Deep Learning Segmentation of Triple-Negative Breast Cancer (TNBC) Patient Derived Tumor Xenograft (PDX) and Sensitivity of Radiomic Pipeline to Tumor Probability Boundary
Abstract
:Simple Summary
Abstract
1. Introduction
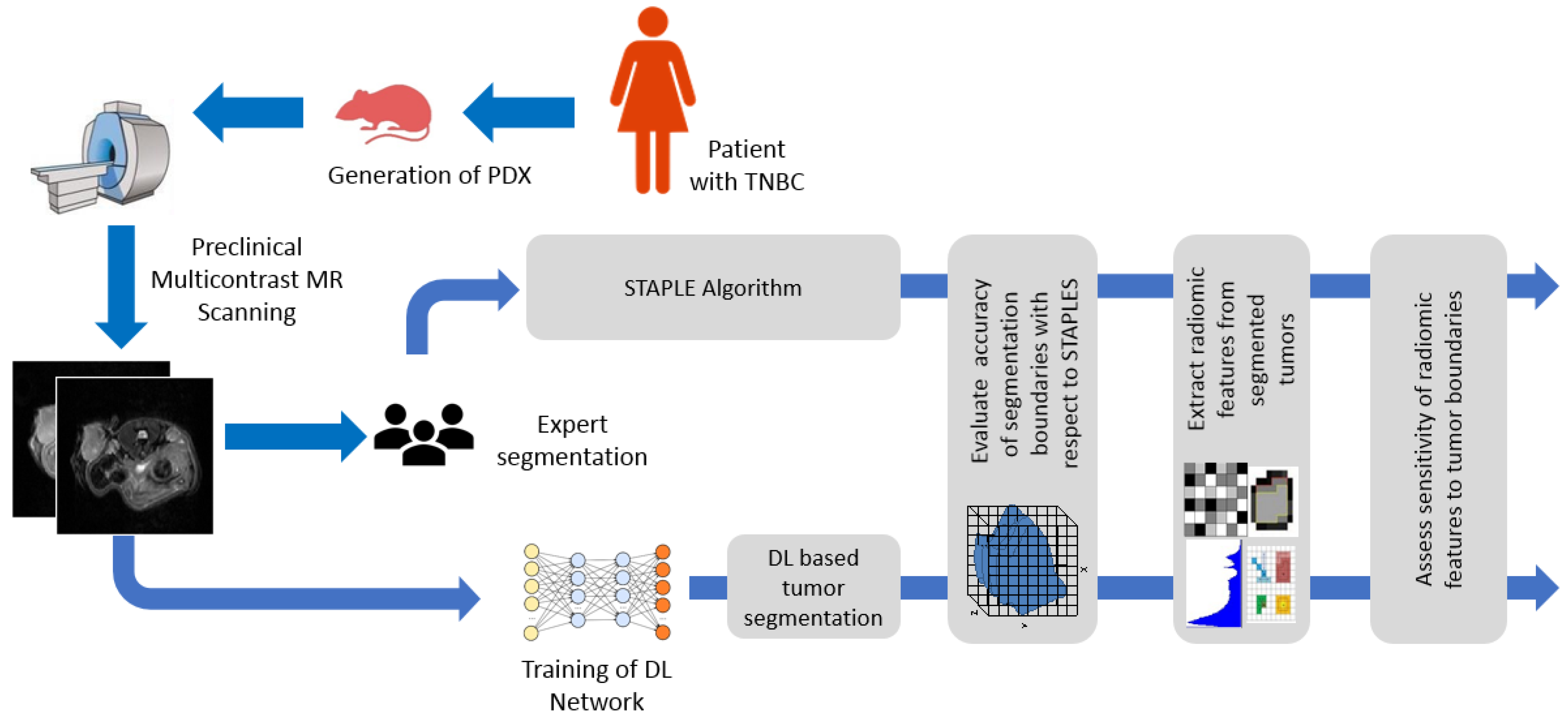
2. Materials and Methods
2.1. Generation of TNBC PDXs
2.2. MR Image Acquisition
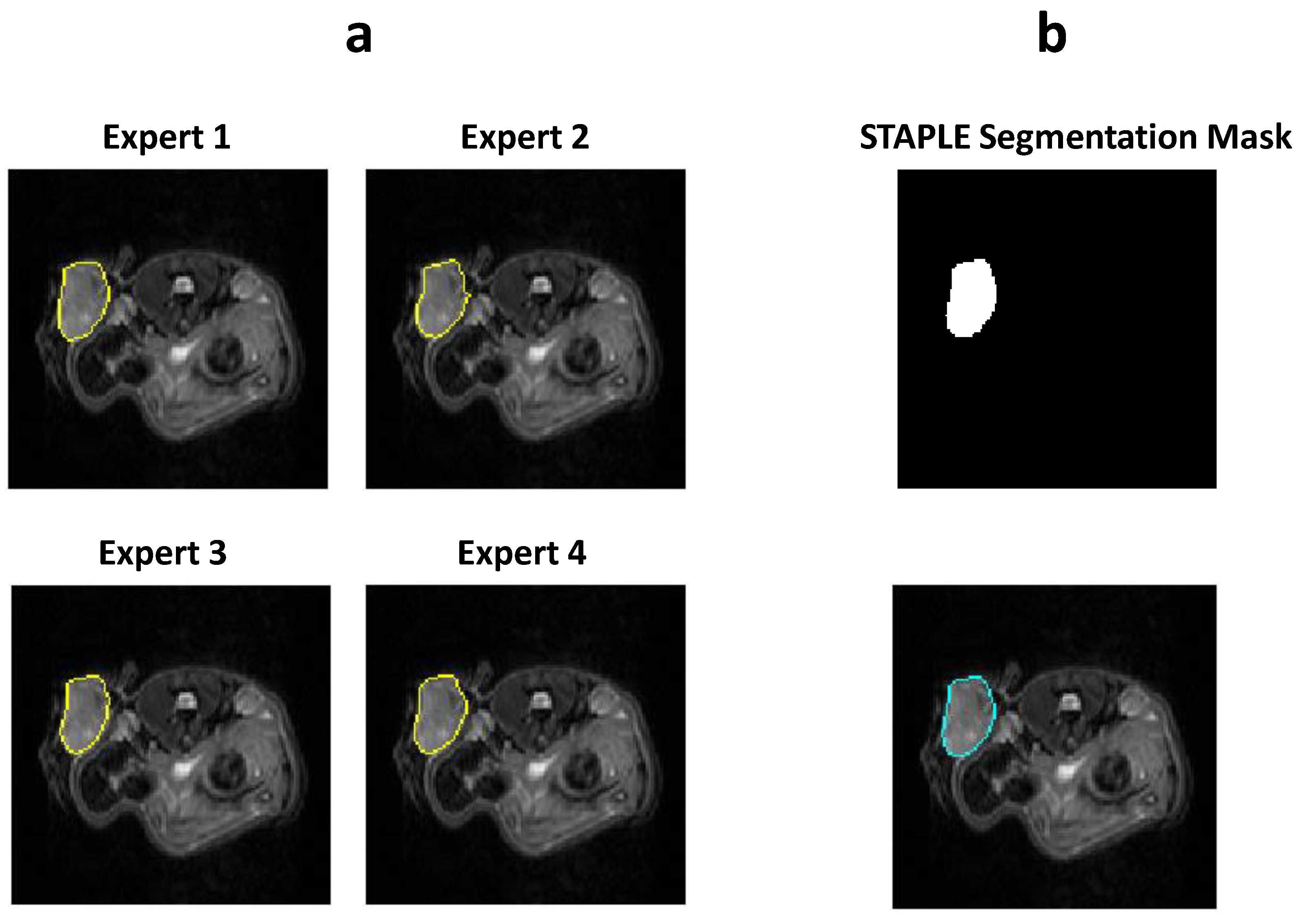
2.3. Manual Segmentation of the MR Images
2.4. CNN Model for Automatic Segmentation
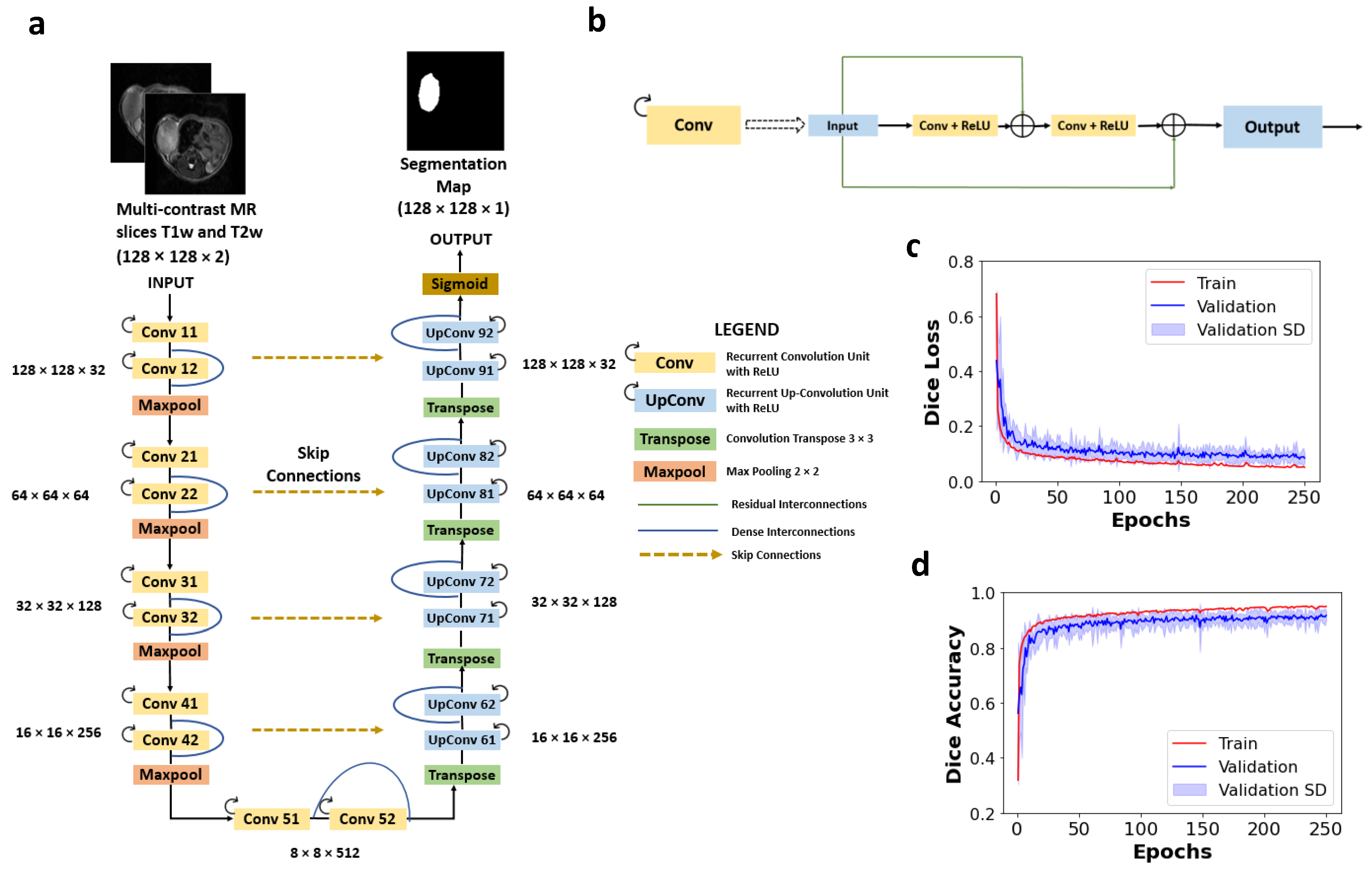
2.4.1. Network Architecture
2.4.2. Preprocessing and Training of the Network
2.4.3. STAPLE Algorithm to Generate Consensus among Experts
2.4.4. Performance Assessment of the Network
- F1-score—the F1-score measures the spatial overlap between the predicted image and the ground truth and is given by Equation (1).
- Precision—precision signifies the fraction of true positives (TP) in relation to that of the segmented tumor region by the algorithm and is given by Equation (2).
- Recall—recall signifies the fraction of true positives (TP) in relation to that of the ground truth segmentation by experts and is given by Equation (3).
- Accuracy—accuracy signifies the fraction of correctly classified voxels in relation to that of the total number of voxels and is given by Equation (4).
