TRAF6 Phosphorylation Prevents Its Autophagic Degradation and Re-Shapes LPS-Triggered Signaling Networks
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and CRISPR-Cas9-Mediated Generation of TRAF6 Knockout Cells
2.2. Characterization of CRISPR-Cas9-Mediated Genomic Indel Mutations
2.3. Induction of Autophagy
2.4. Cell Transfection and Generation of Stable Cell Lines
2.5. Lambda Protein Phosphatase Assays
2.6. Denaturing Lysis and Enrichment of His-Tagged Proteins on Ni-NTA Columns
2.7. Native Cell Lysis and Co-Immunoprecipitation
2.8. Mass Spectrometry for Detection of TRAF6 Phosphorylation Sites
2.9. Phospho-Proteomic Analysis of TRAF6-Mediated Signaling Pathways
2.10. LC-MS/MS Analysis
2.11. Statistical Rationale and Bioinformatic Analysis
2.12. Antibodies, Plasmids and Reagents
3. Results
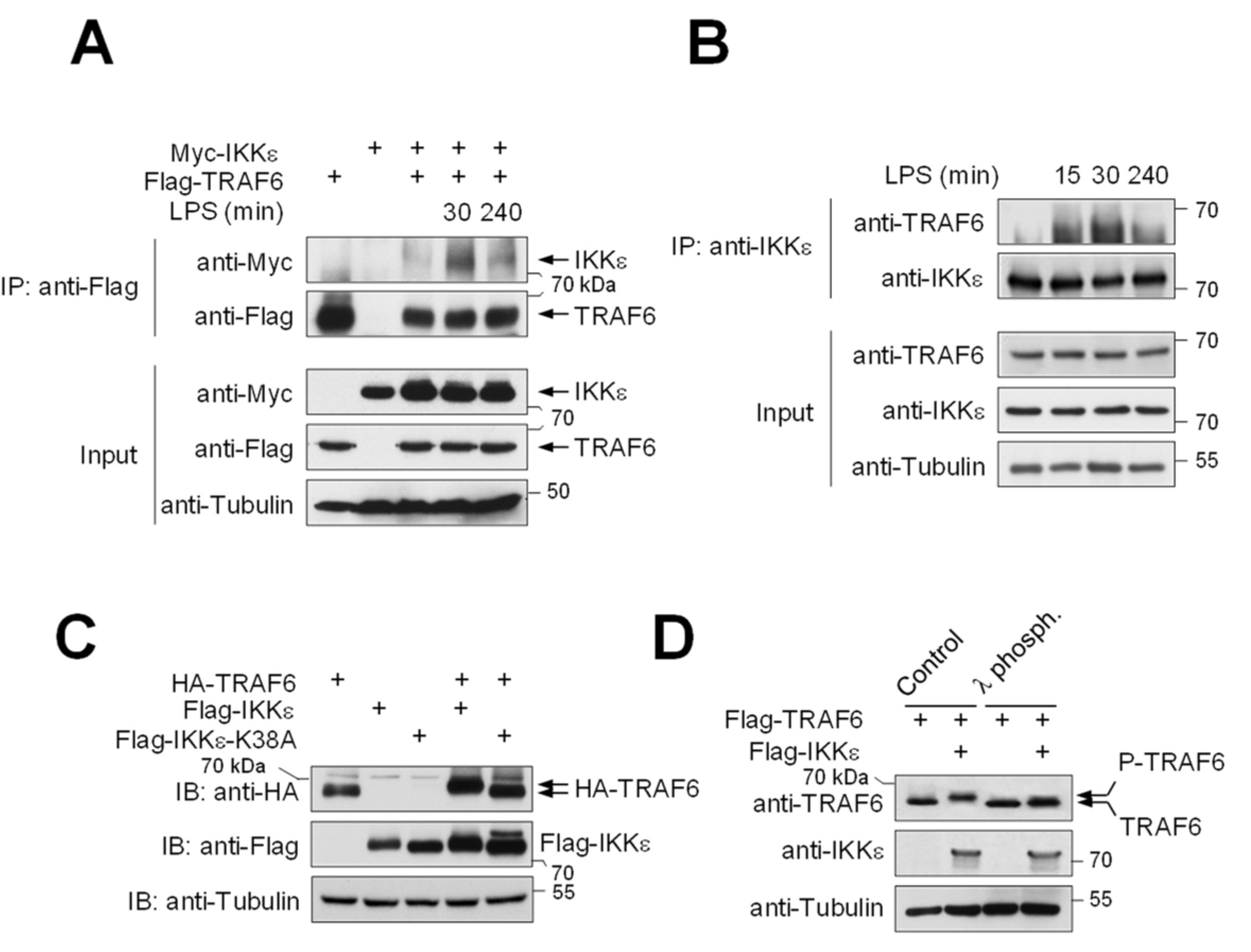
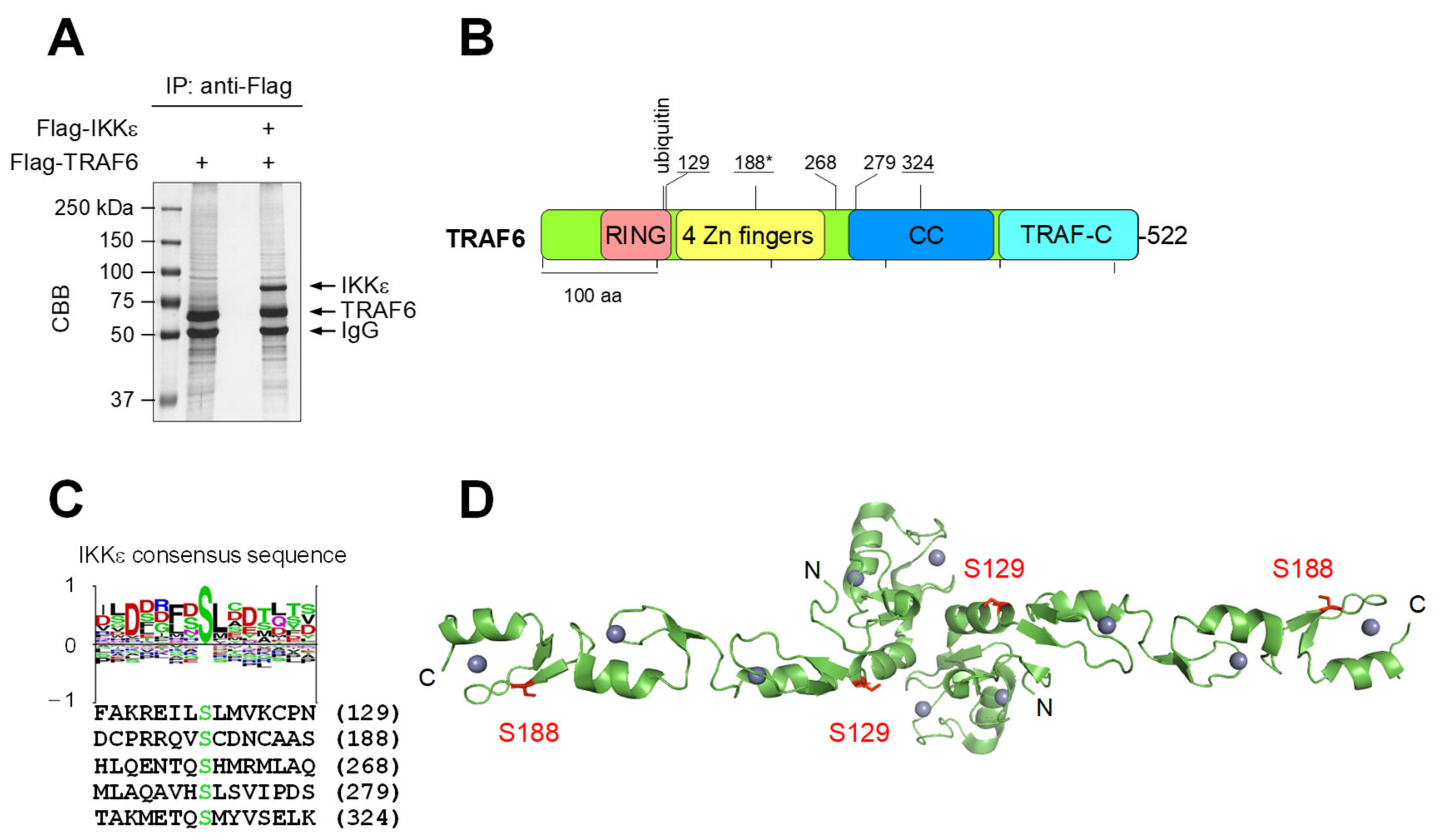
3.1. IKKε Shows LPS-Triggered Interaction with TRAF6 and Phosphorylates TRAF6 at Five Different Serines
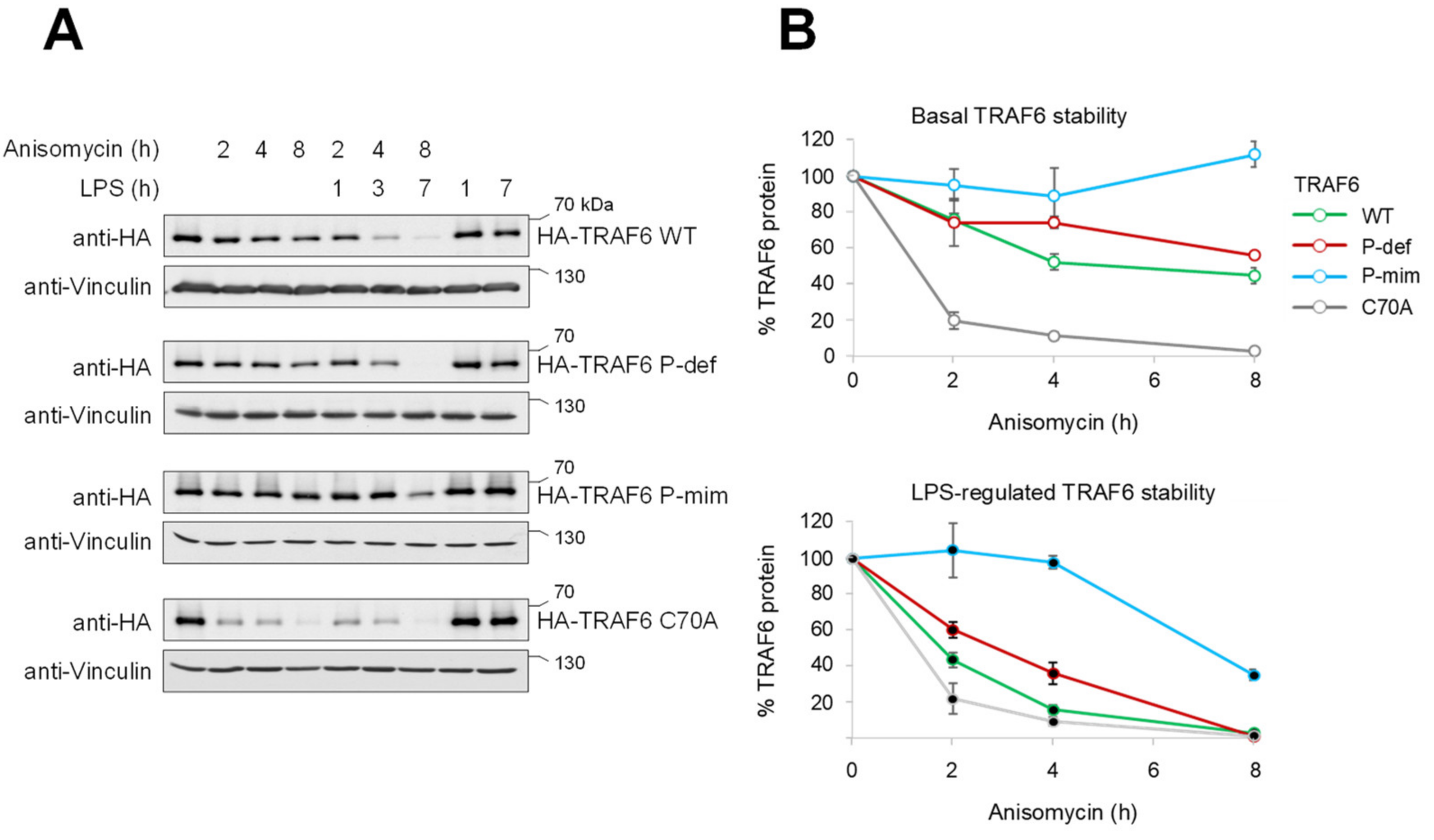
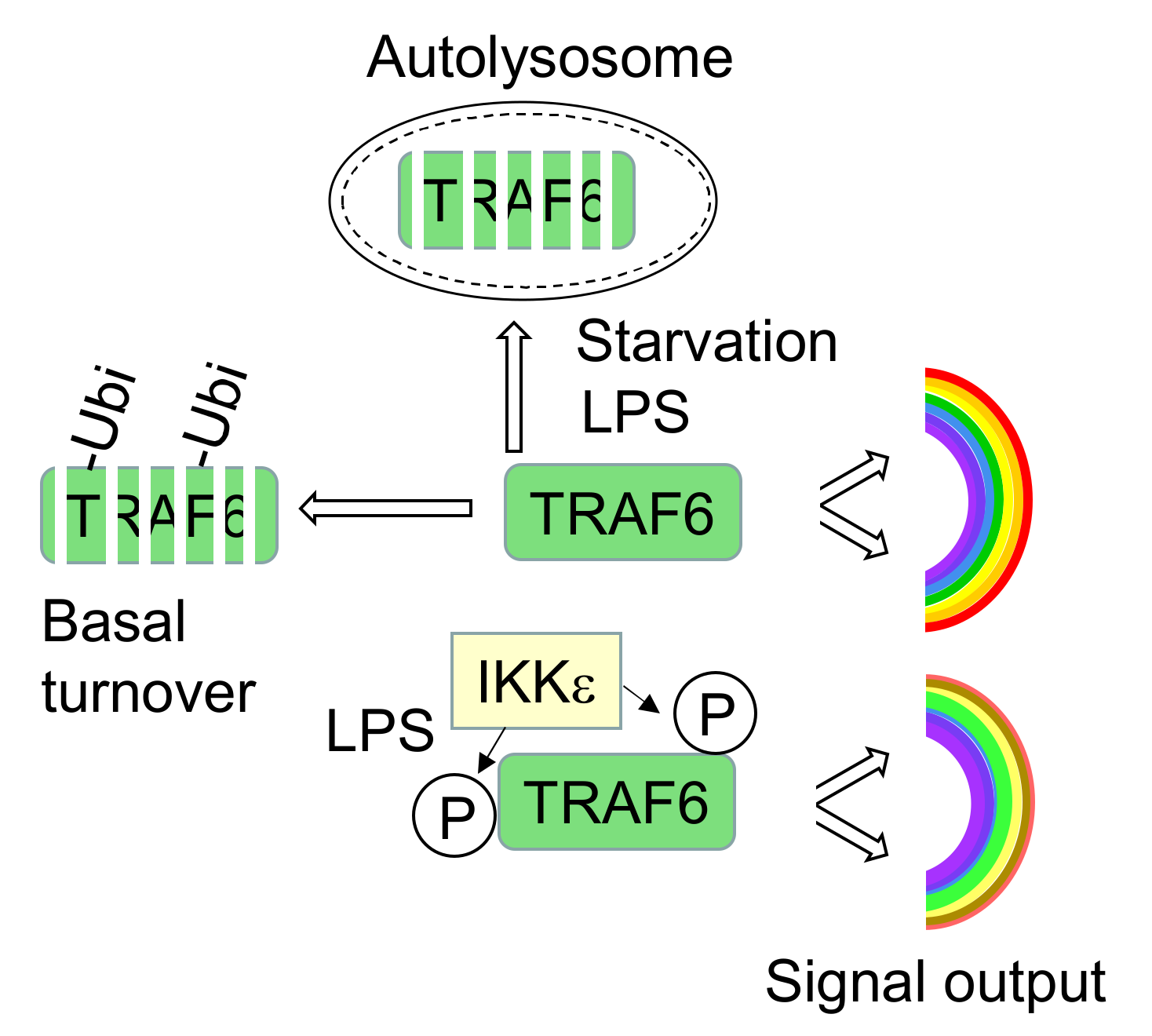
3.2. TRAF6 Phosphorylation Counteracts Constitutive and LPS-Inducible Decay
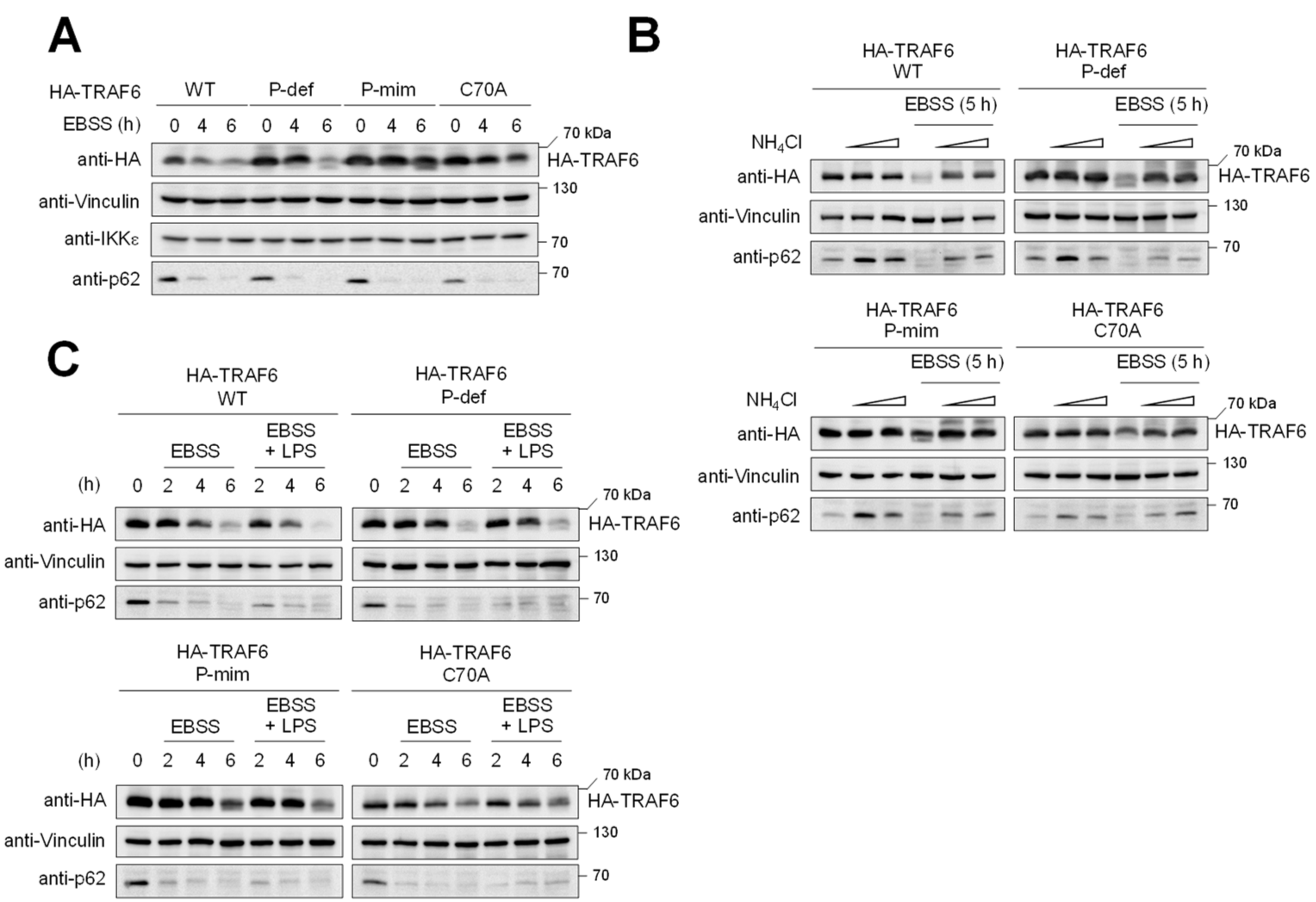
3.3. TRAF6 Phosphorylation Counteracts TRAF6 Elimination by Autophagy
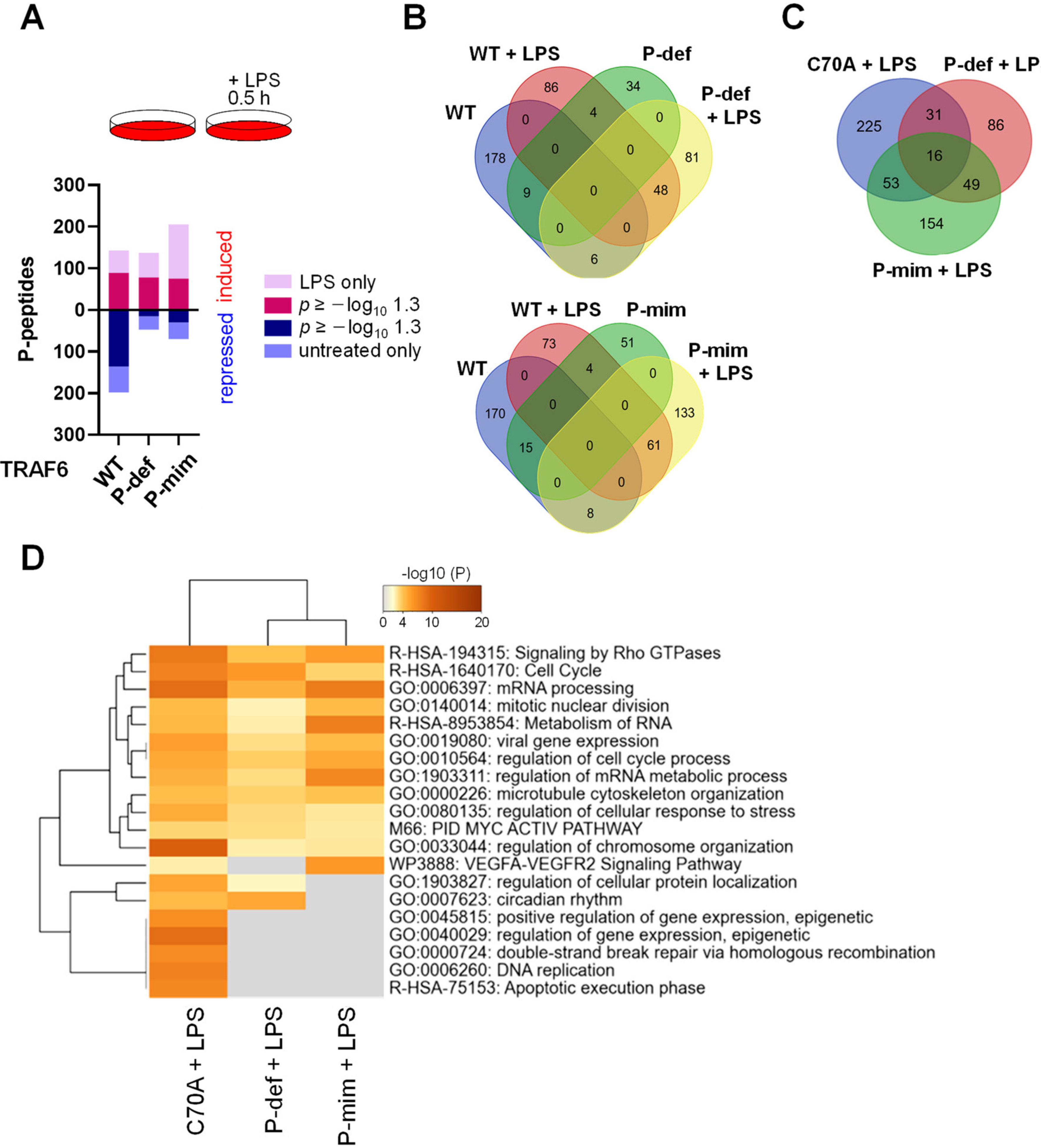
3.4. The E3 Ligase Function of TRAF6 and Its Phosphorylation Differentially Shape LPS-Triggered Signaling Networks
3.5. TRAF6-Dependent Signaling Networks Are Regulated by Nutrient Availability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, H.; Sun, S.-C. Ubiquitin signaling in immune responses. Cell Res. 2016, 26, 457–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newton, K.; Dixit, V.M. Signaling in Innate Immunity and Inflammation. Cold Spring Harb. Perspect. Biol. 2012, 4, a006049. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Smits, A.H.; van Tilburg, G.B.; Jansen, P.W.; Makowski, M.M.; Ovaa, H.; Vermeulen, M. An Interaction Landscape of Ubiquitin Signaling. Mol. Cell 2017, 65, 941–955.e8. [Google Scholar] [CrossRef] [Green Version]
