Comprehensive Library Generation for Identification and Quantification of Endometrial Cancer Protein Biomarkers in Cervico-Vaginal Fluid
Abstract
Simple Summary
Abstract
1. Introduction
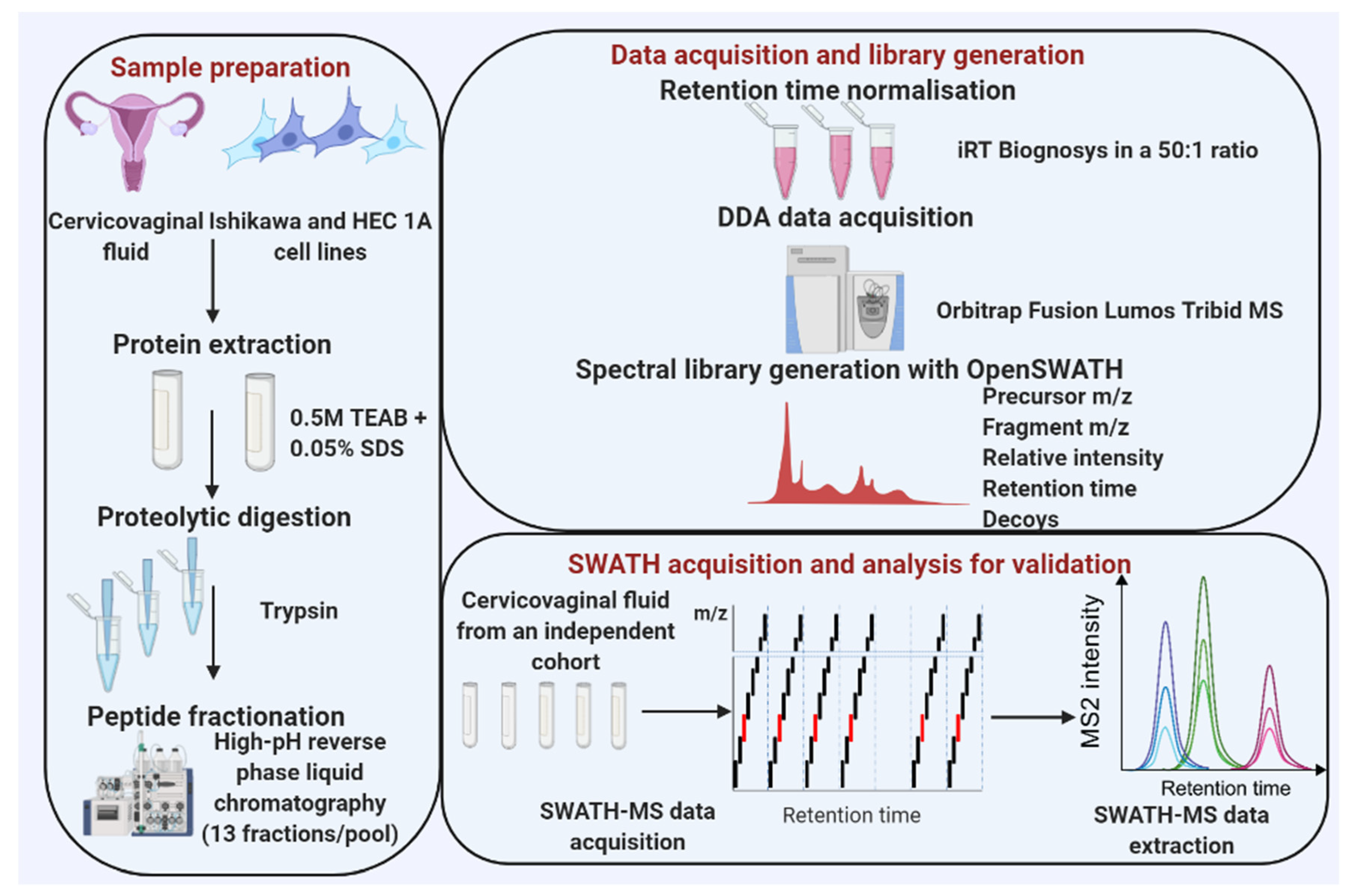
2. Methods
2.1. Research Ethics and Approval
2.2. Sample Collection
2.3. Cell Culture
2.4. Cervico-Vaginal Fluid Supernatant Preparation
2.5. Cell Lysis/Protein Extraction
2.6. Protein Digestion
2.7. High-pH Fractionation of Peptides
2.8. DDA Mass Spectrometry for Spectral Library Generation
2.9. Building the SWATH Spectral Library
2.10. Assay Library False Discovery Rate Control
2.11. SWATH-MS Acquisition and Library Validation
3. Results
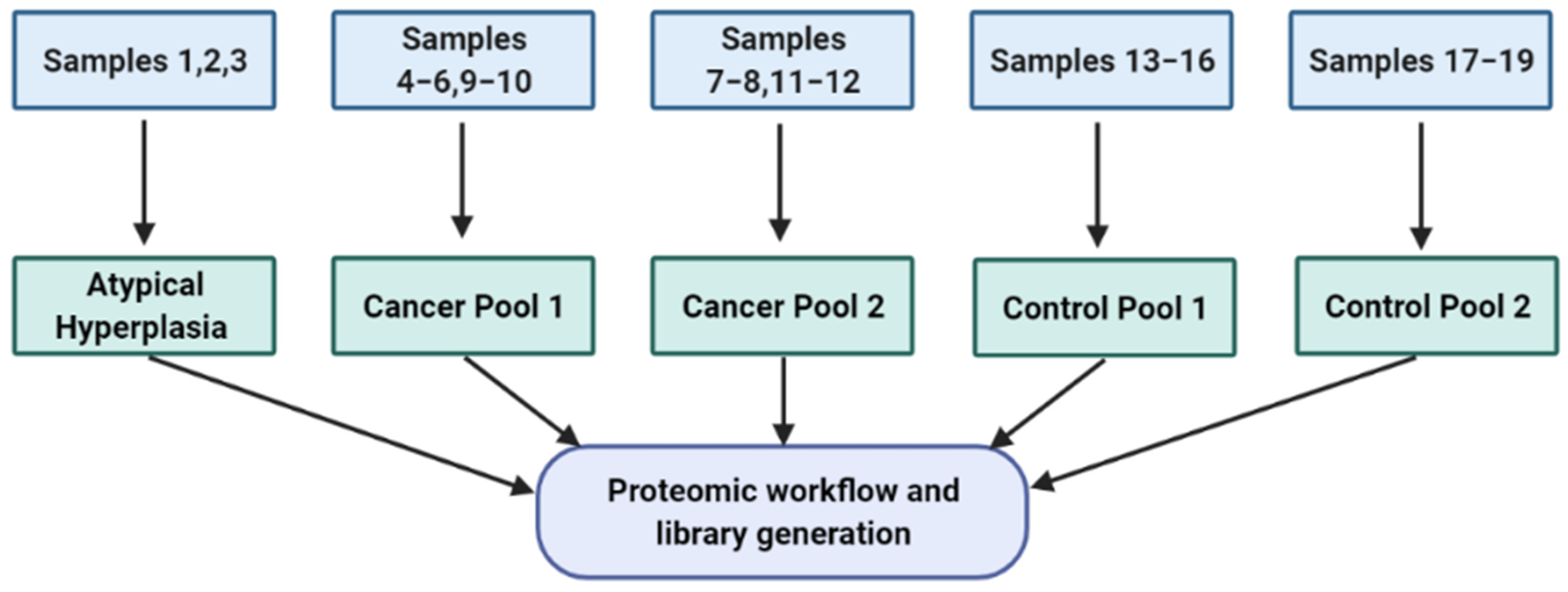
3.1. Descriptive Characteristics of the Study Population
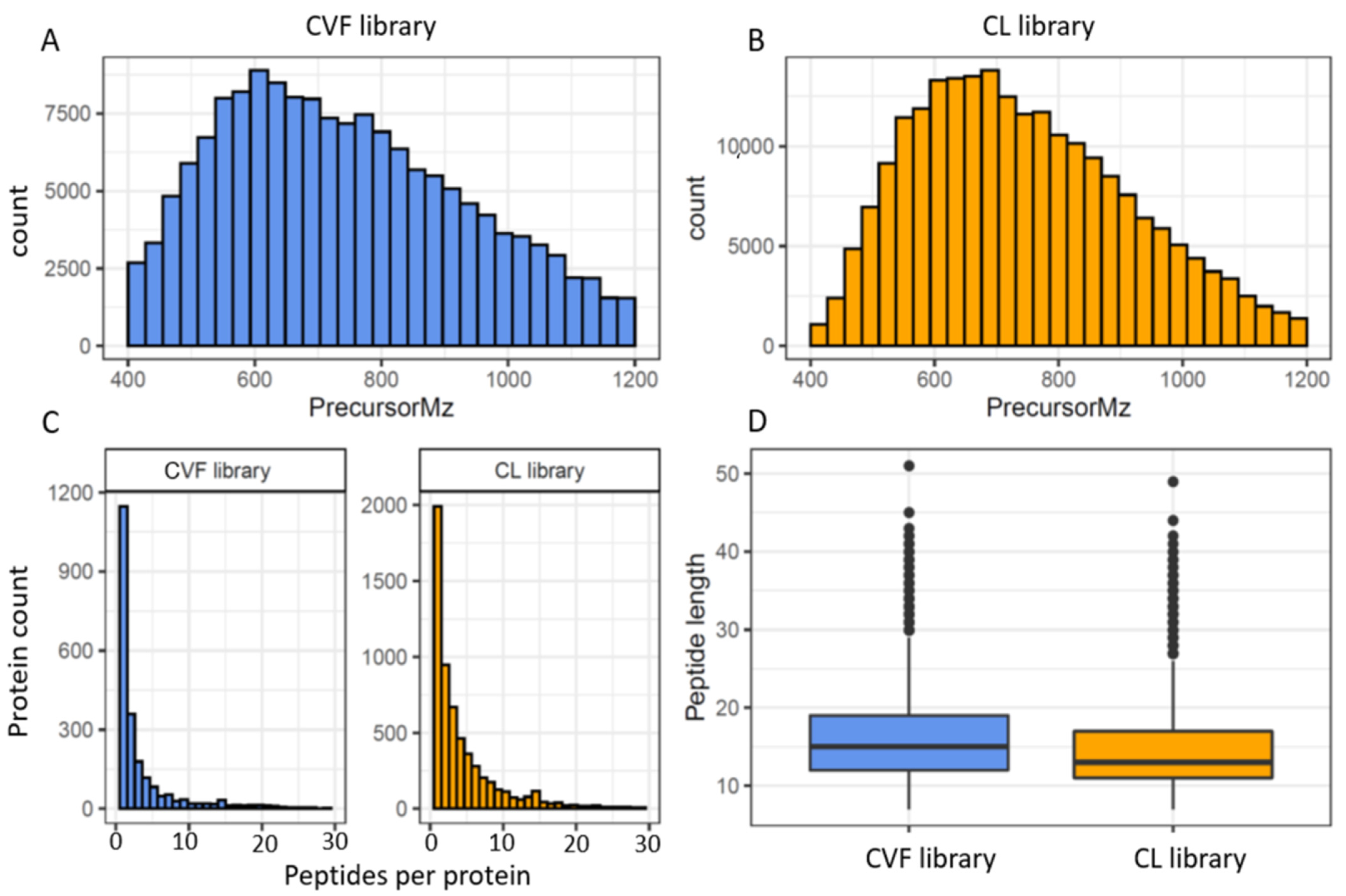
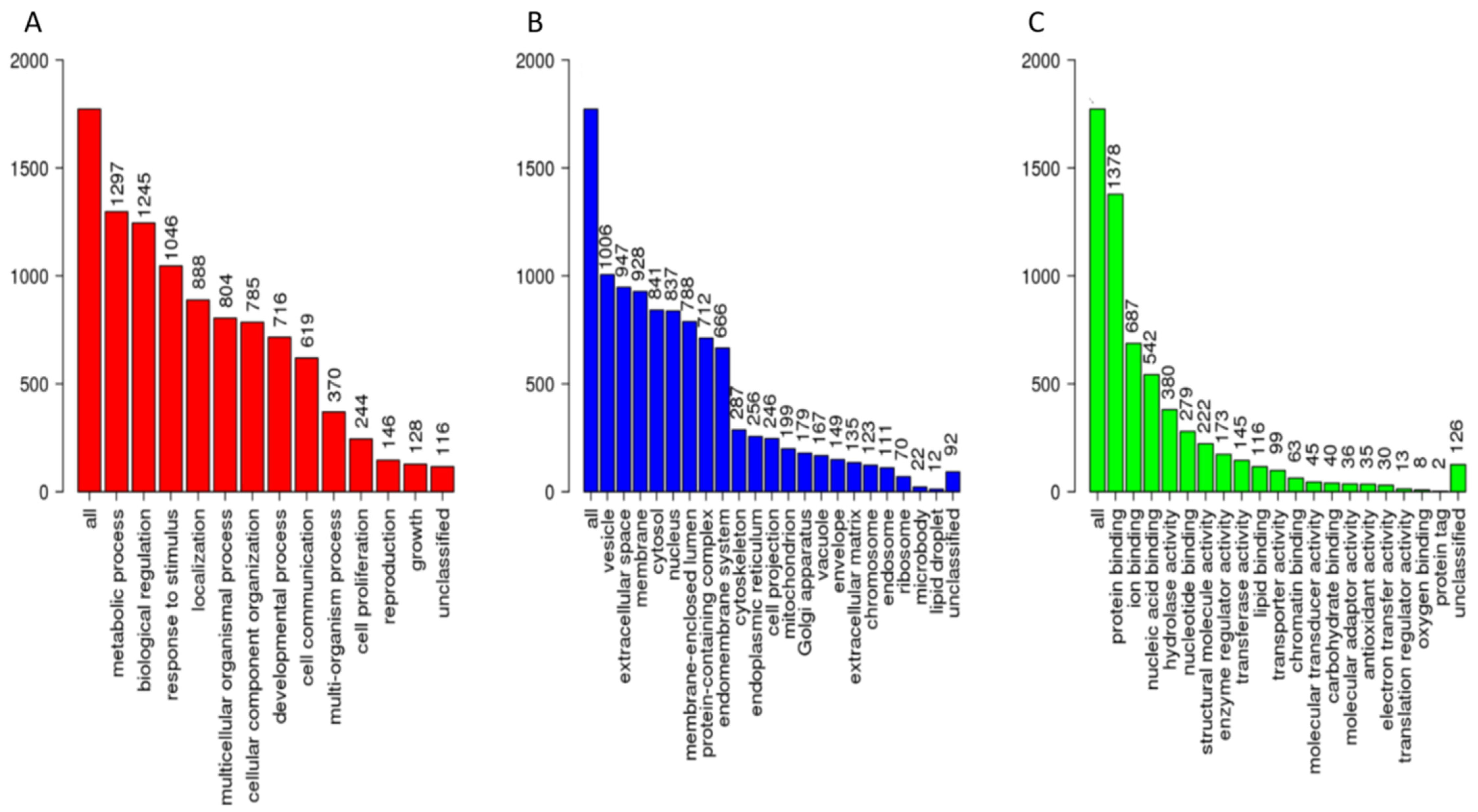
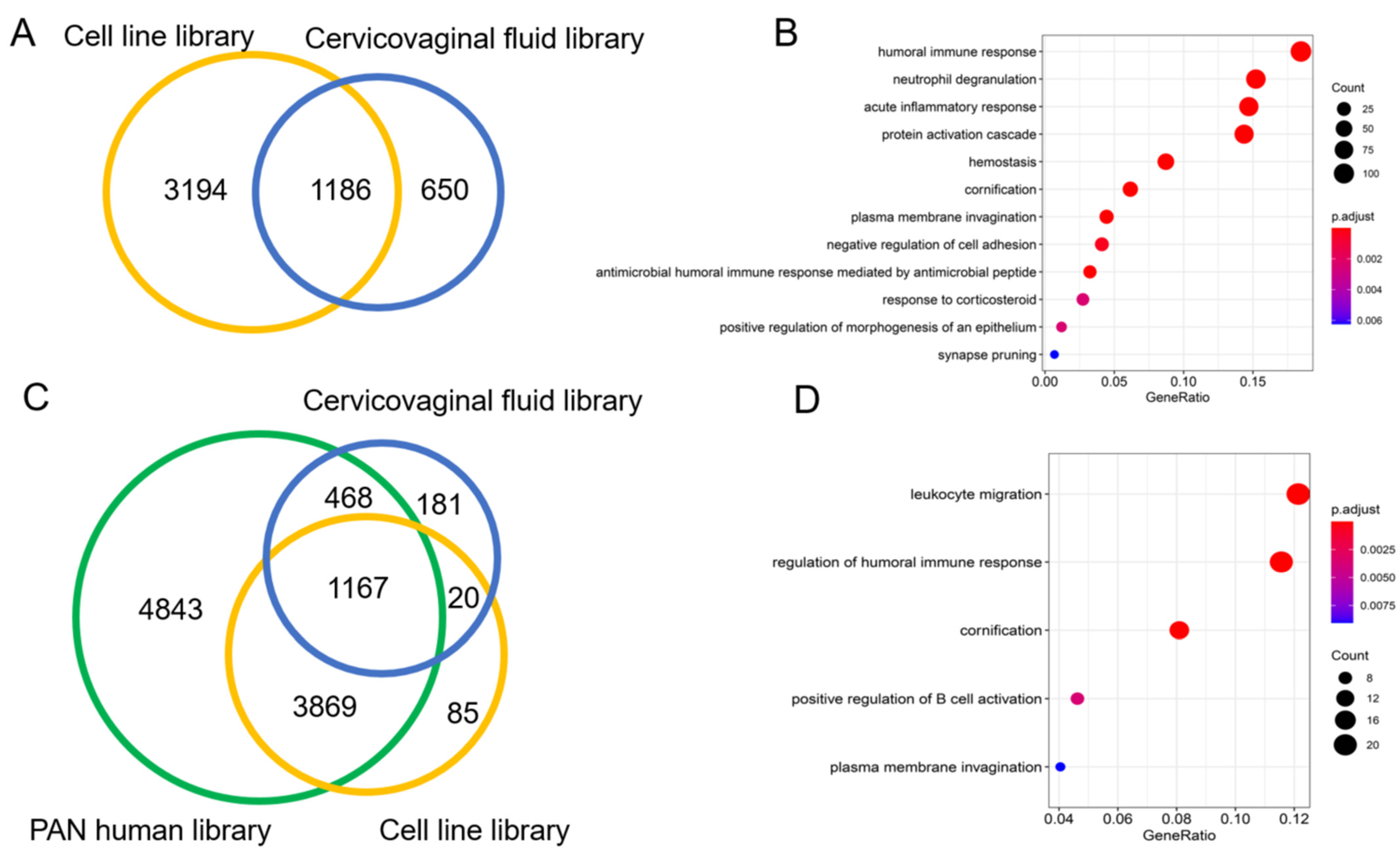
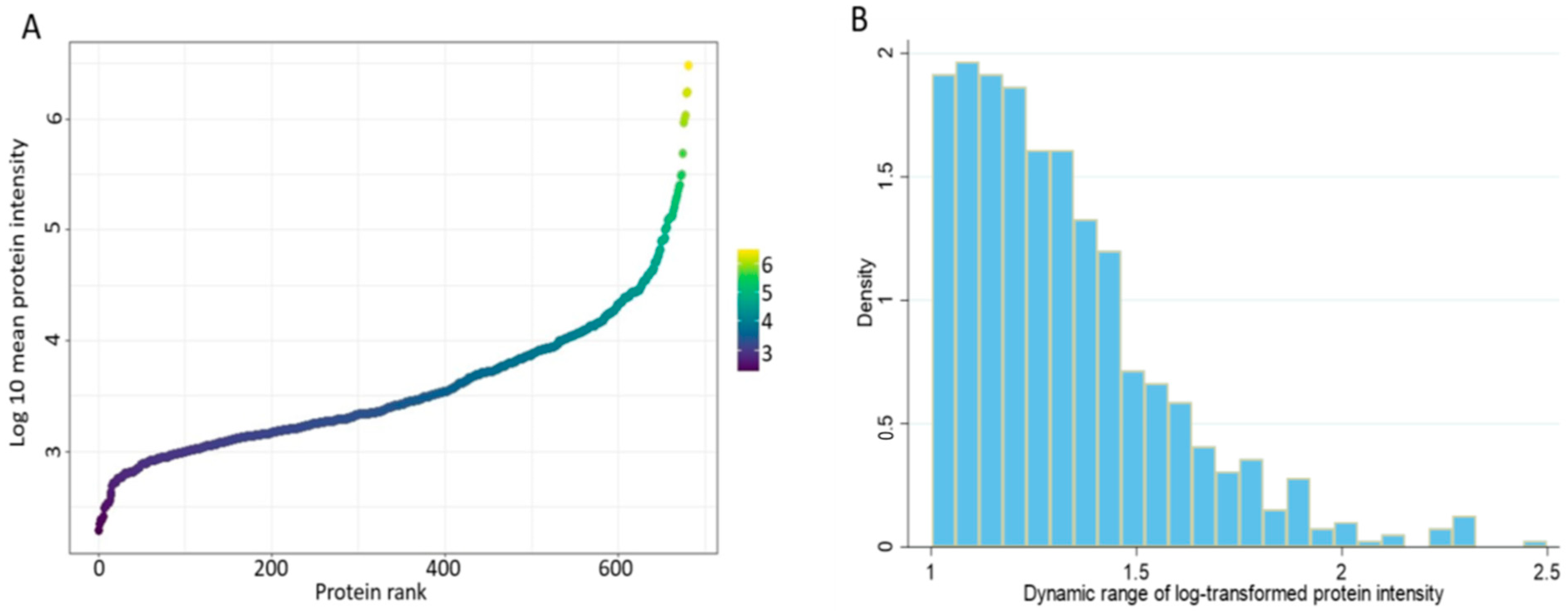
3.2. The Spectral Libraries
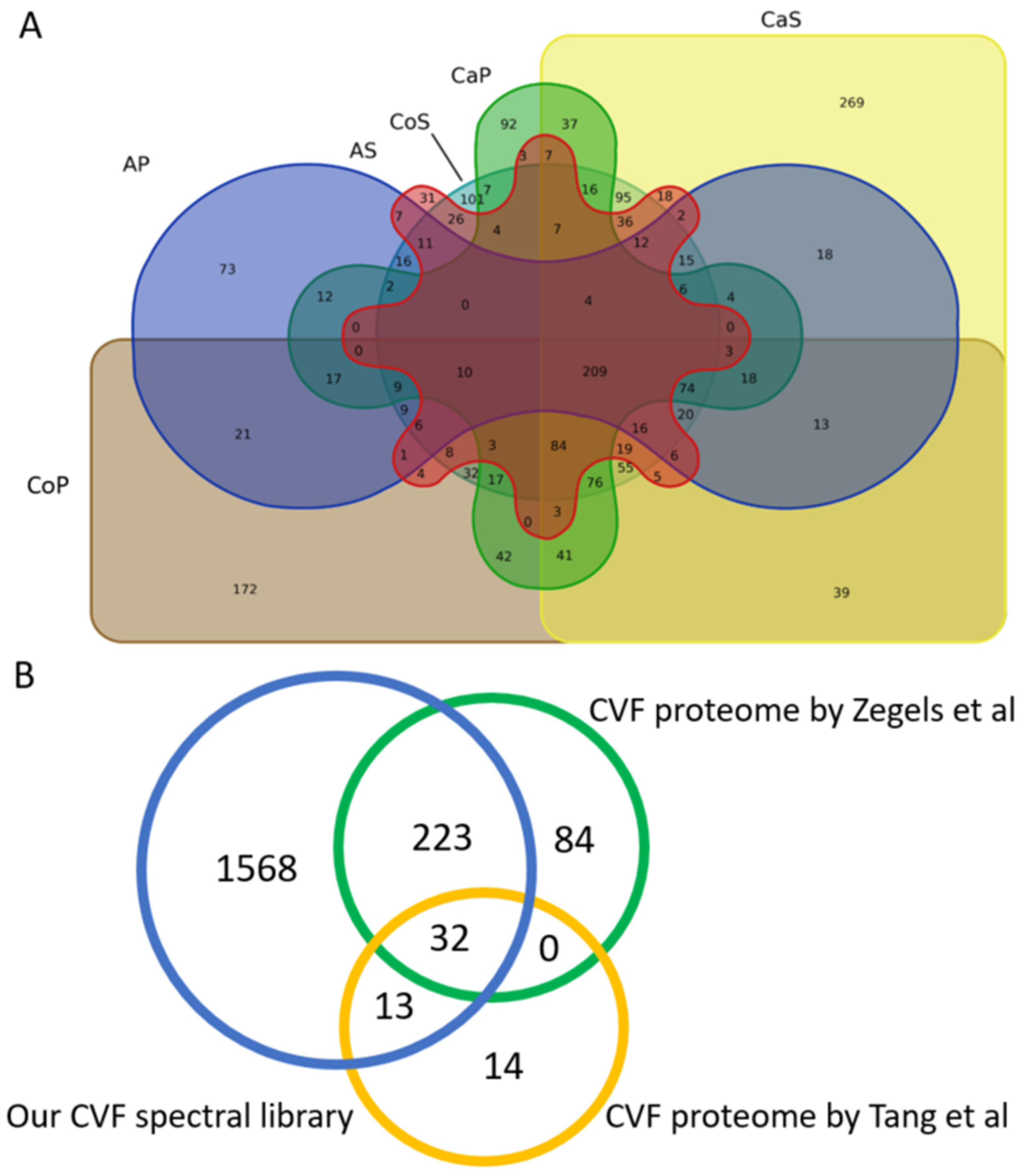
3.3. Spectral Library Validation
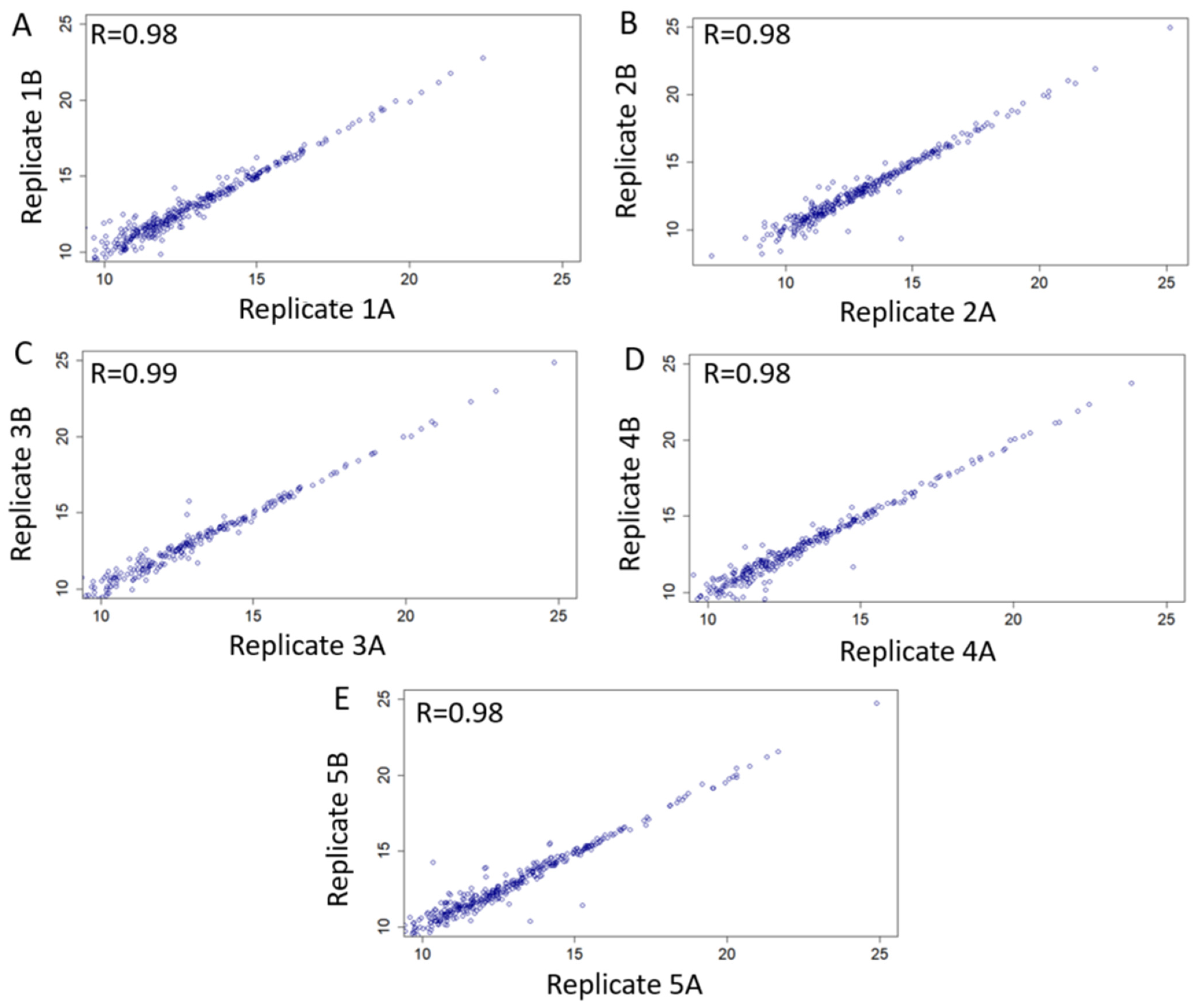
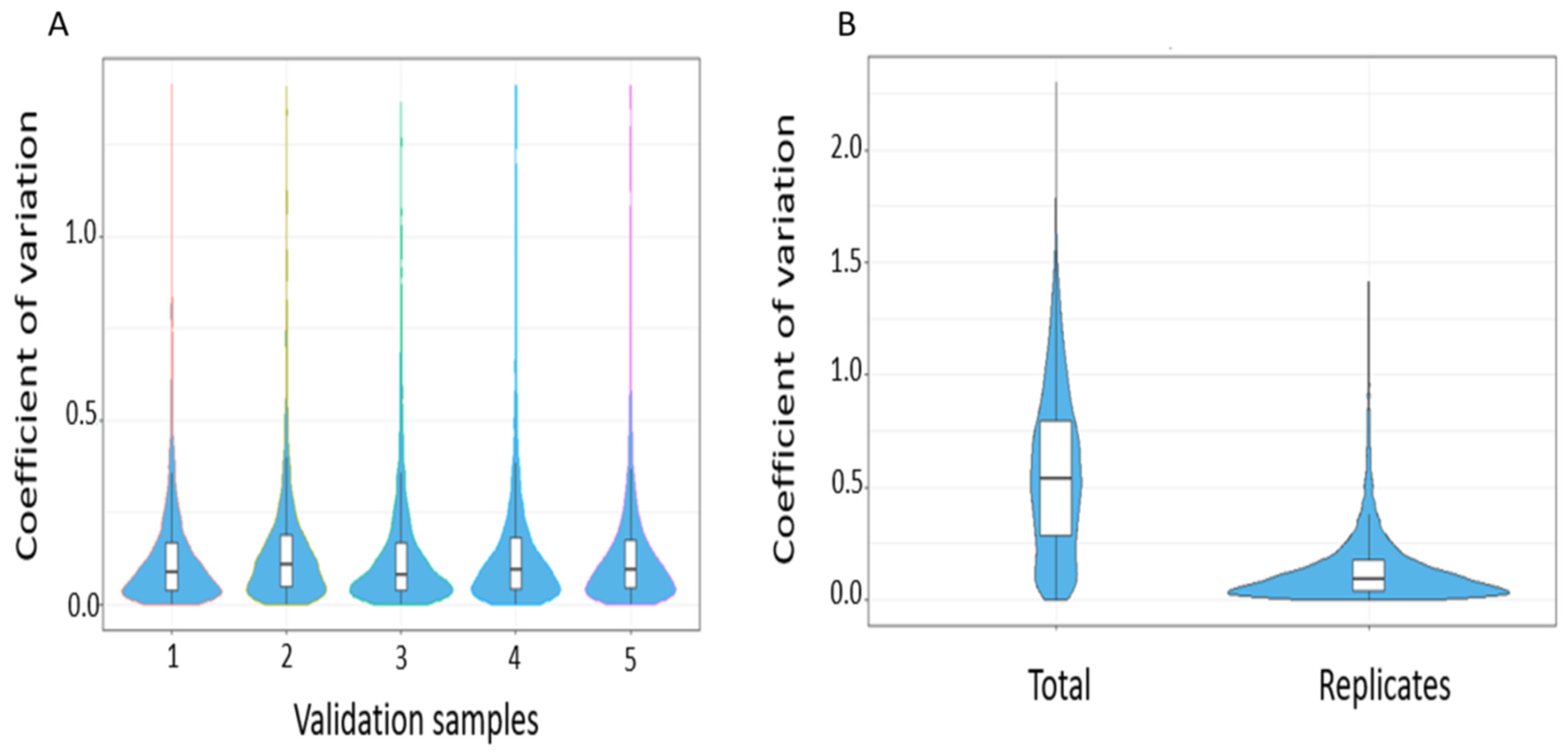
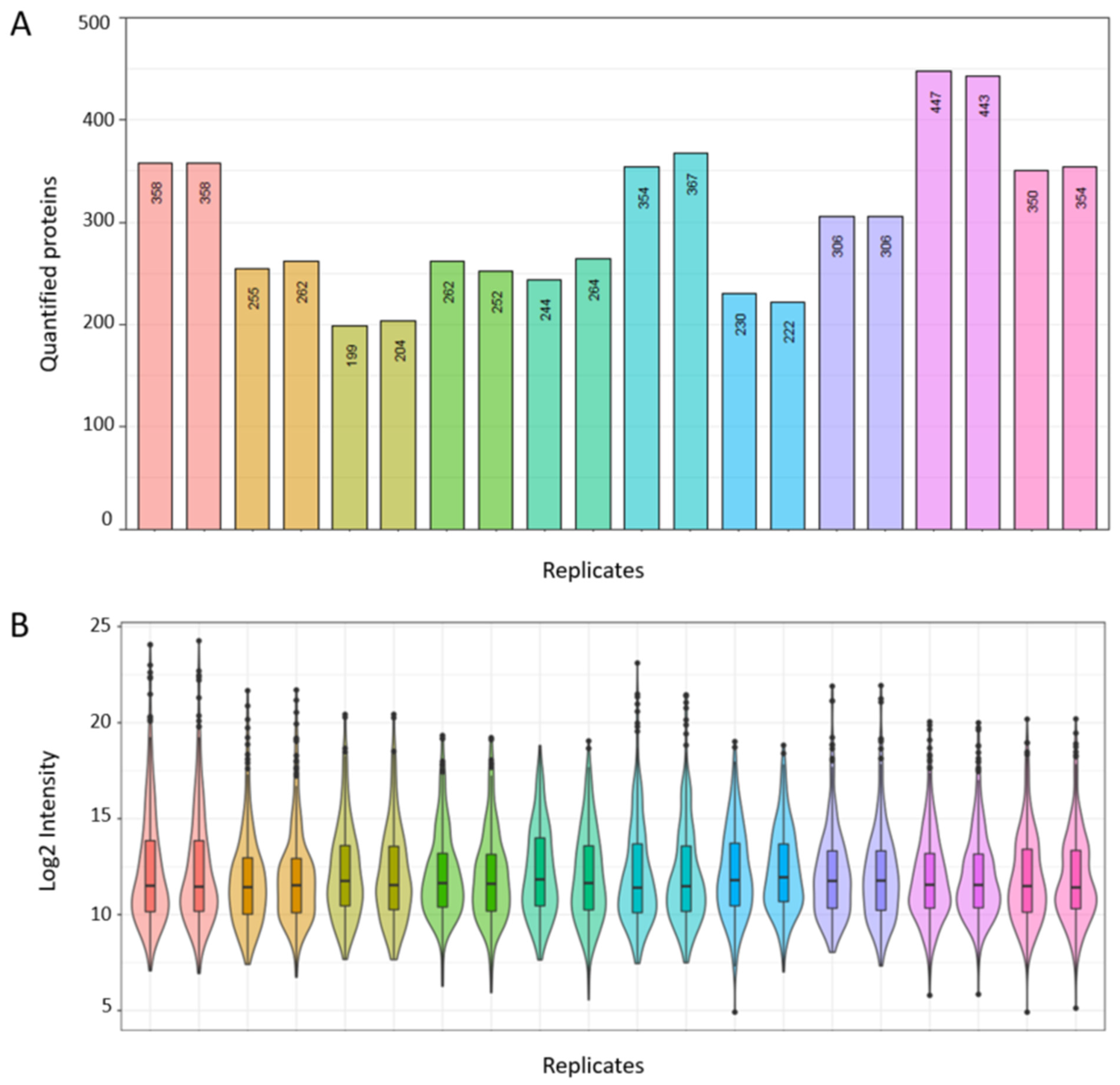
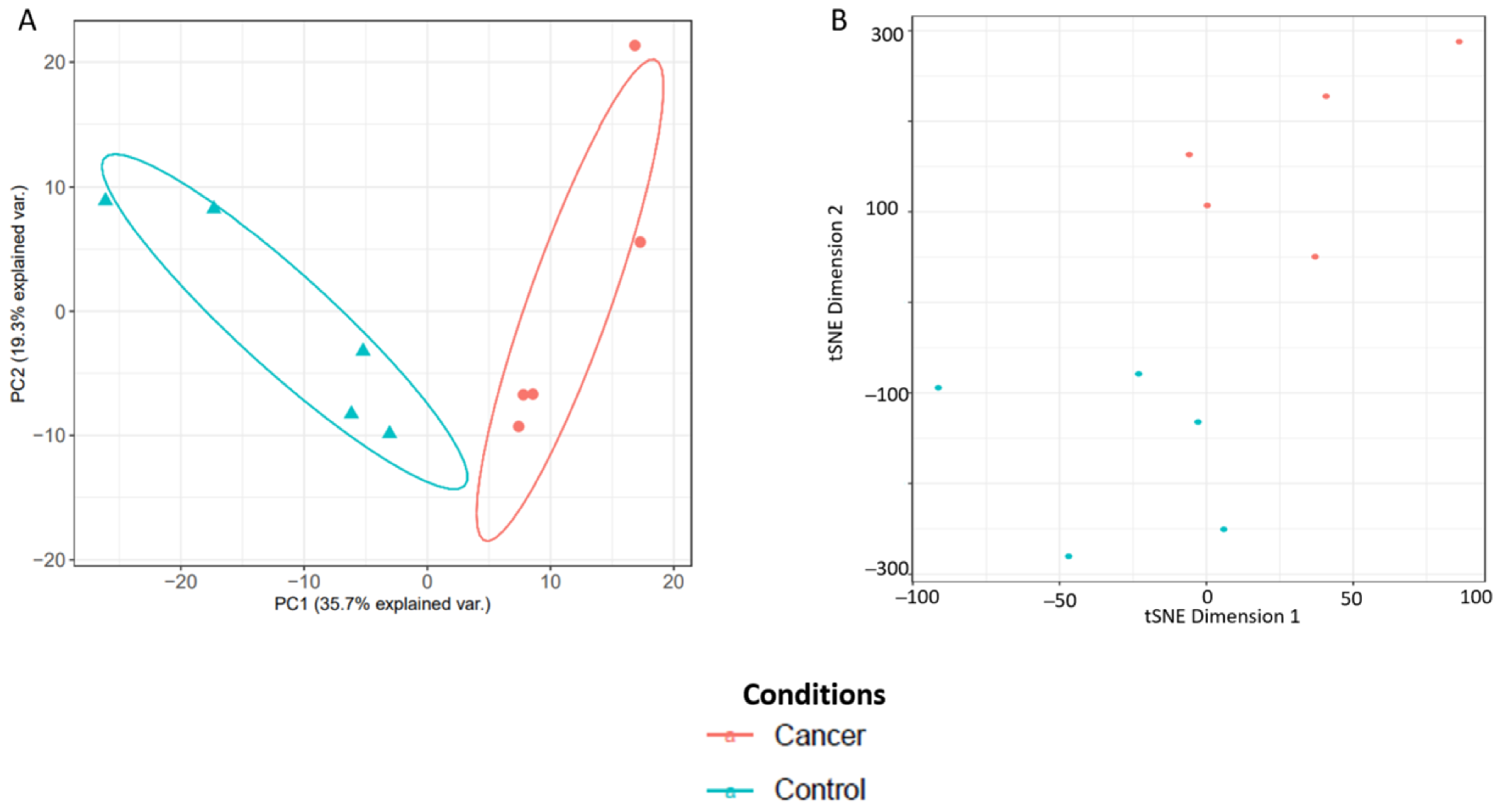
3.3.1. Technical Validation and Applicability for SWATH-MS Data Analysis
3.3.2. Real World Application of Spectral Library for Biomarker Discovery
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Lortet-Tieulent, J.; Ferlay, J.; Bray, F.; Jemal, A. International patterns and trends in endometrial cancer incidence 1978–2013. J. Natl. Cancer Inst. 2018, 110, 354–361. [Google Scholar] [CrossRef]
- Crosbie, E.; Morrison, J. The emerging epidemic of endometrial cancer: Time to take action. Cochrane Database Syst. Rev. 2014, ED000095. [Google Scholar] [CrossRef]
- Bokhman, J.V. Two pathogenetic types of endometrial carcinoma. Gynecol. Oncol. 1983, 15, 10–17. [Google Scholar] [CrossRef]
- Suarez, A.A.; Felix, A.S.; Cohn, D.E. Bokhman redux: Endometrial cancer “types” in the 21st century. Gynecol. Oncol. 2017, 144, 243–249. [Google Scholar] [CrossRef]
- Stelloo, E.; Bosse, T.; Nout, R.A.; MacKay, H.J.; Church, D.N.; Nijman, H.W.; Leary, A.; Edmondson, R.J.; Powell, M.E.; Crosbie, E.J. Refining prognosis and identifying targetable pathways for high-risk endometrial cancer; a TransPORTEC initiative. Mod. Pathol. 2015, 28, 836. [Google Scholar] [CrossRef] [PubMed]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Yang, W.; Lum, A.; Senz, J.; Boyd, N.; Pike, J.; Anglesio, M.; Kwon, J.S. Confirmation of ProMisE: A simple, genomics-based clinical classifier for endometrial cancer. Cancer 2017, 123, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Funston, G.; O’Flynn, H.; Ryan, N.A.; Hamilton, W.; Crosbie, E.J. Recognizing Gynecological Cancer in Primary Care: Risk Factors, Red Flags, and Referrals. Adv. Ther. 2018, 35, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.R.; O’Flynn, H.; Njoku, K.; Crosbie, E.J. Detecting endometrial cancer. Obstet. Gynaecol. 2021, 23, 103–112. [Google Scholar]
