EVI2B Is a New Prognostic Biomarker in Metastatic Melanoma with IFNgamma Associated Immune Infiltration
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Survival Analysis with EVI2B mRNA Level in the Public Database
2.2. Estimated Fractions of TILs
2.3. Patial Pattern of TIL Analysis
2.4. Correlation with Sets of Immunomodulatory Genes
2.5. Statistical Analysis
3. Results
3.1. Prognostic Impact of EVI2B Gene Expression in Metastatic Melanoma
3.2. Spatial Pattern of Infiltrating Immune Cells by EVI2B Gene Expression
3.3. Infiltrating Immune Cells with EVI2B mRNA Level
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schadendorf, D.; van Akkooi, A.C.J.; Berking, C.; Griewank, K.G.; Gutzmer, R.; Hauschild, A.; Stang, A.; Roesch, A.; Ugurel, S. Melanoma. Lancet 2018, 392, 971–984. [Google Scholar] [CrossRef]
- Phillips, M.; Marsden, H.; Jaffe, W.; Matin, R.N.; Wali, G.N.; Greenhalgh, J.; McGrath, E.; James, R.; Ladoyanni, E.; Bewley, A.; et al. Assessment of accuracy of an artificial intelligence algorithm to detect melanoma in images of skin lesions. JAMA Netw. Open 2019, 2, e1913436. [Google Scholar] [CrossRef] [PubMed]
- Braeuer, R.R.; Watson, I.R.; Wu, C.J.; Mobley, A.K.; Kamiya, T.; Shoshan, E.; Bar-Eli, M. Why is melanoma so metastatic? Pigment Cell Melanoma Res. 2014, 27, 19–36. [Google Scholar] [CrossRef]
- Antohe, M.; Nedelcu, R.I.; Nichita, L.; Popp, C.G.; Cioplea, M.; Brinzea, A.; Hodorogea, A.; Calinescu, A.; Balaban, M.; Ion, D.A.; et al. Tumor infiltrating lymphocytes: The regulator of melanoma evolution. Oncol. Lett. 2019, 17, 4155–4161. [Google Scholar] [CrossRef]
- Azimi, F.; Scolyer, R.A.; Rumcheva, P.; Moncrieff, M.; Murali, R.; McCarthy, S.W.; Saw, R.P.; Thompson, J.F. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J. Clin. Oncol. 2012, 30, 2678–2683. [Google Scholar] [CrossRef]
