A Personal Breast Cancer Risk Stratification Model Using Common Variants and Environmental Risk Factors in Japanese Females
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Evaluation of Environmental Risk Factors
2.3. Statistical Analysis
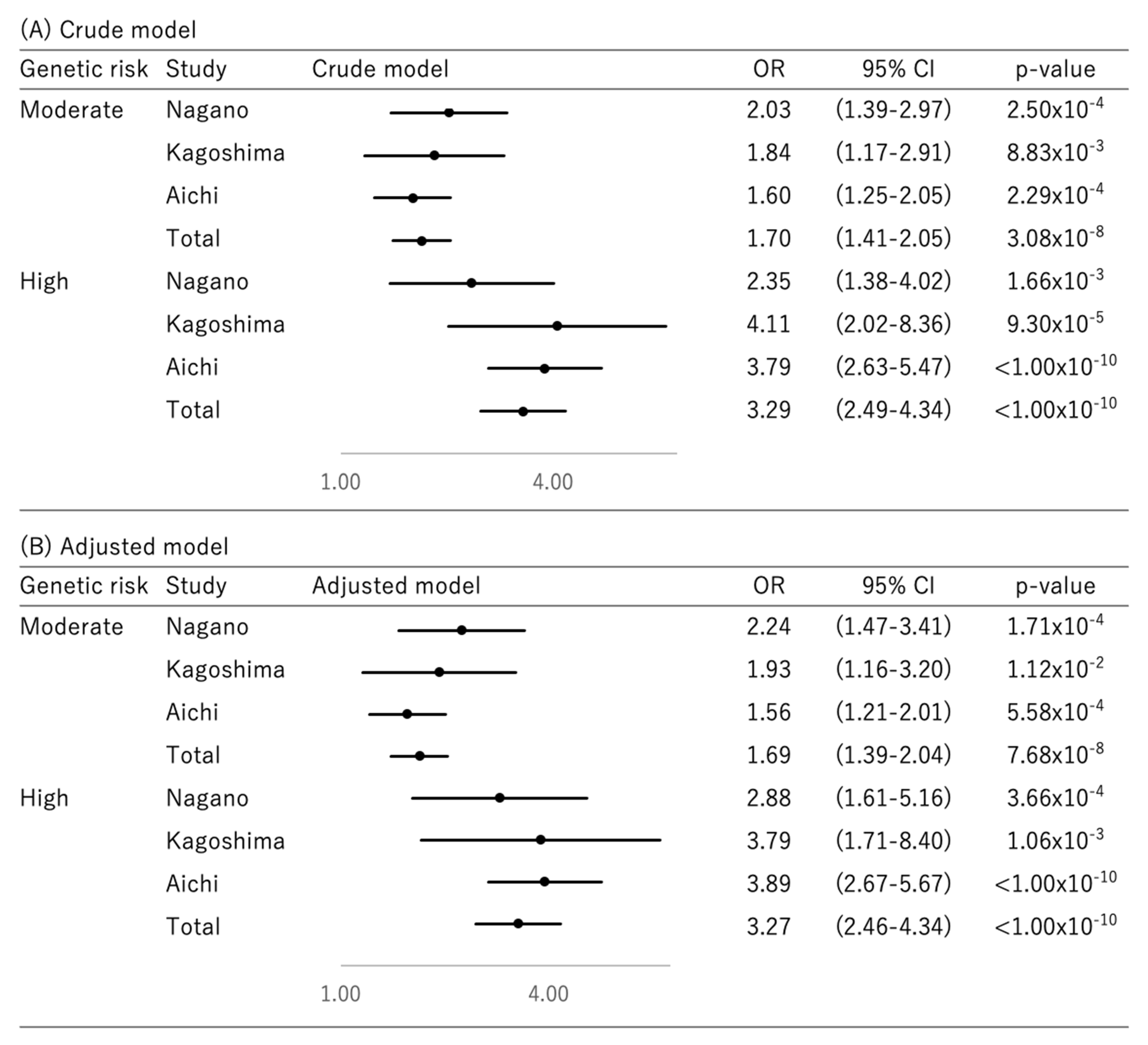
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Zanetti, R.; Ferlay, J. Cancer Incidence in Five Continents, Vol. XI (Electronic Version). Available online: https://ci5.iarc.fr (accessed on 10 May 2021).