- Berndsen, C.; Wolberger, C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 2014, 21, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Walsh, M.C.; Lee, J.; Choi, Y. Tumor necrosis factor receptor- associated factor 6 (TRAF6) regulation of development, function, and homeostasis of the immune system. Immunol. Rev. 2015, 266, 72–92. [Google Scholar] [CrossRef] [PubMed]
- Guven-Maiorov, E.; Keskin, O.; Gursoy, A.; VanWaes, C.; Chen, Z.; Tsai, C.-J.; Nussinov, R. The Architecture of the TIR Domain Signalosome in the Toll-like Receptor-4 Signaling Pathway. Sci. Rep. 2015, 5, srep13128. [Google Scholar] [CrossRef] [Green Version]
- Abbott, D.W.; Yang, Y.; Hutti, J.E.; Madhavarapu, S.; Kelliher, M.A.; Cantley, L.C. Coordinated Regulation of Toll-Like Receptor and NOD2 Signaling by K63-Linked Polyubiquitin Chains. Mol. Cell. Biol. 2007, 27, 6012–6025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanayama, A.; Seth, R.B.; Sun, L.; Ea, C.-K.; Hong, M.; Shaito, A.; Chiu, Y.-H.; Deng, L.; Chen, Z.J. TAB2 and TAB3 Activate the NF-κB Pathway through Binding to Polyubiquitin Chains. Mol. Cell 2004, 15, 535–548. [Google Scholar] [CrossRef]
- Arthur, J.S.C.; Ley, S.C. Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 2013, 13, 679–692. [Google Scholar] [CrossRef]
- Du, M.; Yuan, L.; Tan, X.; Huang, D.; Wang, X.; Zheng, Z.; Mao, X.; Li, X.; Yang, L.; Huang, K.; et al. The LPS-inducible lncRNA Mirt2 is a negative regulator of inflammation. Nat. Commun. 2017, 8, 2049. [Google Scholar] [CrossRef] [Green Version]
- Dai, Y.; Jia, P.; Fang, Y.; Liu, H.; Jiao, X.; He, J.C.; Ding, X. miR-146a is essential for lipopolysaccharide (LPS)-induced cross-tolerance against kidney ischemia/reperfusion injury in mice. Sci. Rep. 2016, 6, 27091. [Google Scholar] [CrossRef] [Green Version]
- Chan, S.T.; Lee, J.; Narula, M.; Ou, J.-H.J. Suppression of Host Innate Immune Response by Hepatitis C Virus via Induction of Autophagic Degradation of TRAF6. J. Virol. 2016, 90, 10928–10935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muroi, M.; Tanamoto, K.-I. IRAK-1-mediated negative regulation of Toll-like receptor signaling through proteasome-dependent downregulation of TRAF6. Biochim. Biophys. Acta (BBA) Bioenerg. 2012, 1823, 255–263. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.; Pang, K.; Park, J.; Hong, E.; Lee, J.; Ooshima, A.; Kim, H.-S.; Cho, J.H.; Han, Y.; Lee, C.; et al. Destablilization of TRAF6 by DRAK1 Suppresses Tumor Growth and Metastasis in Cervical Cancer Cells. Cancer Res. 2020, 80, 2537–2549. [Google Scholar] [CrossRef] [Green Version]
