Molecular Properties and Therapeutic Targeting of the EBV-Encoded Receptor BILF1
Abstract
Simple Summary
Abstract
1. Introduction
2. Epstein–Barr Virus
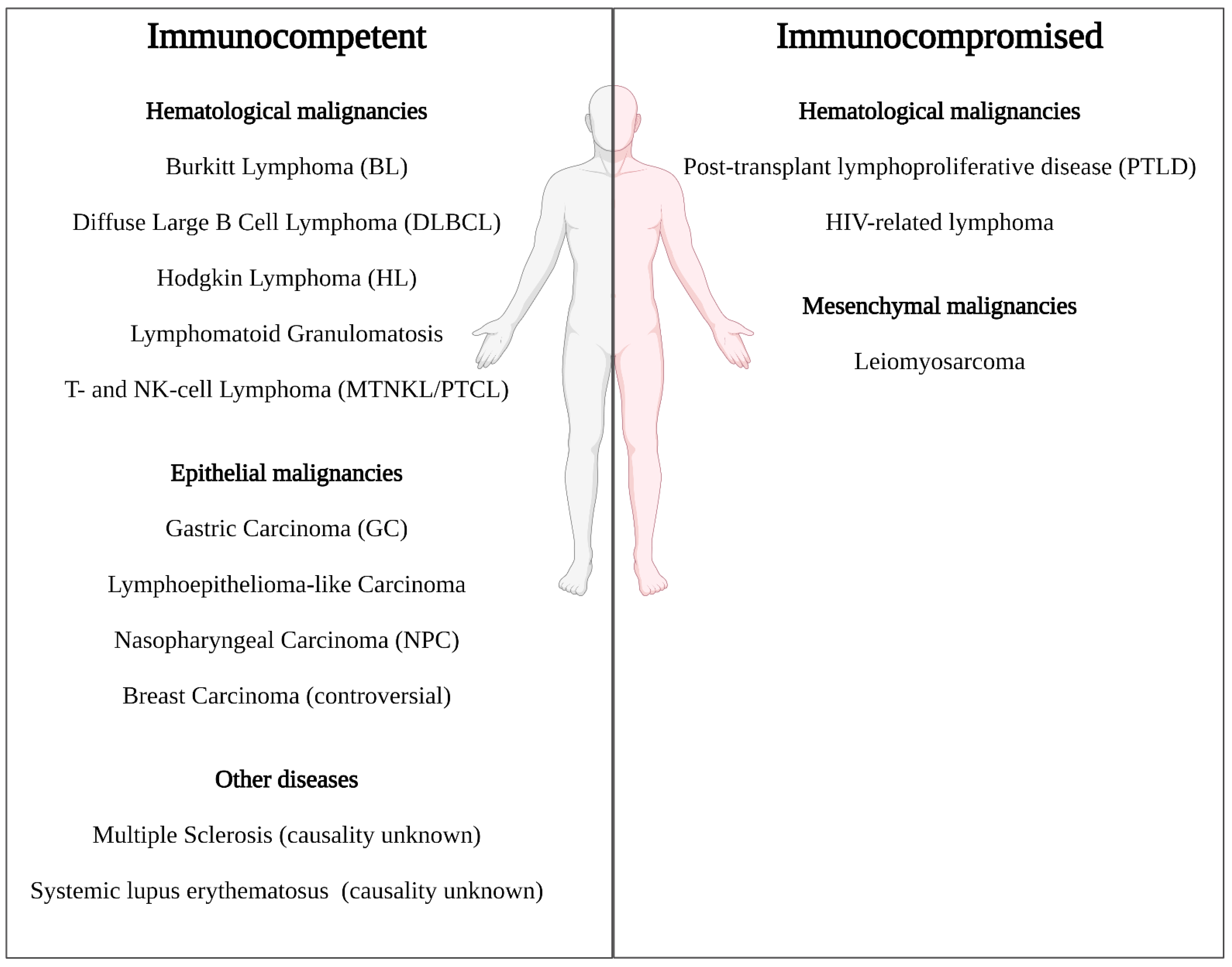
3. EBV Cancers and Standard of Care
4. G Protein–Coupled Receptors
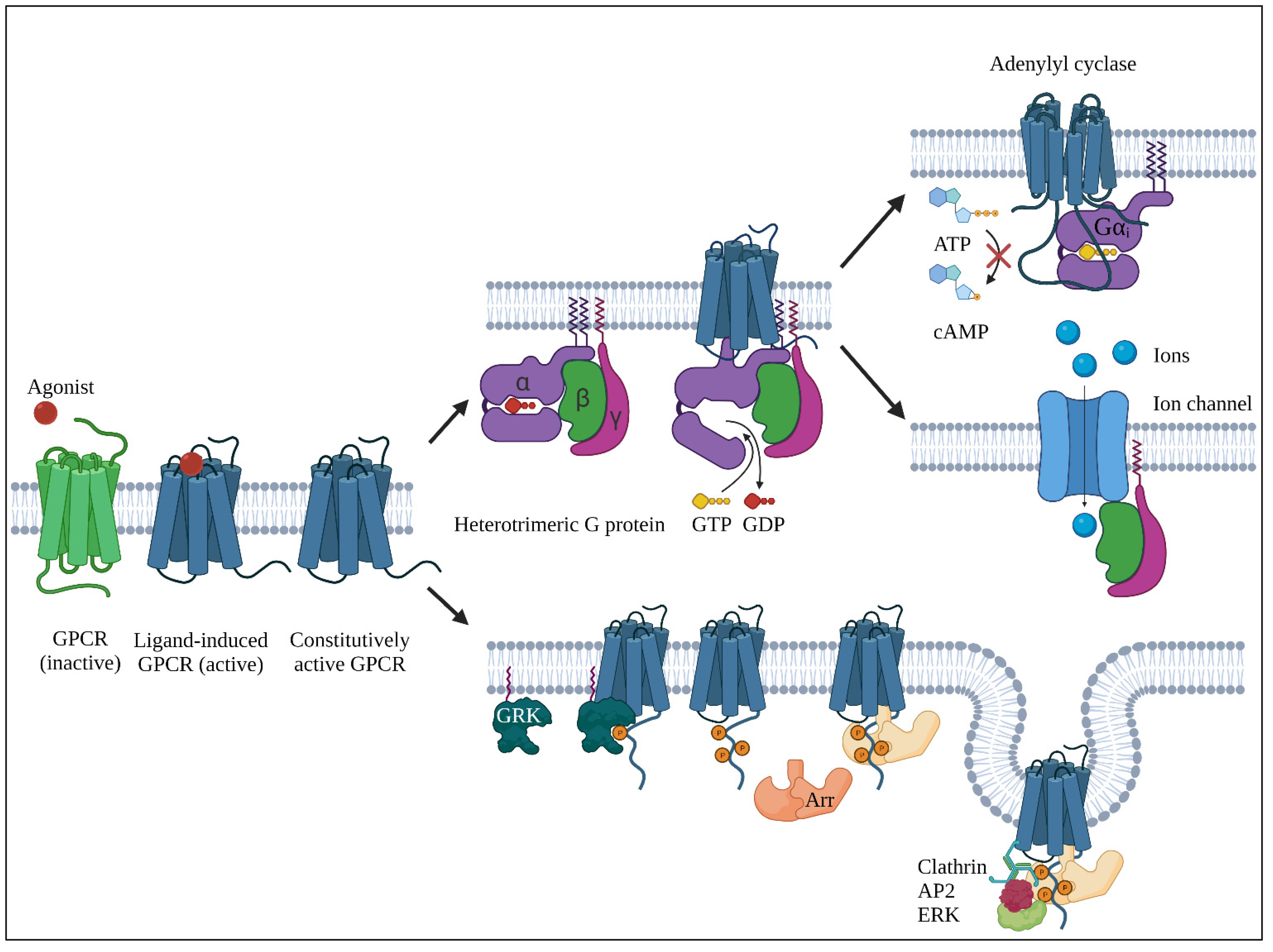
4.1. Basics
4.2. BILF1; a Conserved G Protein–Coupled Receptor in γ1-Herpesviruses
4.2.1. General
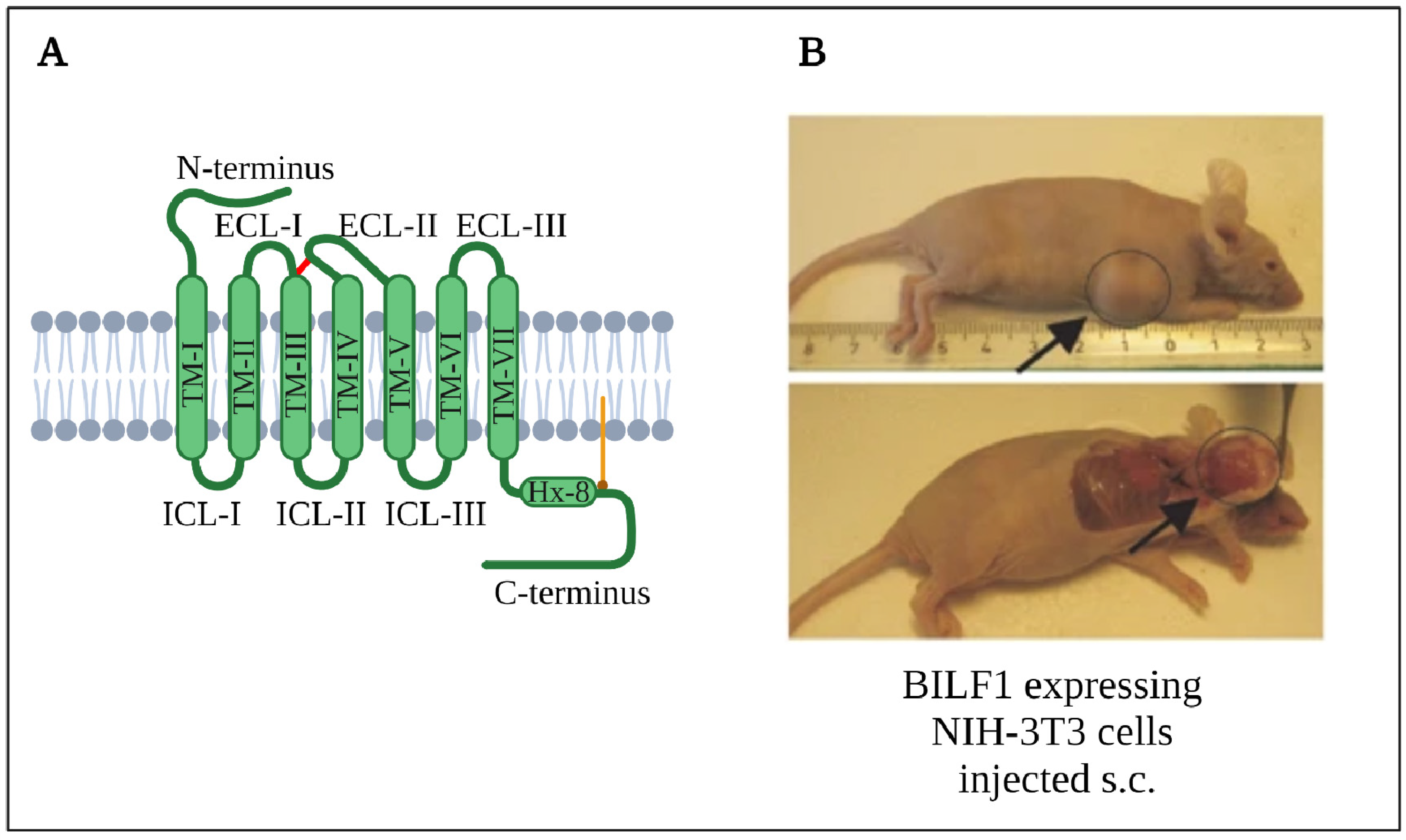
4.2.2. Structure
4.2.3. Expression Patterns
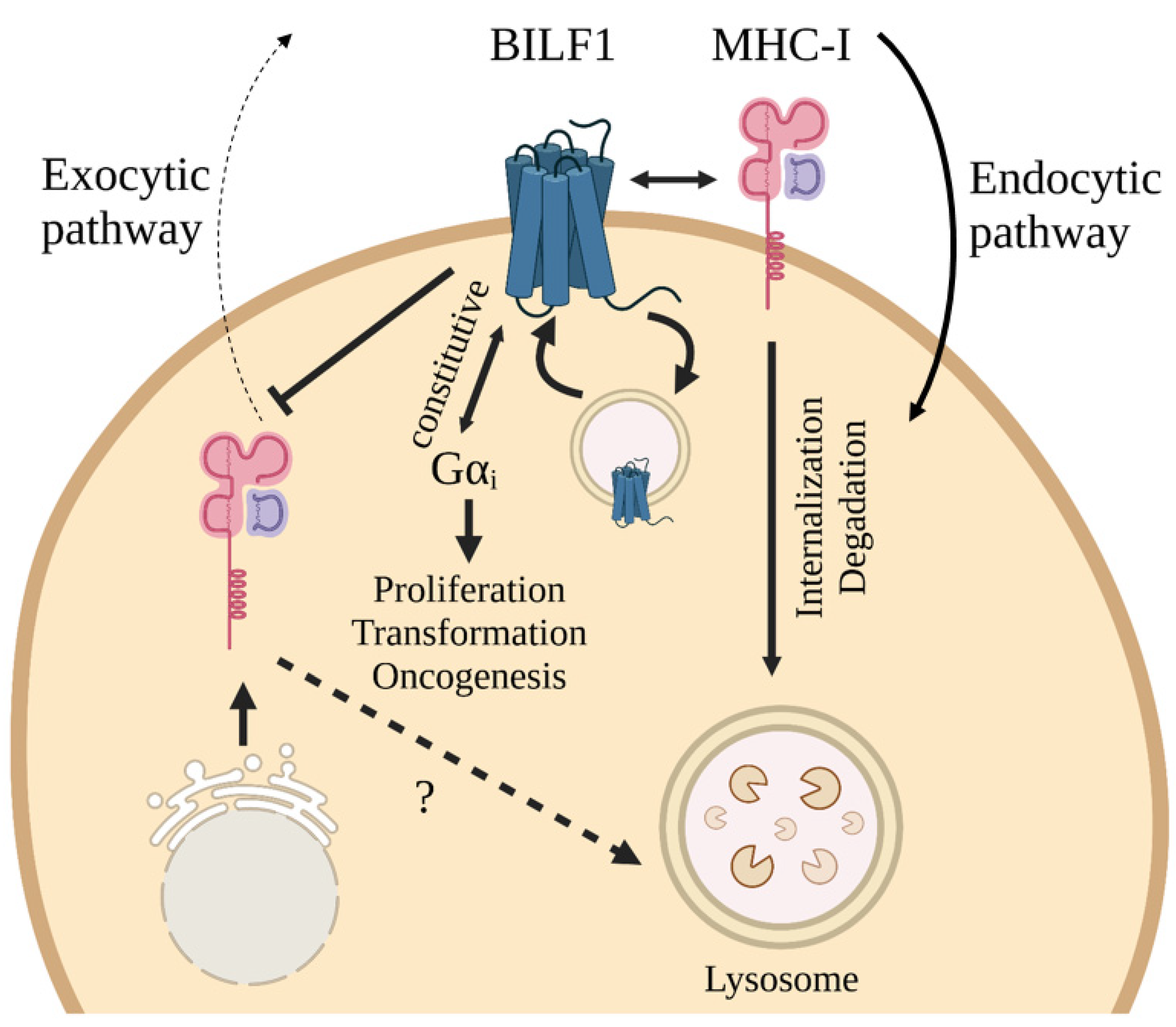
4.2.4. Cellular Effects
4.2.5. Oncogenesis
4.3. Druggability of GPCRs
- Excess of circulating agonists driving GPCR signaling, which promotes tumor progression (e.g., neuropeptides in small cell lung cancer) [155];
- Mutations in GPCRs or Gα subunit leading to aberrant signaling (e.g., G stimulatory protein (gsp), thyroid-stimulating hormone receptor (TSHR) [156]);
- Overexpression of certain GPCRs (e.g., among many others, CXCR4, CCR7 or CXCR1) resulting in increased cancer metastasis, proliferation, cell survival or angiogenesis [157]
5. EBV Drug Pipeline
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schiöth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef]
- Paulsen, S.J.; Rosenkilde, M.M.; Eugen-Olsen, J.; Kledal, T.N. Epstein-Barr Virus-Encoded BILF1 Is a Constitutively Active G Protein-Coupled Receptor. J. Virol. 2005, 79, 536–546. [Google Scholar] [CrossRef]
- Beisser, P.S.; Verzijl, D.; Gruijthuijsen, Y.K.; Beuken, E.; Smit, M.J.; Leurs, R.; Bruggeman, C.A.; Vink, C. The Epstein-Barr Virus BILF1 Gene Encodes a G Protein-Coupled Receptor That Inhibits Phosphorylation of RNA-Dependent Protein Kinase. J. Virol. 2005, 79, 441–449. [Google Scholar] [CrossRef]
- Cohen, J.I. Epstein-Barr Virus Infection. N. Engl. J. Med. 2000, 343, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Ambinder, R.F.; Cesarman, E. Clinical and Pathological Aspects of EBV and KSHV Infection; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University Press: Cambridge, UK, 2007; ISBN 9780521827140. [Google Scholar]
- Wang, F.; Rivailler, P.; Rao, P.; Cho, Y. Simian homologues of Epstein–Barr virus. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2001, 356, 489–497. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Epstein, M.; Achong, B.; Barr, Y. Virus Particles in Cultured Lymphoblasts from Burkitt’s Lymphoma. Lancet 1964, 283, 702–703. [Google Scholar] [CrossRef]
- Arfelt, K.N.; Fares, S.; Rosenkilde, M.M. EBV, the human host, and the 7TM receptors: Defense or offense? In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2015; Volume 129, pp. 395–427. [Google Scholar]
- Babcock, G.J.; Decker, L.L.; Volk, M.; Thorley-Lawson, D.A. EBV Persistence in Memory B Cells In Vivo. Immunity 1998, 9, 395–404. [Google Scholar] [CrossRef]
- Chesnokova, L.S.; Hutt-Fletcher, L.M. Fusion of Epstein-Barr Virus with Epithelial Cells Can Be Triggered by αvβ5 in Addition to αvβ6 and αvβ8, and Integrin Binding Triggers a Conformational Change in Glycoproteins gHgL. J. Virol. 2011, 85, 13214–13223. [Google Scholar] [CrossRef]
- Hutt-Fletcher, L.M. Epstein-Barr Virus Entry. J. Virol. 2007, 81, 7825–7832. [Google Scholar] [CrossRef]
- Tanner, J.; Weis, J.; Fearon, D.; Whang, Y.; Kieff, E. Epstein-barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell 1987, 50, 203–213. [Google Scholar] [CrossRef]
- Kenney, S.C. Reactivation and lytic replication of EBV. In Human Herpesviruses; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University Press: Cambridge, UK, 2007; pp. 403–433. [Google Scholar]
- Murata, T. Encyclopedia of EBV-encoded lytic genes: An update. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2018; Volume 1045, pp. 395–412. [Google Scholar]
- Zuo, J.; Currin, A.; Griffin, B.D.; Shannon-Lowe, C.; Thomas, W.A.; Ressing, M.E.; Wiertz, E.J.H.J.; Rowe, M. The Epstein-Barr Virus G-Protein-Coupled Receptor Contributes to Immune Evasion by Targeting MHC Class I Molecules for Degradation. PLoS Pathog. 2009, 5, e1000255. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Quinn, L.L.; Tamblyn, J.; Thomas, W.A.; Feederle, R.; Delecluse, H.-J.; Hislop, A.D.; Rowe, M. The Epstein-Barr Virus-Encoded BILF1 Protein Modulates Immune Recognition of Endogenously Processed Antigen by Targeting Major Histocompatibility Complex Class I Molecules Trafficking on both the Exocytic and Endocytic Pathways. J. Virol. 2011, 85, 1604–1614. [Google Scholar] [CrossRef]
- Quinn, L.L.; Zuo, J.; Abbott, R.J.M.; Shannon-Lowe, C.; Tierney, R.J.; Hislop, A.D.; Rowe, M. Cooperation between Epstein-Barr Virus Immune Evasion Proteins Spreads Protection from CD8+ T Cell Recognition across All Three Phases of the Lytic Cycle. PLoS Pathog. 2014, 10, e1004322. [Google Scholar] [CrossRef] [PubMed]
- Quinn, L.L.; Williams, L.R.; White, C.; Forrest, C.; Zuo, J.; Rowe, M. The Missing Link in Epstein-Barr Virus Immune Evasion: The BDLF3 Gene Induces Ubiquitination and Downregulation of Major Histocompatibility Complex Class I (MHC-I) and MHC-II. J. Virol. 2016, 90, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T. EBV-encoded latent genes. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2018; Volume 1045, pp. 377–394. [Google Scholar]
- Ambrosio, M.R.; Leoncini, L. Epidemiology of Epstein-Barr Virus and Mechanisms of Carcinogenesis. In Tropical Hemato-Oncology; Springer International Publishing: Cham, Switzerland, 2015; pp. 127–141. ISBN 9783319182575. [Google Scholar]
- Young, L.S.; Rickinson, A.B. Epstein–Barr virus: 40 years on. Nat. Rev. Cancer 2004, 4, 757–768. [Google Scholar] [CrossRef]
- Chakravorty, S.; Yan, B.; Wang, C.; Wang, L.; Quaid, J.T.; Lin, C.F.; Briggs, S.D.; Majumder, J.; Canaria, D.A.; Chauss, D.; et al. Integrated Pan-Cancer Map of EBV-Associated Neoplasms Reveals Functional Host-Virus Interactions. Cancer Res. 2019, 79, 6010–6023. [Google Scholar] [CrossRef]
- Frappier, L. Epstein-Barr virus: Current questions and challenges. Tumour Virus Res. 2021, 12, 200218. [Google Scholar] [CrossRef]
- Hjalgrim, H.; Friborg, J.; Melbye, M. The Epidemiology of EBV and Its Association with Malignant Disease; Cambridge University Press: Cambridge, UK, 2007; ISBN 9780521827140. [Google Scholar]
- Siaghani, P.J.; Wong, J.T.; Chan, J.; Weisenburger, D.D.; Song, J.Y. Epidemiology and Pathology of T- and NK-Cell Lymphomas. In Cancer Treatment and Research; Springer International Publishing: New York, NY, USA, 2019; Volume 176, pp. 1–29. [Google Scholar]
- Houen, G.; Trier, N.H.; Frederiksen, J.L. Epstein-Barr Virus and Multiple Sclerosis. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Hassani, A.; Corboy, J.R.; Al-Salam, S.; Khan, G. Epstein-Barr virus is present in the brain of most cases of multiple sclerosis and may engage more than just B cells. PLoS ONE 2018, 13, e0192109. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Jakimovski, D.; Ramanathan, M.; Weinstock-Guttman, B.; Zivadinov, R. The role of Epstein-Barr virus in multiple sclerosis: From molecular pathophysiology to in vivo imaging. Neural Regen. Res. 2019, 14, 373. [Google Scholar] [CrossRef]
