Risk Score Generated from CT-Based Radiomics Signatures for Overall Survival Prediction in Non-Small Cell Lung Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. Patient’s Clinical Characteristics
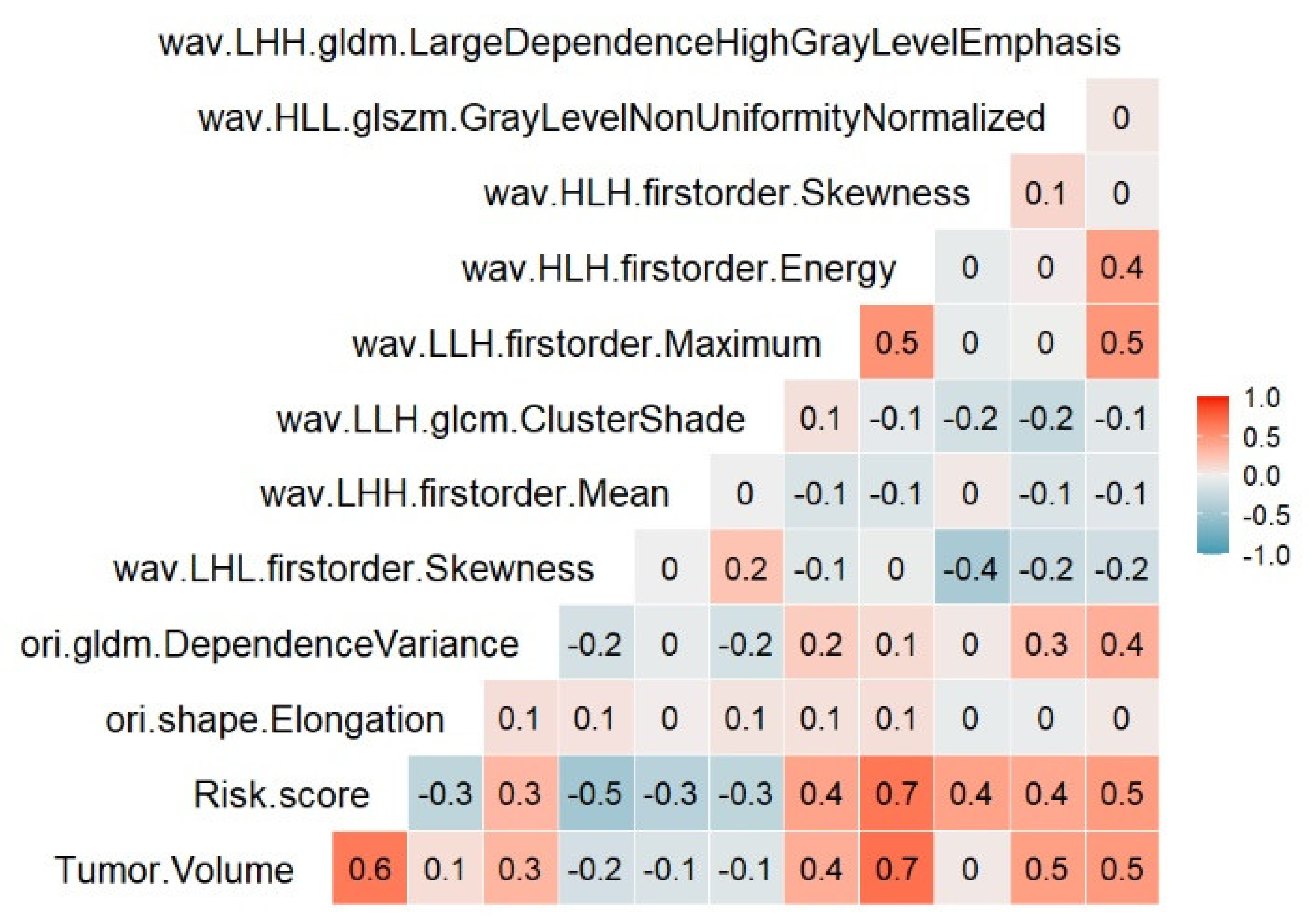
2.2. Feature Selection
2.3. Construction of