- Njoku, K.; Abiola, J.; Russell, J.; Crosbie, E.J. Endometrial cancer prevention in high-risk women. Best Pract. Res. Clin. Obstet. Gynaecol. 2020, 65, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Sundar, S.; Balega, J.; Crosbie, E.; Drake, A.; Edmondson, R.; Fotopoulou, C.; Gallos, I.; Ganesan, R.; Gupta, J.; Johnson, N. BGCS uterine cancer guidelines: Recommendations for practice. Eur. J.Obstet. Gynecol. Reprod. Biol. 2017, 213, 71–97. [Google Scholar] [CrossRef]
- Njoku, K.; Chiasserini, D.; Jones, E.R.; Barr, C.E.; O’flynn, H.; Whetton, A.D.; Crosbie, E.J. Urinary biomarkers and their potential for the non-invasive detection of endometrial cancer. Front. Oncol. 2020, 10, 2420. [Google Scholar] [CrossRef]
- Costas, L.; Frias-Gomez, J.; Guardiola, M.; Benavente, Y.; Pineda, M.; Pavón, M.Á.; Martínez, J.M.; Climent, M.; Barahona, M.; Canet, J. New perspectives on screening and early detection of endometrial cancer. Int. J. Cancer 2019, 145, 3194–3206. [Google Scholar] [CrossRef] [PubMed]
- Njoku, K.; Chiasserini, D.; Whetton, A.D.; Crosbie, E.J. Proteomic biomarkers for the detection of endometrial cancer. Cancers 2019, 11, 1572. [Google Scholar] [CrossRef]
- Banach, P.; Suchy, W.; Derezinski, P.; Matysiak, J.; Kokot, Z.J.; Nowak-Markwitz, E. Mass spectrometry as a tool for biomarkers searching in gynecological oncology. Biomed. Pharmacother. 2017, 92, 836–842. [Google Scholar] [CrossRef] [PubMed]
- Banno, K.; Kisu, I.; Yanokura, M.; Tsuji, K.; Masuda, K.; Ueki, A.; Kobayashi, Y.; Yamagami, W.; Nomura, H.; Tominaga, E. Biomarkers in endometrial cancer: Possible clinical applications. Oncol. Lett. 2012, 3, 1175–1180. [Google Scholar] [CrossRef] [PubMed]
- Njoku, K.; Campbell, A.E.; Geary, B.; MacKintosh, M.L.; Derbyshire, A.E.; Kitson, S.J.; Sivalingam, V.N.; Pierce, A.; Whetton, A.D.; Crosbie, E.J. Metabolomic Biomarkers for the Detection of Obesity-Driven Endometrial Cancer. Cancers 2021, 13, 718. [Google Scholar] [CrossRef] [PubMed]
- Guest, P.C.; Gottschalk, M.G.; Bahn, S. Proteomics: Improving biomarker translation to modern medicine? Genome Med. 2013, 5, 17. [Google Scholar] [CrossRef][Green Version]
- Ludwig, C.; Gillet, L.; Rosenberger, G.; Amon, S.; Collins, B.C.; Aebersold, R. Data-independent acquisition-based SWATH-MS for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2018, 14, e8126. [Google Scholar] [CrossRef]
- Röst, H.L.; Rosenberger, G.; Navarro, P.; Gillet, L.; Miladinović, S.M.; Schubert, O.T.; Wolski, W.; Collins, B.C.; Malmström, J.; Malmström, L. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 2014, 32, 219. [Google Scholar] [CrossRef]