- Aygun, D.; Kuskucu, M.A.; Sahin, S.; Adrovic, A.; Barut, K.; Yıldız, M.; Sharifova, S.; Midilli, K.; Cokugras, H.; Camcıoglu, Y.; et al. Epstein–Barr virus, cytomegalovirus and BK polyomavirus burden in juvenile systemic lupus erythematosus: Correlation with clinical and laboratory indices of disease activity. Lupus 2020, 29, 1263–1269. [Google Scholar] [CrossRef]
- Li, Z.-X.; Zeng, S.; Wu, H.-X.; Zhou, Y. The risk of systemic lupus erythematosus associated with Epstein–Barr virus infection: A systematic review and meta-analysis. Clin. Exp. Med. 2019, 19, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Draborg, A.H.; Duus, K.; Houen, G. Epstein-Barr Virus and Systemic Lupus Erythematosus. Clin. Dev. Immunol. 2012, 2012, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Farahmand, M.; Monavari, S.H.; Shoja, Z.; Ghaffari, H.; Tavakoli, M.; Tavakoli, A. Epstein–Barr virus and risk of breast cancer: A systematic review and meta-analysis. Future Oncol. 2019, 15, 2873–2885. [Google Scholar] [CrossRef]
- Hu, H.; Luo, M.-L.; Desmedt, C.; Nabavi, S.; Yadegarynia, S.; Hong, A.; Konstantinopoulos, P.A.; Gabrielson, E.; Hines-Boykin, R.; Pihan, G.; et al. Epstein–Barr Virus Infection of Mammary Epithelial Cells Promotes Malignant Transformation. EBioMedicine 2016, 9, 148–160. [Google Scholar] [CrossRef]
- Sinclair, A.J.; Moalwi, M.H.; Amoaten, T. Is EBV Associated with Breast Cancer in Specific Geographic Locations? Cancers 2021, 13, 819. [Google Scholar] [CrossRef]
- Jin, Q.; Su, J.; Yan, D.; Wu, S. Epstein-Barr Virus Infection and Increased Sporadic Breast Carcinoma Risk: A Meta-Analysis. Med. Princ. Pract. 2020, 29, 195–200. [Google Scholar] [CrossRef]
- Rosenkilde, M.; Kledal, T. Targeting Herpesvirus Reliance of the Chemokine System. Curr. Drug Targets 2006, 7, 103–118. [Google Scholar] [CrossRef]
- Rezk, S.A.; Zhao, X.; Weiss, L.M. Epstein-Barr virus (EBV)-associated lymphoid proliferations, a 2018 update. Hum. Pathol. 2018, 79, 18–41. [Google Scholar] [CrossRef] [PubMed]
- Cuccaro, A.; Bartolomei, F.; Cupelli, E.; Hohaus, S. Prognostic factors in Hodgkin Lymphoma. Mediterr. J. Hematol. Infect. Dis. 2014, 6, e2014053. [Google Scholar] [CrossRef]
- Kimura, H. EBV in T-/NK-Cell Tumorigenesis. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2018; Volume 1045, pp. 459–475. [Google Scholar]
- Kimura, H.; Fujiwara, S. Overview of EBV-Associated T/NK-Cell Lymphoproliferative Diseases. Front. Pediatr. 2019, 6, 417. [Google Scholar] [CrossRef]
- Cohen, J.I. Vaccine Development for Epstein-Barr Virus. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2018; Volume 1045, pp. 477–493. [Google Scholar]
- WHO. Hodgkin Lymphoma in 2018; GLOBOCAN 2018; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Casulo, C.; Friedberg, J. Treating Burkitt Lymphoma in Adults. Curr. Hematol. Malig. Rep. 2015, 10, 266–271. [Google Scholar] [CrossRef]
- Burkitt, D. A “Tumour Safari” in East and Central Africa. Br. J. Cancer 1962, 16, 379–386. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Miyazaki, K. Current treatment approaches for NK/T-cell lymphoma. J. Clin. Exp. Hematop. 2017, 57, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.-T.; Wang, D.; Luo, H.; Xiao, M.; Zhou, H.-S.; Liu, D.; Ling, S.-P.; Wang, N.; Hu, X.-L.; Luo, Y.; et al. Aggressive NK-cell leukemia: Clinical subtypes, molecular features, and treatment outcomes. Blood Cancer J. 2017, 7, 660. [Google Scholar] [CrossRef]
- Lunning, M.A.; Vose, J.M. Angioimmunoblastic T-cell lymphoma: The many-faced lymphoma. Blood 2017, 129, 1095–1102. [Google Scholar] [CrossRef]
- Querfeld, C.; Zain, J.; Rosen, S.T. T-Cell and NK-Cell Lymphomas; Cancer Treatment and Research; Springer International Publishing: Cham, Switzerland, 2019; Volume 176, ISBN 978-3-319-99715-5. [Google Scholar]
- Gru, A.A.; Haverkos, B.H.; Freud, A.G.; Hastings, J.; Nowacki, N.B.; Barrionuevo, C.; Vigil, C.E.; Rochford, R.; Natkunam, Y.; Baiocchi, R.A.; et al. The Epstein-Barr Virus (EBV) in T Cell and NK Cell Lymphomas: Time for a Reassessment. Curr. Hematol. Malig. Rep. 2015, 10, 456–467. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; IARC: Lyon, France, 2017; Volume 2, ISBN 978-92-832-4494-3. [Google Scholar]
- WHO. Non-Hodgkin Lymphoma in 2018; GLOBOCAN 2018; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Dunleavy, K.; Roschewski, M.; Wilson, W.H. Lymphomatoid Granulomatosis and Other Epstein-Barr Virus Associated Lymphoproliferative Processes. Curr. Hematol. Malig. Rep. 2012, 7, 208–215. [Google Scholar] [CrossRef]
- Gutiérrez-Domingo, Á.; Gutiérrez-Domingo, I.; Gallardo-Rodríguez, K.M. Lymphomatoid Granulomatosis: A Rare Tumor with Poor Prognosis. Arch. Bronconeumol. 2018, 54, 108–109. [Google Scholar] [CrossRef] [PubMed]
- Roschewski, M.; Wilson, W.H. Lymphomatoid Granulomatosis. Cancer J. 2012, 18, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Song, J.Y.; Pittaluga, S.; Dunleavy, K.; Grant, N.; White, T.; Jiang, L.; Davies-Hill, T.; Raffeld, M.; Wilson, W.H.; Jaffe, E.S. Lymphomatoid Granulomatosis—A Single Institute Experience. Am. J. Surg. Pathol. 2015, 39, 141–156. [Google Scholar] [CrossRef]
- Chen, Y.-P.; Chan, A.T.C.; Le, Q.-T.; Blanchard, P.; Sun, Y.; Ma, J. Nasopharyngeal carcinoma. Lancet 2019, 394, 64–80. [Google Scholar] [CrossRef]
- Houldcroft, C.J.; Kellam, P. Host genetics of Epstein-Barr virus infection, latency and disease. Rev. Med. Virol. 2015, 25, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Khan, G.; Hashim, M.J. Global burden of deaths from Epstein-Barr virus attributable malignancies 1990–2010. Infect. Agent. Cancer 2014, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Farrell, P.J. Epstein-Barr Virus and Cancer. Annu. Rev. Pathol. Mech. Dis. 2019, 14, 29–53. [Google Scholar] [CrossRef] [PubMed]
- WHO. Nasopharyngeal Carcinoma in 2018; GLOBOCAN 2018; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Nishikawa, J.; Iizasa, H.; Yoshiyama, H.; Shimokuri, K.; Kobayashi, Y.; Sasaki, S.; Nakamura, M.; Yanai, H.; Sakai, K.; Suehiro, Y.; et al. Clinical Importance of Epstein–Barr Virus-Associated Gastric Cancer. Cancers 2018, 10, 167. [Google Scholar] [CrossRef] [PubMed]
- Naseem, M.; Barzi, A.; Brezden-Masley, C.; Puccini, A.; Berger, M.D.; Tokunaga, R.; Battaglin, F.; Soni, S.; McSkane, M.; Zhang, W.; et al. Outlooks on Epstein-Barr virus associated gastric cancer. Cancer Treat. Rev. 2018, 66, 15–22. [Google Scholar] [CrossRef]
- Kang, B.W.; Baek, D.W.; Kang, H.; Baek, J.H.; Kim, J.G. Novel Therapeutic Approaches for Epstein-Barr Virus Associated Gastric Cancer. Anticancer Res. 2019, 39, 4003–4010. [Google Scholar] [CrossRef]
- Sitarz, R.; Skierucha, M.; Mielko, J.; Offerhaus, J.; Maciejewski, R.; Polkowski, W. Gastric cancer: Epidemiology, prevention, classification, and treatment. Cancer Manag. Res. 2018, 10, 239–248. [Google Scholar] [CrossRef]
- Fukayama, M. Epstein-Barr virus and gastric carcinoma. Pathol. Int. 2010, 60, 337–350. [Google Scholar] [CrossRef]