- Meng, Y.; Liu, C.; Shen, L.; Zhou, M.; Liu, W.; Kowolik, C.; Campbell, J.L.; Zheng, L.; Shen, B. TRAF6 mediates human DNA2 polyubiquitination and nuclear localization to maintain nuclear genome integrity. Nucleic Acids Res. 2019, 47, 7564–7579. [Google Scholar] [CrossRef]
- Tenekeci, U.; Poppe, M.; Beuerlein, K.; Buro, C.; Müller, H.; Weiser, H.; Kettner-Buhrow, D.; Porada, K.; Newel, D.; Xu, M.; et al. K63-Ubiquitylation and TRAF6 Pathways Regulate Mammalian P-Body Formation and mRNA Decapping. Mol. Cell 2016, 62, 943–957. [Google Scholar] [CrossRef] [Green Version]
- Naito, A.; Azuma, S.; Tanaka, S.; Miyazaki, T.; Takaki, S.; Takatsu, K.; Nakao, K.; Nakamura, K.; Katsuki, M.; Yamamoto, T.; et al. Severe osteopetrosis, defective interleukin-1 signalling and lymph node organogenesis inTRAF6-deficient mice. Genes Cells 1999, 4, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The Role of Atg Proteins in Autophagosome Formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef]
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef]
- Nazio, F.; Strappazzon, F.; Antonioli, M.; Bielli, P.; Cianfanelli, V.; Bordi, M.; Gretzmeier, C.; Dengjel, J.; Piacentini, M.; Fimia, G.M.; et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 2013, 15, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.-S.; Kehrl, J.H. TRAF6 and A20 Regulate Lysine 63-Linked Ubiquitination of Beclin-1 to Control TLR4-Induced Autophagy. Sci. Signal. 2010, 3, ra42. [Google Scholar] [CrossRef]
- Antonioli, M.; Di Rienzo, M.; Piacentini, M.; Fimia, G.M. Emerging Mechanisms in Initiating and Terminating Autophagy. Trends Biochem. Sci. 2017, 42, 28–41. [Google Scholar] [CrossRef]
- Oh, S.Y.; Kang, N.; Kang, J.Y.; Kim, K.W.; Choi, J.-H.; Yang, Y.-M.; Shin, D.M. Sestrin2 Regulates Osteoclastogenesis via the p62-TRAF6 Interaction. Front. Cell Dev. Biol. 2021, 9, 646803. [Google Scholar] [CrossRef]
- Cadwell, K. Crosstalk between autophagy and inflammatory signalling pathways: Balancing defence and homeostasis. Nat. Rev. Immunol. 2016, 16, 661–675. [Google Scholar] [CrossRef]