- Prieto, G.; Vázquez, J. Calculation of False Discovery Rate for Peptide and Protein Identification. In Mass Spectrometry Data Analysis in Proteomics; Springer: Cham, Switzerland, 2020; pp. 145–159. [Google Scholar]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Zegels, G.; Van Raemdonck, G.A.; Coen, E.P.; Tjalma, W.A.; Van Ostade, X.W. Comprehensive proteomic analysis of human cervical-vaginal fluid using colposcopy samples. Proteome Sci. 2009, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.-J.; De Seta, F.; Odreman, F.; Venge, P.; Piva, C.; Guaschino, S.; Garcia, R.C. Proteomic analysis of human cervical-vaginal fluids. J. Proteome Res. 2007, 6, 2874–2883. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.L.; Smith, C.R.; Diamandis, E.P. Proteomic analysis of human cervico-vaginal fluid. J. Proteome Res. 2007, 6, 2859–2865. [Google Scholar] [CrossRef] [PubMed]
- Lazar, C.; Gatto, L.; Ferro, M.; Bruley, C.; Burger, T. Accounting for the multiple natures of missing values in label-free quantitative proteomics data sets to compare imputation strategies. J. Proteome Res. 2016, 15, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- Stead, D.A.; Paton, N.W.; Missier, P.; Embury, S.M.; Hedeler, C.; Jin, B.; Brown, A.J.; Preece, A. Information quality in proteomics. Briefings Bioinform. 2008, 9, 174–188. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Costa, C.; Martínez-Bartolomé, S.; McClatchy, D.B.; Saviola, A.J.; Yu, N.-K.; Yates, J.R., III. Impact of the Identification Strategy on the Reproducibility of the DDA and DIA Results. J. Proteome Res. 2020, 19, 3153–3161. [Google Scholar] [CrossRef]
- Albrecht, D.; Kniemeyer, O.; Brakhage, A.A.; Guthke, R. Missing values in gel-based proteomics. Proteomics 2010, 10, 1202–1211. [Google Scholar] [CrossRef]
- Nanjappa, V.; Thomas, J.K.; Marimuthu, A.; Muthusamy, B.; Radhakrishnan, A.; Sharma, R.; Ahmad Khan, A.; Balakrishnan, L.; Sahasrabuddhe, N.A.; Kumar, S. Plasma Proteome Database as a resource for proteomics research: 2014 update. Nucleic Acids Res. 2014, 42, D959–D965. [Google Scholar] [CrossRef]
- Huang, L.; Shao, D.; Wang, Y.; Cui, X.; Li, Y.; Chen, Q.; Cui, J. Human body-fluid proteome: Quantitative profiling and computational prediction. Briefings Bioinform. 2021, 22, 315–333. [Google Scholar] [CrossRef]
- Zegels, G.; Van Raemdonck, G.A.; Tjalma, W.A.; Van Ostade, X.W. Use of cervicovaginal fluid for the identification of biomarkers for pathologies of the female genital tract. Proteome Sci. 2010, 8, 1–23. [Google Scholar] [CrossRef]
- Cole, A.M. Innate Host Defense of Human Vaginal and CervicalMucosae. Curr. Top. Microbiol. Immunol. 2006, 306, 199–230. [Google Scholar]
- Valore, E.V.; Park, C.H.; Igreti, S.L.; Ganz, T. Antimicrobial components of vaginal fluid. Am. J. Obstet. Gynecol. 2002, 187, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.; Reddy, A.P.; Jacob, T.; Thomas, A.; Schneider, K.A.; Dasari, S.; Lapidus, J.A.; Lu, X.; Rodland, M.; Roberts, C.T. Identification of Novel protein biomarkers of preterm birth in human cervical—Vaginal fluid. J. Proteome Res. 2007, 6, 1269–1276. [Google Scholar] [CrossRef]