- WHO. Stomach Cancer in 2018; GLOBOCAN 2018; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Cheng, N.; Hui, D.; Liu, Y.; Zhang, N.; Jiang, Y.; Han, J.; Li, H.; Ding, Y.; Du, H.; Chen, J.; et al. Is gastric lymphoepithelioma-like carcinoma a special subtype of EBV-associated gastric carcinoma? New insight based on clinicopathological features and EBV genome polymorphisms. Gastric Cancer 2015, 18, 246–255. [Google Scholar] [CrossRef]
- Iezzoni, J.C.; Gaffey, M.J.; Weiss, L.M. The Role of Epstein-Barr Virus in Lymphoepithelioma-like Carcinomas. Am. J. Clin. Pathol. 1995, 103, 308–315. [Google Scholar] [CrossRef]
- Díaz del Arco, C.; Esteban Collazo, F.; Fernández Aceñero, M.J. Lymphoepithelioma-like carcinoma of the large intestine: A case report and literature review. Rev. Esp. Patol. 2018, 51, 18–22. [Google Scholar] [CrossRef]
- Labgaa, I.; Stueck, A.; Ward, S.C. Lymphoepithelioma-Like Carcinoma in Liver. Am. J. Pathol. 2017, 187, 1438–1444. [Google Scholar] [CrossRef] [PubMed]
- Koufopoulos, N.; Syrios, J.; Papanikolaou, A.; Misitzis, I.; Kapatou, K.A.; Dimas, D.; Khaldi, L. Lymphoepithelioma-like breast carcinoma. Pol. J. Pathol. 2018, 69, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-J.; Qiao, Y.-W.; Zhao, X.-Q.; Liu, J. Clinicopathological features of Epstein-Barr virus-associated gastric carcinoma: A systematic review and meta-analysis. J. Balk. Union Oncol. 2019, 24, 1092–1099. [Google Scholar]
- Bittar, Z.; Fend, F.; Quintanilla-Martinez, L. Lymphoepithelioma-like carcinoma of the stomach: A case report and review of the literature. Diagn. Pathol. 2013, 8, 184. [Google Scholar] [CrossRef]
- Bai, Y.; Gao, Q.; Ren, G.; Wang, B.; Xiang, H. Epstein-Barr virus-associated lymphoepithelioma-like gastric carcinoma located on gastric high body: Two case reports. Indian J. Pathol. Microbiol. 2014, 57, 463. [Google Scholar] [CrossRef] [PubMed]
- Bedri, S.; Sultan, A.A.; Alkhalaf, M.; Al Moustafa, A.-E.; Vranic, S. Epstein-Barr virus (EBV) status in colorectal cancer: A mini review. Hum. Vaccin. Immunother. 2019, 15, 603–610. [Google Scholar] [CrossRef]
- WHO. Breast Cancer in 2018; GLOBOCAN 2018; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Münz, C. Epstein Barr Virus Volume 1; Current Topics in Microbiology and Immunology; Springer International Publishing: Cham, Switzerland, 2015; Volume 390, ISBN 978-3-319-22821-1. [Google Scholar]
- Li, S.; Young, K.H.; Medeiros, L.J. Diffuse large B-cell lymphoma. Pathology 2018, 50, 74–87. [Google Scholar] [CrossRef]
- Castillo, J.J.; Beltran, B.E.; Miranda, R.N.; Young, K.H.; Chavez, J.C.; Sotomayor, E.M. EBV-positive diffuse large B-cell lymphoma, not otherwise specified: 2018 update on diagnosis, risk-stratification and management. Am. J. Hematol. 2018, 93, 953–962. [Google Scholar] [CrossRef]
- Lu, T.-X.; Liang, J.-H.; Miao, Y.; Fan, L.; Wang, L.; Qu, X.-Y.; Cao, L.; Gong, Q.-X.; Wang, Z.; Zhang, Z.-H.; et al. Epstein-Barr virus positive diffuse large B-cell lymphoma predict poor outcome, regardless of the age. Sci. Rep. 2015, 5, 12168. [Google Scholar] [CrossRef]
- Liu, Y.; Barta, S.K. Diffuse large B-cell lymphoma: 2019 update on diagnosis, risk stratification, and treatment. Am. J. Hematol. 2019, 94, 604–616. [Google Scholar] [CrossRef]
- Green, M.; Michaels, M.G. Epstein-Barr Virus Infection and Posttransplant Lymphoproliferative Disorder. Am. J. Transplant. 2013, 13, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Montoto, S.; Wilson, J.; Shaw, K.; Heath, M.; Wilson, A.; McNamara, C.; Orkin, C.; Nelson, M.; Johnson, M.; Bower, M.; et al. Excellent immunological recovery following CODOX-M/IVAC, an effective intensive chemotherapy for HIV-associated Burkitt’s lymphoma. AIDS 2010, 24, 851–856. [Google Scholar] [CrossRef]
- Jacobson, C.A.; Abramson, J.S. HIV-Associated Hodgkin’s Lymphoma: Prognosis and Therapy in the Era of cART. Adv. Hematol. 2012, 2012, 1–8. [Google Scholar] [CrossRef]
- Lopez, A.; Abrisqueta, P. Plasmablastic lymphoma: Current perspectives. Blood Lymphat. Cancer Targets Ther. 2018, 8, 63–70. [Google Scholar] [CrossRef]
- Tchernonog, E.; Faurie, P.; Coppo, P.; Monjanel, H.; Bonnet, A.; Algarte Génin, M.; Mercier, M.; Dupuis, J.; Bijou, F.; Herbaux, C.; et al. Clinical characteristics and prognostic factors of plasmablastic lymphoma patients: Analysis of 135 patients from the LYSA group. Ann. Oncol. 2017, 28, 843–848. [Google Scholar] [CrossRef]
- Gravelle, P.; Péricart, S.; Tosolini, M.; Fabiani, B.; Coppo, P.; Amara, N.; Traverse-Gléhen, A.; Van Acker, N.; Brousset, P.; Fournie, J.-J.; et al. EBV infection determines the immune hallmarks of plasmablastic lymphoma. Oncoimmunology 2018, 7, e1486950. [Google Scholar] [CrossRef] [PubMed]
- Grogg, K.L.; Miller, R.F.; Dogan, A. HIV infection and lymphoma. J. Clin. Pathol. 2006, 60, 1365–1372. [Google Scholar] [CrossRef] [PubMed]
- UNAIDS. Global HIV & AIDS Statistics—2020 Fact Sheet. Available online: https://www.unaids.org/en/resources/fact-sheet (accessed on 27 May 2021).
- Bibas, M.; Antinori, A. EBV and HIV-Related Lymphoma. Mediterr. J. Hematol. Infect. Dis. 2009, 1, e2009032. [Google Scholar] [CrossRef] [PubMed]
- GODT. WHO International Report on Organ Donation and Transplantation Activities 2019; Executive Summary 2019; WHO: Geneva, Switzerland, 2021; pp. 1–12. [Google Scholar]
- Dharnidharka, V.R.; Webster, A.C.; Martinez, O.M.; Preiksaitis, J.K.; Leblond, V.; Choquet, S. Post-transplant lymphoproliferative disorders. Nat. Rev. Dis. Prim. 2016, 2, 15088. [Google Scholar] [CrossRef]
- Lee, B.; Bower, M.; Newsom-Davis, T.; Nelson, M. HIV-related lymphoma. HIV Ther. 2010, 4, 649–659. [Google Scholar] [CrossRef]
- Rosen, S.T. HIV/AIDS-Associated Viral Oncogenesis; Meyers, C., Ed.; Cancer Treatment and Research; Springer International Publishing: Cham, Switzerland, 2019; Volume 177, ISBN 978-3-030-03501-3. [Google Scholar]
- Wu, D.; Chen, C.; Zhang, M.; Li, Z.; Wang, S.; Shi, J.; Zhang, Y.; Yao, D.; Hu, S. The clinical features and prognosis of 100 AIDS-related lymphoma cases. Sci. Rep. 2019, 9, 5381. [Google Scholar] [CrossRef]
- Gopal, S.; Patel, M.R.; Yanik, E.L.; Cole, S.R.; Achenbach, C.J.; Napravnik, S.; Burkholder, G.A.; Reid, E.G.; Rodriguez, B.; Deeks, S.G.; et al. Temporal Trends in Presentation and Survival for HIV-Associated Lymphoma in the Antiretroviral Therapy Era. J. Natl. Cancer Inst. 2013, 105, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Meister, A.; Hentrich, M.; Wyen, C.; Hübel, K. Malignant lymphoma in the HIV-positive patient. Eur. J. Haematol. 2018, 101, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Nowalk, A.; Green, M. Epstein-Barr Virus. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Gross, T.G.; Orjuela, M.A.; Perkins, S.L.; Park, J.R.; Lynch, J.C.; Cairo, M.S.; Smith, L.M.; Hayashi, R.J. Low-Dose Chemotherapy and Rituximab for Posttransplant Lymphoproliferative Disease (PTLD): A Children’s Oncology Group Report. Am. J. Transplant. 2012, 12, 3069–3075. [Google Scholar] [CrossRef] [PubMed]