- Global Burden of Disease Cancer Collaboration; Fitzmaurice, C.; Akinyemiju, T.F.; Al Lami, F.H.; Alam, T.; Alizadeh-Navaei, R.; Allen, C.; Alsharif, U.; Alvis-Guzman, N.; Amini, E.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2018, 4, 1553–1568. [Google Scholar] [CrossRef] [PubMed]
- Hori, M.; Matsuda, T.; Shibata, A.; Katanoda, K.; Sobue, T.; Nishimoto, H.; Japan Cancer Surveillance Research, G. Cancer incidence and incidence rates in Japan in 2009: A study of 32 population-based cancer registries for the Monitoring of Cancer Incidence in Japan (MCIJ) project. Jpn. J. Clin. Oncol. 2015, 45, 884–891. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.G.; Howell, S.J.; Howell, A. Personalized prevention in high risk individuals: Managing hormones and beyond. Breast 2018, 39, 139–147. [Google Scholar] [CrossRef]
- Chowkwanyun, M.; Bayer, R.; Galea, S. “Precision” Public Health—Between Novelty and Hype. N. Engl. J. Med. 2018, 379, 1398–1400. [Google Scholar] [CrossRef]
- Vineis, P.; Wild, C.P. The science of precision prevention of cancer. Lancet Oncol. 2017, 18, 997–998. [Google Scholar] [CrossRef]
- Gail, M.H. Twenty-five years of breast cancer risk models and their applications. J. Natl. Cancer Inst. 2015, 107. [Google Scholar] [CrossRef]
- Park, B.; Ma, S.H.; Shin, A.; Chang, M.C.; Choi, J.Y.; Kim, S.; Han, W.; Noh, D.Y.; Ahn, S.H.; Kang, D.; et al. Korean risk assessment model for breast cancer risk prediction. PLoS ONE 2013, 8, e76736. [Google Scholar] [CrossRef]
- Gail, M.H.; Costantino, J.P.; Pee, D.; Bondy, M.; Newman, L.; Selvan, M.; Anderson, G.L.; Malone, K.E.; Marchbanks, P.A.; McCaskill-Stevens, W.; et al. Projecting individualized absolute invasive breast cancer risk in African American women. J. Natl. Cancer Inst. 2007, 99, 1782–1792. [Google Scholar] [CrossRef] [PubMed]
- Costantino, J.P.; Gail, M.H.; Pee, D.; Anderson, S.; Redmond, C.K.; Benichou, J.; Wieand, H.S. Validation studies for models projecting the risk of invasive and total breast cancer incidence. J. Natl. Cancer Inst. 1999, 91, 1541–1548. [Google Scholar] [CrossRef] [PubMed]
- Gail, M.H.; Brinton, L.A.; Byar, D.P.; Corle, D.K.; Green, S.B.; Schairer, C.; Mulvihill, J.J. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J. Natl. Cancer Inst. 1989, 81, 1879–1886. [Google Scholar] [CrossRef] [PubMed]
- Saslow, D.; Boetes, C.; Burke, W.; Harms, S.; Leach, M.O.; Lehman, C.D.; Morris, E.; Pisano, E.; Schnall, M.; Sener, S.; et al. American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography. CA Cancer J. Clin. 2007, 57, 75–89. [Google Scholar] [CrossRef]
- Michailidou, K.; Beesley, J.; Lindstrom, S.; Canisius, S.; Dennis, J.; Lush, M.J.; Maranian, M.J.; Bolla, M.K.; Wang, Q.; Shah, M.; et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat. Genet. 2015, 47, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Zhang, B.; Sung, H.; Low, S.K.; Kweon, S.S.; Lu, W.; Shi, J.; Long, J.; Wen, W.; Choi, J.Y.; et al. Genome-wide association analysis in East Asians identifies breast cancer susceptibility loci at 1q32.1, 5q14.3 and 15q26.1. Nat. Genet. 2014, 46, 886–890. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.C.; Lee, J.Y.; Sung, H.; Choi, J.Y.; Park, S.K.; Lee, K.M.; Kim, Y.J.; Go, M.J.; Li, L.; Cho, Y.S.; et al. A genome-wide association study identifies a breast cancer risk variant in ERBB4 at 2q34: Results from the Seoul Breast Cancer Study. Breast Cancer Res. BCR 2012, 14, R56. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Cai, Q.; Sung, H.; Shi, J.; Zhang, B.; Choi, J.Y.; Wen, W.; Delahanty, R.J.; Lu, W.; Gao, Y.T.; et al. Genome-wide association study in east Asians identifies novel susceptibility loci for breast cancer. PLoS Genet. 2012, 8, e1002532. [Google Scholar] [CrossRef]
