R273H) breast cancer cells [84]. Wei et al., also reported recently that a combination of gemcitabine with a protein arginine methyltransferase 5 (PRMT5) inhibitor yielded a synergistic lethal effect on a patient-derived xenograft PDAC model [85]. Thus, mutp53 and G2/M checkpoints for mitotic inhibition and the synthetic lethality of p53 loss are crucial for understanding cancer development.
7. Immunoregulation of the Microenvironment of PDAC
7.1. PD-1/PD-L1 in PDAC
Previous studies showed that immunotherapy with PD-1/PD-L1 failed to exert any effects on PDAC. The microenvironment of PDAC is not immunogenic or immunosuppressive [86]. In general, pancreatic cancer produces a local and systemic immune dysfunction or immunosuppression to avoid recognition and attachment by effective immune-competent cells. The microenvironment of cancers exhibits a lower number of tumor-infiltrating lymphocytes, and dendritic cells and many suppressor T cells [87]. Tumor cells use mechanisms that act via the PD-L1 or CTLA-4 program, a blockage of co-stimulation to activate T cells, and the recruitment of tumor-associated macrophages and marrow-derived suppressor cells (MDSCs), to achieve immune suppression [88]. Recently, Amin et al., reported that the addition of immunotherapies to adjuvant chemotherapy improved survival compared with chemotherapy alone after curative-intent resection of PDAC [89]. Cao et al., reported that p53-mediated PD-1 activation is involved in tumor suppression in an immunity-independent manner [90].
7.2. Mutant p53 GOF Mechanisms via the Shedding of the Tumor-Promoting Secretome (Including Exosomes)
Mutp53-harboring cancer cells secrete numerous extracellular factors (secretomes) to create a supportive microenvironment for their progression. These factors can be either soluble or contained within vesicles (such as exosomes). Cooks et al., reported that a mutp53-bearing colon cancer secreted exosomes enriched in miR-1246. These exosomes were taken up by nearby macrophages and reprogrammed them into a tumor-promoting M2 status [91]. Tran et al., isolated tumor-infiltrating lymphocytes (TILs) from a patient with metastatic cholangiocarcinoma containing CD4+ helper T cells that could specifically recognize a patient-specific mutant protein, the erbb2-interacting protein (ERBB2IPE805G). This immune response was presented by HLA-DQB1*0601, and the minimal neoepitope was located within the following 13-amino-acid sequence: NSKEETGHLENGN (where E is Glu; G, Gly; H, His; K, Lys; L, Leu; N, Asn; S, Ser; and T, Thr). The patient received two courses of this patient-mutant-specific helper-T cell therapy and exhibited tumor shrinkage. Three lung metastases, which were resected nearly a half year after this immunotherapy, were infiltrated by the ERBB2IP mutation-reactive T cells, suggesting that they contributed to cancer regression and stabilization of disease [92].
7.3. Neoepitopes from mutp53 Proteins Are Recognized by TCRs on CD8+ T Cells
To date, no ideal tumor-associated antigens have been identified, either localized within cells or mounted on the surfaces of normal cells. Small peptides (neoantigens) degraded from the mutp53 protein are secreted (as secretomes) from cancer cells, to be taken up by surrounding T cells. Previous experiments have shown that these small mutant peptides (neoantigens, about 10–15-peptide-containing mutated neoepitopes) are highly immunogenic and trigger the killing of cancer cells via both CD8+ and CD4+ immune responses [91]. These small peptides (neoantigens) are taken up by antigen-presenting cells. and formed an HLA/MHC1-peptide complex that is recognized by T-cell receptors (TCRs) expressed on the surface of CD8+ T cells. Interestingly, small peptides derived from hot-spot-mutp53 proteins can become “public neoantigens” [91]; i.e., hotspots carrying mutp53 genes and encoding these hotspot-mutant peptides frequently occur in many human cancers. These neoepitopes derived from hot-spot-mutp53 proteins can become “public” neoantigens [93].
7.4. CAR-T Cells Promote T-Cell Expansion to Promote Anti-Tumor Function
A comprehensive picture of the GOF of p53 is necessary to achieve personalized cancer treatment. Using the NGS method, physicians can now determine the landscape of the whole-genome data of patients. Pavlakis and Stieve [94] proposed that mutp53-harboring cancer cells secrete numerous secretomes (including miRNA-enriched exosomes) to recruit different immune/stellate cells and remodel the microenvironment to favor tumor promotion. In contrast, these recruited inflammatory and immune cells can also affect and shape the GOF activities of mutant p53 within cancer cells [95]. In the past decade, adoptive T-cell therapies (ACTs) have yielded excellent results for chronic lymphocytic leukemia [96]. Those authors isolated endogenous TILs for ex vivo expansion, genetically engineered TCRs, or chimeric antigen receptors (CARs) against cancer-specific antigens. Fifth-generation CAR-T cells were designed to activate the JAK–STAT pathway and promote T-cell expansion, which afforded a better antitumor effect [97]. A phase 1 clinical trial has demonstrated that anti-EGFR CAR-T treatment is safe and effective in patients with metastatic PDAC (median overall survival, 4.9 months; range, 2.9–30 months) [98]. The potential obstacles for ACT in PDAC include appropriate ways to deliver CAR-T cells to fibrotic, immunosuppressive cancer environments. However, whether mutp53 neoantigen-specific T cells can cause solid tumor regression has yet to be determined. These trials of cancer immunotherapies are also critical for developing new therapeutics for PDACs.