- Dekate, J.; Chetty, R. Epstein-Barr Virus-Associated Smooth Muscle Tumor. Arch. Pathol. Lab. Med. 2016, 140, 718–722. [Google Scholar] [CrossRef]
- Hussein, K.; Rath, B.; Ludewig, B.; Kreipe, H.; Jonigk, D. Clinico-pathological characteristics of different types of immunodeficiency-associated smooth muscle tumours. Eur. J. Cancer 2014, 50, 2417–2424. [Google Scholar] [CrossRef]
- Prockop, S.E.; Doubrovina, E.; Boulad, F.; Kernan, N.A.; Kobos, R.; Scaradavou, A.; Abramson, S.J.; Laquaglia, M.; Price, A.; O’Reilly, R.J. Adoptive Treatment Of EBV-Associated Leiomyosarcoma in Immunodeficient Patients With EBV Specific Cytotoxic T Cells. Blood 2013, 122, 3267. [Google Scholar] [CrossRef]
- Jonigk, D.; Laenger, F.; Maegel, L.; Izykowski, N.; Rische, J.; Tiede, C.; Klein, C.; Maecker-Kolhoff, B.; Kreipe, H.; Hussein, K. Molecular and Clinicopathological Analysis of Epstein-Barr Virus-Associated Posttransplant Smooth Muscle Tumors. Am. J. Transplant. 2012, 12, 1908–1917. [Google Scholar] [CrossRef]
- Magg, T.; Schober, T.; Walz, C.; Ley-Zaporozhan, J.; Facchetti, F.; Klein, C.; Hauck, F. Epstein-Barr Virus+ Smooth Muscle Tumors as Manifestation of Primary Immunodeficiency Disorders. Front. Immunol. 2018, 9, 368. [Google Scholar] [CrossRef]
- Pender, M.P.; Csurhes, P.A.; Smith, C.; Douglas, N.L.; Neller, M.A.; Matthews, K.K.; Beagley, L.; Rehan, S.; Crooks, P.; Hopkins, T.J.; et al. Epstein-Barr virus–specific T cell therapy for progressive multiple sclerosis. JCI Insight 2018, 3, e124714. [Google Scholar] [CrossRef]
- Dunleavy, K.; Pittaluga, S.; Shovlin, M.; Steinberg, S.M.; Cole, D.; Grant, C.; Widemann, B.; Staudt, L.M.; Jaffe, E.S.; Little, R.F.; et al. Low-Intensity Therapy in Adults with Burkitt’s Lymphoma. N. Engl. J. Med. 2013, 369, 1915–1925. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.A.; Faderl, S.; O’Brien, S.; Bueso-Ramos, C.; Cortes, J.; Garcia-Manero, G.; Giles, F.J.; Verstovsek, S.; Wierda, W.G.; Pierce, S.A.; et al. Chemoimmunotherapy with hyper-CVAD plus rituximab for the treatment of adult Burkitt and Burkitt-type lymphoma or acute lymphoblastic leukemia. Cancer 2006, 106, 1569–1580. [Google Scholar] [CrossRef] [PubMed]
- Sweetenham, J.W.; Pearce, R.; Taghipour, G.; Blaise, D.; Gisselbrecht, C.; Goldstone, A.H. Adult Burkitt’s and Burkitt-like non-Hodgkin’s lymphoma—Outcome for patients treated with high-dose therapy and autologous stem-cell transplantation in first remission or at relapse: Results from the European Group for Blood and Marrow Transplantation. J. Clin. Oncol. 1996, 14, 2465–2472. [Google Scholar] [CrossRef]
- Oriol, A.; Ribera, J.; Bergua, J.; Giménez Mesa, E.; Grande, C.; Esteve, J.; Brunet, S.; Moreno, M.; Escoda, L.; Hernandez-Rivas, J.; et al. High-dose chemotherapy and immunotherapy in adult Burkitt lymphoma. Cancer 2008, 113, 117–125. [Google Scholar] [CrossRef]
- Haahr, S.; Höllsberg, P. Multiple sclerosis is linked to Epstein-Barr virus infection. Rev. Med. Virol. 2006, 16, 297–310. [Google Scholar] [CrossRef] [PubMed]
- WHO. Atlas: Multiple Sclerosis in the World in 2008; WHO: Geneva, Switzerland, 2008. [Google Scholar]
- Fernández-Menéndez, S.; Fernández-Morán, M.; Fernández-Vega, I.; Pérez-Álvarez, A.; Villafani-Echazú, J. Epstein-Barr virus and multiple sclerosis. From evidence to therapeutic strategies. J. Neurol. Sci. 2016, 361, 213–219. [Google Scholar] [CrossRef]
- Laurence, M.; Benito-León, J. Epstein-Barr virus and multiple sclerosis: Updating Pender’s hypothesis. Mult. Scler. Relat. Disord. 2017, 16, 8–14. [Google Scholar] [CrossRef]
- Lucas, R.M.; Hughes, A.M.; Lay, M.-L.J.; Ponsonby, A.-L.; Dwyer, D.E.; Taylor, B.V.; Pender, M.P. Epstein-Barr virus and multiple sclerosis. J. Neurol. Neurosurg. Psychiatry 2011, 82, 1142–1148. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, R.; Rashid, W. Multiple sclerosis. BMJ Clin. Evid. 2012, 10, 1202. [Google Scholar]
- Alexander, S.P.H.; Davenport, A.P.; Kelly, E.; Marrion, N.; Peters, J.A.; Benson, H.E.; Faccenda, E.; Pawson, A.J.; Sharman, J.L.; Southan, C.; et al. The Concise Guide to Pharmacology 2015/16: G protein-coupled receptors. Br. J. Pharmacol. 2015, 172, 5744–5869. [Google Scholar] [CrossRef]
- Weis, W.I.; Kobilka, B.K. The Molecular Basis of G Protein-Coupled Receptor Activation. Annu. Rev. Biochem. 2018, 87, 897–919. [Google Scholar] [CrossRef]
- Mirzadegan, T.; Benko, G.; Filipek, S.; Palczewski, K. Sequence Analyses of G-Protein-Coupled Receptors: Similarities to Rhodopsin. Biochemistry 2003, 42, 2759. [Google Scholar] [CrossRef] [PubMed]
- Rovati, G.E.; Capra, V.; Neubig, R.R. The Highly Conserved DRY Motif of Class A G Protein-Coupled Receptors: Beyond the Ground State. Mol. Pharmacol. 2007, 71, 959–964. [Google Scholar] [CrossRef]
- Trzaskowski, B.; Latek, D.; Yuan, S.; Ghoshdastider, U.; Debinski, A.; Filipek, S. Action of Molecular Switches in GPCRs—Theoretical and Experimental Studies. Curr. Med. Chem. 2012, 19, 1090–1109. [Google Scholar] [CrossRef]
- Wacker, D.; Stevens, R.C.; Roth, B.L. How Ligands Illuminate GPCR Molecular Pharmacology. Cell 2017, 170, 414–427. [Google Scholar] [CrossRef]
- Lyngaa, R.; Nørregaard, K.; Kristensen, M.; Kubale, V.; Rosenkilde, M.M.; Kledal, T.N. Cell transformation mediated by the Epstein–Barr virus G protein-coupled receptor BILF1 is dependent on constitutive signaling. Oncogene 2010, 29, 4388–4398. [Google Scholar] [CrossRef]
- Downes, G.B.; Gautam, N. The G Protein Subunit Gene Families. Genomics 1999, 62, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Dhanasekaran, N.; Dermott, J.M. Signaling by the G12 class of G proteins. Cell. Signal. 1996, 8, 235–245. [Google Scholar] [CrossRef]
- Suzuki, N.; Hajicek, N.; Kozasa, T. Regulation and physiological functions of G12/13-mediated signaling pathways. NeuroSignals 2009, 17, 55–70. [Google Scholar] [CrossRef]
- Komolov, K.E.; Du, Y.; Duc, N.M.; Betz, R.M.; Rodrigues, J.P.G.L.M.; Leib, R.D.; Patra, D.; Skiniotis, G.; Adams, C.M.; Dror, R.O.; et al. Structural and Functional Analysis of a β2-Adrenergic Receptor Complex with GRK5. Cell 2017, 169, 407–421.e16. [Google Scholar] [CrossRef]
- Spiess, K.; Fares, S.; Sparre-Ulrich, A.H.; Hilgenberg, E.; Jarvis, M.A.; Ehlers, B.; Rosenkilde, M.M. Identification and Functional Comparison of Seven-Transmembrane G-Protein-Coupled BILF1 Receptors in Recently Discovered Nonhuman Primate Lymphocryptoviruses. J. Virol. 2015, 89, 2253–2267. [Google Scholar] [CrossRef]
- Rummel, P.C.; Thiele, S.; Hansen, L.S.; Petersen, T.P.; Sparre-Ulrich, A.H.; Ulven, T.; Rosenkilde, M.M. Extracellular Disulfide Bridges Serve Different Purposes in Two Homologous Chemokine Receptors, CCR1 and CCR5. Mol. Pharmacol. 2013, 84, 335–345. [Google Scholar] [CrossRef]
- Mavri, M.; Spiess, K.; Rosenkilde, M.M.; Rutland, C.S.; Vrecl, M.; Kubale, V. Methods for Studying Endocytotic Pathways of Herpesvirus Encoded G Protein-Coupled Receptors. Molecules 2020, 25, 5710. [Google Scholar] [CrossRef]