- Shah, S.J.; Yu, K.H.; Sangar, V.; Parry, S.I.; Blair, I.A. Identification and quantification of preterm birth biomarkers in human cervicovaginal fluid by liquid chromatography/tandem mass spectrometry. J. Proteome Res. 2009, 8, 2407–2417. [Google Scholar] [CrossRef] [PubMed]
- Amabebe, E.; Reynolds, S.; Stern, V.L.; Parker, J.L.; Stafford, G.P.; Paley, M.N.; Anumba, D.O. Identifying metabolite markers for preterm birth in cervicovaginal fluid by magnetic resonance spectroscopy. Metabolomics 2016, 12, 67. [Google Scholar] [CrossRef]
- Park, S.; You, Y.-A.; Yun, H.; Choi, S.-J.; Hwang, H.-S.; Choi, S.-K.; Lee, S.M.; Kim, Y.J. Cervicovaginal fluid cytokines as predictive markers of preterm birth in symptomatic women. Obstet. Gynecol. Sci. 2020, 63, 455–463. [Google Scholar] [CrossRef]
- Ferreira, C.S.T.; da Silva, M.G.; de Pontes, L.G.; Dos Santos, L.D.; Marconi, C. Protein content of cervicovaginal fluid is altered during bacterial vaginosis. J. Low. Genit. Tract Dis. 2018, 22, 147–151. [Google Scholar] [CrossRef]
- Noda-Nicolau, N.M.; Bastos, L.B.; Bolpetti, A.N.; Pinto, G.V.S.; Marcolino, L.D.; Marconi, C.; Ferreira, C.S.T.; Polettini, J.; Vieira, E.P.; Da Silva, M.G. Cervicovaginal Levels of Human β-Defensin 1, 2, 3, and 4 of Reproductive-Aged Women With Chlamydia trachomatis Infection. J. Low. Genit. Tract Dis. 2017, 21, 189–192. [Google Scholar] [CrossRef] [PubMed]
- Njoku, K.; Crosbie, E. Does the vaginal microbiome drive cervical carcinogenesis? BJOG An Int. J. Obstet. Gynaecol. 2020, 127, 181. [Google Scholar] [CrossRef] [PubMed]
- Starodubtseva, N.L.; Brzhozovskiy, A.G.; Bugrova, A.E.; Kononikhin, A.S.; Indeykina, M.I.; Gusakov, K.I.; Chagovets, V.V.; Nazarova, N.M.; Frankevich, V.E.; Sukhikh, G.T. Label-free cervicovaginal fluid proteome profiling reflects the cervix neoplastic transformation. J. Mass Spectrom. 2019, 54, 693–703. [Google Scholar] [CrossRef]
- Prilepskaya, V.N.; Mheryan, A.N.; Akopian, A.S.; Nazarova, N.M.; Dovletkhanova, E.R.; Abakarova, P.R.; Starodubtseva, N.L. Biomarkers of cervicovaginal fluid for the diagnosis of cervical diseases associated with human papilloma virus (literature review). Gynecology 2019, 21, 6–11. [Google Scholar] [CrossRef]
- Njoku, K.; Sutton, C.J.; Whetton, A.D.; Crosbie, E.J. Metabolomic Biomarkers for Detection, Prognosis and Identifying Recurrence in Endometrial Cancer. Metabolites 2020, 10, 314. [Google Scholar] [CrossRef]
- O’Flynn, H.; Ryan, N.A.; Narine, N.; Shelton, D.; Rana, D.; Crosbie, E.J. Diagnostic accuracy of cytology for the detection of endometrial cancer in urine and vaginal samples. Nat. Commun. 2021, 12, 1–8. [Google Scholar] [CrossRef]
- Di Quinzio, M.K.; Oliva, K.; Holdsworth, S.J.; Ayhan, M.; Walker, S.P.; Rice, G.E.; Georgiou, H.M.; Permezel, M. Proteomic analysis and characterisation of human cervico-vaginal fluid proteins. Aust. N. Z. J. Obstet. Gynaecol. 2007, 47, 9–15. [Google Scholar] [CrossRef]