- Long, J.; Cai, Q.; Shu, X.O.; Qu, S.; Li, C.; Zheng, Y.; Gu, K.; Wang, W.; Xiang, Y.B.; Cheng, J.; et al. Identification of a functional genetic variant at 16q12.1 for breast cancer risk: Results from the Asia Breast Cancer Consortium. PLoS Genet. 2010, 6, e1001002. [Google Scholar] [CrossRef]
- Barnholtz-Sloan, J.S.; Shetty, P.B.; Guan, X.; Nyante, S.J.; Luo, J.; Brennan, D.J.; Millikan, R.C. FGFR2 and other loci identified in genome-wide association studies are associated with breast cancer in African-American and younger women. Carcinogenesis 2010, 31, 1417–1423. [Google Scholar] [CrossRef]
- Turnbull, C.; Ahmed, S.; Morrison, J.; Pernet, D.; Renwick, A.; Maranian, M.; Seal, S.; Ghoussaini, M.; Hines, S.; Healey, C.S.; et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 2010, 42, 504–507. [Google Scholar] [CrossRef]
- Thomas, G.; Jacobs, K.B.; Kraft, P.; Yeager, M.; Wacholder, S.; Cox, D.G.; Hankinson, S.E.; Hutchinson, A.; Wang, Z.; Yu, K.; et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat. Genet. 2009, 41, 579–584. [Google Scholar] [CrossRef]
- Ahmed, S.; Thomas, G.; Ghoussaini, M.; Healey, C.S.; Humphreys, M.K.; Platte, R.; Morrison, J.; Maranian, M.; Pooley, K.A.; Luben, R.; et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat. Genet. 2009, 41, 585–590. [Google Scholar] [CrossRef]
- Udler, M.S.; Meyer, K.B.; Pooley, K.A.; Karlins, E.; Struewing, J.P.; Zhang, J.; Doody, D.R.; MacArthur, S.; Tyrer, J.; Pharoah, P.D.; et al. FGFR2 variants and breast cancer risk: Fine-scale mapping using African American studies and analysis of chromatin conformation. Hum. Mol. Genet. 2009, 18, 1692–1703. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Long, J.; Gao, Y.T.; Li, C.; Zheng, Y.; Xiang, Y.B.; Wen, W.; Levy, S.; Deming, S.L.; Haines, J.L.; et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 2009, 41, 324–328. [Google Scholar] [CrossRef]
- Stacey, S.N.; Manolescu, A.; Sulem, P.; Thorlacius, S.; Gudjonsson, S.A.; Jonsson, G.F.; Jakobsdottir, M.; Bergthorsson, J.T.; Gudmundsson, J.; Aben, K.K.; et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2008, 40, 703–706. [Google Scholar] [CrossRef] [PubMed]
- Gold, B.; Kirchhoff, T.; Stefanov, S.; Lautenberger, J.; Viale, A.; Garber, J.; Friedman, E.; Narod, S.; Olshen, A.B.; Gregersen, P.; et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc. Natl. Acad. Sci. USA 2008, 105, 4340–4345. [Google Scholar] [CrossRef] [PubMed]
- Stacey, S.N.; Manolescu, A.; Sulem, P.; Rafnar, T.; Gudmundsson, J.; Gudjonsson, S.A.; Masson, G.; Jakobsdottir, M.; Thorlacius, S.; Helgason, A.; et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2007, 39, 865–869. [Google Scholar] [CrossRef]
- Hunter, D.J.; Kraft, P.; Jacobs, K.B.; Cox, D.G.; Yeager, M.; Hankinson, S.E.; Wacholder, S.; Wang, Z.; Welch, R.; Hutchinson, A.; et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007, 39, 870–874. [Google Scholar] [CrossRef]
- Easton, D.F.; Pooley, K.A.; Dunning, A.M.; Pharoah, P.D.; Thompson, D.; Ballinger, D.G.; Struewing, J.P.; Morrison, J.; Field, H.; Luben, R.; et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 2007, 447, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.; Dunning, A.M.; Garcia-Closas, M.; Balasubramanian, S.; Reed, M.W.; Pooley, K.A.; Scollen, S.; Baynes, C.; Ponder, B.A.; Chanock, S.; et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat. Genet. 2007, 39, 352–358. [Google Scholar] [CrossRef]
- Yanes, T.; Young, M.A.; Meiser, B.; James, P.A. Clinical applications of polygenic breast cancer risk: A critical review and perspectives of an emerging field. Breast Cancer Res. BCR 2020, 22, 21. [Google Scholar] [CrossRef]