- Rosenkilde, M.M.; Smit, M.J.; Waldhoer, M. Structure, function and physiological consequences of virally encoded chemokine seven transmembrane receptors. Br. J. Pharmacol. 2008, 153, 154–166. [Google Scholar] [CrossRef] [PubMed]
- Rosenkilde, M.M.; Waldhoer, M.; Lüttichau, H.R.; Schwartz, T.W. Virally encoded 7TM receptors. Oncogene 2001, 20, 1582–1593. [Google Scholar] [CrossRef] [PubMed]
- Casarosa, P.; Bakker, R.A.; Verzijl, D.; Navis, M.; Timmerman, H.; Leurs, R.; Smiti, M.J. Constitutive signaling of the human cytomegalovirus-encoded chemokine receptor US28. J. Biol. Chem. 2001, 276, 1133–1137. [Google Scholar] [CrossRef]
- Geras-Raaka, E.; Arvanitakis, L.; Bais, C.; Cesarman, E.; Mesri, E.A.; Gershengorn, M.C. Inhibition of Constitutive Signaling of Kaposi’s Sarcoma-Associated Herpesvirus G Protein-Coupled Receptor by Protein Kinases in Mammalian Cells in Culture. J. Exp. Med. 1998, 187, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Smit, M.J.; Verzijl, D.; Casarosa, P.; Navis, M.; Timmerman, H.; Leurs, R. Kaposi’s Sarcoma-Associated Herpesvirus-Encoded G Protein-Coupled Receptor ORF74 Constitutively Activates p44/p42 MAPK and Akt via Gi and Phospholipase C-Dependent Signaling Pathways. J. Virol. 2002, 76, 1744–1752. [Google Scholar] [CrossRef][Green Version]
- Couty, J.P.; Geras-Raaka, E.; Weksler, B.B.; Gershengorn, M.C. Kaposi’s Sarcoma-associated Herpesvirus G Protein-coupled Receptor Signals through Multiple Pathways in Endothelial Cells. J. Biol. Chem. 2001, 276, 33805–33811. [Google Scholar] [CrossRef]
- Griffin, B.D.; Gram, A.M.; Mulder, A.; Van Leeuwen, D.; Claas, F.H.J.; Wang, F.; Ressing, M.E.; Wiertz, E. EBV BILF1 Evolved to Downregulate Cell Surface Display of a Wide Range of HLA Class I Molecules through Their Cytoplasmic Tail. J. Immunol. 2013, 190, 1672–1684. [Google Scholar] [CrossRef]
- Tsutsumi, N.; Qu, Q.; Mavri, M.; Baggesen, M.S.; Maeda, S.; Waghray, D.; Berg, C.; Kobilka, B.K.; Rosenkilde, M.M.; Skiniotis, G.; et al. Structural basis for the constitutive activity and immunomodulatory properties of the Epstein-Barr virus-encoded G protein-coupled receptor BILF1. Immunity 2021, 54, 1405–1416. [Google Scholar] [CrossRef]
- Tierney, R.J.; Shannon-Lowe, C.D.; Fitzsimmons, L.; Bell, A.I.; Rowe, M. Unexpected patterns of Epstein-Barr virus transcription revealed by a High throughput PCR array for absolute quantification of viral mRNA. Virology 2015, 474, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Gao, J.; Cheng, L.; Yang, X.; Li, F.; Jiang, G. The Epstein-Barr virus-encoded G protein-coupled receptor BILF1 upregulates ICAM-1 through a mechanism involving the NF-κB pathway. Biosci. Biotechnol. Biochem. 2020, 84, 1810–1819. [Google Scholar] [CrossRef]
- Bayda, N.; Tilloy, V.; Chaunavel, A.; Bahri, R.; Halabi, M.A.; Feuillard, J.; Jaccard, A.; Ranger-Rogez, S. Comprehensive Epstein-Barr Virus Transcriptome by RNA-Sequencing in Angioimmunoblastic T Cell Lymphoma (AITL) and Other Lymphomas. Cancers 2021, 13, 610. [Google Scholar] [CrossRef] [PubMed]
- Borozan, I.; Zapatka, M.; Frappier, L.; Ferretti, V. Analysis of Epstein-Barr Virus Genomes and Expression Profiles in Gastric Adenocarcinoma. J. Virol. 2017, 92. [Google Scholar] [CrossRef]
- Fares, S.; Spiess, K.; Olesen, E.T.B.; Zuo, J.; Jackson, S.; Kledal, T.N.; Wills, M.R.; Rosenkilde, M.M. Distinct Roles of Extracellular Domains in the Epstein-Barr Virus-Encoded BILF1 Receptor for Signaling and Major Histocompatibility Complex Class I Downregulation. mBio 2019, 10, e01707-18. [Google Scholar] [CrossRef]
- Vischer, H.F.; Nijmeijer, S.; Smit, M.J.; Leurs, R. Viral hijacking of human receptors through heterodimerization. Biochem. Biophys. Res. Commun. 2008, 377, 93–97. [Google Scholar] [CrossRef]
- Nijmeijer, S.; Leurs, R.; Smit, M.J.; Vischer, H.F. The Epstein-Barr Virus-encoded G Protein-coupled Receptor BILF1 Hetero-oligomerizes with Human CXCR4, Scavenges Gαi Proteins, and Constitutively Impairs CXCR4 Functioning. J. Biol. Chem. 2010, 285, 29632–29641. [Google Scholar] [CrossRef] [PubMed]
- Paydas, S.; Ergin, M.; Erdogan, S.; Seydaoglu, G. Prognostic significance of EBV-LMP1 and VEGF-A expressions in non-Hodgkin’s lymphomas. Leuk. Res. 2008, 32, 1424–1430. [Google Scholar] [CrossRef] [PubMed]
- Krishna, S.M.; James, S.; Balaram, P. Expression of VEGF as prognosticator in primary nasopharyngeal cancer and its relation to EBV status. Virus Res. 2006, 115, 85–90. [Google Scholar] [CrossRef]
- Hayes, S.H.; Seigel, G.M. Immunoreactivity of ICAM-1 in human tumors, metastases and normal tissues. Int. J. Clin. Exp. Pathol. 2009, 2, 553–560. [Google Scholar] [PubMed]
- Lindberg, J.S.; Moe, S.M.; Goodman, W.G.; Coburn, J.W.; Sprague, S.M.; Liu, W.; Blaisdell, P.W.; Brenner, R.M.; Turner, S.A.; Martin, K.J. The calcimimetic AMG 073 reduces parathyroid hormone and calcium x phosphorus in secondary hyperparathyroidism. Kidney Int. 2003, 63, 248–254. [Google Scholar] [CrossRef][Green Version]
- Ritter, K.; Buning, C.; Halland, N.; Pöverlein, C.; Schwink, L. G Protein-Coupled Receptor 119 (GPR119) Agonists for the Treatment of Diabetes: Recent Progress and Prevailing Challenges. J. Med. Chem. 2016, 59, 3579–3592. [Google Scholar] [CrossRef]
- Irving, C.B.; Adams, C.E.; Lawrie, S. Haloperidol versus placebo for schizophrenia. In Cochrane Database of Systematic Reviews; Irving, C.B., Ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2006. [Google Scholar]
- Brinkmann, V.; Davis, M.D.; Heise, C.E.; Albert, R.; Cottens, S.; Hof, R.; Bruns, C.; Prieschl, E.; Baumruker, T.; Hiestand, P.; et al. The Immune Modulator FTY720 Targets Sphingosine 1-Phosphate Receptors. J. Biol. Chem. 2002, 277, 21453–21457. [Google Scholar] [CrossRef] [PubMed]
- Nieto Gutierrez, A.; Mcdonald, P.H. GPCRs: Emerging anti-cancer drug targets. Cell. Signal. 2017, 41, 65–74. [Google Scholar] [CrossRef]
- Perez Almeria, C.V.; Setiawan, I.M.; Siderius, M.; Smit, M.J. G protein-coupled receptors as promising targets in cancer. Curr. Opin. Endocr. Metab. Res. 2021, 16, 119–127. [Google Scholar] [CrossRef]
- Dorr, P.; Westby, M.; Dobbs, S.; Griffin, P.; Irvine, B.; Macartney, M.; Mori, J.; Rickett, G.; Smith-Burchnell, C.; Napier, C.; et al. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 2005, 49, 4721–4732. [Google Scholar] [CrossRef] [PubMed]
- Heasley, L.E. Autocrine and paracrine signaling through neuropeptide receptors in human cancer. Oncogene 2001, 20, 1563–1569. [Google Scholar] [CrossRef] [PubMed]
- Trejo, J. Dysregulation of G Protein-Coupled Receptor Signaling in Cancer. In Signal Transduction: Pathways, Mechanisms and Diseases; Springer: Berlin/Heidelberg, Germany, 2010; pp. 83–98. ISBN 9783642021114. [Google Scholar]
- Kashyap, M.K.; Kumar, D.; Jones, H.; Amaya-Chanaga, C.I.; Choi, M.Y.; Melo-Cardenas, J.; Ale-Ali, A.; Kuhne, M.R.; Sabbatini, P.; Cohen, L.J.; et al. Ulocuplumab (BMS-936564/MDX1338): A fully human anti- CXCR4 antibody induces cell death in chronic lymphocytic leukemia mediated through a reactive oxygen speciesdependent pathway. Oncotarget 2016, 7, 2809–2822. [Google Scholar] [CrossRef] [PubMed]