8. MDM2–MDMX(MDM4)–p53 Axis
Normal cells contain a lower level of p53 proteins because of their short half-life, which is modulated by a ring finger E3 ubiquitin ligase termed MDM2. Moreover, MDMX(MDM4), an analog of MDM2, enhances the activity of MDM2 further to polyubiquitinate p53 by forming a complex with MDM2 [99]. The mutual dependence of MDM2 and MDMX regarding their p53-inactivation function, as well as their essential roles in controlling p53 levels and activity in vivo, have been reported [100]. The target genes under the control of the MDM2–MDMX–p53 loop are primarily critical for carcinogenesis. Recently, small molecules, such as protoporphyrin IX (PpIX), have been used to target the p53/MDM2 and p53/MDMX interactions and promote alternatives to target wt-p53-bearing tumors, such as pancreatic carcinoma [101]. Therefore, the MDM2–MDMX–p53 pathway is subjected to multiple layers of control in response to various stress signals and cancers, including PDACs.
9. Future Perspectives
One of the possible interesting technologies for the treatment of solid tumors, including PDACs, is the liquid biopsy (LB) to obtain information for diagnostic, prognostic, and predictive purposes in the near future [102,103]. However, to date, this approach has been used in breast cancer, colorectal cancer, and melanoma exclusively. This new technique is emerging as an identification clinical tool. In current clinical practice, LB is used for the identification of driver mutations in the circulating DNA derived from both tumors and circulating neoplastic cells. Liquid derivatives other than LBs, such as circulating tumor cells, circulating tumor RNA/DNA, microRNA, platelets, and extracellular vesicles, as well as other biofluids, such as urine and cerebrospinal fluid, may be adopted to detect mutations of tumor suppressor genes and their related genes in the future.
10. Conclusions
This review focused on novel therapies to treat patients carrying mutated TSGs. However, the success of these treatments remains low. Collisson et al. [104] showed that there is a human QM-PDAC (quasi-mesenchymal) subtype in which the genetic alterations were similar to those of cell lines obtained from genetically engineered p53Lox/+ mice (lacking the tumor suppressor p53 gene). Bailey et al., reported that the “squamous” subtypes of PDAC are more aggressive, metastatic, and undifferentiated tumors, because enriched TP53 mutations were detected in this subtype [105].
The laser-capture microdissection technique was used to dissect purified PDAC cancer tissues, to study whole genomes and perform transcriptome analysis. Chan-Seng-Yue et al. [106] proposed two subtypes: the classical and basal subtypes. The basal-like-A phenotype (high squamous signature) is linked to major K-ras imbalances in late-stage disease. Conversely, the classical subtype is believed to have a better response to a chemotherapy regimen (mFOLFIRINOX), as well as a better prognosis. It is currently undergoing a clinical trial known as NCT04683315.
Genetic studies of patients with breast cancer yielded similar results. Most mutTP53 clusters were detected in the basal-like subtype of breast cancer, which is chemoresistant and has the worst prognosis [107]. Mutant p53 activates lipid metabolism in tumors; moreover, it binds to and activates a series of transcription factors, the sterol regulatory element-binding proteins, and induces the expression of many genes in the mevalonate pathway, which leads to the disruption of breast cancer cell architecture in a 3D culture model and contributes to increased proliferation, survival, invasion, and metastasis [107].
p53 is the master regulator of tumor suppressor genes, and the GOF mutp53 functions not only to generate cancer cells, but also to promote a higher proliferation capability by triggering EMT and promoting chemoradio resistance. Furthermore, its secretome remodels the extracellular matrix and creates an immune-suppressive microenvironment that promotes the migration and metastasis of cancer cells. A subtype of cancer cells acquire the GOF mutp53-specific characteristics of “cancer stem cells,” which is thought to be responsible for carcinogenesis and chemoradioresistance. Therefore, approaches aimed at eradicating these frequently encountered “mut-TP53-harboring cancer cells” are an important clinical issue for future studies. Understanding these issues is crucial for improving the current cancer treatments.