- Liong, S.; Di Quinzio, M.K.; Heng, Y.J.; Fleming, G.; Permezel, M.; Rice, G.E.; Georgiou, H.M. Proteomic analysis of human cervicovaginal fluid collected before preterm premature rupture of the fetal membranes. Reproduction 2013, 145, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Parry, S.; Leite, R.; Esplin, M.S.; Bukowski, R.; Zhang, H.; Varner, M.; Andrews, W.W.; Saade, G.R.; Ilekis, J.; Reddy, U.M. Cervicovaginal fluid proteomic analysis to identify potential biomarkers for preterm birth. Am. J. Obstet. Gynecol. 2020, 222, 493.e1–493.e13. [Google Scholar] [CrossRef] [PubMed]
- Ciocca, D.R.; Calderwood, S.K. Heat shock proteins in cancer: Diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 2005, 10, 86. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.C.; Guo, J.; Diehl, G.; DeSouza, L.; Rodrigues, M.J.; Romaschin, A.D.; Colgan, T.J.; Siu, K.M. Protein expression profiling of endometrial malignancies reveals a new tumor marker: Chaperonin 10. J. Proteome Res. 2004, 3, 636–643. [Google Scholar] [CrossRef] [PubMed]
- DeSouza, L.V.; Krakovska, O.; Darfler, M.M.; Krizman, D.B.; Romaschin, A.D.; Colgan, T.J.; Siu, K.W.M. mTRAQ-based quantification of potential endometrial carcinoma biomarkers from archived formalin-fixed paraffin-embedded tissues. Proteomics 2010, 10, 3108–3116. [Google Scholar] [CrossRef]
- Dube, V.; Grigull, J.; DeSouza, L.V.; Ghanny, S.; Colgan, T.J.; Romaschin, A.D.; Michael Siu, K.W. Verification of endometrial tissue biomarkers previously discovered using mass spectrometry-based proteomics by means of immunohistochemistry in a tissue microarray format. J. Proteome Res. 2007, 6, 2648–2655. [Google Scholar] [CrossRef] [PubMed]
- Dombrauckas, J.D.; Santarsiero, B.D.; Mesecar, A.D. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry 2005, 44, 9417–9429. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Min, W.; Huang, C.; Bai, S.; Tang, M.; Zhao, X. Proteomics-based approach identified differentially expressed proteins with potential roles in endometrial carcinoma. Int. J. Gynecol. Cancer 2010, 20, 9–15. [Google Scholar] [CrossRef]
- Njoku, K.; Barr, C.E.; Hotchkies, L.; Quille, N.; Louise Wan, Y.; Crosbie, E.J. Impact of socioeconomic deprivation on endometrial cancer survival in the North West of England: A prospective database analysis. BJOG Int. J. Obstet. Gynaecol. 2020, 128, 1215–1224. [Google Scholar] [CrossRef] [PubMed]
- Njoku, K.; O’Flynn, H.; Jones, E.; Ramchander, N.C.; White, H.; Macey, R.; Crosbie, E.J. Screening tests for endometrial cancer in the general population. Cochrane Database Syst. Rev. 2021, 128, 1215–1224. [Google Scholar]