- Struck, T.J.; Mannakee, B.K.; Gutenkunst, R.N. The impact of genome-wide association studies on biomedical research publications. Hum. Genom. 2018, 12, 38. [Google Scholar] [CrossRef]
- Britt, K.L.; Cuzick, J.; Phillips, K.A. Key steps for effective breast cancer prevention. Nat. Rev. Cancer 2020, 20, 417–436. [Google Scholar] [CrossRef]
- Sugrue, L.P.; Desikan, R.S. What Are Polygenic Scores and Why Are They Important? JAMA 2019, 321, 1820–1821. [Google Scholar] [CrossRef]
- Lee, A.; Mavaddat, N.; Wilcox, A.N.; Cunningham, A.P.; Carver, T.; Hartley, S.; Babb de Villiers, C.; Izquierdo, A.; Simard, J.; Schmidt, M.K.; et al. BOADICEA: A comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet. Med. 2019, 21, 1708–1718. [Google Scholar] [CrossRef]
- Shieh, Y.; Hu, D.; Ma, L.; Huntsman, S.; Gard, C.C.; Leung, J.W.T.; Tice, J.A.; Vachon, C.M.; Cummings, S.R.; Kerlikowske, K.; et al. Breast cancer risk prediction using a clinical risk model and polygenic risk score. Breast Cancer Res. Treat. 2016, 159, 513–525. [Google Scholar] [CrossRef]
- Maas, P.; Barrdahl, M.; Joshi, A.D.; Auer, P.L.; Gaudet, M.M.; Milne, R.L.; Schumacher, F.R.; Anderson, W.F.; Check, D.; Chattopadhyay, S.; et al. Breast Cancer Risk From Modifiable and Nonmodifiable Risk Factors Among White Women in the United States. JAMA Oncol. 2016, 2, 1295–1302. [Google Scholar] [CrossRef]
- Kuchiba, A.; Iwasaki, M.; Ono, H.; Kasuga, Y.; Yokoyama, S.; Onuma, H.; Nishimura, H.; Kusama, R.; Tsugane, S.; Yoshida, T. Global methylation levels in peripheral blood leukocyte DNA by LUMA and breast cancer: A case-control study in Japanese women. Br. J. Cancer 2014, 110, 2765–2771. [Google Scholar] [CrossRef] [PubMed]
- Hamajima, N.; Matsuo, K.; Saito, T.; Hirose, K.; Inoue, M.; Takezaki, T.; Kuroishi, T.; Tajima, K. Gene-environment Interactions and Polymorphism Studies of Cancer Risk in the Hospital-based Epidemiologic Research Program at Aichi Cancer Center II (HERPACC-II). Asian Pac. J. Cancer Prev. 2001, 2, 99–107. [Google Scholar] [PubMed]
- Tajima, K.; Hirose, K.; Inoue, M.; Takezaki, T.; Hamajima, N.; Kuroishi, T. A Model of Practical Cancer Prevention for Out-patients Visiting a Hospital: The Hospital-based Epidemiologic Research Program at Aichi Cancer Center (HERPACC). Asian Pac. J. Cancer Prev. 2000, 1, 35–47. [Google Scholar] [PubMed]
- Sueta, A.; Ito, H.; Kawase, T.; Hirose, K.; Hosono, S.; Yatabe, Y.; Tajima, K.; Tanaka, H.; Iwata, H.; Iwase, H.; et al. A genetic risk predictor for breast cancer using a combination of low-penetrance polymorphisms in a Japanese population. Breast Cancer Res. Treat. 2012, 132, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Mavaddat, N.; Pharoah, P.D.P.; Michailidou, K.; Tyrer, J.; Brook, M.N.; Bolla, M.K.; Wang, Q.; Dennis, J.; Dunning, A.M.; Shah, M.; et al. Prediction of breast cancer risk based on profiling with common genetic variants. J. Natl. Cancer Inst. 2015, 107, djv036. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Hosmer, D.W.; Lemeshow, S. Applied Logistic Regression, 2nd ed.; Wiley: New York, NY, USA, 2000. [Google Scholar]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Lemeshow, S.; Hosmer, D.W., Jr. A review of goodness of fit statistics for use in the development of logistic regression models. Am. J. Epidemiol. 1982, 115, 92–106. [Google Scholar] [CrossRef]
- Manolio, T.A. Bringing genome-wide association findings into clinical use. Nat. Rev. Genet. 2013, 14, 549–558. [Google Scholar] [CrossRef]