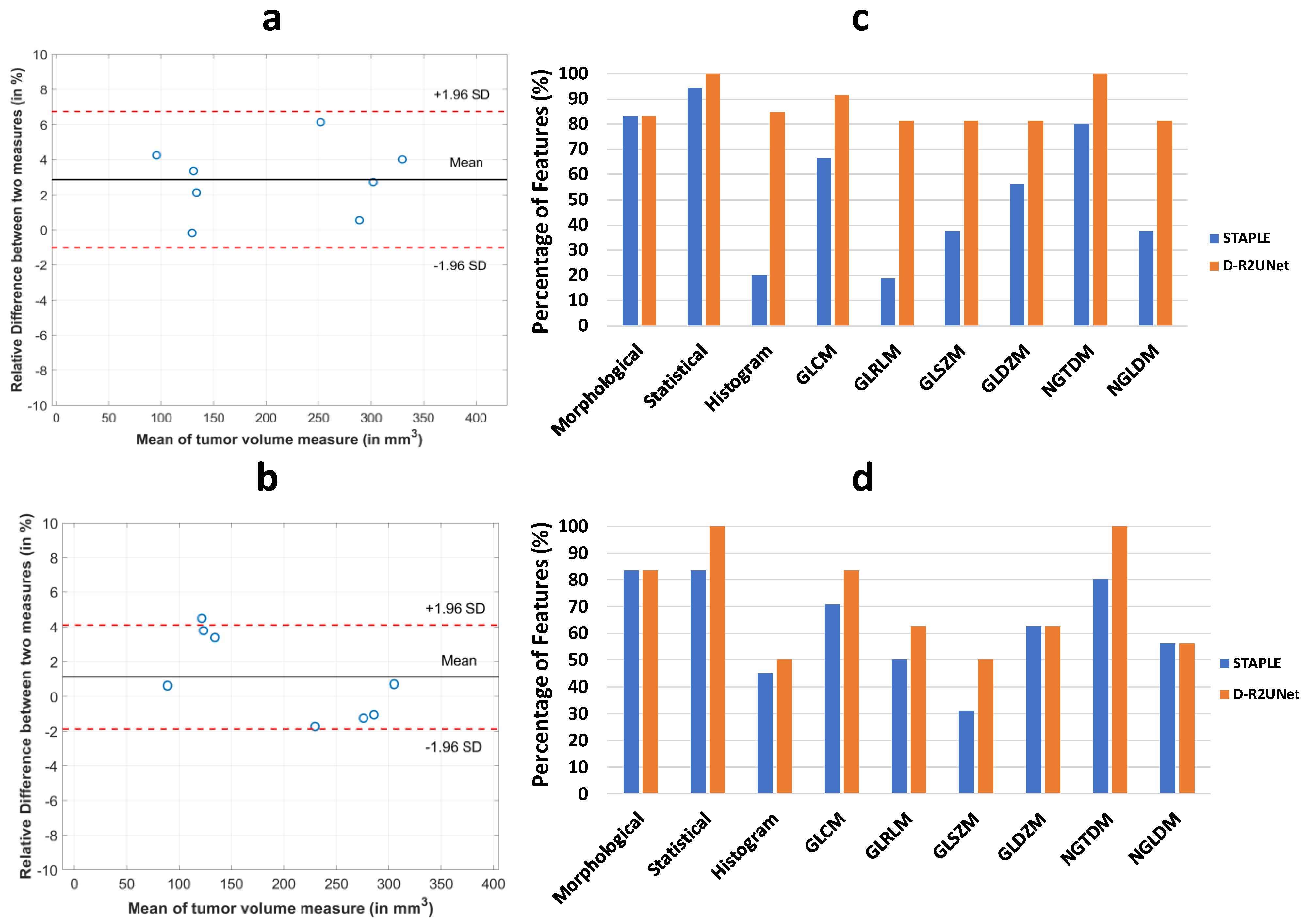
2.5. Extraction of Radiomic Features and Correlation between STAPLE and D-R2UNet
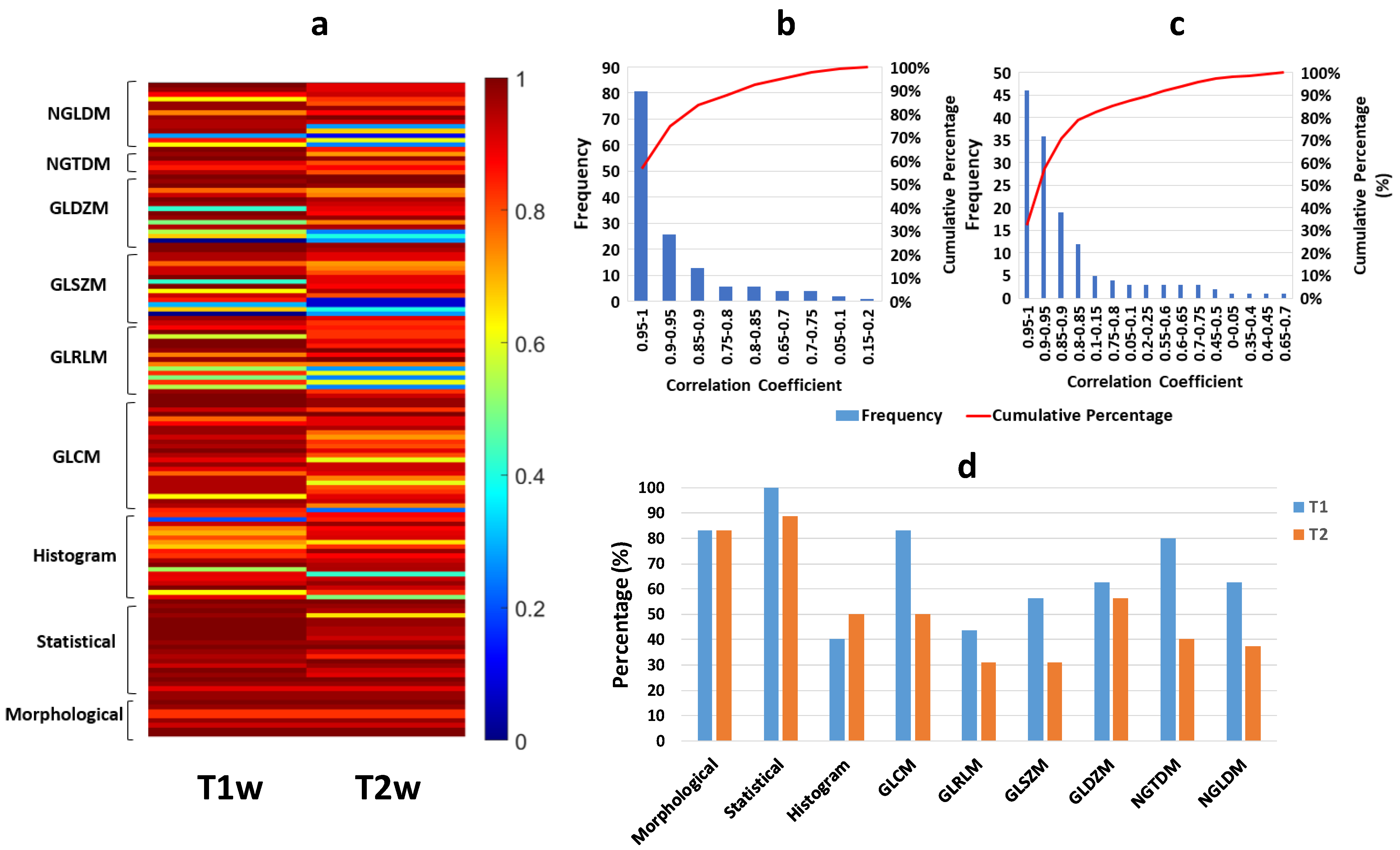
2.6. Reproducibility of Radiomic Features by Experts
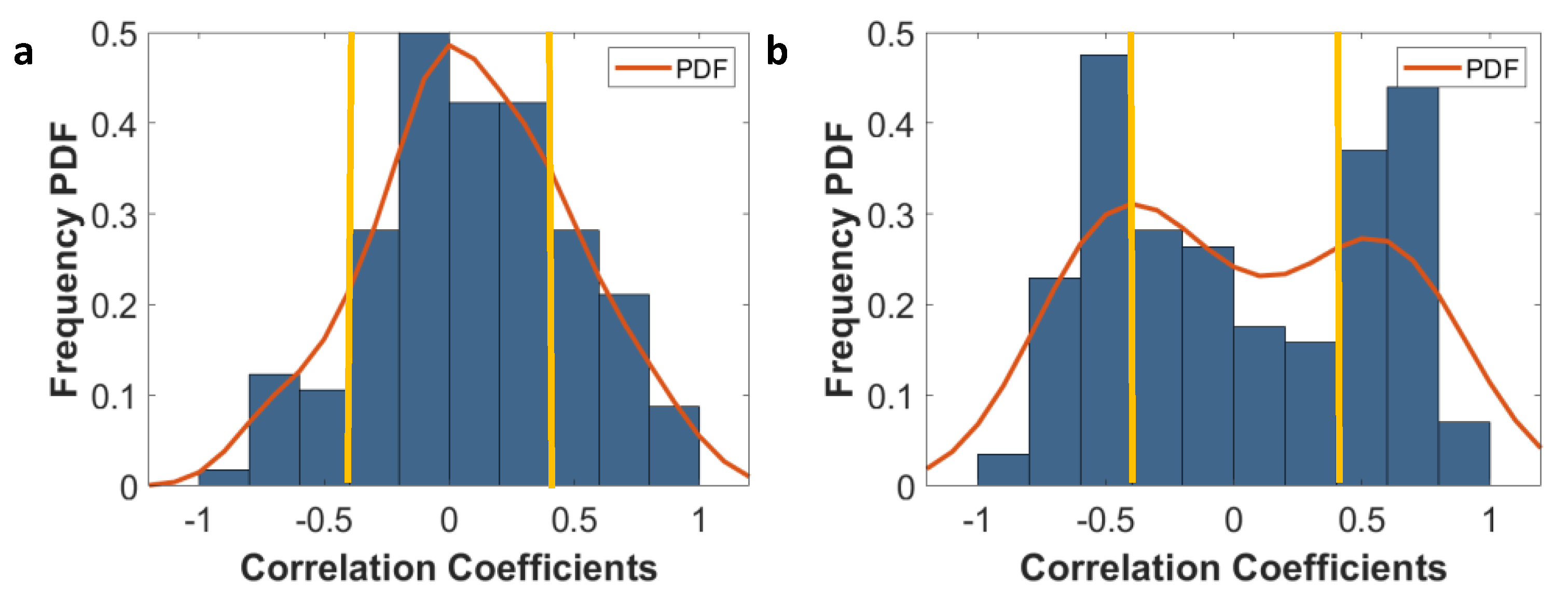
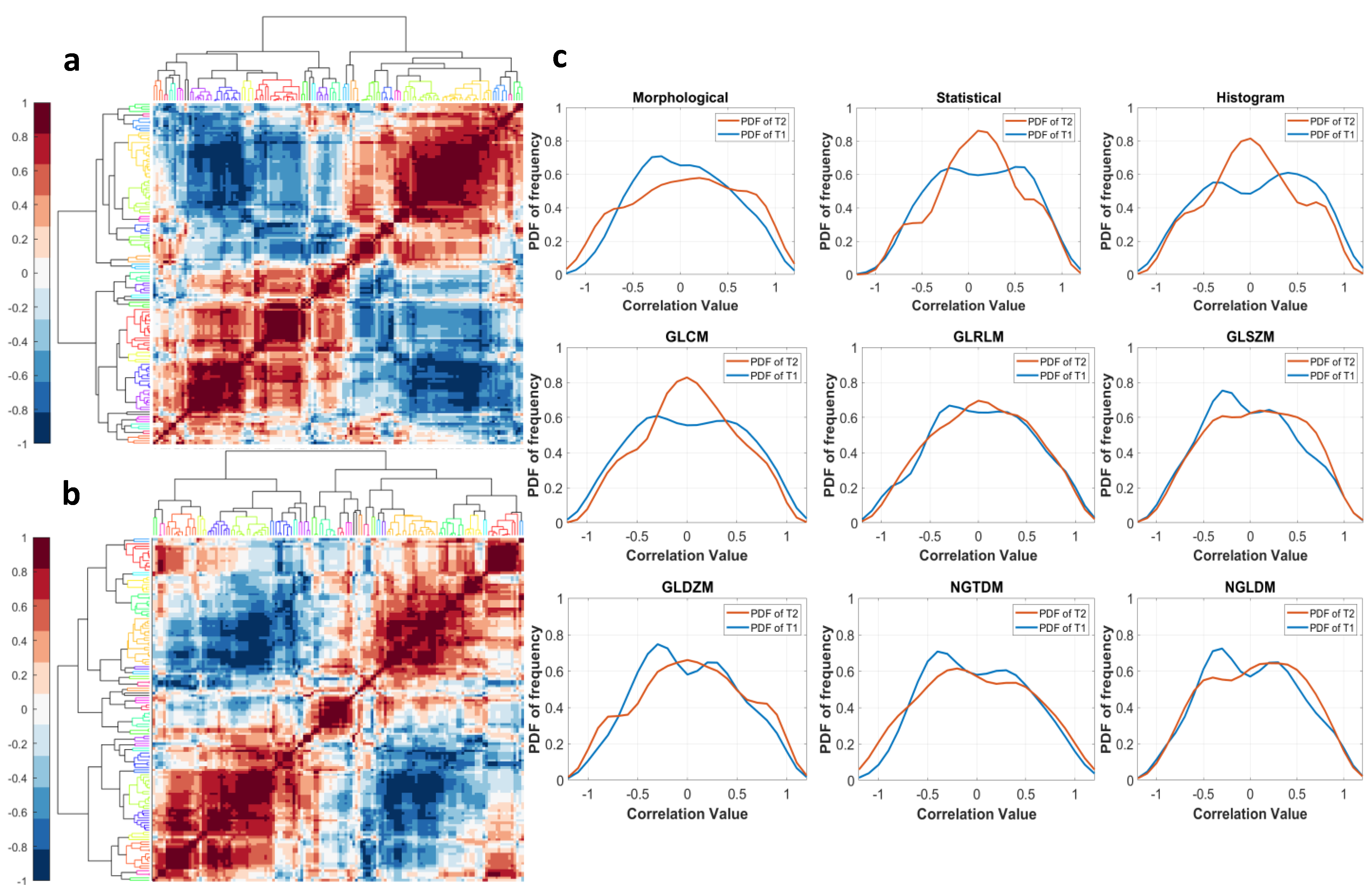
2.7. Sensitivity of Features to Tumor Probability Boundaries
3. Results
3.1. Performance of CNN Segmentation
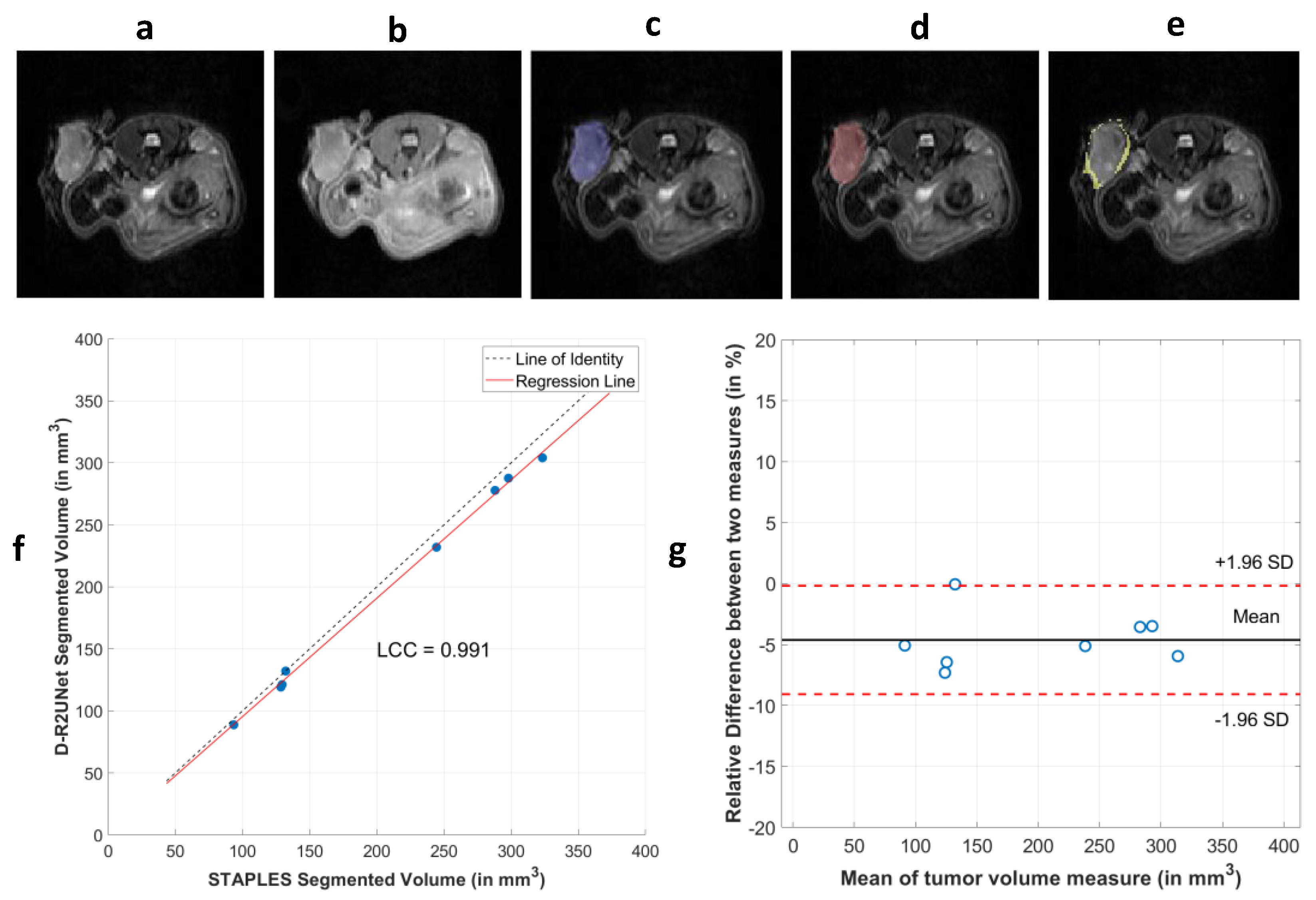
3.2. Robustness of Radiomic Parameters Extracted from the D-R2UNet Algorithm
3.3. Reproducibility Analysis of the Radiomic Parameters
3.4. Sensitivity of Radiomic Features to Tumor Boundaries
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Z.; Akbay, E.; Mikse, O.; Tupper, T.; Cheng, K.; Wang, Y.; Tan, X.; Altabef, A.; Woo, S.A.; Chen, L.; et al. Co-clinical trials demonstrate superiority of crizotinib to chemotherapy in ALK-rearranged non-small cell lung cancer and predict strategies to overcome resistance. Clin. Cancer Res. 2014, 20, 1204–1211. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.R.; Kang, H.N.; Shim, H.S.; Kim, E.Y.; Kim, J.; Kim, D.J.; Lee, J.G.; Lee, C.Y.; Hong, M.H.; Kim, S.M.; et al. Co-clinical trials demonstrate predictive biomarkers for dovitinib, an FGFR inhibitor, in lung squamous cell carcinoma. Ann. Oncol. 2017, 28, 1250–1259. [Google Scholar] [CrossRef]
- Kwong, L.N.; Boland, G.M.; Frederick, D.T.; Helms, T.L.; Akid, A.T.; Miller, J.P.; Jiang, S.; Cooper, Z.A.; Song, X.; Seth, S.; et al. Co-clinical assessment identifies patterns of BRAF inhibitor resistance in melanoma. J. Clin. Investig. 2015, 125, 1459–1470. [Google Scholar] [CrossRef] [Green Version]
- Lunardi, A.; Ala, U.; Epping, M.T.; Salmena, L.; Clohessy, J.G.; Webster, K.A.; Wang, G.; Mazzucchelli, R.; Bianconi, M.; Stack, E.C.; et al. A co-clinical approach identifies mechanisms and potential therapies for androgen deprivation resistance in prostate cancer. Nat. Genet. 2013, 45, 747–755. [Google Scholar] [CrossRef]
- Nishino, M.; Sacher, A.G.; Gandhi, L.; Chen, Z.; Akbay, E.; Fedorov, A.; Westin, C.F.; Hatabu, H.; Johnson, B.E.; Hammerman, P.; et al. Co-clinical quantitative tumor volume imaging in ALK-rearranged NSCLC treated with crizotinib. Eur. J. Radiol. 2017, 88, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Owonikoko, T.K.; Zhang, G.; Kim, H.S.; Stinson, R.M.; Bechara, R.; Zhang, C.; Chen, Z.; Saba, N.F.; Pakkala, S.; Pillai, R.; et al. Patient-derived xenografts faithfully replicated clinical outcome in a phase II co-clinical trial of arsenic trioxide in relapsed small cell lung cancer. J. Transl. Med. 2016, 14, 111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sia, D.; Moeini, A.; Labgaa, I.; Villanueva, A. The future of patient-derived tumor xenografts in cancer treatment. Pharmacogenomics 2015, 16, 1671–1683. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, A.; Wang, L. Bridging the divide: Preclinical research discrepancies between triple-negative breast cancer cell lines and patient tumors. Oncotarget 2017, 8, 113269–113281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeRose, Y.S.; Wang, G.; Lin, Y.C.; Bernard, P.S.; Buys, S.S.; Ebbert, M.T.; Factor, R.; Matsen, C.; Milash, B.A.; Nelson, E.; et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med. 2011, 17, 1514–1520. [Google Scholar] [CrossRef]
- Krepler, C.; Xiao, M.; Spoesser, K.; Brafford, P.A.; Shannan, B.; Beqiri, M.; Liu, Q.; Xu, W.; Garman, B.; Nathanson, K.L.; et al. Personalized pre-clinical trials in BRAF inhibitor resistant patient derived xenograft models identify second line combination therapies. Clin. Cancer Res. 2015, 22, 1592–1602. [Google Scholar] [CrossRef] [Green Version]
- Shoghi, K.I.; Badea, C.T.; Blocker, S.J.; Chenevert, T.L.; Laforest, R.; Lewis, M.T.; Luker, G.D.; Manning, H.C.; Marcus, D.S.; Mowery, Y.M.; et al. Co-Clinical Imaging Resource Program (CIRP): Bridging the Translational Divide to Advance Precision Medicine. Tomography 2020, 6, 273–287. [Google Scholar] [CrossRef]
- Sardanelli, F.; Boetes, C.; Borisch, B.; Decker, T.; Federico, M.; Gilbert, F.J.; Helbich, T.; Heywang-Köbrunner, S.H.; Kaiser, W.A.; Kerin, M.J.; et al. Magnetic resonance imaging of the breast: Recommendations from the EUSOMA working group. Eur. J. Cancer 2010, 46, 1296–1316. [Google Scholar] [CrossRef] [PubMed]
- Uematsu, T. MR imaging of triple-negative breast cancer. Breast Cancer 2011, 18, 161–164. [Google Scholar] [CrossRef]
- Uematsu, T.; Kasami, M.; Yuen, S. Triple-Negative Breast Cancer: Correlation between MR Imaging and Pathologic Findings. Radiology 2009, 250, 638–647. [Google Scholar] [CrossRef]
- Cui, S.; Mao, L.; Jiang, J.; Liu, C.; Xiong, S. Automatic Semantic Segmentation of Brain Gliomas from MRI Images Using a Deep Cascaded Neural Network. J. Healthc. Eng. 2018, 2018, 1–14. [Google Scholar] [CrossRef]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.-M.; Larochelle, H. Brain tumor segmentation with Deep Neural Networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trebeschi, S.; van Griethuysen, J.J.M.; Lambregts, D.M.J.; Lahaye, M.J.; Parmar, C.; Bakers, F.C.H.; Peters, N.H.G.M.; Beets-Tan, R.G.H.; Aerts, H.J.W.L. Deep Learning for Fully-Automated Localization and Segmentation of Rectal Cancer on Multiparametric MR. Sci. Rep. 2017, 7, 5301. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. Lect. Notes Comput. Sci. 2015, 9351, 234–241. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Liu, Q.; Wang, Y. Road Extraction by Deep Residual U-Net. IEEE Geosci. Remote Sens. Lett. 2018, 15, 749–753. [Google Scholar] [CrossRef] [Green Version]