- Wan, Y.L.; Beverley-Stevenson, R.; Carlisle, D.; Clarke, S.; Edmondson, R.J.; Glover, S.; Holland, J.; Hughes, C.; Kitchener, H.C.; Kitson, S. Working together to shape the endometrial cancer research agenda: The top ten unanswered research questions. Gynecol. Oncol. 2016, 143, 287–293. [Google Scholar] [CrossRef]
- Badrick, E.; Cresswell, K.; Ellis, P.; Crosbie, P.; Hall, P.S.; O’Flynn, H.; Martin, R.; Leighton, J.; Brown, L.; Makin, D. Top ten research priorities for detecting cancer early. Lancet Public Health 2019, 4, e551. [Google Scholar] [CrossRef]
- Snowhite, I.; Jones, W.; Dumestre, J.; Dunlap, K.; Braly, P.; Hagensee, M. Comparative analysis of methods for collection and measurement of cytokines and immunoglobulins in cervical and vaginal secretions of HIV and HPV infected women. J. Immunol. Methods 2002, 263, 85–95. [Google Scholar] [CrossRef]
- Andréoletti, L.; Grésenguet, G.; Chomont, N.; Matta, M.; Quiniou, Y.; Si-Mohamed, A.; Bélec, L. Comparison of washing and swabbing procedures for collecting genital fluids to assess shedding of human immunodeficiency virus type 1 (HIV-1) RNA in asymptomatic HIV-1-infected women. J. Clin. Microbiol. 2003, 41, 449–452. [Google Scholar] [CrossRef][Green Version]
- Igidbashian, S.; Boveri, S.; Spolti, N.; Radice, D.; Sandri, M.T.; Sideri, M. Self-collected human papillomavirus testing acceptability: Comparison of two self-sampling modalities. J. Women’s Health 2011, 20, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Escher, C.; Reiter, L.; MacLean, B.; Ossola, R.; Herzog, F.; Chilton, J.; MacCoss, M.J.; Rinner, O. Using i RT, a normalized retention time for more targeted measurement of peptides. Proteomics 2012, 12, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Parker, S.J.; Rost, H.; Rosenberger, G.; Collins, B.C.; Malmström, L.; Amodei, D.; Venkatraman, V.; Raedschelders, K.; Van Eyk, J.E.; Aebersold, R. Identification of a set of conserved eukaryotic internal retention time standards for data-independent acquisition mass spectrometry. Mol. Cell. Proteom. 2015, 14, 2800–2813. [Google Scholar] [CrossRef] [PubMed]
- Toprak, U.H.; Gillet, L.C.; Maiolica, A.; Navarro, P.; Leitner, A.; Aebersold, R. Conserved peptide fragmentation as a benchmarking tool for mass spectrometers and a discriminating feature for targeted proteomics. Mol. Cell. Proteom. 2014, 13, 2056–2071. [Google Scholar] [CrossRef] [PubMed]
| Cases | Controls |
|---|---|
| Referred with postmenopausal bleeding | Referred with postmenopausal bleeding |
| Biopsy proven AH or EC | AH and EC excluded following routine diagnostic investigations, including TVS, biopsy and/or hysteroscopy |
| Able to give informed consent | Able to give informed consent |
| Sample taken prior to commencement of any treatment, including surgery, hormone therapy or chemotherapy | Sample taken prior to commencement of any treatment, including surgery or hormone therapy |
| Can include those with benign pathologies such as benign polyp or atrophic vaginitis |
| Serial Number | Age (Years) | BMI (kg/m2) | Diagnosis | Grade | Stage |