- Rosenkilde, M.M.; Kledal, T.N.; Bräuner-Osborne, H.; Schwartz, T.W. Agonists and Inverse Agonists for the Herpesvirus 8-encoded Constitutively Active Seven-transmembrane Oncogene Product, ORF-74. J. Biol. Chem. 1999, 274, 956–961. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, D.; Chandran, B.; Berger, E.A. Selective killing of Kaposi’s sarcoma-associated herpesvirus lytically infected cells with a recombinant immunotoxin targeting the viral gpK8.1A envelope glycoprotein. mAbs 2012, 4, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Berger, E.A. An immunotoxin targeting the gH glycoprotein of KSHV for selective killing of cells in the lytic phase of infection. Antivir. Res. 2011, 90, 143–150. [Google Scholar] [CrossRef]
- Lee, S.; Chung, Y.H.; Lee, C. US28, a Virally-Encoded GPCR as an Antiviral Target for Human Cytomegalovirus Infection. Biomol. Ther. 2017, 25, 69–79. [Google Scholar] [CrossRef]
- Kralj, A.; Wetzel, A.; Mahmoudian, S.; Stamminger, T.; Tschammer, N.; Heinrich, M.R. Identification of novel allosteric modulators for the G-protein coupled US28 receptor of human cytomegalovirus. Bioorg. Med. Chem. Lett. 2011, 21, 5446–5450. [Google Scholar] [CrossRef]
- Kralj, A.; Nguyen, M.T.; Tschammer, N.; Ocampo, N.; Gesiotto, Q.; Heinrich, M.R.; Phanstiel, O. Development of flavonoid-based inverse agonists of the key signaling receptor US28 of human cytomegalovirus. J. Med. Chem. 2013, 56, 5019–5032. [Google Scholar] [CrossRef] [PubMed]
- Kralj, A.; Kurt, E.; Tschammer, N.; Heinrich, M.R. Synthesis and Biological Evaluation of Biphenyl Amides That Modulate the US28 Receptor. ChemMedChem 2014, 9, 151–168. [Google Scholar] [CrossRef]
- Lückmann, M.; Amarandi, R.-M.; Papargyri, N.; Jakobsen, M.H.; Christiansen, E.; Jensen, L.J.; Pui, A.; Schwartz, T.W.; Rosenkilde, M.M.; Frimurer, T.M. Structure-based discovery of novel US28 small molecule ligands with different modes of action. Chem. Biol. Drug Des. 2017, 89, 289–296. [Google Scholar] [CrossRef]
- Amărandi, R.-M.; Lückmann, M.; Melynis, M.; Jakobsen, M.H.; Fallah, Z.; Spiess, K.; Hjortø, G.M.; Pui, A.; Frimurer, T.M.; Rosenkilde, M.M. Ligand-selective small molecule modulators of the constitutively active vGPCR US28. Eur. J. Med. Chem. 2018, 155, 244–254. [Google Scholar] [CrossRef]
- Heukers, R.; Fan, T.S.; de Wit, R.H.; van Senten, J.R.; De Groof, T.W.M.; Bebelman, M.P.; Lagerweij, T.; Vieira, J.; de Munnik, S.M.; Smits-de Vries, L.; et al. The constitutive activity of the virally encoded chemokine receptor US28 accelerates glioblastoma growth. Oncogene 2018, 37, 4110–4121. [Google Scholar] [CrossRef] [PubMed]
- De Groof, T.W.M.; Mashayekhi, V.; Fan, T.S.; Bergkamp, N.D.; Sastre Toraño, J.; van Senten, J.R.; Heukers, R.; Smit, M.J.; Oliveira, S. Nanobody-Targeted Photodynamic Therapy Selectively Kills Viral GPCR-Expressing Glioblastoma Cells. Mol. Pharm. 2019, 16, 3145–3156. [Google Scholar] [CrossRef]
- De Groof, T.; Elder, E.; Heukers, R.; Lim, E.; Wills, M.; Sinclair, J.; Smit, M. Targeting the latent human cytomegalovirus reservoir with virus specific nanobodies. bioRxiv 2020. [Google Scholar] [CrossRef]
- Spiess, K.; Jeppesen, M.G.; Malmgaard-Clausen, M.; Krzywkowski, K.; Dulal, K.; Cheng, T.; Hjortø, G.M.; Larsen, O.; Burg, J.S.; Jarvis, M.A.; et al. Rationally designed chemokine-based toxin targeting the viral G protein-coupled receptor US28 potently inhibits cytomegalovirus infection in vivo. Proc. Natl. Acad. Sci. USA 2015, 112, 8427–8432. [Google Scholar] [CrossRef]
- Krishna, B.A.; Spiess, K.; Poole, E.L.; Lau, B.; Voigt, S.; Kledal, T.N.; Rosenkilde, M.M.; Sinclair, J.H. Targeting the latent cytomegalovirus reservoir with an antiviral fusion toxin protein. Nat. Commun. 2017, 8, 14321. [Google Scholar] [CrossRef] [PubMed]
- Spiess, K.; Jeppesen, M.G.; Malmgaard-Clausen, M.; Krzywkowski, K.; Kledal, T.N.; Rosenkilde, M.M. Novel Chemokine-Based Immunotoxins for Potent and Selective Targeting of Cytomegalovirus Infected Cells. J. Immunol. Res. 2017, 2017, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Zhang, D.; Mao, Y.; Zhu, J.; Ming, H.; Wen, J.; Ma, J.; Cao, Q.; Lin, H.; Tang, Q.; et al. A Human Fab-Based Immunoconjugate Specific for the LMP1 Extracellular Domain Inhibits Nasopharyngeal Carcinoma Growth In Vitro and In Vivo. Mol. Cancer Ther. 2012, 11, 594–603. [Google Scholar] [CrossRef]
- Zhu, S.; Chen, J.; Xiong, Y.; Kamara, S.; Gu, M.; Tang, W.; Chen, S.; Dong, H.; Xue, X.; Zheng, Z.-M.; et al. Novel EBV LMP-2-affibody and affitoxin in molecular imaging and targeted therapy of nasopharyngeal carcinoma. PLoS Pathog. 2020, 16, e1008223. [Google Scholar] [CrossRef] [PubMed]
| EBV-Associated Malignancy | EBV Association | EBV Association—Dependent Upon Type | Incidence with EBV Association | Geography | Cellular Background | Localization | Prognosis | |
|---|---|---|---|---|---|---|---|---|
| Immunocompetent patients | Hodgkin lymphoma (latency II) [37,38,39,40,41] | Developed cnt: 30–50% Developing cnt: 80–90% | Mixed cellularity: 60–80% Nodular sclerosis: 20–40% | 29,000/y [41] Up to 56,500 cases and 20,500 deaths in 2018 * [42] | WW | B cells (Reed–Sternberg), (T cells <2%) | Nodal: state 1–3 Extranodal: state 4 | CRR: 80–90% [38] 15–20% are resistant/relapse |
| Burkitt lymphoma (latency I) [7,37,41,43,44] | Africa: 85%. USA: 15% | Endemic: 95%, sporadic: 25%. | 7000/y, 20/100,000 children between 5–9y in sub-Saharan Africa [41] | Endemic: Equa. Africa, New Guinea; Sporadic: WW HIV: WW | B cells | Germinal centers, jaw (young children), breast and abdomen (older children) | Developed cnt: Overall Cure > 90% | |
| 3y OS for chemo-resistant: 7% | ||||||||
| MTNKL/PTCL (latency II) [25,39,45,46,47,48,49,50] | 40–50% [49] | AITL: >90%, ANKL: >90%, ENKTCL-NT: 100%, (PTCL-NOS): 30%, SEBV+LOC: 100% [39] 10–30% of all NHL are PTCL/MTNKL [48] | Up to 76,400 cases and 37,500 deaths in 2018 * [48,49,51] | (East) Asia, America, Europe, America | NK cells, T cells | Systemic (AITL, ANKL, SEBV+TLOC); midline nasal/oral cavity, pharynx (ENKTCL-NT), | ENKTCL: 5y OS < 50%. ANKL: Med. OS time 55 days. AITL: Med. 5y OS 32%. PTCL-NOS: Med.5y OS: 20–30% SEBV+LOC: Death within few weeks | |
| Lymphomatoid granulomatosis (latency I-II) [39,52,53,54,55] | 100% | Very rare Prevalence unknown | Western countries | B cells | Lungs, kidneys, skin, CNS | 5y OS rate: 40% 50–60% mortality rate | ||
| NPC (latency II) [41,56,57,58,59] | 95–100% | Type 1: Squamous (low EBV assoc.) Type 2: Non-keratinizing (high EBV assoc.) Type 3: Undifferentiated (high EBV assoc.) | 78,000/y, 80/100,000 mean > 40 years old in Southern China [41] Up to 129,000 cases and 73,000 deaths in 2018 * [56,60] | Asia (Southern China), Africa (north, northwest, central west) | Epithelial cells | Nasal/oral cavity, pharynx | 3y OS 86%, 5y OS 79% | |
| Gastric carcinomas (latency I) [41,61,62,63,64,65] | 9–10% | Gastric lymphoepithelioma: 90%, Moderately diff. adenocarcinomas: 7%, Poorly diff. gastric adenocarcinomas: 6% | 84,000/y [41] Up to 100,000 cases and 78,000 deaths in 2018 * [65,66] | WW, male predominance | Epithelial cells (Gastric pit cell) | Stomach | Overall general GC: median survival time < 12 months, EBV+ 5y OS: 71% | |