- Alom, M.Z.; Hasan, M.; Yakopcic, C.; Taha, T.M.; Asari, V.K. Recurrent Residual Convolutional Neural Network based on U-Net (R2U-Net) for Medical Image Segmentation. arXiv 2018, arXiv:1802.06955. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. 2016, 2016, 770–778. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef] [Green Version]
- Kolařík, M.; Burget, R.; Uher, V.; Říha, K.; Dutta, M.K. Optimized high resolution 3D dense-U-Net network for brain and spine segmentation. Appl. Sci. 2019, 9, 404. [Google Scholar] [CrossRef] [Green Version]
- Dutta, K. Densely Connected Recurrent Residual (Dense R2UNet) Convolutional Neural Network for Segmentation of Lung CT Images. arXiv 2021, arXiv:2102.00663. [Google Scholar]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [Green Version]
- Trebeschi, S.; Drago, S.G.; Birkbak, N.J.; Kurilova, I.; Calin, A.M.; Pizzi, A.D.; Lalezari, F.; Lambregts, D.M.J.; Rohaan, M.W.; Parmar, C.; et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann. Oncol. 2019, 30, 998–1004. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [Green Version]
- Li, S.Q.; Shen, D.; Shao, J.Y.; Crowder, R.; Liu, W.B.; Prat, A.; He, X.P.; Liu, S.Y.; Hoog, J.; Lu, C.; et al. Endocrine-Therapy-Resistant ESR1 Variants Revealed by Genomic Characterization of Breast-Cancer-Derived Xenografts. Cell Rep. 2013, 4, 1116–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drozdzal, M.; Vorontsov, E.; Chartrand, G.; Kadoury, S.; Pal, C. The importance of skip connections in biomedical image segmentation. Lect. Notes Comput. Sci. 2016, 10008 LNCS, 179–187. [Google Scholar] [CrossRef] [Green Version]
- Hinton, G.E.; Srivastava, N.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R.R. Improving neural networks by preventing co-adaptation of feature detectors. arXiv 2012, arXiv:1207.0580. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the thirteenth international conference on artificial intelligence and statistics, Sardinia, Italy, 13–15 May 2010; pp. 249–256. [Google Scholar]
- Warfield, S.K.; Zou, K.H.; Wells, W.M. Simultaneous Truth and Performance Level Estimation (STAPLE): An Algorithm for the Validation of Image Segmentation. IEEE Trans. Med. Imaging 2004, 23, 903–921. [Google Scholar] [CrossRef] [Green Version]