- Nagata, C.; Mizoue, T.; Tanaka, K.; Tsuji, I.; Wakai, K.; Inoue, M.; Tsugane, S.; Research Group for the Development; Evaluation of Cancer Prevention Strategies in Japan. Tobacco smoking and breast cancer risk: An evaluation based on a systematic review of epidemiological evidence among the Japanese population. Jpn J. Clin. Oncol. 2006, 36, 387–394. [Google Scholar] [CrossRef]
- Nagata, C.; Mizoue, T.; Tanaka, K.; Tsuji, I.; Wakai, K.; Inoue, M.; Tsugane, S.; Research Group for the Development; Evaluation of Cancer Prevention Strategies in Japan. Alcohol drinking and breast cancer risk: An evaluation based on a systematic review of epidemiologic evidence among the Japanese population. Jpn J. Clin. Oncol. 2007, 37, 568–574. [Google Scholar] [CrossRef][Green Version]
- Wada, K.; Nagata, C.; Tamakoshi, A.; Matsuo, K.; Oze, I.; Wakai, K.; Tsuji, I.; Sugawara, Y.; Mizoue, T.; Tanaka, K.; et al. Body mass index and breast cancer risk in Japan: A pooled analysis of eight population-based cohort studies. Ann. Oncol. 2014, 25, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Iwase, M.; Matsuo, K.; Koyanagi, Y.N.Y.; Ito, H.; Tamakoshi, A.; Wang, C.; Utada, M.; Ozasa, K.; Sugawara, Y.; Tsuji, I.; et al. Alcohol consumption and breast cancer risk in Japan: A pooled analysis of eight population-based cohort studies. Int. J. Cancer 2021. [Google Scholar] [CrossRef]
- Shieh, Y.; Ziv, E.; Eklund, M.; Sabacan, L.; Firouzian, R.; Madlensky, L.; Anton-Culver, H.; Borowsky, A.; LaCroix, A.; Naeim, A.; et al. Abstract P3-09-02: Risk stratification using clinical risk factors and genetic variants in a personalized screening trial. Cancer Res. 2018, 78. [Google Scholar] [CrossRef]
- Kim, J.O.; Schaid, D.J.; Cooke, A.; Kim, C.; Goldenberg, B.A.; Highsmith, W.E.; Grenier, D.; Sinnwell, J.P.; Degnim, A.C.; Couch, F.; et al. Impact of a breast cancer (BC) polygenic risk score (PRS) on the decision to take preventive endocrine therapy (ET): The Genetic Risk Estimate (GENRE) trial. J. Clin. Oncol. 2019, 37, 1501. [Google Scholar] [CrossRef]
- Hamashima, C.; Japanese Research Group for the Development of Breast Cancer Screening Guidelines; Hamashima, C.C.; Hattori, M.; Honjo, S.; Kasahara, Y.; Katayama, T.; Nakai, M.; Nakayama, T.; Morita, T.; et al. The Japanese Guidelines for Breast Cancer Screening. Jpn. J. Clin. Oncol. 2016, 46, 482–492. [Google Scholar] [CrossRef]
- Harvie, M.; Howell, A.; Evans, D.G. Can diet and lifestyle prevent breast cancer: What is the evidence? Am. Soc. Clin. Oncol. Educ. Book 2015, e66–e73. [Google Scholar] [CrossRef]
- Arthur, R.S.; Wang, T.; Xue, X.; Kamensky, V.; Rohan, T.E. Genetic factors, adherence to healthy lifestyle behavior, and risk of invasive breast cancer among women in the UK Biobank. J. Natl. Cancer Inst. 2020, djz241. [Google Scholar] [CrossRef] [PubMed]
- Mavaddat, N.; Michailidou, K.; Dennis, J.; Lush, M.; Fachal, L.; Lee, A.; Tyrer, J.P.; Chen, T.H.; Wang, Q.; Bolla, M.K.; et al. Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. Am. J. Hum. Genet. 2019, 104, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Low, S.K.; Chin, Y.M.; Ito, H.; Matsuo, K.; Tanikawa, C.; Matsuda, K.; Saito, H.; Sakurai-Yageta, M.; Nakaya, N.; Shimizu, A.; et al. Identification of two novel breast cancer loci through large-scale genome-wide association study in the Japanese population. Sci. Rep. 2019, 9, 17332. [Google Scholar] [CrossRef]
- Low, S.K.; Takahashi, A.; Ashikawa, K.; Inazawa, J.; Miki, Y.; Kubo, M.; Nakamura, Y.; Katagiri, T. Genome-wide association study of breast cancer in the Japanese population. PLoS ONE 2013, 8, e76463. [Google Scholar] [CrossRef]
- Elgazzar, S.; Zembutsu, H.; Takahashi, A.; Kubo, M.; Aki, F.; Hirata, K.; Takatsuka, Y.; Okazaki, M.; Ohsumi, S.; Yamakawa, T.; et al. A genome-wide association study identifies a genetic variant in the SIAH2 locus associated with hormonal receptor-positive breast cancer in Japanese. J. Hum. Genet. 2012, 57, 766–771. [Google Scholar] [CrossRef]