Author Contributions
K.-K.K., Y.-H.Y., D.-C.W., and K.K.Y. wrote the draft of the manuscript. P.-J.H., W.-T.C., S.-C.C., K.W., C.-C.K., J.-B.P., C.-P.L., K.K., and C.-J.L. critically reviewed the manuscript. All authors approved the final version of the manuscript and agreed to be accountable for all aspects of the work by ensuring that questions related to the accuracy or integrity of any part of the work were appropriately investigated and resolved. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Ministry of Science and Technology (MOST 109-2314-B-037-119; MOST 109-2320-B-037-033), the National Health Research Institutes (NHRI-EX109-10720SI), Kaohsiung Medical University Hospital (SA10803C), and Kaohsiung Medical University Research Center (KMU-TC109A02; KMU-TC109A02-0, KMU-TC109A02-3).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cer statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galvano, A.; Castiglia, M.; Rizzo, S.; Silvestris, N.; Brunetti, O.; Vaccaro, G.; Gristina, V.; Barraco, N.; Bono, M.; Guercio, G.; et al. Moving the Target on the Optimal Adjuvant Strategy for Resected Pancreatic Cancers: A Systematic Review with Meta-Analysis. Cancers 2020, 12, 534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Roessel, S.; van Veldhuisen, E.; Klompmaker, S.; Janssen, Q.P.; Abu Hilal, M.; Alseidi, A.; Balduzzi, A.; Balzano, G.; Bassi, C.; Berrevoet, F.; et al. Evaluation of Adjuvant Chemotherapy in Patients with Resected Pancreatic Cancer After Neoadjuvant FOLFIRINOX Treatment. JAMA Oncol. 2020, 6, 1733–1740. [Google Scholar] [CrossRef]
- Gamboa, A.C.; Rupji, M.; Switchenko, J.M.; Lee, R.M.; Turgeon, M.K.; Meyer, B.I.; Russell, M.C.; Cardona, K.; Kooby, D.A.; Maithel, S.K.; et al. Optimal timing and treatment strategy for pancreatic cancer. J. Surg. Oncol. 2020, 122, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, G.; Daye, D.; Wilson, N.M.; Noman, R.; Mahendra, A.M.; Doshi, M.H. Ablation in Pancreatic Cancer: Past, Present and Future. Cancers 2021, 13, 2511. [Google Scholar] [CrossRef]
- Hingorani, S.R.; Wang, L.; Multani, A.S.; Combs, C.; Deramaudt, T.B.; Hruban, R.H.; Rustgi, A.K.; Chang, S.; Tuveson, D.A. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005, 7, 469–483. [Google Scholar] [CrossRef] [Green Version]
- Ponz-Sarvise, M.; Tuveson, D.A.; Yu, K.H. Mouse Models of Pancreatic Ductal Adenocarcinoma. Hematol. Oncol. Clin. N. Am. 2015, 29, 609–617. [Google Scholar] [CrossRef]
- Kuo, K.K.; Kuo, C.J.; Chiu, C.Y.; Liang, S.S.; Huang, C.H.; Chi, S.W.; Tsai, K.B.; Chen, C.Y.; Hsi, E.; Cheng, K.H.; et al. Quantitative Proteomic Analysis of Differentially Expressed Protein Profiles Involved in Pancreatic Ductal Adenocarcinoma. Pancreas 2016, 45, 71–83. [Google Scholar] [CrossRef] [Green Version]
- Zhen, D.B.; Rabe, K.G.; Gallinger, S.; Syngal, S.; Schwartz, A.G.; Goggins, M.G.; Hruban, R.H.; Cote, M.L.; McWilliams, R.R.; Roberts, N.J.; et al. BRCA1, BRCA2, PALB2, and CDKN2A mutations in familial pancreatic cancer: A PACGENE study. Genet. Med. 2015, 17, 569–577. [Google Scholar] [CrossRef] [Green Version]
- Axilbund, J.E.; Wiley, E.A. Genetic testing by cancer site: Pancreas. Cancer J. 2012, 18, 350–354. [Google Scholar] [CrossRef]
- Klein, A.P. Genetic susceptibility to pancreatic cancer. Mol. Carcinog. 2012, 51, 14–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, C.; Hong, S.M.; Lim, P.; Kamiyama, H.; Khan, M.; Anders, R.A.; Goggins, M.; Hruban, R.H.; Eshleman, J.R. KRAS2 mutations in human pancreatic acinar-ductal metaplastic lesions are limited to those with PanIN: Implications for the human pancreatic cancer cell of origin. Mol. Cancer Res. 2009, 7, 230–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mueller, S.; Engleitner, T.; Maresch, R.; Zukowska, M.; Lange, S.; Kaltenbacher, T.; Konukiewitz, B.; Ollinger, R.; Zwiebel, M.; Strong, A.; et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature 2018, 554, 62–68. [Google Scholar] [CrossRef]
- Habbe, N.; Shi, G.; Meguid, R.A.; Fendrich, V.; Esni, F.; Chen, H.; Feldmann, G.; Stoffers, D.A.; Konieczny, S.F.; Leach, S.D.; et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc. Natl. Acad. Sci. USA 2008, 105, 18913–18918. [Google Scholar] [CrossRef] [Green Version]
- Furukawa, T.; Sunamura, M.; Horii, A. Molecular mechanisms of pancreatic carcinogenesis. Cancer Sci. 2006, 97, 1–7. [Google Scholar] [CrossRef]
- Furukawa, T.; Sunamura, M.; Motoi, F.; Matsuno, S.; Horii, A. Potential tumor suppressive pathway involving DUSP6/MKP-3 in pancreatic cancer. Am. J. Pathol. 2003, 162, 1807–1815. [Google Scholar] [CrossRef] [Green Version]