- Birmingham, C.L.; Smith, A.C.; Bakowski, M.A.; Yoshimori, T.; Brumell, J.H. Autophagy Controls Salmonella Infection in Response to Damage to the Salmonella-containing Vacuole. J. Biol. Chem. 2006, 281, 11374–11383. [Google Scholar] [CrossRef] [Green Version]
- Delgado, M.A.; Elmaoued, R.A.; Davis, A.S.; Kyei, G.; Deretic, V. Toll-like receptors control autophagy. EMBO J. 2008, 27, 1110–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, C.-S.; Kehrl, J.H. MyD88 and Trif Target Beclin 1 to Trigger Autophagy in Macrophages. J. Biol. Chem. 2008, 283, 33175–33182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, Z.; Xie, X.; Cao, H.; Zhou, X.; Zhang, X.D.; Fan, H.; Liu, Z. Autophagy facilitates TLR4- and TLR3-triggered migration and invasion of lung cancer cells through the promotion of TRAF6 ubiquitination. Autophagy 2014, 10, 257–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inokuchi-Shimizu, S.; Park, E.J.; Roh, Y.S.; Yang, L.; Zhang, B.; Song, J.; Liang, S.; Pimienta, M.; Taniguchi, K.; Wu, X.; et al. TAK1-mediated autophagy and fatty acid oxidation prevent hepatosteatosis and tumorigenesis. J. Clin. Investig. 2014, 124, 3566–3578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, M.; Sato, K.; Tomura, K.; Kosako, H.; Sato, K. The autophagy receptor ALLO-1 and the IKKE-1 kinase control clearance of paternal mitochondria in Caenorhabditis elegans. Nat. Cell Biol. 2018, 20, 81–91. [Google Scholar] [CrossRef]
- Saul, V.V.; Seibert, M.; Krüger, M.; Jeratsch, S.; Kracht, M.; Schmitz, M.L. ULK1/2 Restricts the Formation of Inducible SINT-Speckles, Membraneless Organelles Controlling the Threshold of TBK1 Activation. iScience 2019, 19, 527–544. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Tenoever, B.R.; Grandvaux, N.; Zhou, G.-P.; Lin, R.; Hiscott, J. Triggering the Interferon Antiviral Response Through an IKK-Related Pathway. Science 2003, 300, 1148–1151. [Google Scholar] [CrossRef]
- Boehm, J.; Zhao, J.J.; Yao, J.; Kim, S.Y.; Firestein, R.; Dunn, I.F.; Sjostrom, S.K.; Garraway, L.A.; Weremowicz, S.; Richardson, A.L.; et al. Integrative Genomic Approaches Identify IKBKE as a Breast Cancer Oncogene. Cell 2007, 129, 1065–1079. [Google Scholar] [CrossRef] [Green Version]