- Yan, Y.; Leontovich, A.A.; Gerdes, M.J.; Desai, K.; Dong, J.; Sood, A.; Santamaria-Pang, A.; Mansfield, A.S.; Chadwick, C.; Zhang, R.; et al. Understanding heterogeneous tumor microenvironment in metastatic melanoma. PLoS ONE 2019, 14, e0216485. [Google Scholar] [CrossRef]
- Cawthon, R.M.; Andersen, L.B.; Buchberg, A.M.; Xu, G.F.; O’Connell, P.; Viskochil, D.; Weiss, R.B.; Wallace, M.R.; Marchuk, D.A.; Culver, M.; et al. cDNA sequence and genomic structure of EV12B, a gene lying within an intron of the neurofibromatosis type 1 gene. Genomics 1991, 9, 446–460. [Google Scholar] [CrossRef][Green Version]
- Buchberg, A.M.; Bedigian, H.G.; Jenkins, N.A.; Copeland, N.G. Evi-2, a common integration site involved in murine myeloid leukemogenesis. Mol. Cell Biol. 1990, 10, 4658–4666. [Google Scholar] [CrossRef]
- Zjablovskaja, P.; Kardosova, M.; Danek, P.; Angelisova, P.; Benoukraf, T.; Wurm, A.A.; Kalina, T.; Sian, S.; Balastik, M.; Delwel, R.; et al. EVI2B is a C/EBPalpha target gene required for granulocytic differentiation and functionality of hematopoietic progenitors. Cell Death Differ. 2017, 24, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.Y.; Ou Yang, T.H.; Anastassiou, D. Biomolecular events in cancer revealed by attractor metagenes. PLoS Comput. Biol. 2013, 9, e1002920. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Chen, J.; Wang, J.; Xu, M.; Zhang, Y.; Sun, P.; Liang, L. Identification hub genes in colorectal cancer by integrating weighted gene co-expression network analysis and clinical validation in vivo and vitro. Front. Oncol. 2020, 10, 638. [Google Scholar] [CrossRef] [PubMed]