Ten Radiomics Signatures Prognostic Model
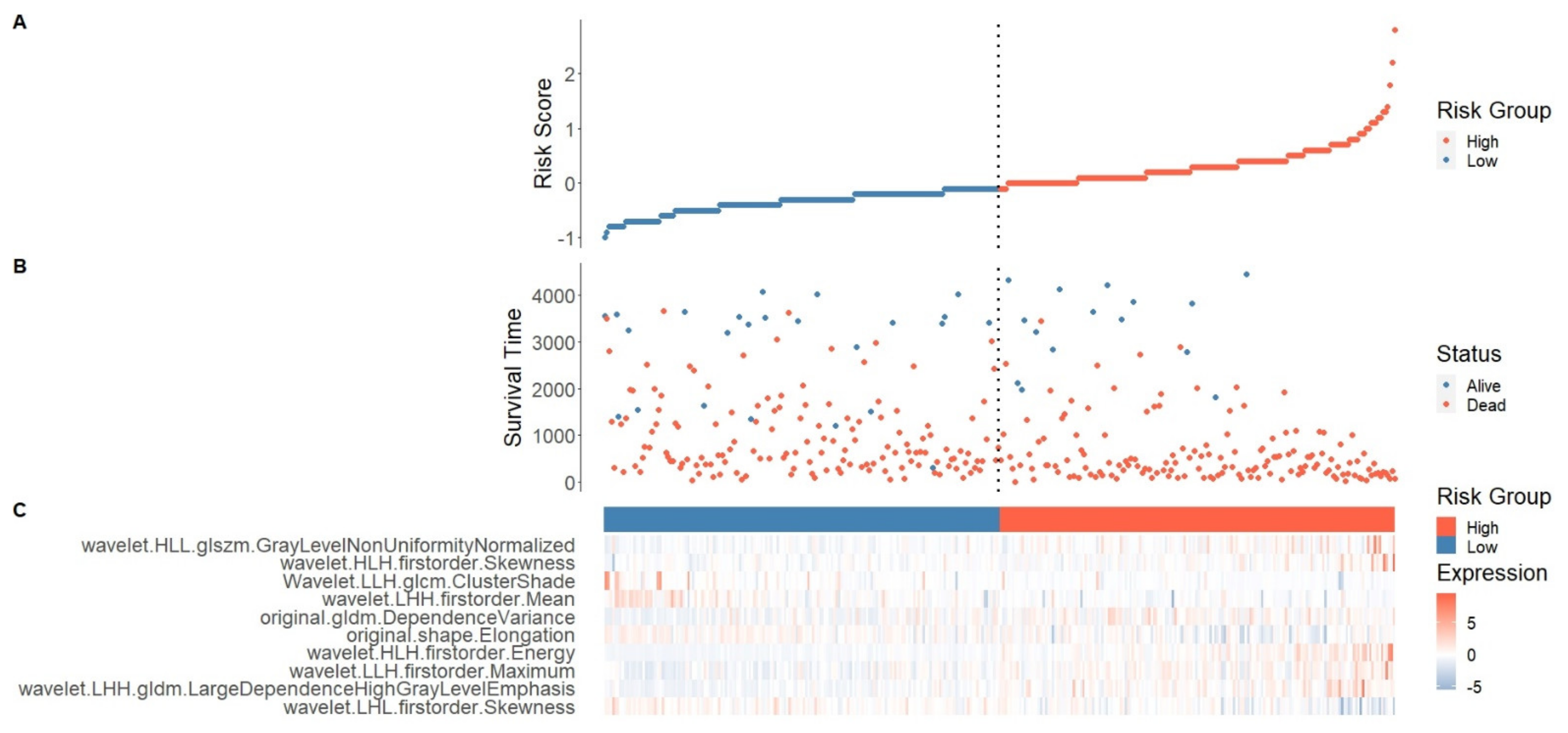
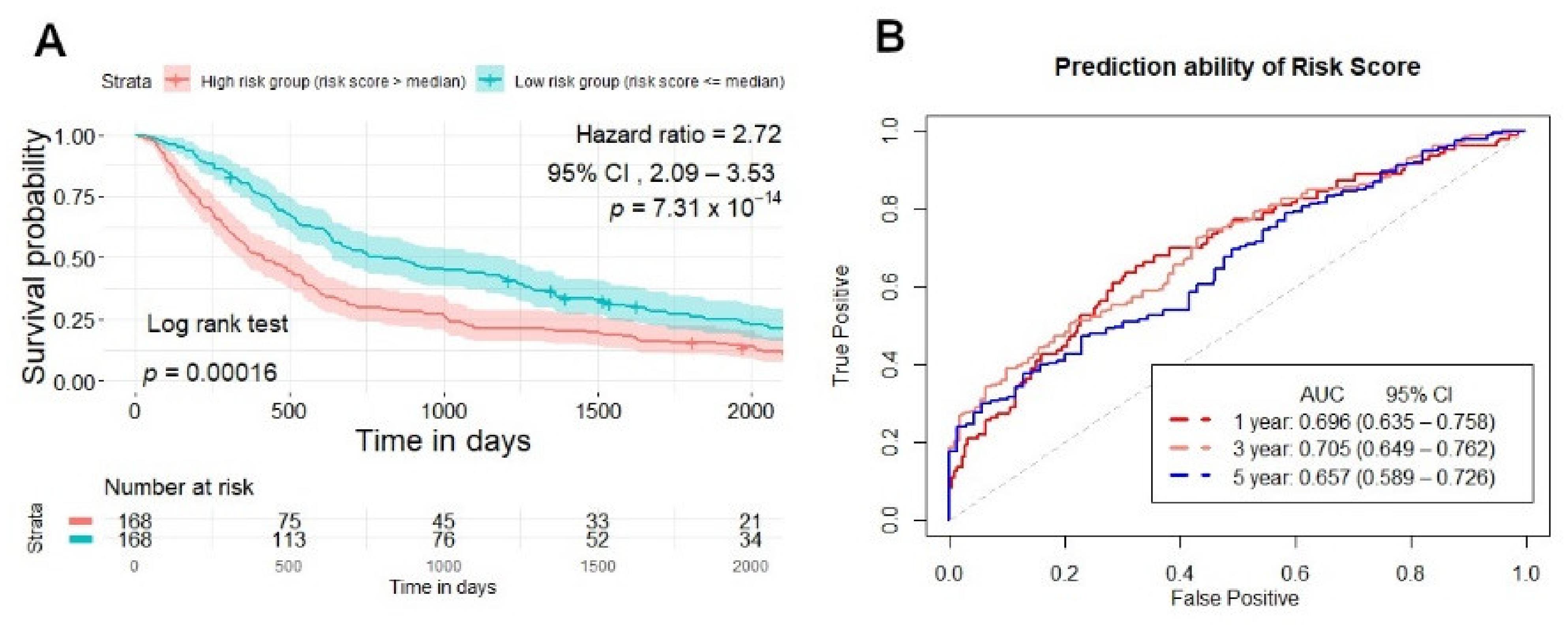