- Bardeesy, N.; Aguirre, A.J.; Chu, G.C.; Cheng, K.H.; Lopez, L.V.; Hezel, A.F.; Feng, B.; Brennan, C.; Weissleder, R.; Mahmood, U.; et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc. Natl. Acad. Sci. USA 2006, 103, 5947–5952. [Google Scholar] [CrossRef] [Green Version]
- Lang, G.A.; Iwakuma, T.; Suh, Y.A.; Liu, G.; Rao, V.A.; Parant, J.M.; Valentin-Vega, Y.A.; Terzian, T.; Caldwell, L.C.; Strong, L.C.; et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 2004, 119, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguirre, A.J.; Bardeesy, N.; Sinha, M.; Lopez, L.; Tuveson, D.A.; Horner, J.; Redston, M.S.; DePinho, R.A. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003, 17, 3112–3126. [Google Scholar] [CrossRef] [Green Version]
- Biankin, A.V.; Waddell, N.; Kassahn, K.S.; Gingras, M.C.; Muthuswamy, L.B.; Johns, A.L.; Miller, D.K.; Wilson, P.J.; Patch, A.M.; Wu, J.; et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012, 491, 399–405. [Google Scholar] [CrossRef]
- Singhi, A.D.; George, B.; Greenbowe, J.R.; Chung, J.; Suh, J.; Maitra, A.; Klempner, S.J.; Hendifar, A.; Milind, J.M.; Golan, T.; et al. Real-Time Targeted Genome Profile Analysis of Pancreatic Ductal Adenocarcinomas Identifies Genetic Alterations That Might Be Targeted with Existing Drugs or Used as Biomarkers. Gastroenterology 2019, 156, 2242–2253. [Google Scholar] [CrossRef] [Green Version]
- Jones, S.; Zhang, X.; Parsons, D.W.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Kamiyama, H.; Jimeno, A.; et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008, 321, 1801–1806. [Google Scholar] [CrossRef] [Green Version]
- Jones, S.; Hruban, R.H.; Kamiyama, M.; Borges, M.; Zhang, X.; Parsons, D.W.; Lin, J.C.; Palmisano, E.; Brune, K.; Jaffee, E.M.; et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science 2009, 324, 217. [Google Scholar] [CrossRef] [Green Version]
- Win, A.K.; Young, J.P.; Lindor, N.M.; Tucker, K.M.; Ahnen, D.J.; Young, G.P.; Buchanan, D.D.; Clendenning, M.; Giles, G.G.; Winship, I.; et al. Colorectal and other cancer risks for carriers and noncarriers from families with a DNA mismatch repair gene mutation: A prospective cohort study. J. Clin. Oncol. 2012, 30, 958–964. [Google Scholar] [CrossRef] [Green Version]
- Grant, R.C.; Selander, I.; Connor, A.A.; Selvarajah, S.; Borgida, A.; Briollais, L.; Petersen, G.M.; Lerner-Ellis, J.; Holter, S.; Gallinger, S. Prevalence of germline mutations in cancer predisposition genes in patients with pancreatic cancer. Gastroenterology 2015, 148, 556–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaffee, K.G.; Oberg, A.L.; McWilliams, R.R.; Majithia, N.; Allen, B.A.; Kidd, J.; Singh, N.; Hartman, A.R.; Wenstrup, R.J.; Petersen, G.M. Prevalence of germ-line mutations in cancer genes among pancreatic cancer patients with a positive family history. Genet. Med. 2018, 20, 119–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, G.M.; Chaffee, K.G.; McWilliams, R.R.; Majithia, N.; Allen, B.; Kidd, J.; Singh, N.; Hartman, A.-R.; Oberg, A.L. Genetic heterogeneity and survival among pancreatic adenocarcinoma (PDAC) patients with positive family history. J. Clin. Oncol. 2016, 34, 4108. [Google Scholar] [CrossRef]
- Miki, Y.; Swensen, J.; Shattuck-Eidens, D.; Futreal, P.A.; Harshman, K.; Tavtigian, S.; Liu, Q.; Cochran, C.; Bennett, L.M.; Ding, W.; et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994, 266, 66–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wooster, R.; Bignell, G.; Lancaster, J.; Swift, S.; Seal, S.; Mangion, J.; Collins, N.; Gregory, S.; Gumbs, C.; Micklem, G. Identification of the breast cancer susceptibility gene BRCA2. Nature 1995, 378, 789–792. [Google Scholar] [CrossRef]
- Hahn, S.A.; Greenhalf, B.; Ellis, I.; Sina-Frey, M.; Rieder, H.; Korte, B.; Gerdes, B.; Kress, R.; Ziegler, A.; Raeburn, J.A.; et al. BRCA2 germline mutations in familial pancreatic carcinoma. J. Natl. Cancer Inst. 2003, 95, 214–221. [Google Scholar] [CrossRef]
- Murphy, K.M.; Brune, K.A.; Griffin, C.; Sollenberger, J.E.; Petersen, G.M.; Bansal, R.; Hruban, R.H.; Kern, S.E. Evaluation of candidate genes MAP2K4, MADH4, ACVR1B, and BRCA2 in familial pancreatic cancer: Deleterious BRCA2 mutations in 17%. Cancer Res. 2002, 62, 3789–3793. [Google Scholar] [PubMed]
- Golan, T.; Hammel, P.; Reni, M.; Van Cutsem, E.; Macarulla, T.; Hall, M.J.; Park, J.O.; Hochhauser, D.; Arnold, D.; Oh, D.Y.; et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N. Engl. J. Med. 2019, 381, 317–327. [Google Scholar] [CrossRef]