- Saltz, J.; Gupta, R.; Hou, L.; Kurc, T.; Singh, P.; Nguyen, V.; Samaras, D.; Shroyer, K.R.; Zhao, T.; Batiste, R.; et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018, 23, 181–193.e187. [Google Scholar] [CrossRef]
- Scott, A.J.; Symons, M.J. Clustering methods based on likelihood ratio criteria. Biometrics 1971, 27, 387–397. [Google Scholar] [CrossRef]
- Hubert, L.; Schultz, J. Quadratic assignment as a general data analysis strategy. Br. J. Math. Stat. Psychol. 1976, 29, 190–241. [Google Scholar] [CrossRef]
- Banfield, J.D.; Raftery, A.E. Model-based gaussian and non-gaussian clustering. Biometrics 1993, 49, 803–821. [Google Scholar] [CrossRef]
- Ball, G.H.; Hall, D.J. A Novel Method of Data Analysis and Pattern Classification; Technical Report; Stanford Research Institute: Menlo Park, CA, USA, 1965. [Google Scholar]
- Finotello, F.; Mayer, C.; Plattner, C.; Laschober, G.; Rieder, D.; Hackl, H.; Krogsdam, A.; Loncova, Z.; Posch, W.; Wilflingseder, D.; et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019, 11, 34. [Google Scholar] [CrossRef]
- Ayers, M.; Lunceford, J.; Nebozhyn, M.; Murphy, E.; Loboda, A.; Kaufman, D.R.; Albright, A.; Cheng, J.D.; Kang, S.P.; Shankaran, V.; et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 2017, 127, 2930–2940. [Google Scholar] [CrossRef]
- Shukla, S.A.; Bachireddy, P.; Schilling, B.; Galonska, C.; Zhan, Q.; Bango, C.; Langer, R.; Lee, P.C.; Gusenleitner, D.; Keskin, D.B.; et al. Cancer-Germline Antigen Expression Discriminates Clinical Outcome to CTLA-4 Blockade. Cell 2018, 173, 624–633.e628. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Q.; Wang, L.; Xie, L.; An, Y.; Zhang, G.; Zhu, W.; Li, Y.; Liu, Z.; Zhang, X.; et al. OSskcm: An online survival analysis webserver for skin cutaneous melanoma based on 1085 transcriptomic profiles. Cancer Cell Int. 2020, 20, 176. [Google Scholar] [CrossRef] [PubMed]