- Rajurkar, M.; Dang, K.; Fernandez-Barrena, M.G.; Liu, X.; Fernandez-Zapico, M.E.; Lewis, B.C.; Mao, J. IKBKE Is Required during KRAS-Induced Pancreatic Tumorigenesis. Cancer Res. 2017, 77, 320–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Axe, E.L.; Walker, S.; Manifava, M.; Chandra, P.; Roderick, H.; Habermann, A.; Griffiths, G.; Ktistakis, N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008, 182, 685–701. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Wittwer, T.; Weber, A.; Schneider, H.; Moreno, R.; Maine, G.N.; Kracht, M.; Schmitz, M.L.; Burstein, E. Regulation of NF-κB activity by competition between RelA acetylation and ubiquitination. Oncogene 2011, 31, 611–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gloeckner, C.J.; Boldt, K.; Ueffing, M. Strep/FLAG Tandem Affinity Purification (SF-TAP) to Study Protein Interactions. Curr. Protoc. Protein Sci. 2009, 57, 19.20.1–19.20.19. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Renner, F.; Moreno, R.; Schmitz, M.L. SUMOylation-Dependent Localization of IKKɛ in PML Nuclear Bodies Is Essential for Protection against DNA-Damage-Triggered Cell Death. Mol. Cell 2010, 37, 503–515. [Google Scholar] [CrossRef]
- Fang, R.; Jiang, Q.; Zhou, X.; Wang, C.; Guan, Y.; Tao, J.; Xi, J.; Feng, J.-M.; Jiang, Z. MAVS activates TBK1 and IKKε through TRAFs in NEMO dependent and independent manner. PLoS Pathog. 2017, 13, e1006720. [Google Scholar] [CrossRef]
- Mertins, P.; Mani, D.R.; Ruggles, K.; Gillette, M.A.; Clauser, K.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; Petralia, F.; et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nat. Cell Biol. 2016, 534, 55–62. [Google Scholar] [CrossRef] [Green Version]
- Fu, T.-M.; Shen, C.; Li, Q.; Zhang, P.; Wu, H. Mechanism of ubiquitin transfer promoted by TRAF6. Proc. Natl. Acad. Sci. USA 2018, 115, 1783–1788. [Google Scholar] [CrossRef] [Green Version]
- Munafó, D.B.; Colombo, M.I. A novel assay to study autophagy: Regulation of autophagosome vacuole size by amino acid deprivation. J. Cell Sci. 2001, 114, 3619–3629. [Google Scholar] [CrossRef]