- Tuli, R.; Shiao, S.L.; Nissen, N.; Tighiouart, M.; Kim, S.; Osipov, A.; Bryant, M.; Ristow, L.; Placencio-Hickok, V.; Hoffman, D.; et al. A phase 1 study of veliparib, a PARP-1/2 inhibitor, with gemcitabine and radiotherapy in locally advanced pancreatic cancer. EBioMedicine 2019, 40, 375–381. [Google Scholar] [CrossRef] [Green Version]
- de Bono, J.; Ramanathan, R.K.; Mina, L.; Chugh, R.; Glaspy, J.; Rafii, S.; Kaye, S.; Sachdev, J.; Heymach, J.; Smith, D.C.; et al. Phase I, Dose-Escalation, Two-Part Trial of the PARP Inhibitor Talazoparib in Patients with Advanced Germline BRCA1/2 Mutations and Selected Sporadic Cancers. Cancer Discov. 2017, 7, 620–629. [Google Scholar] [CrossRef] [Green Version]
- Shroff, R.T.; Hendifar, A.; McWilliams, R.R.; Geva, R.; Epelbaum, R.; Rolfe, L.; Goble, S.; Lin, K.K.; Biankin, A.V.; Giordano, H.; et al. Rucaparib Monotherapy in Patients with Pancreatic Cancer and a Known Deleterious BRCA Mutation. JCO Precis. Oncol. 2018, 2, 1–15. [Google Scholar] [CrossRef]
- Aubrey, B.J.; Kelly, G.L.; Janic, A.; Herold, M.J.; Strasser, A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018, 25, 104–113. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Hao, Q.; Lu, H. Mutant p53 in cancer therapy-the barrier or the path. J. Mol. Cell Biol. 2019, 11, 293–305. [Google Scholar] [CrossRef] [Green Version]
- Malkin, D.; Li, F.P.; Strong, L.C.; Fraumeni, J.F., Jr.; Nelson, C.E.; Kim, D.H.; Kassel, J.; Gryka, M.A.; Bischoff, F.Z.; Tainsky, M.A.; et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 1990, 250, 1233–1238. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Zou, Z.Q.; Pirollo, K.; Blattner, W.; Chang, E.H. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature 1990, 348, 747–749. [Google Scholar] [CrossRef] [PubMed]
- Donehower, L.A.; Soussi, T.; Korkut, A.; Liu, Y.; Schultz, A.; Cardenas, M.; Li, X.; Babur, O.; Hsu, T.K.; Lichtarge, O.; et al. Integrated Analysis of TP53 Gene and Pathway Alterations in The Cancer Genome Atlas. Cell Rep. 2019, 28, 3010. [Google Scholar] [CrossRef]
- Di Agostino, S.; Strano, S.; Emiliozzi, V.; Zerbini, V.; Mottolese, M.; Sacchi, A.; Blandino, G.; Piaggio, G. Gain of function of mutant p53: The mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell 2006, 10, 191–202. [Google Scholar] [CrossRef] [Green Version]
- Shetzer, Y.; Molchadsky, A.; Rotter, V. Oncogenic Mutant p53 Gain of Function Nourishes the Vicious Cycle of Tumor Development and Cancer Stem-Cell Formation. Cold Spring Harb. Perspect. Med. 2016, 6, a026203. [Google Scholar] [CrossRef]
- Synnott, N.C.; Murray, A.; McGowan, P.M.; Kiely, M.; Kiely, P.A.; O’Donovan, N.; O’Connor, D.P.; Gallagher, W.M.; Crown, J.; Duffy, M.J. Mutant p53: A novel target for the treatment of patients with triple-negative breast cancer? Int. J. Cancer 2017, 140, 234–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baugh, E.H.; Ke, H.; Levine, A.J.; Bonneau, R.A.; Chan, C.S. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 2018, 25, 154–160. [Google Scholar] [CrossRef]
- Mello, S.S.; Attardi, L.D. Not all p53 gain-of-function mutants are created equal. Cell Death Differ. 2013, 20, 855–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bullock, A.N.; Fersht, A.R. Rescuing the function of mutant p53. Nat. Rev. Cancer 2001, 1, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Takimoto, R.; Wang, W.; Dicker, D.T.; Rastinejad, F.; Lyssikatos, J.; el-Deiry, W.S. The mutant p53-conformation modifying drug, CP-31398, can induce apoptosis of human cancer cells and can stabilize wild-type p53 protein. Cancer Biol. Ther. 2002, 1, 47–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rippin, T.M.; Bykov, V.J.; Freund, S.M.; Selivanova, G.; Wiman, K.G.; Fersht, A.R. Characterization of the p53-rescue drug CP-31398 in vitro and in living cells. Oncogene 2002, 21, 2119–2129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zache, N.; Lambert, J.M.; Rokaeus, N.; Shen, J.; Hainaut, P.; Bergman, J.; Wiman, K.G.; Bykov, V.J. Mutant p53 targeting by the low molecular weight compound STIMA-1. Mol. Oncol. 2008, 2, 70–80. [Google Scholar] [CrossRef] [Green Version]