- Cirenajwis, H.; Ekedahl, H.; Lauss, M.; Harbst, K.; Carneiro, A.; Enoksson, J.; Rosengren, F.; Werner-Hartman, L.; Torngren, T.; Kvist, A.; et al. Molecular stratification of metastatic melanoma using gene expression profiling: Prediction of survival outcome and benefit from molecular targeted therapy. Oncotarget 2015, 6, 12297–12309. [Google Scholar] [CrossRef] [PubMed]
- Bogunovic, D.; O’Neill, D.W.; Belitskaya-Levy, I.; Vacic, V.; Yu, Y.L.; Adams, S.; Darvishian, F.; Berman, R.; Shapiro, R.; Pavlick, A.C.; et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc. Natl. Acad. Sci. USA 2009, 106, 20429–20434. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic. Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Li, B.; Severson, E.; Pignon, J.C.; Zhao, H.; Li, T.; Novak, J.; Jiang, P.; Shen, H.; Aster, J.C.; Rodig, S.; et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016, 17, 174. [Google Scholar] [CrossRef] [PubMed]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautes-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef]
- Benjamini, Y.; Krieger, A.M.; Yekutieli, D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika 2006, 93, 491–507. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic. Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, D.H.; Shankaran, V.; Dighe, A.S.; Stockert, E.; Aguet, M.; Old, L.J.; Schreiber, R.D. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc. Natl. Acad. Sci. USA 1998, 95, 7556–7561. [Google Scholar] [CrossRef]