- Fujita, K.-I.; Srinivasula, S.M. TLR4-mediated autophagy in macrophages is a p62-dependent type of selective autophagy of aggresome-like induced structures (ALIS). Autophagy 2011, 7, 552–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lomaga, M.A.; Yeh, W.-C.; Sarosi, I.; Duncan, G.S.; Furlonger, C.; Ho, A.; Morony, S.; Capparelli, C.; Van, G.; Kaufman, S.; et al. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 1999, 13, 1015–1024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwandner, R.; Yamaguchi, K.; Cao, Z. Requirement of Tumor Necrosis Factor Receptor–Associated Factor (Traf)6 in Interleukin 17 Signal Transduction. J. Exp. Med. 2000, 191, 1233–1240. [Google Scholar] [CrossRef] [Green Version]
- Davies, C.; Mak, T.W.; Young, L.S.; Eliopoulos, A. TRAF6 Is Required for TRAF2-Dependent CD40 Signal Transduction in Nonhemopoietic Cells. Mol. Cell. Biol. 2005, 25, 9806–9819. [Google Scholar] [CrossRef] [Green Version]
- Mostowy, S.; Shenoy, A.R. The cytoskeleton in cell-autonomous immunity: Structural determinants of host defence. Nat. Rev. Immunol. 2015, 15, 559–573. [Google Scholar] [CrossRef] [Green Version]
- Shi, C.-S.; Qi, H.-Y.; Boularan, C.; Huang, N.-N.; Abu-Asab, M.; Shelhamer, J.H.; Kehrl, J.H. SARS-Coronavirus Open Reading Frame-9b Suppresses Innate Immunity by Targeting Mitochondria and the MAVS/TRAF3/TRAF6 Signalosome. J. Immunol. 2014, 193, 3080–3089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, C.; An, R.; Yu, Y.; Dai, H.; Qu, Z.; Gao, M.; Wang, J. BICP0 Negatively Regulates TRAF6-Mediated NF-κB and Interferon Activation by Promoting K48-Linked Polyubiquitination of TRAF6. Front. Microbiol. 2019, 10, 3040. [Google Scholar] [CrossRef]
- Jang, H.-D.; Hwang, H.Z.; Kim, H.-S.; Lee, S.Y. C-Cbl negatively regulates TRAF6-mediated NF-κB activation by promoting K48-linked polyubiquitination of TRAF6. Cell. Mol. Biol. Lett. 2019, 24, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.K.; Gupta, S.K.; Bhatnagar, S.; Panguluri, S.K.; Darnay, B.G.; Choi, Y.; Kumar, A. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J. Cell Biol. 2010, 191, 1395–1411. [Google Scholar] [CrossRef] [Green Version]