- Lambert, J.M.; Moshfegh, A.; Hainaut, P.; Wiman, K.G.; Bykov, V.J. Mutant p53 reactivation by PRIMA-1MET induces multiple signaling pathways converging on apoptosis. Oncogene 2010, 29, 1329–1338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bykov, V.J.; Wiman, K.G. Mutant p53 reactivation by small molecules makes its way to the clinic. FEBS Lett. 2014, 588, 2622–2627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinmann, L.; Wischhusen, J.; Demma, M.J.; Naumann, U.; Roth, P.; Dasmahapatra, B.; Weller, M. A novel p53 rescue compound induces p53-dependent growth arrest and sensitises glioma cells to Apo2L/TRAIL-induced apoptosis. Cell Death Differ. 2008, 15, 718–729. [Google Scholar] [CrossRef] [PubMed]
- Bromley, D.; Bauer, M.R.; Fersht, A.R.; Daggett, V. An in silico algorithm for identifying stabilizing pockets in proteins: Test case, the Y220C mutant of the p53 tumor suppressor protein. Protein Eng. Des. Sel. 2016, 29, 377–390. [Google Scholar] [CrossRef] [Green Version]
- Joerger, A.C.; Ang, H.C.; Fersht, A.R. Structural basis for understanding oncogenic p53 mutations and designing rescue drugs. Proc. Natl. Acad. Sci. USA 2006, 103, 15056–15061. [Google Scholar] [CrossRef] [Green Version]
- Boeckler, F.M.; Joerger, A.C.; Jaggi, G.; Rutherford, T.J.; Veprintsev, D.B.; Fersht, A.R. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc. Natl. Acad Sci. USA 2008, 105, 10360–10365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Wilcken, R.; Joerger, A.C.; Chuckowree, I.S.; Amin, J.; Spencer, J.; Fersht, A.R. Small molecule induced reactivation of mutant p53 in cancer cells. Nucleic Acids Res. 2013, 41, 6034–6044. [Google Scholar] [CrossRef]
- Burmakin, M.; Shi, Y.; Hedstrom, E.; Kogner, P.; Selivanova, G. Dual targeting of wild-type and mutant p53 by small molecule RITA results in the inhibition of N-Myc and key survival oncogenes and kills neuroblastoma cells in vivo and in vitro. Clin. Cancer Res. 2013, 19, 5092–5103. [Google Scholar] [CrossRef] [Green Version]
- Hiraki, M.; Hwang, S.Y.; Cao, S.; Ramadhar, T.R.; Byun, S.; Yoon, K.W.; Lee, J.H.; Chu, K.; Gurkar, A.U.; Kolev, V.; et al. Small-Molecule Reactivation of Mutant p53 to Wild-Type-like p53 through the p53-Hsp40 Regulatory Axis. Chem. Biol. 2015, 22, 1206–1216. [Google Scholar] [CrossRef] [Green Version]
- Kung, A.L.; Zabludoff, S.D.; France, D.S.; Freedman, S.J.; Tanner, E.A.; Vieira, A.; Cornell-Kennon, S.; Lee, J.; Wang, B.; Wang, J.; et al. Small molecule blockade of transcriptional coactivation of the hypoxia-inducible factor pathway. Cancer Cell 2004, 6, 33–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staab, A.; Loeffler, J.; Said, H.M.; Diehlmann, D.; Katzer, A.; Beyer, M.; Fleischer, M.; Schwab, F.; Baier, K.; Einsele, H.; et al. Effects of HIF-1 inhibition by chetomin on hypoxia-related transcription and radiosensitivity in HT 1080 human fibrosarcoma cells. BMC Cancer 2007, 7, 213. [Google Scholar] [CrossRef] [Green Version]
- Kessler, J.; Hahnel, A.; Wichmann, H.; Rot, S.; Kappler, M.; Bache, M.; Vordermark, D. HIF-1alpha inhibition by siRNA or chetomin in human malignant glioma cells: Effects on hypoxic radioresistance and monitoring via CA9 expression. BMC Cancer 2010, 10, 605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aggarwal, M.; Saxena, R.; Sinclair, E.; Fu, Y.; Jacobs, A.; Dyba, M.; Wang, X.; Cruz, I.; Berry, D.; Kallakury, B.; et al. Reactivation of mutant p53 by a dietary-related compound phenethyl isothiocyanate inhibits tumor growth. Cell Death Differ. 2016, 23, 1615–1627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pintus, S.S.; Ivanisenko, N.V.; Demenkov, P.S.; Ivanisenko, T.V.; Ramachandran, S.; Kolchanov, N.A.; Ivanisenko, V.A. The substitutions G245C and G245D in the Zn(2+)-binding pocket of the p53 protein result in differences of conformational flexibility of the DNA-binding domain. J. Biomol. Struct. Dyn. 2013, 31, 78–86. [Google Scholar] [CrossRef]
- Margalit, O.; Simon, A.J.; Yakubov, E.; Puca, R.; Yosepovich, A.; Avivi, C.; Jacob-Hirsch, J.; Gelernter, I.; Harmelin, A.; Barshack, I.; et al. Zinc supplementation augments in vivo antitumor effect of chemotherapy by restoring p53 function. Int. J. Cancer 2012, 131, E562–E568. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Vazquez, A.; Levine, A.J.; Carpizo, D.R. Allele-specific p53 mutant reactivation. Cancer Cell 2012, 21, 614–625. [Google Scholar] [CrossRef] [Green Version]
- Lindemann, A.; Patel, A.A.; Silver, N.L.; Tang, L.; Liu, Z.; Wang, L.; Tanaka, N.; Rao, X.; Takahashi, H.; Maduka, N.K.; et al. COTI-2, A Novel Thiosemicarbazone Derivative, Exhibits Antitumor Activity in HNSCC through p53-dependent and -independent Mechanisms. Clin. Cancer Res. 2019, 25, 5650–5662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarado-Ortiz, E.; de la Cruz-Lopez, K.G.; Becerril-Rico, J.; Sarabia-Sanchez, M.A.; Ortiz-Sanchez, E.; Garcia-Carranca, A. Mutant p53 Gain-of-Function: Role in Cancer Development, Progression, and Therapeutic Approaches. Front. Cell Dev. Biol. 2020, 8, 607670. [Google Scholar] [CrossRef] [PubMed]