- Bhat, P.; Leggatt, G.; Waterhouse, N.; Frazer, I.H. Interferon-gamma derived from cytotoxic lymphocytes directly enhances their motility and cytotoxicity. Cell Death Dis. 2017, 8, e2836. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, C.; Uekusa, Y.; Iwasaki, M.; Yamaguchi, N.; Mukai, T.; Gao, P.; Tomura, M.; Ono, S.; Tsujimura, T.; Fujiwara, H.; et al. A role of interferon-gamma (IFN-gamma) in tumor immunity: T cells with the capacity to reject tumor cells are generated but fail to migrate to tumor sites in IFN-gamma-deficient mice. Cancer Res. 2001, 61, 3399–3405. [Google Scholar]
- Cursons, J.; Souza-Fonseca-Guimaraes, F.; Foroutan, M.; Anderson, A.; Hollande, F.; Hediyeh-Zadeh, S.; Behren, A.; Huntington, N.D.; Davis, M.J. A gene signature predicting natural killer cell infiltration and improved survival in melanoma patients. Cancer Immunol. Res. 2019, 7, 1162–1174. [Google Scholar] [CrossRef]
- Lee, H.; Da Silva, I.P.; Palendira, U.; Scolyer, R.A.; Long, G.V.; Wilmott, J.S. Targeting NK cells to enhance melanoma response to immunotherapies. Cancers 2021, 13, 1363. [Google Scholar] [CrossRef]
- Cogdill, A.P.; Andrews, M.C.; Wargo, J.A. Hallmarks of response to immune checkpoint blockade. Br. J. Cancer 2017, 117, 1–7. [Google Scholar] [CrossRef] [PubMed]
- de Assis, L.V.M.; Kinker, G.S.; Moraes, M.N.; Markus, R.P.; Fernandes, P.A.; Castrucci, A.M.L. Expression of the circadian clock gene BMAL1 Positively correlates with antitumor immunity and patient survival in metastatic melanoma. Front. Oncol. 2018, 8, 185. [Google Scholar] [CrossRef]
- Venet, D.; Dumont, J.E.; Detours, V. Most random gene expression signatures are significantly associated with breast cancer outcome. PLoS Comput. Biol. 2011, 7, e1002240. [Google Scholar] [CrossRef]
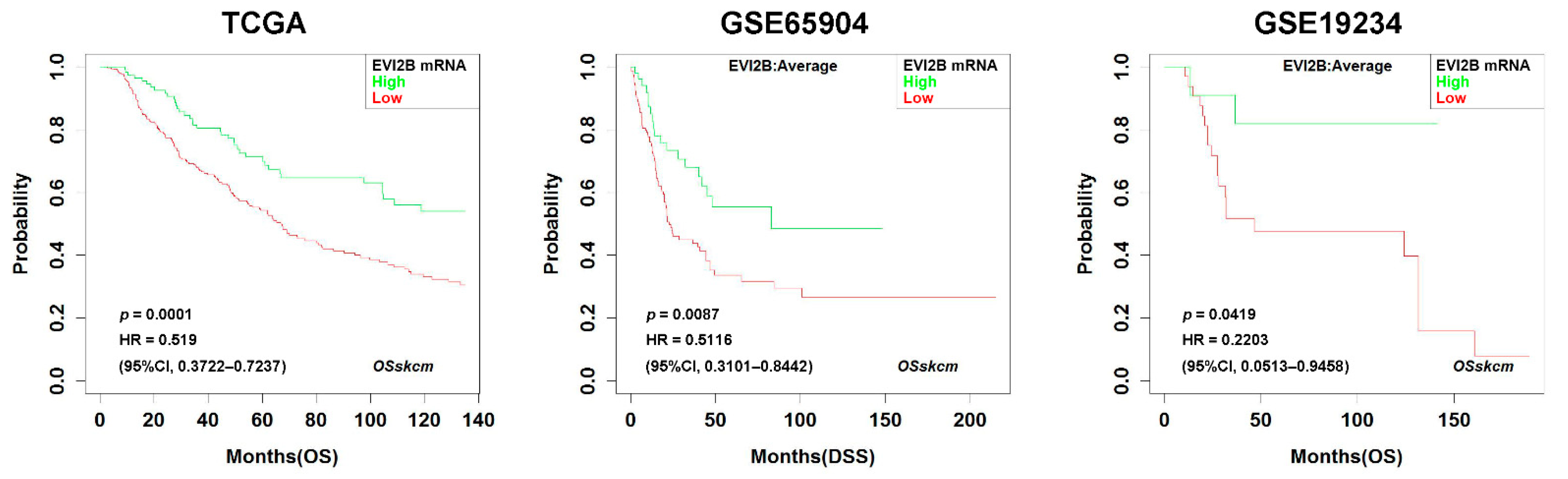
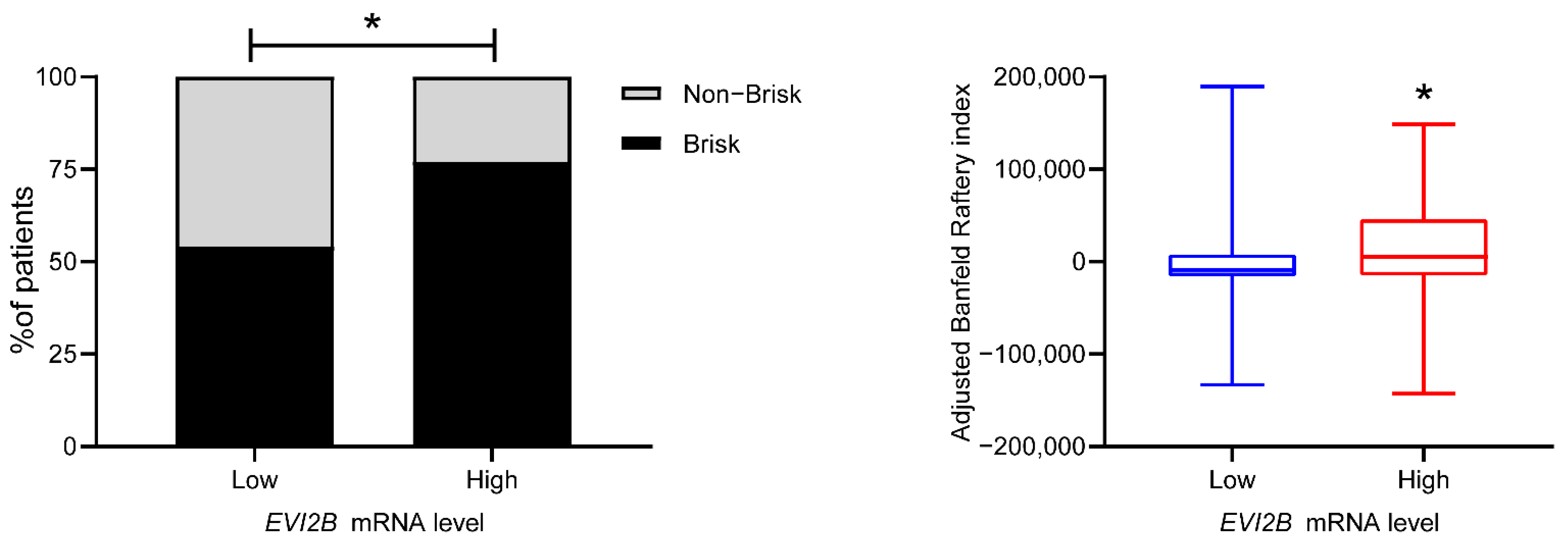
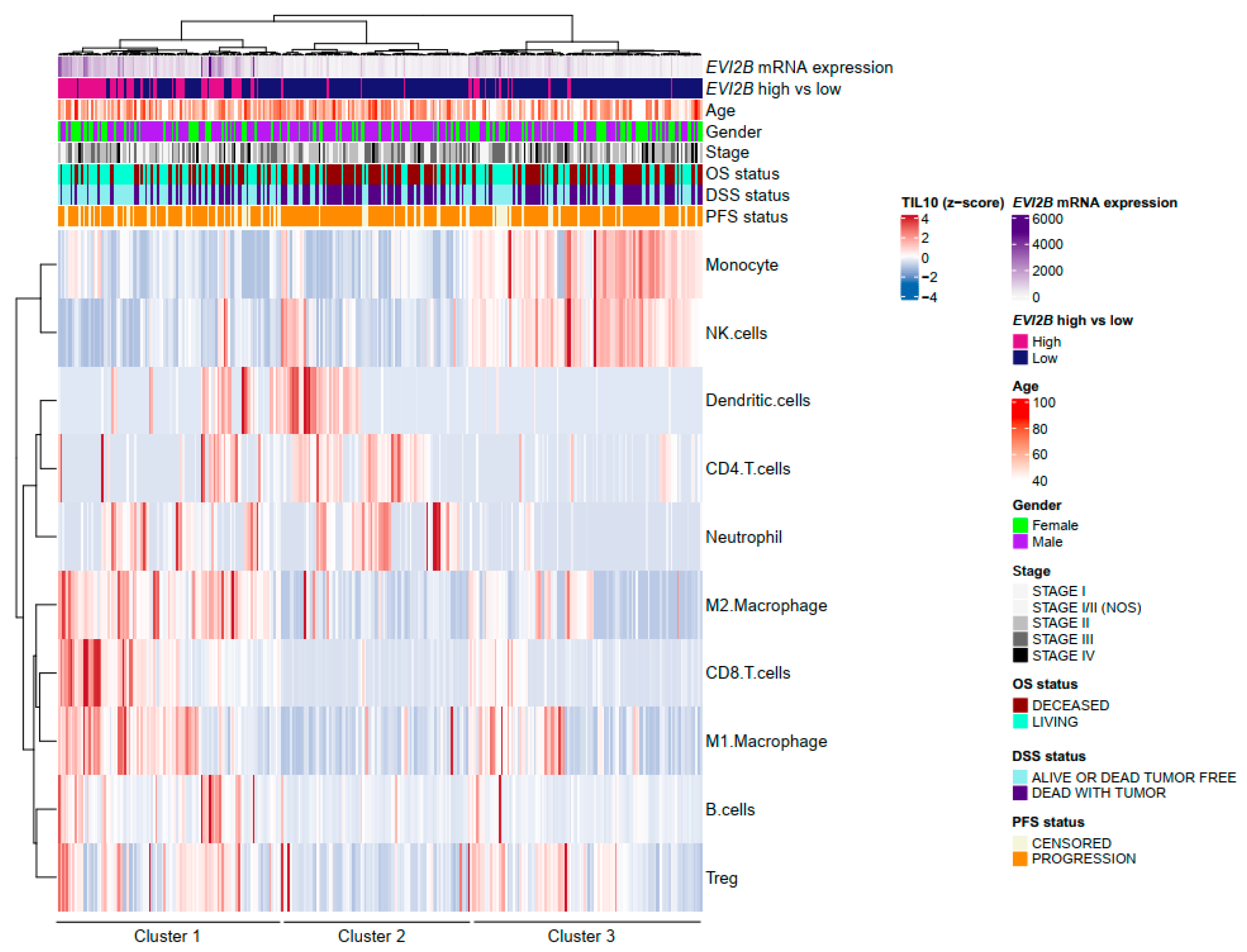
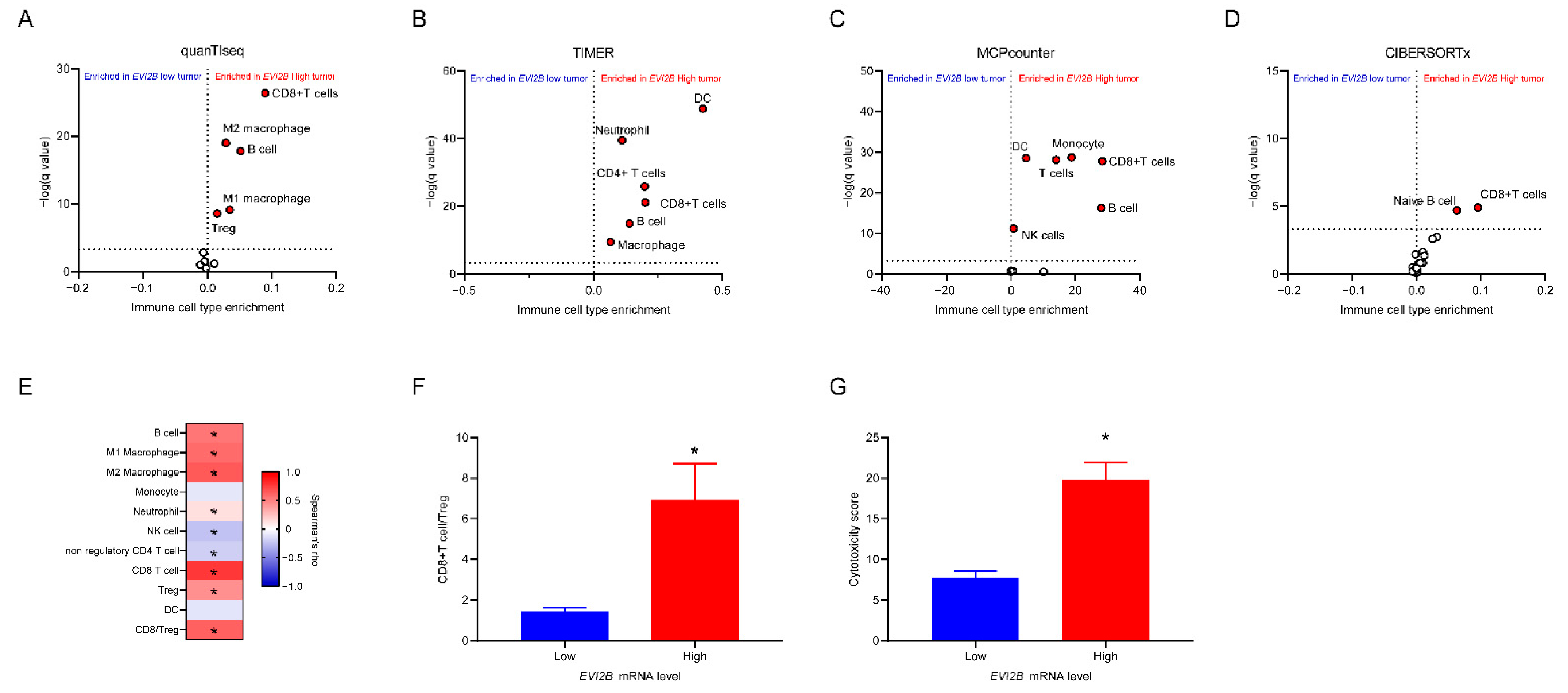

| Variables | EVI2B Low | EVI2B High | p Value * |
|---|---|---|---|