- Vallières, M.; Freeman, C.R.; Skamene, S.R.; El Naqa, I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys. Med. Biol. 2015, 60, 5471–5496. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, S.P. Least-Squares Quantization in Pcm. IEEE Trans. Inf. Theory 1982, 28, 129–137. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, L.I. A Concordance Correlation-Coefficient to Evaluate Reproducibility. Biometrics 1989, 45, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Tunali, I.; Hall, L.O.; Napel, S.; Cherezov, D.; Guvenis, A.; Gillies, R.J.; Schabath, M.B. Stability and reproducibility of computed tomography radiomic features extracted from peritumoral regions of lung cancer lesions. Med. Phys. 2019, 46, 5075–5085. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.H. Biostatistics 304. Cluster analysis. Singap. Med. J. 2005, 46, 153–160. [Google Scholar]
- Balagurunathan, Y.; Gu, Y.; Wang, H.; Kumar, V.; Grove, O.; Hawkins, S.; Kim, J.; Goldgof, D.B.; Hall, L.O.; Gatenby, R.A.; et al. Reproducibility and Prognosis of Quantitative Features Extracted from CT Images. Transl. Oncol. 2014, 7, 72–87. [Google Scholar] [CrossRef] [Green Version]
- Fried, D.V.; Tucker, S.L.; Zhou, S.; Liao, Z.; Mawlawi, O.; Ibbott, G.; Court, L.E. Prognostic value and reproducibility of pretreatment CT texture features in stage III non-small cell lung cancer. Int. J. Radiat. Oncol. Biol. Phys. 2014, 90, 834–842. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.; Shoghi, K.I. Computer-Aided Tumor Segmentation from T2-Weighted MR Images of Patient-Derived Tumor Xenografts; Springer: Cham, Switzerlands, 2019; pp. 159–171. [Google Scholar] [CrossRef]
- Holbrook, M.D.; Blocker, S.J.; Mowery, Y.M.; Badea, A.; Qi, Y.; Xu, E.S.; Kirsch, D.G.; Johnson, G.A.; Badea, C.T. MRI-Based Deep Learning Segmentation and Radiomics of Sarcoma in Mice. Tomography 2020, 6, 23–33. [Google Scholar] [CrossRef]
- Narayana, P.A.; Coronado, I.; Sujit, S.J.; Sun, X.; Wolinsky, J.S.; Gabr, R.E. Are multi-contrast magnetic resonance images necessary for segmenting multiple sclerosis brains? A large cohort study based on deep learning. Magn. Reson. Imaging 2020, 65, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Ashton, E.A.; Takahashi, C.; Berg, M.J.; Goodman, A.; Totterman, S.; Ekholm, S. Accuracy and reproducibility of manual and semiautomated quantification of MS lesions by MRI. J. Magn. Reson. Imaging 2003, 17, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Hurtz, S.; Chow, N.; Watson, A.E.; Somme, J.H.; Goukasian, N.; Hwang, K.S.; Morra, J.; Elashoff, D.; Gao, S.; Petersen, R.C.; et al. Automated and manual hippocampal segmentation techniques: Comparison of results, reproducibility and clinical applicability. Neuroimage Clin. 2019, 21, 101574. [Google Scholar] [CrossRef] [PubMed]