- Escoll, M.; Gargini, R.; Cuadrado, A.; Anton, I.M.; Wandosell, F. Mutant p53 oncogenic functions in cancer stem cells are regulated by WIP through YAP/TAZ. Oncogene 2017, 36, 3515–3527. [Google Scholar] [CrossRef]
- Hassan, Z.; Schneeweis, C.; Wirth, M.; Muller, S.; Geismann, C.; Neuss, T.; Steiger, K.; Kramer, O.H.; Schmid, R.M.; Rad, R.; et al. Important role of Nfkb2 in the Kras(G12D)-driven carcinogenesis in the pancreas. Pancreatology 2021. [Google Scholar] [CrossRef]
- Kim, M.P.; Li, X.; Deng, J.; Zhang, Y.; Dai, B.; Allton, K.L.; Hughes, T.G.; Siangco, C.; Augustine, J.J.; Kang, Y.; et al. Oncogenic KRAS recruits an expansive transcriptional network through mutant p53 to drive pancreatic cancer metastasis. Cancer Discov. 2021. [Google Scholar] [CrossRef]
- Capaci, V.; Bascetta, L.; Fantuz, M.; Beznoussenko, G.V.; Sommaggio, R.; Cancila, V.; Bisso, A.; Campaner, E.; Mironov, A.A.; Wisniewski, J.R.; et al. Mutant p53 induces Golgi tubulo-vesiculation driving a prometastatic secretome. Nat. Commun. 2020, 11, 3945. [Google Scholar] [CrossRef]
- Eliyahu, D.; Raz, A.; Gruss, P.; Givol, D.; Oren, M. Participation of p53 cellular tumour antigen in transformation of normal embryonic cells. Nature 1984, 312, 646–649. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, J.R.; Rudge, K.; Currie, G.A. Cellular immortalization by a cDNA clone encoding the transformation-associated phosphoprotein p53. Nature 1984, 312, 651–654. [Google Scholar] [CrossRef]
- Parada, L.F.; Land, H.; Weinberg, R.A.; Wolf, D.; Rotter, V. Cooperation between gene encoding p53 tumour antigen and ras in cellular transformation. Nature 1984, 312, 649–651. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, I.B.; Joe, A.K. Mechanisms of disease: Oncogene addiction--a rationale for molecular targeting in cancer therapy. Nat. Clin. Pract. Oncol. 2006, 3, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Marchenko, N.D.; Schulz, R.; Fischer, V.; Velasco-Hernandez, T.; Talos, F.; Moll, U.M. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol. Cancer Res. 2011, 9, 577–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blagosklonny, M.V.; Toretsky, J.; Neckers, L. Geldanamycin selectively destabilizes and conformationally alters mutated p53. Oncogene 1995, 11, 933–939. [Google Scholar]
- Alexandrova, E.M.; Yallowitz, A.R.; Li, D.; Xu, S.; Schulz, R.; Proia, D.A.; Lozano, G.; Dobbelstein, M.; Moll, U.M. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature 2015, 523, 352–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, Z.; Oleinick, N.L.; Zhang, J. ATR/CHK1 inhibitors and cancer therapy. Radiother. Oncol. 2018, 126, 450–464. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Bi, J.; Li, Y.; Yang, S.; Zhang, Y.; Li, M.; Liu, H.; Li, Y.; McDonald, M.E.; Thiel, K.W.; et al. AZD1775 Increases Sensitivity to Olaparib and Gemcitabine in Cancer Cells with p53 Mutations. Cancers 2018, 10, 149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leijen, S.; van Geel, R.M.; Sonke, G.S.; de Jong, D.; Rosenberg, E.H.; Marchetti, S.; Pluim, D.; van Werkhoven, E.; Rose, S.; Lee, M.A.; et al. Phase II Study of WEE1 Inhibitor AZD1775 Plus Carboplatin in Patients with TP53-Mutated Ovarian Cancer Refractory or Resistant to First-Line Therapy Within 3 Months. J. Clin. Oncol. 2016, 34, 4354–4361. [Google Scholar] [CrossRef] [Green Version]
- Hartman, S.J.; Bagby, S.M.; Yacob, B.W.; Simmons, D.M.; MacBeth, M.; Lieu, C.H.; Davis, S.L.; Leal, A.D.; Tentler, J.J.; Diamond, J.R.; et al. WEE1 Inhibition in Combination with Targeted Agents and Standard Chemotherapy in Preclinical Models of Pancreatic Ductal Adenocarcinoma. Front. Oncol. 2021, 11, 642328. [Google Scholar] [CrossRef]
- Xiao, G.; Lundine, D.; Annor, G.K.; Canar, J.; Ellison, V.; Polotskaia, A.; Donabedian, P.L.; Reiner, T.; Khramtsova, G.F.; Olopade, O.I.; et al. Gain-of-Function Mutant p53 R273H Interacts with Replicating DNA and PARP1 in Breast Cancer. Cancer Res. 2020, 80, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Yang, J.; Adair, S.J.; Ozturk, H.; Kuscu, C.; Lee, K.Y.; Kane, W.J.; O’Hara, P.E.; Liu, D.; Demirlenk, Y.M.; et al. Targeted CRISPR screening identifies PRMT5 as synthetic lethality combinatorial target with gemcitabine in pancreatic cancer cells. Proc. Natl. Acad. Sci. USA 2020, 117, 28068–28079. [Google Scholar] [CrossRef]