| No. of patients (%) | 272 (75%) | 91 (25%) | |
| Age (years) (median, range) | 57 (15–87) | 54 (18–86) | 0.491 |
| Sex | 0.211 | ||
| Male | 176 | 52 | |
| Female | 96 | 39 | |
| Race | 0.169 | ||
| White | 253 | 86 | |
| Others (Asian or African American) | 3 | 3 | |
| NA | 16 | 2 | |
| TNM stage | 0.375 | ||
| Stage 0 | 6 | 1 | |
| Stage I | 55 | 20 | |
| Stage I/II (NOS) | 10 | 3 | |
| Stage II | 60 | 12 | |
| Stage III | 101 | 40 | |
| Stage IV | 18 | 3 | |
| NA | 22 | 12 |
| Variable | HR | 95% CI | p-Value |
|---|---|---|---|
| Age at diagnosis | 1.021 | 1.010–1.032 | <0.0001 |
| Male (ref: Female) | 0.885 | 0.629–1.245 | 0.483 |
| Stage at initial diagnosis (ref: Stage 0/I) | |||
| II | 0.998 | 0.633–1.574 | 0.993 |
| III | 1.594 | 1.056–2.404 | 0.026 |
| IV | 2.967 | 1.355–6.498 | 0.007 |
| Tumor purity | 0.777 | 0.294–2.056 | 0.611 |
| EVI2B mRNA level | 0.716 | 0.610–0.840 | <0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yonekura, S.; Ueda, K. EVI2B Is a New Prognostic Biomarker in Metastatic Melanoma with IFNgamma Associated Immune Infiltration. Cancers 2021, 13, 4110. https://doi.org/10.3390/cancers13164110
Yonekura S, Ueda K. EVI2B Is a New Prognostic Biomarker in Metastatic Melanoma with IFNgamma Associated Immune Infiltration. Cancers. 2021; 13(16):4110. https://doi.org/10.3390/cancers13164110
Chicago/Turabian StyleYonekura, Satoru, and Kosuke Ueda. 2021. "EVI2B Is a New Prognostic Biomarker in Metastatic Melanoma with IFNgamma Associated Immune Infiltration" Cancers 13, no. 16: 4110. https://doi.org/10.3390/cancers13164110
APA StyleYonekura, S., & Ueda, K. (2021). EVI2B Is a New Prognostic Biomarker in Metastatic Melanoma with IFNgamma Associated Immune Infiltration. Cancers, 13(16), 4110. https://doi.org/10.3390/cancers13164110