- Ministry of Health, Labour and Welfare, Japan. The National Health and Nutrition Survey. Available online: https://www.mhlw.go.jp/bunya/kenkou/kenkou_eiyou_chousa.html (accessed on 26 April 2021).

| Cases | Controls | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nagano (%) | Kaghoshima (%) | Aichi (%) | Total (%) | Nagano (%) | Kaghoshima (%) | Aichi (%) | Total (%) | ||||||||||
| Age | |||||||||||||||||
| −39 | 33 | (8.48) | 13 | (5.58) | 105 | (15.06) | 151 | (11.45) | 26 | (6.68) | 79 | (23.87) | 204 | (14.63) | 309 | (14.62) | |
| 40–49 | 112 | (28.79) | 43 | (18.45) | 197 | (28.26) | 352 | (26.69) | 107 | (27.51) | 96 | (29.00) | 416 | (29.84) | 619 | (29.28) | |
| 50–59 | 122 | (31.36) | 56 | (24.03) | 222 | (31.85) | 400 | (30.33) | 135 | (34.70) | 86 | (25.98) | 429 | (30.77) | 650 | (30.75) | |
| 60–69 | 88 | (22.62) | 55 | (23.61) | 132 | (18.94) | 275 | (20.85) | 96 | (24.68) | 41 | (12.39) | 271 | (19.44) | 408 | (19.30) | |
| 70+ | 34 | (8.74) | 62 | (26.61) | 41 | (5.88) | 137 | (10.39) | 25 | (6.43) | 29 | (8.76) | 74 | (5.31) | 128 | (6.05) | |
| UK | 0 | (0.00) | 4 | (1.72) | 0 | (0.00) | 4 | (0.30) | 0 | (0.00) | 0 | (0.00) | 0 | (0.00) | 0 | (0.00) | |
| BMI | |||||||||||||||||
| <18.5 | 30 | (7.71) | 11 | (4.72) | 56 | (8.03) | 97 | (7.35) | 13 | (3.34) | 32 | (9.67) | 116 | (8.32) | 161 | (7.62) | |
| 18.5–24.9 | 275 | (70.69) | 145 | (62.23) | 498 | (71.45) | 918 | (69.60) | 284 | (73.01) | 249 | (75.23) | 1021 | (73.24) | 1554 | (73.51) | |
| ≥25 | 84 | (21.59) | 66 | (28.33) | 143 | (20.52) | 293 | (22.21) | 92 | (23.65) | 50 | (15.11) | 245 | (17.58) | 387 | (18.31) | |
| UK | 0 | (0.00) | 11 | (4.72) | 0 | (0.00) | 11 | (0.83) | 0 | (0.00) | 0 | (0.00) | 12 | (0.86) | 12 | (0.57) | |
| Ethanol intake | |||||||||||||||||
| Never | 288 | (74.04) | 182 | (78.11) | 509 | (73.03) | 979 | (74.22) | 272 | (69.92) | 248 | (74.92) | 1027 | (73.67) | 1547 | (73.18) | |
| <23 g/day | 72 | (18.51) | 37 | (15.88) | 148 | (21.23) | 257 | (19.48) | 88 | (22.62) | 68 | (20.54) | 299 | (21.45) | 455 | (21.52) | |
| ≥23 g/day | 27 | (6.94) | 9 | (3.86) | 33 | (4.73) | 69 | (5.23) | 29 | (7.46) | 14 | (4.23) | 51 | (3.66) | 94 | (4.45) | |
| UK | 2 | (0.51) | 5 | (2.15) | 7 | (1.00) | 14 | (1.06) | 0 | (0.00) | 1 | (0.30) | 17 | (1.22) | 18 | (0.85) | |
| Smoking | |||||||||||||||||
| Never | 307 | (78.92) | 183 | (78.54) | 585 | (83.93) | 1075 | (81.50) | 358 | (92.03) | 283 | (85.50) | 1113 | (79.84) | 1754 | (82.97) | |
| Ever | 78 | (20.05) | 46 | (19.74) | 110 | (15.78) | 234 | (17.74) | 30 | (7.71) | 48 | (14.50) | 278 | (19.94) | 356 | (16.84) | |
| UK | 4 | (1.03) | 4 | (1.72) | 2 | (0.29) | 10 | (0.76) | 1 | (0.26) | 0 | (0.00) | 3 | (0.22) | 4 | (0.19) | |
| Physical activity | |||||||||||||||||
| No | 337 | (86.63) | 128 | (54.94) | 416 | (59.68) | 881 | (66.79) | 324 | (83.29) | 176 | (53.17) | 862 | (61.84) | 1362 | (64.43) | |
| Yes | 47 | (12.08) | 95 | (40.77) | 281 | (40.32) | 423 | (32.07) | 64 | (16.45) | 153 | (46.22) | 532 | (38.16) | 749 | (35.43) | |
| UK | 5 | (1.29) | 10 | (4.29) | 0 | (0.00) | 5 | (0.38) | 1 | (0.26) | 2 | (0.60) | 0 | (0.00) | 3 | (0.14) | |
| Family history of breast cancer | |||||||||||||||||
| No | 323 | (83.03) | 195 | (83.69) | 632 | (90.67) | 1150 | (87.19) | 350 | (89.97) | 296 | (89.43) | 1305 | (93.62) | 1951 | (92.29) | |
| Yes | 40 | (10.28) | 30 | (12.88) | 65 | (9.33) | 135 | (10.24) | 25 | (6.43) | 30 | (9.06) | 89 | (6.38) | 144 | (6.81) | |
| UK | 26 | (6.68) | 8 | (3.43) | 0 | (0.00) | 34 | (2.58) | 14 | (3.60) | 5 | (1.51) | 0 | (0.00) | 19 | (0.90) | |