- Vallieres, M.; Zwanenburg, A.; Badic, B.; Cheze Le Rest, C.; Visvikis, D.; Hatt, M. Responsible Radiomics Research for Faster Clinical Translation. J. Nucl. Med. 2018, 59, 189–193. [Google Scholar] [CrossRef]
- Pavic, M.; Bogowicz, M.; Wurms, X.; Glatz, S.; Finazzi, T.; Riesterer, O.; Roesch, J.; Rudofsky, L.; Friess, M.; Veit-Haibach, P.; et al. Influence of inter-observer delineation variability on radiomics stability in different tumor sites. Acta Oncol. 2018, 57, 1070–1074. [Google Scholar] [CrossRef] [Green Version]
- Park, J.E.; Park, S.Y.; Kim, H.J.; Kim, H.S. Reproducibility and Generalizability in Radiomics Modeling: Possible Strategies in Radiologic and Statistical Perspectives. Korean J. Radiol. 2019, 20, 1124–1137. [Google Scholar] [CrossRef]
- Traverso, A.; Wee, L.; Dekker, A.; Gillies, R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1143–1158. [Google Scholar] [CrossRef] [Green Version]
- Haarburger, C.; Muller-Franzes, G.; Weninger, L.; Kuhl, C.; Truhn, D.; Merhof, D. Radiomics feature reproducibility under inter-rater variability in segmentations of CT images. Sci. Rep. 2020, 10, 12688. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Leger, S.; Agolli, L.; Pilz, K.; Troost, E.G.C.; Richter, C.; Lock, S. Assessing robustness of radiomic features by image perturbation. Sci. Rep. 2019, 9, 614. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Tan, Y.; Tsai, W.Y.; Qi, J.; Xie, C.; Lu, L.; Schwartz, L.H. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci. Rep. 2016, 6, 23428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, P.; Wang, J.; Zhong, H.; Zhou, Z.; Shen, L.; Hu, W.; Zhang, Z. Reproducibility with repeat CT in radiomics study for rectal cancer. Oncotarget 2016, 7, 71440–71446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Timmeren, J.E.; Leijenaar, R.T.H.; van Elmpt, W.; Wang, J.; Zhang, Z.; Dekker, A.; Lambin, P. Test-Retest Data for Radiomics Feature Stability Analysis: Generalizable or Study-Specific? Tomography 2016, 2, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. J. Chiropr. Chiropr. Med. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Cicchetti, D.V. Guidelines, criteria, and rules of thumb for evaluating normed and standardized assessment instruments in psychology. Psychol. Assess. 1994, 6, 284. [Google Scholar] [CrossRef]
| Total No. of Mice Used for Study | No. of Mice Used for Training and Validation of the CNN | No. of Mice Used for Testing the Performance of the CNN | No. of MR Slices for Training and Validation | No. of MR Slices Used for Testing |
|---|---|---|---|---|
| 49 | 41 | 8 | 255 | 39 |
| Input Data | Network | F1-Score | Recall | Precision | AUC |
|---|---|---|---|---|---|
| T2w and T1w | U-Net | 0.929 ± 0.072 | 0.928 ± 0.040 | 0.935 ± 0.098 | 0.962 ± 0.019 |