| Lymphoepithelioma-like carcinomas (latency I ?) [67,68,69,70,71,72,73,74] | Varies | Liver: rare Stomach: >80% Colon: rare Salivary gland a: 90% Lungs a: 64% Thymus a: 44% | Very rare | Mainly Asia | Epithelia | Varies | N/A | |
| Colorectal carcinomas [75] | Controversial | Controversial | Controversial | WW | Epithelia | Colon | N/A | |
| Breast carcinoma (latency II) [32,33,34,35] | Controversial [32] | N/A | Potentially Up to 520,000 cases and 162,500 deaths in 2018 * [32,76] | WW, highest EBV Assoc. Asia and America | Mammary epithelia | Breast | 5y OS > 90% | |
| Diffuse large B cell lymphomas NOS (DLBCL) (latency I-III) [50,77,78,79,80,81] | ~10% [79] | 30–40% of all NHL are DLBCL [78] | Up to 2000 cases in 2018 * [51,78,79] | WW, 10–15% developing cnt. 5% developed cnt. | B cells | Nodal, extranodal (lungs, gastrointestinal tract) | EBV+ DLBCL: 5y OS 25–54%. | |
| Immunocompromised patients | Lymphomas (latency I-III) [39,77,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99] | PTLD: 50–80% [92] HIV: 40–50% [39,88] | Hodgkin (HIV): 100% DLBCL (HIV): 30–90%, Burkitt (HIV): 50–60%, PbL (HIV): 80% PbL (PTLD): 30% PTLD in transplant patients: SOT: 1–20%, HSCT: <2% [92] | PTLD: up to 24,640 in 2019 ** [91,92]. HIV: up to 60,000 of newly HIV infected in 2020 will develop EBV+ lymphoma *** [89]; 1–6% of HIV+ patients develop lymphomas each year [90], 50% are EBV+ [88] | WW | B cells (90%), T cells (seldom), | Extranodal, CNS, gastrointestinal | Highly variable (see Table 2) PTLD; 2y OS 83% |
| Leiomyosarcomas/smooth muscle tumors (latency I-III) [100,101,102,103,104] | HIV: 85% PT: 98% (B cell > 90%, T cell > 70%) CI: 100% [101] | HIV, PT, CI: Tumor manifestation in <1–5% of each group [101] | WW | Smooth muscle cells | CNS, gut/liver, skin, lungs, larynx, pharynx, adrenal glands, spleen | 2y OS 66%, 5y OS 50% |
| EBV-Associated Malignancy | Treatment | Prognosis | |
|---|---|---|---|
| Immunocompetent patients | Hodgkin lymphoma (latency II) [37,38,39,40,41] | Chemotherapy, radiation therapy, stem cell transplant | CCRR: 80–90% Resistant/relapse: 15–20% |
| Burkitt lymphoma (latency I) [7,37,41,43,44,106,107,108,109] | Multiple drug chemotherapy | Overall cure rate in dev. Countries >90%, worse in low-income. 3y OS is 7% for chemoresistant patients | |
| Combined chemotherapy and immunotherapy (rituximab, α-CD20) | 100% overall survival and 95% progression-free survival at 86 months. 3y OS: 89%, 2y OS: 82% | ||
| Mature T- and NK-cell neoplasms/Peripheral T-cell Lymphoma (latency II) [25,39,45,46,47,48,49,50] | ENKTCL-NT: Chemotherapy, radiotherapy | General: 5y OS < 50%. Stage 1 and 2 diseases: 5y PFS 70–72%; 5y OS 61–63%. Stage 1–2: CRR 87%; 5y OS 73%. Stage 3–4: CRR 45%, 5y OS 47% 1y PFS 80%; advances stages: 5y OS 24%, PFS 16% | |
| ANKL: chemotherapy, HSCT | Median OS: 55 days. 1y OS: 4.4%. Up to median OS 300 days with allo-HSCT and 43% 2y OS (“subacute ANKL”) | ||
| AITL: chemotherapy (CHOP), immunotherapy (CHOP + rituximab/alemtuzumab), high-dose therapy and autologous stem cell rescue (HDT-ASCR) | AITL: Median 5y OS 32% IT(Rituximab)+SCT: ORR 80%, CRR 44%, 2y OS 62% IT(azmab)+SCT: ORR 66–100%, CRR 13–65%, 2y OS < 50% HDT-ASCR: 5y OS 52% | ||
| PTCL-NOS: chemotherapy, HDT-ASCT, no established SOC for relapse/refractory patients | PTCL-NOS: Median 5y OS: 20–30% (< 50% with ASCT) ORR: 50–60%, CRR: 20–30% | ||
| Systemic EBV-positive T-cell lymphoma of childhood: chemotherapy, HSCT | SEBV+TLOC: death within days or weeks of diagnosis | ||
| Lymphomatoid granulomatosis (latency I-II) [39,52,53,54,55] | SOC: corticosteroids, chemotherapy, IFN-α, immunotherapy (rituximab) | 5y OS: 40% (SOC), Grade I-II: PFS 5y 56%, Grade III: PFS 4y 40%, CRR 66%, 50–60% mortality rate | |
| In trials: IFN-α (p with CNS involment), HSCT | 80–90% complete remission | ||
| Nasopharyngeal carcinomas (latency II) [41,56,57,58,59] | Surgery, chemotherapy, radiotherapy, 1st line | Phase 2 and 3 trials (n = 7) comparing induction chemotherapy and concurrent chemoradiotherapy vs. concurrent chemoradiotherapy: avg 3y (n = 5) OS 86% vs. 75% | PFS 76% vs. 64% avg 5y (n = 2) OS 79% vs. 73% | PFS 69% vs. 58% | |
| Immunotherapy (CPI: α-PD1) in recurrent or metastatic disease | Phase 1/2 trials (n = 3): avg ORR 27% | avg 1y OS (n = 2) 61%, median 16.8 months | avg 1y PFS (n = 3) 26%, median 5 months | ||
| CPI α-PD1 with chemotherapy | Phase 1 trial: ORR 91%, 1y PFS 61% | ||
| Gastric carcinomas (latency I) [41,61,62,63,64,65] | General GC: surgical resection with lymphadenectomy, radiotherapy, chemotherapy EBV+ possibly resistant to current chemotherapy options (incl. docetaxel, 5-Fluorouracil) | Overall general GC: OS 20%, median survival time <12 months Recurrence rates (EBV+GC, stages): 0% (I), 21% (II), 33% (III), 83% (IV) | |
| Immunotherapy (CPI: α-PD1, α-PDL1) | 2nd–3rd line of treatment, phase II/III trials over 100 patients (n = 2): avg ORR 11.3%, OS time 5.43 months (vs. placebo 0%, 4.14 months) α-PDL1 vs. chemotherapy trial: RR 2.2% vs. 4.3%, OS time 4.6 vs. 5 months | ||
| DNA methylation inhibitors/Demethylating agents | Phase I trial: Significant epigenetic and clinical responses of epigenetic priming with 5-azacytidine (prior to chemotherapy) in patients with locally advanced esophageal/gastric adenocarcinoma | ||
| PI3K inhibitors | Phase III trial: no significant improvement in OS for advanced GC of everlimos in 3rd line treatment Phase I trial: prolonged stable disease with continuous dosing of PX-866 | ||
| Lymphoepithelioma-like carcinomas (latency I ?) [67,68,69,70,71,72,73,74] | Surgery, chemotherapy | Varies | |
| Breast carcinoma (latency II) [32,33,34,35] | Surgery, radiotherapy, chemotherapy, immunotherapy | Generally 5y OS > 90% | |
| EBV+ diffuse large B cell lymphoma, NOS (latency I-III) [50,77,78,79,80,81] | Antiviral chemotherapy R-CHOP immunotherapy rituximab, durvalumab, nivolumab (α-CD20/PDL1/PD1) EBV CTL, ASCT | 5y OS 25–54% >45 y/o: median survival 2 years <45 y/o: CRR > 80% | |
| Multiple sclerosis [26,27,28,77,105,110,111,112,113,114,115] | Immunotherapy, EBV-specific T-cell immunotherapy in trials | Life expectancy not greatly affected, irreversible disabilities possible, 90% relapsing, remitting MS 10% progressive MS [115] | |
| Immunocompromised patients | Lymphomas (latency I-III) [39,77,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,106,109] | Reduction/cessatation of immune suppression (1st line) | PTLD: ORR 0–73% (biggest study: CRR 37%) |
| Chemotherapy | HIV: Burkitt’s lymphoma HAART + chemotherapy: ORR 70%, 3y OS 52% HIV: Hodgkin’s lymphoma cART + chemotherapies: avg 3y OS 51% | 5y OS 76% HIV: Burkitt lymphoma chemotherapy: 4y OS 72% PbL-HIV: median OS 6–19 months PbL-PTLD: median OS 7 months | ||