| Age at menarche | |||||||||||||||||
| ≤12 y.o. | 139 | (35.73) | 63 | (27.04) | 216 | (30.99) | 418 | (31.69) | 145 | (37.28) | 115 | (34.74) | 439 | (31.49) | 699 | (33.07) | |
| 13–14 y.o. | 164 | (42.16) | 106 | (45.49) | 340 | (48.78) | 610 | (46.25) | 175 | (44.99) | 155 | (46.83) | 648 | (46.48) | 978 | (46.26) | |
| ≥15 y.o. | 85 | (21.85) | 57 | (24.46) | 133 | (19.08) | 275 | (20.85) | 69 | (17.74) | 59 | (17.82) | 277 | (19.87) | 405 | (19.16) | |
| UK | 1 | (0.26) | 7 | (3.00) | 8 | (1.15) | 16 | (1.21) | 0 | (0.00) | 2 | (0.60) | 30 | (2.15) | 32 | (1.51) | |
| Parity | |||||||||||||||||
| No | 15 | (3.86) | 32 | (13.73) | 107 | (15.35) | 154 | (11.68) | 4 | (1.03) | 69 | (20.85) | 203 | (14.56) | 276 | (13.06) | |
| Yes | 334 | (85.86) | 175 | (75.11) | 589 | (84.51) | 1098 | (83.24) | 336 | (86.38) | 249 | (75.23) | 1188 | (85.22) | 1773 | (83.87) | |
| UK | 40 | (10.28) | 26 | (11.16) | 1 | (0.14) | 67 | (5.08) | 49 | (12.60) | 13 | (3.93) | 3 | (0.22) | 65 | (3.07) | |
| Number of births | |||||||||||||||||
| nonparous | 15 | (3.86) | 32 | (13.73) | 107 | (15.35) | 154 | (11.68) | 4 | (1.03) | 69 | (20.85) | 203 | (14.56) | 276 | (13.06) | |
| 1 or 2 | 237 | (60.93) | 115 | (49.36) | 452 | (64.85) | 804 | (60.96) | 213 | (54.76) | 155 | (46.83) | 867 | (62.20) | 1235 | (58.42) | |
| ≥3 | 97 | (24.94) | 60 | (25.75) | 137 | (19.66) | 294 | (22.29) | 123 | (31.62) | 94 | (28.40) | 315 | (22.60) | 532 | (25.17) | |
| UK | 40 | (10.28) | 26 | (11.16) | 1 | (0.14) | 67 | (5.08) | 49 | (12.60) | 13 | (3.93) | 9 | (0.65) | 71 | (3.36) | |
| Age at first birth | |||||||||||||||||
| <30 y.o. | 137 | (35.22) | 78 | (33.48) | 270 | (38.74) | 485 | (36.77) | 148 | (38.05) | 89 | (26.89) | 653 | (46.84) | 890 | (42.10) | |
| ≥30 y.o. | 197 | (50.64) | 95 | (40.77) | 316 | (45.34) | 608 | (46.10) | 188 | (48.33) | 160 | (48.34) | 522 | (37.45) | 870 | (41.15) | |
| nonparous | 15 | (3.86) | 32 | (13.73) | 108 | (15.49) | 155 | (11.75) | 4 | (1.03) | 69 | (20.85) | 207 | (14.85) | 280 | (13.25) | |
| UK | 40 | (10.28) | 28 | (12.02) | 3 | (0.43) | 71 | (5.38) | 49 | (12.60) | 13 | (3.93) | 12 | (0.86) | 74 | (3.50) | |
| Breastfeeding | |||||||||||||||||
| No | 72 | (18.51) | 17 | (7.30) | 131 | (18.79) | 220 | (16.68) | 67 | (17.22) | 21 | (6.34) | 260 | (18.65) | 348 | (16.46) | |
| Yes | 306 | (78.66) | 158 | (67.81) | 558 | (80.06) | 1022 | (77.48) | 322 | (82.78) | 229 | (69.18) | 1117 | (80.13) | 1668 | (78.90) | |
| UK | 11 | (2.83) | 58 | (24.89) | 8 | (1.15) | 77 | (5.84) | 0 | (0.00) | 81 | (24.47) | 17 | (1.22) | 98 | (4.64) | |
| Hormone therapy | |||||||||||||||||
| No | 327 | (84.06) | 203 | (87.12) | 603 | (86.51) | 1133 | (85.90) | 331 | (85.09) | 265 | (80.06) | 1141 | (81.85) | 1737 | (82.17) | |
| Yes | 55 | (14.14) | 22 | (9.44) | 88 | (12.63) | 165 | (12.51) | 55 | (14.14) | 63 | (19.03) | 229 | (16.43) | 347 | (16.41) | |
| UK | 7 | (1.80) | 8 | (3.43) | 6 | (0.86) | 21 | (1.59) | 3 | (0.77) | 3 | (0.91) | 24 | (1.72) | 30 | (1.42) | |
| Total | Nagano Study | Kagoshima Study | Aichi Study | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chromosome | Risk/Reference Allele | OR | 95% CI | p | OR | 95% CI | p | OR | 95% CI | p | OR | 95% CI | p |
| rs4849887 | 2q14.2 | C/T | 1.16 | (1.00–1.34) | 0.046 | 0.97 | (0.73–1.28) | 0.820 | 1.38 | (0.93–2.06) | 0.111 | 1.21 | (1.00–1.46) | 0.047 |
| rs10931936 | 2q33.1 | T/C | 1.11 | (1.00–1.23) | 0.047 | 1.15 | (0.93–1.40) | 0.192 | 1.20 | (0.91–1.58) | 0.200 | 1.08 | (0.94–1.23) | 0.278 |
| rs16857609 | 2q35 | T/C | 1.14 | (1.03–1.26) | 0.012 | 0.97 | (0.80–1.19) | 0.803 | 1.29 | (1.00–1.67) | 0.052 | 1.18 | (1.03–1.35) | 0.015 |