- Marchiando, A.M.; Ramanan, D.; Ding, Y.; Gomez, L.E.; Hubbard-Lucey, V.M.; Maurer, K.; Wang, C.; Ziel, J.W.; van Rooijen, N.; Nuñez, G.; et al. A Deficiency in the Autophagy Gene Atg16L1 Enhances Resistance to Enteric Bacterial Infection. Cell Host Microbe 2013, 14, 216–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Q.; Yokoyama, C.C.; Williams, J.; Baldridge, M.; Jin, X.; DesRochers, B.; Bricker, T.; Wilen, C.; Bagaitkar, J.; Loginicheva, E.; et al. Homeostatic Control of Innate Lung Inflammation by Vici Syndrome Gene Epg5 and Additional Autophagy Genes Promotes Influenza Pathogenesis. Cell Host Microbe 2016, 19, 102–113. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.J.; Ye, L.; Huang, W.F.; Guo, L.J.; Xu, Z.G.; Wu, H.L.; Yang, C.; Liu, H.F. p62 links the autophagy pathway and the ubiqutin–proteasome system upon ubiquitinated protein degradation. Cell. Mol. Biol. Lett. 2016, 21, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Pawson, T.; Scott, J.D. Protein phosphorylation in signaling—50 years and counting. Trends Biochem. Sci. 2005, 30, 286–290. [Google Scholar] [CrossRef]
- Das, A.; Middleton, A.J.; Padala, P.; Ledgerwood, E.C.; Mace, P.D.; Day, C.L. The Structure and Ubiquitin Binding Properties of TRAF RING Heterodimers. J. Mol. Biol. 2021, 433, 166844. [Google Scholar] [CrossRef]
- Zucchelli, S.; Codrich, M.; Marcuzzi, F.; Pinto, M.; Vilotti, S.; Biagioli, M.; Ferrer, I.; Gustincich, S. TRAF6 promotes atypical ubiquitination of mutant DJ-1 and alpha-synuclein and is localized to Lewy bodies in sporadic Parkinson’s disease brains. Hum. Mol. Genet. 2010, 19, 3759–3770. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Macartney, T.; Peggie, M.; Cohen, P. Interleukin-1 and TRAF6-dependent activation of TAK1 in the absence of TAB2 and TAB3. Biochem. J. 2017, 474, 2235–2248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balic, J.J.; Albargy, H.; Luu, K.; Kirby, F.J.; Jayasekara, W.S.N.; Mansell, F.; Garama, D.J.; De Nardo, D.; Baschuk, N.; Louis, C.; et al. STAT3 serine phosphorylation is required for TLR4 metabolic reprogramming and IL-1β expression. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chariot, A.; Leonardi, A.; Muller, J.; Bonif, M.; Brown, K.; Siebenlist, U. Association of the adaptor TANK with the I kappa B kinase (IKK) regulator NEMO connects IKK complexes with IKK epsilon and TBK1 kinases. J. Biol. Chem. 2002, 277, 37029–37036. [Google Scholar] [CrossRef] [Green Version]