- Feng, M.; Xiong, G.; Cao, Z.; Yang, G.; Zheng, S.; Song, X.; You, L.; Zheng, L.; Zhang, T.; Zhao, Y. PD-1/PD-L1 and immunotherapy for pancreatic cancer. Cancer Lett. 2017, 407, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Nummer, D.; Suri-Payer, E.; Schmitz-Winnenthal, H.; Bonertz, A.; Galindo, L.; Antolovich, D.; Koch, M.; Buchler, M.; Weitz, J.; Schirrmacher, V.; et al. Role of tumor endothelium in CD4+ CD25+ regulatory T cell infiltration of human pancreatic carcinoma. J. Natl. Cancer Inst. 2007, 99, 1188–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat. Rev. Immunol. 2004, 4, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.; Baine, M.J.; Meza, J.L.; Lin, C. Association of Immunotherapy with Survival Among Patients With Brain Metastases Whose Cancer Was Managed With Definitive Surgery of the Primary Tumor. JAMA Netw. Open 2020, 3, e2015444. [Google Scholar] [CrossRef]
- Cao, Z.; Kon, N.; Liu, Y.; Xu, W.; Wen, J.; Yao, H.; Zhang, M.; Wu, Z.; Yan, X.; Zhu, W.G.; et al. An unexpected role for p53 in regulating cancer cell-intrinsic PD-1 by acetylation. Sci. Adv. 2021, 7, eabf4148. [Google Scholar] [CrossRef]
- Cooks, T.; Pateras, I.S.; Jenkins, L.M.; Patel, K.M.; Robles, A.I.; Morris, J.; Forshew, T.; Appella, E.; Gorgoulis, V.G.; Harris, C.C. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat. Commun. 2018, 9, 771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, E.; Turcotte, S.; Gros, A.; Robbins, P.F.; Lu, Y.C.; Dudley, M.E.; Wunderlich, J.R.; Somerville, R.P.; Hogan, K.; Hinrichs, C.S.; et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 2014, 344, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Lugli, E.; Kvistborg, P.; Galletti, G. Cancer neoantigens targeted by adoptive T cell transfer: Private no more. J. Clin. Investig. 2019, 129, 949–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavlakis, E.; Stiewe, T. p53’s Extended Reach: The Mutant p53 Secretome. Biomolecules 2020, 10, 307. [Google Scholar] [CrossRef] [Green Version]
- Amelio, I.; Melino, G. Context is everything: Extrinsic signalling and gain-of-function p53 mutants. Cell Death Discov. 2020, 6, 16. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Chen, Y.; Francisco, N.M.; Zhang, Y.; Wu, M. The application of CAR-T cell therapy in hematological malignancies: Advantages and challenges. Acta Pharm. Sin. B 2018, 8, 539–551. [Google Scholar] [CrossRef]
- Umut, Ö.; Gottschlich, A.; Endres, S.; Kobold, S. CAR T cell therapy in solid tumors: A short review. Memo Mag. Eur. Med. Oncol. 2021. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, Y.; Wu, Z.; Feng, K.; Tong, C.; Wang, Y.; Dai, H.; Shi, F.; Yang, Q.; Han, W. Anti-EGFR chimeric antigen receptor-modified T cells in metastatic pancreatic carcinoma: A phase I clinical trial. Cytotherapy 2020, 22, 573–580. [Google Scholar] [CrossRef]
- Wade, M.; Li, Y.C.; Wahl, G.M. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 2013, 13, 83–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.; Yan, Z.; Liao, X.; Li, Y.; Yang, J.; Wang, Z.G.; Zuo, Y.; Kawai, H.; Shadfan, M.; Ganapathy, S.; et al. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 12001–12006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Zawacka-Pankau, J. The p53/MDM2/MDMX-targeted therapies-a clinical synopsis. Cell Death Dis. 2020, 11, 237. [Google Scholar] [CrossRef] [PubMed]
- Said, R.; Guibert, N.; Oxnard, G.R.; Tsimberidou, A.M. Circulating tumor DNA analysis in the era of precision oncology. Oncotarget 2020, 11, 188–211. [Google Scholar] [CrossRef] [Green Version]
- Russo, A.; Incorvaia, L.; Del Re, M.; Malapelle, U.; Capoluongo, E.; Gristina, V.; Castiglia, M.; Danesi, R.; Fassan, M.; Giuffrè, G.; et al. The molecular profiling of solid tumors by liquid biopsy: A position paper of the AIOM-SIAPEC-IAP-SIBioC-SIC-SIF Italian Scientific Societies. ESMO Open 2021, 6, 100164. [Google Scholar] [CrossRef] [PubMed]
- Collisson, E.A.; Sadanandam, A.; Olson, P.; Gibb, W.J.; Truitt, M.; Gu, S.; Cooc, J.; Weinkle, J.; Kim, G.E.; Jakkula, L.; et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011, 17, 500–503. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.M.; Gingras, M.C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Chan-Seng-Yue, M.; Kim, J.C.; Wilson, G.W.; Ng, K.; Figueroa, E.F.; O’Kane, G.M.; Connor, A.A.; Denroche, R.E.; Grant, R.C.; McLeod, J.; et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat. Genet. 2020, 52, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).