|---|---|---|---|---|---|
| Library Generation Cohort | |||||
| 1 | 57 | 38 | Atypical Hyperplasia | - | - |
| 2 | 78 | 45 | Atypical Hyperplasia | - | - |
| 3 | 61 | 37 | Atypical Hyperplasia | - | - |
| 4 | 65 | 27 | Endometrioid Endometrial cancer | 1 | 1A |
| 5 | 60 | 43 | Endometrioid Endometrial cancer | 1 | 1A |
| 6 | 73 | 29 | Endometrioid Endometrial cancer | 2 | 1A |
| 7 | 52 | 30 | Endometrioid Endometrial cancer | 2 | 1A |
| 8 | 62 | 27 | Clear Cell Endometrial cancer | 3 | 1A |
| 9 | 52 | 30 | Serous Endometrial cancer | 3 | 1A |
| 10 | 74 | 27 | Carcinosarcoma | 3 | 1B |
| 11 | 72 | 27 | Mixed (Clear Cell/Endometrioid) cancer | 3 | 2 |
| 12 | 84 | 28 | Endometrioid Endometrial cancer | 3 | 4B |
| 13 | 54 | 29 | Control (No endometrial pathology) | - | - |
| 14 | 81 | 24 | Control (No endometrial pathology) | - | - |
| 15 | 56 | 24 | Control (Atrophic vaginitis) | - | - |
| 16 | 56 | 40 | Control (No endometrial pathology) | - | - |
| 17 | 52 | 44 | Control (No endometrial pathology) | - | - |
| 18 | 50 | 24 | Control (No endometrial pathology) | - | - |
| 19 | 56 | 24 | Control (No endometrial pathology) | - | - |
| Assays | Proteotypic | Proteotypic and Shared |
|---|---|---|
| Consensus cervico-vaginal fluid spectral library | ||
| Proteins | 1836 | 2425 |
| Peptides | 18,247 | 19,394 |
| Precursors | 22,455 | 23,886 |
| Transitions | 144,060 | 154,206 |
| Endometrial cancer cell-line based spectral library | ||
| Proteins | 5140 | 6003 |
| Peptides | 27,972 | 29,384 |
| Precursors | 29,509 | 30,876 |
| Transitions | 209,988 | 221,844 |
| Study Characteristics | Our CVF Spectral Library | CVF Proteome by Zegel et al. [23] | CVF Proteome by Tang et al. [24] | CVF Proteome by Shaw et al. [25] |
|---|---|---|---|---|
| Sample size | 19 | 7 | 29 | 5 |
| Age range | 50–67 years | 37–45 years | 24–48 years | 20–40 years |
| Menopausal status | Post-menopausal | Pre-menopausal | Pre-menopausal | Pre-menopausal |
| Clinical diagnoses | 9 EC cases, 3 AH and 7 symptomatic women with no EC | All had cervical pre-cancer | Asymptomatic women, 7 had candida | Healthy female volunteers |
| Sample collection | Delphi screener (saline based wash) | Colposcopy (5% acetic acid wash) | Syringe (saline based wash) | Gauze (inserted in vagina for 1 h) |
| Sample description | Supernatant and pellets | Supernatant only | Supernatant only | Whole fluid |
| Sample analysis | Orbitrap Fusion Lumos Tribrid LC-MS | MALDI-TOF-TOF MS/MS | MALDI-TOF-TOF MS/MS | 1D-SDS-PAGE, cation exchange, LS-MS/MS |
| Protein identification | X!Tandem | MASCOT | MASCOT | MASCOT & X!Tandem |
| Spectral library protein count | 2425 | 339 | 147 (59 unique) | 685 |
| Serial Number | Age (Years) | BMI (kg/m2) | Diagnosis | Grade | Stage |
|---|---|---|---|---|---|
| Technical Validation Cohort | |||||
| 20 | 50 | 19 | Control (No endometrial pathology) | ||