- Moreno, R.; Sobotzik, J.M.; Schultz, C.; Schmitz, M.L. Specification of the NF-kappaB transcriptional response by p65 phosphorylation and TNF-induced nuclear translocation of IKK epsilon. Nucleic Acids Res. 2010, 38, 6029–6044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamothe, B.; Besse, A.; Campos, A.D.; Webster, W.K.; Wu, H.; Darnay, B.G. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. J. Biol. Chem. 2007, 282, 4102–4112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choudhury, A.D.; Xu, H.; Baer, R. Ubiquitination and proteasomal degradation of the BRCA1 tumor suppressor is regulated during cell cycle progression. J. Biol. Chem. 2004, 279, 33909–33918. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Busch, J.; Moreno, R.; de la Vega, L.; Saul, V.V.; Bacher, S.; von Zweydorf, F.; Ueffing, M.; Weber, A.; Gloeckner, C.J.; Linne, U.; et al. TRAF6 Phosphorylation Prevents Its Autophagic Degradation and Re-Shapes LPS-Triggered Signaling Networks. Cancers 2021, 13, 3618. https://doi.org/10.3390/cancers13143618
Busch J, Moreno R, de la Vega L, Saul VV, Bacher S, von Zweydorf F, Ueffing M, Weber A, Gloeckner CJ, Linne U, et al. TRAF6 Phosphorylation Prevents Its Autophagic Degradation and Re-Shapes LPS-Triggered Signaling Networks. Cancers. 2021; 13(14):3618. https://doi.org/10.3390/cancers13143618
Chicago/Turabian StyleBusch, Julia, Rita Moreno, Laureano de la Vega, Vera Vivian Saul, Susanne Bacher, Felix von Zweydorf, Marius Ueffing, Axel Weber, Christian Johannes Gloeckner, Uwe Linne, and et al. 2021. "TRAF6 Phosphorylation Prevents Its Autophagic Degradation and Re-Shapes LPS-Triggered Signaling Networks" Cancers 13, no. 14: 3618. https://doi.org/10.3390/cancers13143618
APA StyleBusch, J., Moreno, R., de la Vega, L., Saul, V. V., Bacher, S., von Zweydorf, F., Ueffing, M., Weber, A., Gloeckner, C. J., Linne, U., Kracht, M., & Schmitz, M. L. (2021). TRAF6 Phosphorylation Prevents Its Autophagic Degradation and Re-Shapes LPS-Triggered Signaling Networks. Cancers, 13(14), 3618. https://doi.org/10.3390/cancers13143618