| rs4973768 | 3p24.1 | T/C | 1.15 | (1.02–1.30) | 0.028 | 0.93 | (0.73–1.18) | 0.552 | 1.40 | (1.03–1.90) | 0.034 | 1.20 | (1.02–1.42) | 0.028 |
| rs7697216 | 4q34.1 | C/T | 1.26 | (1.10–1.44) | 0.001 | 1.30 | (0.99–1.71) | 0.056 | 1.26 | (0.91–1.74) | 0.171 | 1.24 | (1.04–1.48) | 0.015 |
| rs1432679 | 5q33.3 | C/T | 1.13 | (1.01–1.25) | 0.028 | 1.15 | (0.93–1.43) | 0.197 | 1.14 | (0.87–1.50) | 0.338 | 1.11 | (0.97–1.28) | 0.127 |
| rs2046210 | 6q25.1 | T/C | 1.23 | (1.11–1.37) | 1.53 × 10−4 | 1.09 | (0.88–1.35) | 0.441 | 1.20 | (0.90–1.60) | 0.203 | 1.30 | (1.13–1.49) | 1.84 × 10−4 |
| rs13365225 | 8p11.23 | A/G | 1.11 | (1.00–1.23) | 0.043 | 1.15 | (0.93–1.41) | 0.188 | 1.23 | (0.95–1.59) | 0.124 | 1.07 | (0.94–1.22) | 0.324 |
| rs13281615 | 8q24.21 | G/A | 1.13 | (1.02–1.25) | 0.022 | 1.00 | (0.81–1.22) | 0.984 | 1.15 | (0.88–1.49) | 0.307 | 1.18 | (1.04–1.35) | 0.014 |
| rs2981579 | 10q26.13 | G/A | 1.22 | (1.10–1.34) | 1.42 × 10−4 | 1.37 | (1.13–1.67) | 0.002 | 1.21 | (0.93–1.56) | 0.152 | 1.15 | (1.01–1.32) | 0.031 |
| rs17271951 | 16q12.1 | C/T | 1.41 | (1.24–1.59) | 4.89 × 10−8 | 1.34 | (1.06–1.70) | 0.016 | 1.58 | (1.14–2.19) | 0.006 | 1.40 | (1.19–1.64) | 3.40 × 10−5 |
| rs4784227 | 16q12.1 | T/C | 1.46 | (1.30–1.64) | 2.47 × 10−10 | 1.52 | (1.20–1.92) | 4.48 × 10−4 | 1.63 | (1.19–2.23) | 0.002 | 1.40 | (1.20–1.62) | 1.07 × 10−5 |
| rs8051542 | 16q12.1 | T/C | 1.21 | (1.07–1.36) | 0.002 | 1.08 | (0.85–1.37) | 0.533 | 1.39 | (1.01–1.92) | 0.043 | 1.23 | (1.05–1.43) | 0.010 |
| rs11075995 | 16q12.2 | A/T | 1.18 | (1.06–1.31) | 0.003 | 1.25 | (1.01–1.56) | 0.041 | 1.11 | (0.85–1.44) | 0.461 | 1.16 | (1.02–1.33) | 0.028 |
| Genetic | Environment | Inclusive | |||||
|---|---|---|---|---|---|---|---|
| C-Statistics | 95% CI | C-Statistics | 95% CI | C-Statistics | 95% CI | p-Value | |
| Nagano | 0.605 | (0.566–0.645) | 0.691 | (0.654–0.728) | 0.721 | (0.685–0.757) | 0.005 |
| Kagoshima | 0.609 | (0.560–0.657) | 0.767 | (0.726–0.808) | 0.789 | (0.750–0.828) | 0.018 |
| Aichi | 0.604 | (0.579–0.630) | 0.581 | (0.555–0.607) | 0.635 | (0.610–0.660) | 7.05 × 10−6 |
| Total | 0.633 | (0.614–0.652) | 0.616 | (0.596–0.636) | 0.659 | (0.640–0.678) | 1.67 × 10−9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oze, I.; Ito, H.; Kasugai, Y.; Yamaji, T.; Kijima, Y.; Ugai, T.; Kasuga, Y.; Ouellette, T.K.; Taniyama, Y.; Koyanagi, Y.N.; et al. A Personal Breast Cancer Risk Stratification Model Using Common Variants and Environmental Risk Factors in Japanese Females. Cancers 2021, 13, 3796. https://doi.org/10.3390/cancers13153796
Oze I, Ito H, Kasugai Y, Yamaji T, Kijima Y, Ugai T, Kasuga Y, Ouellette TK, Taniyama Y, Koyanagi YN, et al. A Personal Breast Cancer Risk Stratification Model Using Common Variants and Environmental Risk Factors in Japanese Females. Cancers. 2021; 13(15):3796. https://doi.org/10.3390/cancers13153796
Chicago/Turabian StyleOze, Isao, Hidemi Ito, Yumiko Kasugai, Taiki Yamaji, Yuko Kijima, Tomotaka Ugai, Yoshio Kasuga, Tomoyo K. Ouellette, Yukari Taniyama, Yuriko N. Koyanagi, and et al. 2021. "A Personal Breast Cancer Risk Stratification Model Using Common Variants and Environmental Risk Factors in Japanese Females" Cancers 13, no. 15: 3796. https://doi.org/10.3390/cancers13153796
APA StyleOze, I., Ito, H., Kasugai, Y., Yamaji, T., Kijima, Y., Ugai, T., Kasuga, Y., Ouellette, T. K., Taniyama, Y., Koyanagi, Y. N., Imoto, I., Tsugane, S., Koriyama, C., Iwasaki, M., & Matsuo, K. (2021). A Personal Breast Cancer Risk Stratification Model Using Common Variants and Environmental Risk Factors in Japanese Females. Cancers, 13(15), 3796. https://doi.org/10.3390/cancers13153796