| 21 | 52 | 37 | Control (Benign endometrial polyp) | ||
| 22 | 55 | 34 | Control (No endometrial pathology) | ||
| 23 | 56 | 19 | Control (No endometrial pathology) | ||
| 24 | 56 | 37 | Control (No endometrial pathology) | ||
| Clinical Validation Cohort | |||||
| 25 | 58 | 36 | Atypical Hyperplasia | - | - |
| 26 | 67 | 30 | Endometrioid endometrial cancer | 1 | 1A |
| 27 | 55 | 34 | Endometrioid endometrial cancer | 1 | 1A |
| 28 | 50 | 54 | Endometrioid endometrial cancer | 1 | 1A |
| 29 | 62 | 27 | Clear cell | 3 | 1A |
| 30 | 60 | 58 | Control (No endometrial pathology) | - | - |
| 31 | 62 | 24 | Control (Atrophic vaginitis) | - | - |
| 32 | 58 | 30 | Control (Benign endometrial polyp) | - | - |
| 33 | 47 | 24 | Control (No endometrial pathology) | - | - |
| 34 | 52 | 33 | Control (No endometrial pathology) | - | - |
| Sample | Protein Count | Missing Values in Replicate 1 | Missing Values in Replicate 2 | Overall Rate |
|---|---|---|---|---|
| Sample 1 | 410 | 21(5.1%) | 32(7.8%) | 12.9% |
| Sample 2 | 389 | 25(6.4%) | 28(7.2%) | 13.6% |
| Sample 3 | 308 | 37(12.0%) | 30(9.7%) | 21.8% |
| Sample 4 | 378 | 29(7.7%) | 33(8.7%) | 16.4% |
| Sample 5 | 426 | 25(5.9%) | 35(8.2%) | 14.1% |
| Biomarker Candidate | Gene Name | Intensity Correlation Coefficient | Coefficient of Variation |
|---|---|---|---|
| Human Epididymis Protein 4 | HE4/WFDC2 | 0.99 | 3.29 |
| Cancer Associated Antigen 15-3 | MUC-1 | 0.99 | 4.88 |
| Matrix Metalloproteinase 9 | MMP9 | 0.99 | 2.01 |
| Fatty Acid Binding Protein-5 | FABP5 | 0.99 | 0.62 |
| Alpha-1B-Glycoprotein | AIBG | 0.99 | 1.36 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Njoku, K.; Chiasserini, D.; Geary, B.; Pierce, A.; Jones, E.R.; Whetton, A.D.; Crosbie, E.J. Comprehensive Library Generation for Identification and Quantification of Endometrial Cancer Protein Biomarkers in Cervico-Vaginal Fluid. Cancers 2021, 13, 3804. https://doi.org/10.3390/cancers13153804
Njoku K, Chiasserini D, Geary B, Pierce A, Jones ER, Whetton AD, Crosbie EJ. Comprehensive Library Generation for Identification and Quantification of Endometrial Cancer Protein Biomarkers in Cervico-Vaginal Fluid. Cancers. 2021; 13(15):3804. https://doi.org/10.3390/cancers13153804
Chicago/Turabian StyleNjoku, Kelechi, Davide Chiasserini, Bethany Geary, Andrew Pierce, Eleanor R. Jones, Anthony D. Whetton, and Emma J. Crosbie. 2021. "Comprehensive Library Generation for Identification and Quantification of Endometrial Cancer Protein Biomarkers in Cervico-Vaginal Fluid" Cancers 13, no. 15: 3804. https://doi.org/10.3390/cancers13153804
APA StyleNjoku, K., Chiasserini, D., Geary, B., Pierce, A., Jones, E. R., Whetton, A. D., & Crosbie, E. J. (2021). Comprehensive Library Generation for Identification and Quantification of Endometrial Cancer Protein Biomarkers in Cervico-Vaginal Fluid. Cancers, 13(15), 3804. https://doi.org/10.3390/cancers13153804