| Dense U-Net | 0.923 ± 0.066 | 0.960 ± 0.025 | 0.897 ± 0.116 | 0.977 ± 0.012 | |
| Res-Net | 0.922 ± 0.074 | 0.947 ± 0.034 | 0.910 ± 0.117 | 0.971 ± 0.017 | |
| R2U-Net | 0.929 ± 0.072 | 0.933 ± 0.037 | 0.937 ± 0.128 | 0.965 ± 0.018 | |
| Dense R2U-Net | 0.948 ± 0.026 | 0.928 ± 0.032 | 0.970 ± 0.042 | 0.963 ± 0.016 | |
| T2w | U-Net | 0.927 ± 0.076 | 0.950 ± 0.031 | 0.919 ± 0.119 | 0.973 ± 0.016 |
| Dense U-Net | 0.910 ± 0.077 | 0.932 ± 0.033 | 0.900 ± 0.130 | 0.963 ± 0.017 | |
| Res-Net | 0.913 ± 0.079 | 0.884 ± 0.079 | 0.943 ± 0.091 | 0.943 ± 0.040 | |
| R2U-Net | 0.924 ± 0.067 | 0.959 ± 0.026 | 0.902 ± 0.116 | 0.977 ± 0.012 | |
| Dense R2U-Net | 0.935 ± 0.064 | 0.954 ± 0.023 | 0.925 ± 0.107 | 0.975 ± 0.011 |
| Network | F1-Score | Recall | Precision | AUC |
|---|---|---|---|---|
| U-Net | 0.906 ± 0.022 | 0.907 ± 0.027 | 0.914 ± 0.015 | 0.961 ± 0.013 |
| Dense U-Net | 0.911 ± 0.016 | 0.903 ± 0.025 | 0.930 ± 0.016 | 0.960 ± 0.012 |
| Res-Net | 0.909 ± 0.010 | 0.902 ± 0.030 | 0.925 ± 0.028 | 0.961 ± 0.012 |
| R2U-Net | 0.917 ± 0.013 | 0.909 ± 0.030 | 0.933 ± 0.008 | 0.960 ± 0.013 |
| Dense R2U-Net | 0.922 ± 0.009 | 0.937 ± 0.005 | 0.928 ± 0.016 | 0.963 ± 0.008 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dutta, K.; Roy, S.; Whitehead, T.D.; Luo, J.; Jha, A.K.; Li, S.; Quirk, J.D.; Shoghi, K.I. Deep Learning Segmentation of Triple-Negative Breast Cancer (TNBC) Patient Derived Tumor Xenograft (PDX) and Sensitivity of Radiomic Pipeline to Tumor Probability Boundary. Cancers 2021, 13, 3795. https://doi.org/10.3390/cancers13153795
Dutta K, Roy S, Whitehead TD, Luo J, Jha AK, Li S, Quirk JD, Shoghi KI. Deep Learning Segmentation of Triple-Negative Breast Cancer (TNBC) Patient Derived Tumor Xenograft (PDX) and Sensitivity of Radiomic Pipeline to Tumor Probability Boundary. Cancers. 2021; 13(15):3795. https://doi.org/10.3390/cancers13153795
Chicago/Turabian StyleDutta, Kaushik, Sudipta Roy, Timothy Daniel Whitehead, Jingqin Luo, Abhinav Kumar Jha, Shunqiang Li, James Dennis Quirk, and Kooresh Isaac Shoghi. 2021. "Deep Learning Segmentation of Triple-Negative Breast Cancer (TNBC) Patient Derived Tumor Xenograft (PDX) and Sensitivity of Radiomic Pipeline to Tumor Probability Boundary" Cancers 13, no. 15: 3795. https://doi.org/10.3390/cancers13153795
APA StyleDutta, K., Roy, S., Whitehead, T. D., Luo, J., Jha, A. K., Li, S., Quirk, J. D., & Shoghi, K. I. (2021). Deep Learning Segmentation of Triple-Negative Breast Cancer (TNBC) Patient Derived Tumor Xenograft (PDX) and Sensitivity of Radiomic Pipeline to Tumor Probability Boundary. Cancers, 13(15), 3795. https://doi.org/10.3390/cancers13153795