| Immunotherapy (rituximab) (+ chemotherapy) | PTLD (SOT) chemotherapy + immunotherapy, age <30y (n = 55): CRR 69%, 2y OS 83% PTLD: Phase II trials rituximab: ORR 55%, 25% relapse HIV: Hodgkin’s lymphoma ASCT + high dose chemotherapy (relapse): 32-month avg OS 61% HIV: Burkitt’s lymphoma EPOCH-R: 90% OS and 100% PFS at 86 months HIV: Burkitt’s 2y OS: 73% | ||
| Cellular immunotherapy | Phase II trial PTLD (HSCT, SOT) allogeneic EBV-specific CTL by best HLA match: 6-month ORR 52%, 42% CRR | ||
| Transplantation (+ medication) | HIV: Hodgkin’s lymphoma ASCT + high dose chemotherapy (relapse): 32-month avg OS 61% | ||
| Leiomyosarcomas/smooth muscle tumors (latency I-III) [100,101,102,103,104] | Chemotherapy, surgery, antiviral therapy, reduced immunosuppression, adoptive T-cell therapy | PT-SMT: 2y OS 66%, median of death post manifestation 5.5 months PT/HIV-SMT: 5y OS 50% |
| Name/EBV-Associated Malignancy | Latency | Type | References |
|---|---|---|---|
| AIDS-related lymphoma (ARL) | I-III | Tissue sample | [140] |
| Angioimmunoblastic T-cell lymphoma (AITL) | II | Tissue sample | [22,140] |
| Anaplastic large cell lymphoma (ALCL) | II ? | Tissue sample | [140] |
| Burkitt lymphoma (BL) | I | Tissue sample | [22,138] |
| Classical Hodgkin lymphoma, nodular sclerosis (cHL-NS) | II | Tissue sample | [140] |
| Cutaneous T-cell lymphoma (CTCL) | II ? | Tissue sample | [140] |
| Diffuse large B-cell lymphoma (DLBCL) | I-III | Tissue sample | [22,140] |
| Gastric carcinoma (GC) | I | Tissue sample | [22,141] |
| Mature T- and NK-cell lymphoma (MTNKL) | II | Tissue sample | [22] |
| Nasopharyngeal carcinoma (NPC) | II | Tissue sample | [22] |
| Nodular lymphocyte predominant Hodgkin lymphoma (NLPHL) | II | Tissue sample | [140] |
| Peripheral T-cell lymphoma, not otherwise specified (PTCL-NOS) | II | Tissue sample | [140] |
| B95-8 (LCL) | III | Cell line | [3,17,140] |
| HH514.c16 (BL) | I | Cell line | [3] |
| Jijoye (BL) | I | Cell line | [140] |
| JY (LCL) | III | Cell line | [3] |
| KREB2 (LCL) | III | Cell line | [140] |
| MEC04 (MTNKL) | II | Cell line | [140] |
| MLEB2 (LCL) | III | Cell line | [140] |
| Namalwa (BL) | I | Cell line | [3,140] |
| P3HR1 (BL) | I | Cell line | [140] |
| Raji (BL) | I | Cell line | [139,140] |
| SNK6 (MTNKL) | II | Cell line | [140] |
| Various BL cell lines | I | Cell line | [138] |
| X50-7 (LCL) | III | Cell line | [3] |
| Indication | Drug Name | Action | Development Stage | Identifier |
|---|---|---|---|---|
| (Non-) Hodgkin lymphoma | ipilimumab + nivolumab | α-CTLA-4 + α-PD-1 | Phase II | NCT01592370 |
| B cell lymphoma | nivolumab | α-PD-1 | Phase I | NCT03097939 |
| B cell lymphoma | Viroprev | α-TK1 | IND application | http://savoypharmaceuticals.com/viroprev.php, accessed on 12 August 2021 |
| Indication | Drug Name | Action | Development Stage | Identifier |
|---|---|---|---|---|
| Nasopharyngeal carcinoma (NPC) | Autologous EBV T Cells | Phase II | NCT00834093 | |
| NPC, first-line in combination with gemcitabine + carboplatin | TT10 EBVSTs | Autologous EBV-CTL (EBNA1, BARF-1, LMP) | Phase III | NCT02578641 |
| NPC, relapse/refractory | YT-E001 | Autologous EBV-CTL (EBNA1, LMP1, LMP2) | Phase I/II | NCT03648697 |
| NPC | LT-C50 | EBV-CTL | Preclinical | https://liontcr.com/pipeline/, accessed on 12 August 2021 |
| Gastric Carcinoma | LT-C60 | EBV-CTL | Preclinical | https://liontcr.com/pipeline/, accessed on 12 August 2021 |
| NPC, recurrent/metastatic (platinum-pretreated) | Tabelecleucel + pembrolizumab | Allogeneic EBV-CTL | Phase Ib/II | NCT03769467 |
| CD30+ EBV-lymphomas | TT11x | Autologous EBV-CTL (EBNA1, BARF-1, LMP) + CD30 CAR | Phase I | NCT04288726 |
| EBV+ PTLD | Viralym-M (ALVR105) | Multi-virus specific allogenic T-Cells | Phase II | NCT04693637 |
| Post HSCT opportunistic infections | Autologous or allogenic EBV CTL | Phase II | NCT03159364 | |
| EBV+ PTLD after SOT or alloHCT (after failure of rituximab/r+chemo) | tabelecleucel | Allogeneic EBV-CTL | Phase III | NCT03394365 |
| EBV+ PTLD after alloHCT (after failure of rituximab) | tabelecleucel | Allogeneic EBV-CTL | Phase III | NCT03392142 |
| Progressive multiple sclerosis | ATA-188 | Allogeneic EBV-CTL | Phase I | NCT03283826 |
| Advanced stage EBV+ malignancies (stage IV gastric carcinoma, NPC, lymphoma after SOC) | Autologous PD-1 knockout EBV-CTL + Fludarabine + Cyclophosphamide + IL-2 | Phase II | NCT03044743 | |
| Systemic Lupus Erythematosus (SLE) | LUPUS CTL EBV | Autologous EBV-CTL | Phase I/II | NCT02677688 |
| ENKTCL, (PTLD, NPC) | VT-EBV-N | Autologous EBV CTL (LMP1, LMP2a) | Phase II | NCT03671850 |
| ENKTCL, 2nd line | EBViNT | Autologous EBV CTL (LMP2a) | Phase I/II | NCT03789617 |
| Reactivation/infection prevention post cord blood transplant | Autologous EBV CTL | Phase I/II | NCT03594981 NCT01923766 |
| Indication | Drug Name | Action | Development Stage | Identifier |
|---|---|---|---|---|
| NPC, 3rd line, locally recurrent or metastatic | apatinib mesylate | Vascular endothelial growth factor receptor 2 (VEGFR-2) inhibitor | Phase II | NCT03130270 |
| EBV+ Lymphoma | Nanatinostat (VRx-3996) | Histone deacetylase (HDAC) inhibitor | Phase I/II | NCT03397706 |
| Viral cancers/EBV diseases | Inhibition of replication | Lead selection | http://www.virostatics.com/research-and-development/, accessed on 12 August 2021 | |
| EBV diseases | BZLF1 activator | Discovery | http://www.vironika.com/pipeline, accessed on 12 August 2021 | |
| EBV diseases | EBNA1 inhibitor | Pre-IND | http://www.vironika.com/pipeline, accessed on 12 August 2021 |
| Indication | Drug Name | Action | Development Stage | Identifier |
|---|---|---|---|---|
| Persistent, recurrent or metastatic NPC | MVA vaccine | Recombinant modified vaccinia Ankara (MVA) EBNA1/LMP2 vaccine | Phase II | NCT01094405 |
| EBV infection | mRNA-1189 | mRNA-based vaccine (gp350, gH/gL/gp42, gH/gL, gB) | Preclinical | https://www.modernatx.com/pipeline, accessed on 12 August 2021 |
| EBV infection | Vaccine | gp350 blocking | Phase I recruitment | NCT04645147 |
| EBV infection | Vaccine | g42, gH/gL blocking | Preclinical | 10.1016/j.immuni.2019.03.010, Patent No. EP3054971, accessed on 12 August 2021 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knerr, J.M.; Kledal, T.N.; Rosenkilde, M.M. Molecular Properties and Therapeutic Targeting of the EBV-Encoded Receptor BILF1. Cancers 2021, 13, 4079. https://doi.org/10.3390/cancers13164079
Knerr JM, Kledal TN, Rosenkilde MM. Molecular Properties and Therapeutic Targeting of the EBV-Encoded Receptor BILF1. Cancers. 2021; 13(16):4079. https://doi.org/10.3390/cancers13164079
Chicago/Turabian StyleKnerr, Julius Maximilian, Thomas Nitschke Kledal, and Mette Marie Rosenkilde. 2021. "Molecular Properties and Therapeutic Targeting of the EBV-Encoded Receptor BILF1" Cancers 13, no. 16: 4079. https://doi.org/10.3390/cancers13164079
APA StyleKnerr, J. M., Kledal, T. N., & Rosenkilde, M. M. (2021). Molecular Properties and Therapeutic Targeting of the EBV-Encoded Receptor BILF1. Cancers, 13(16), 4079. https://doi.org/10.3390/cancers13164079