2.4. Risk Score Establishment for Overall Survival Prediction in the Training Set
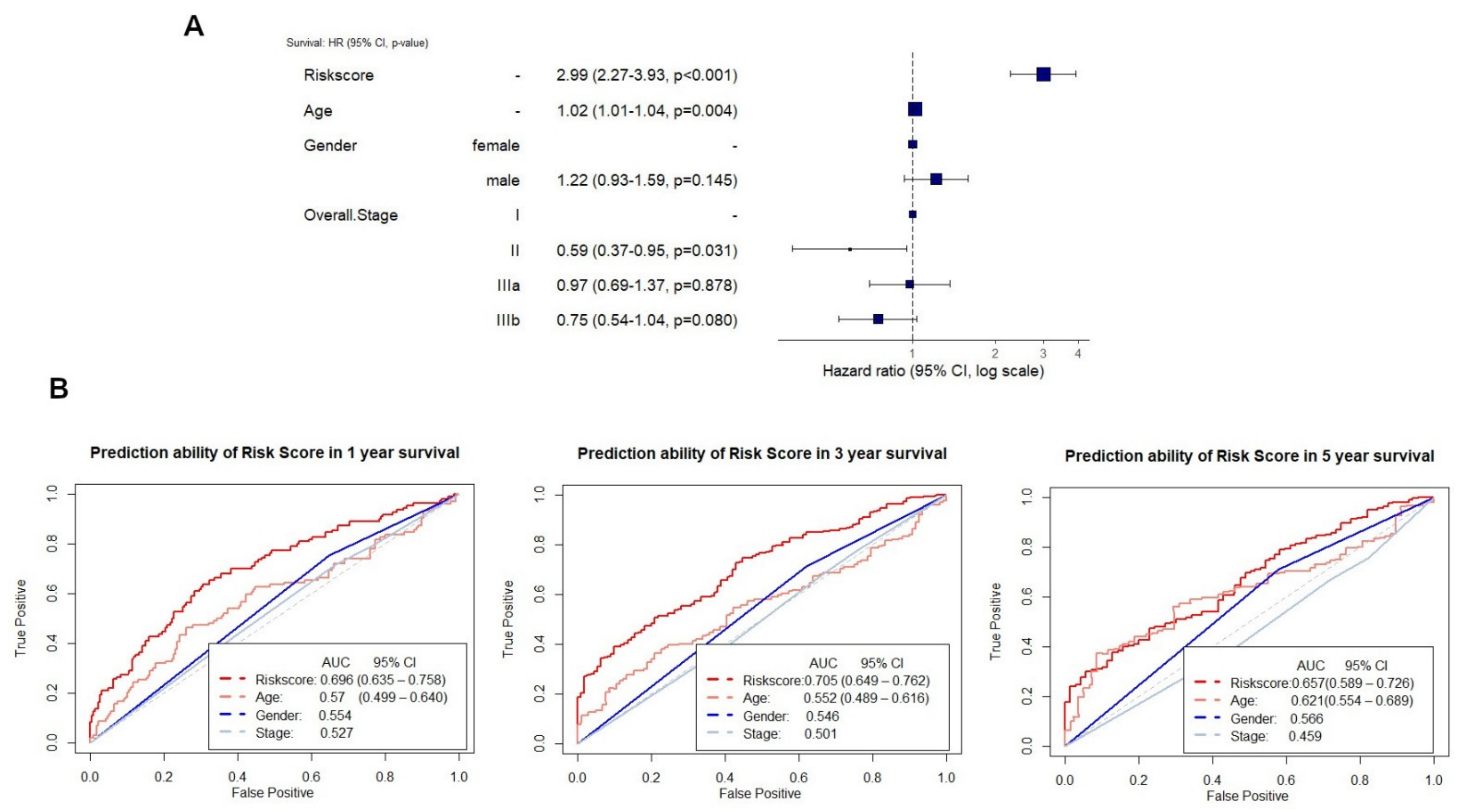
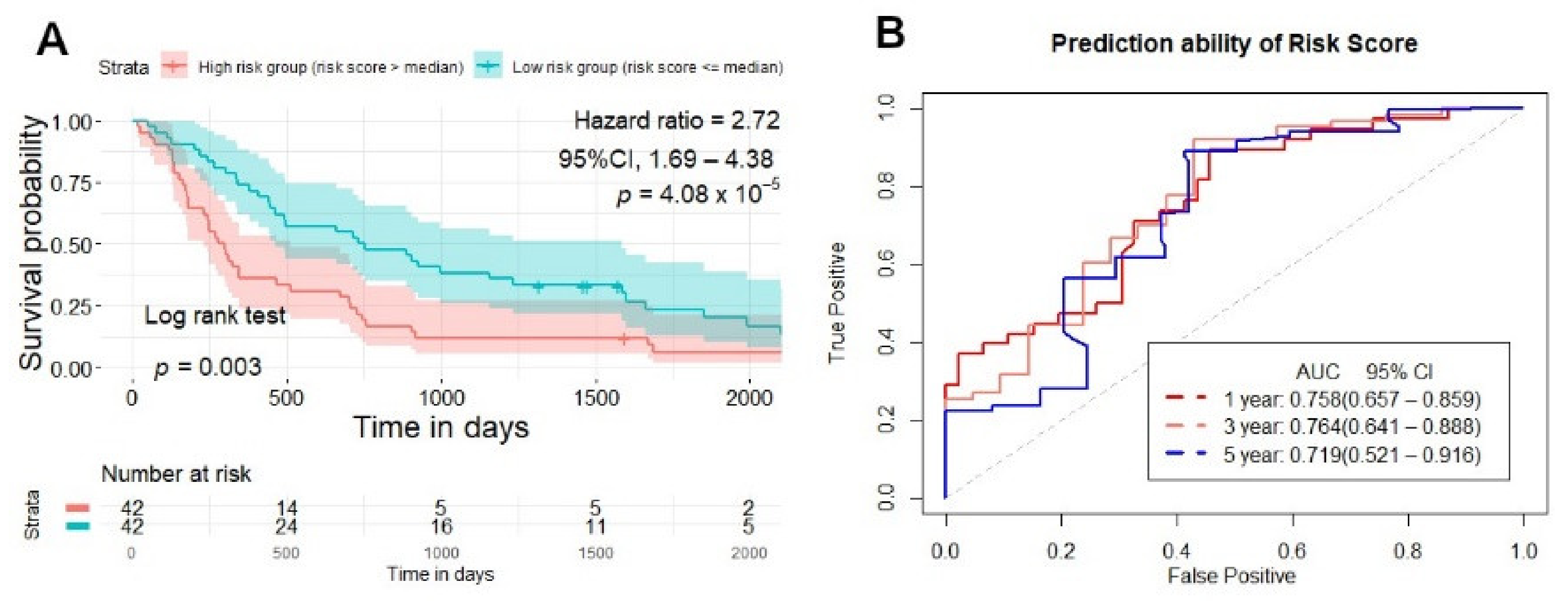
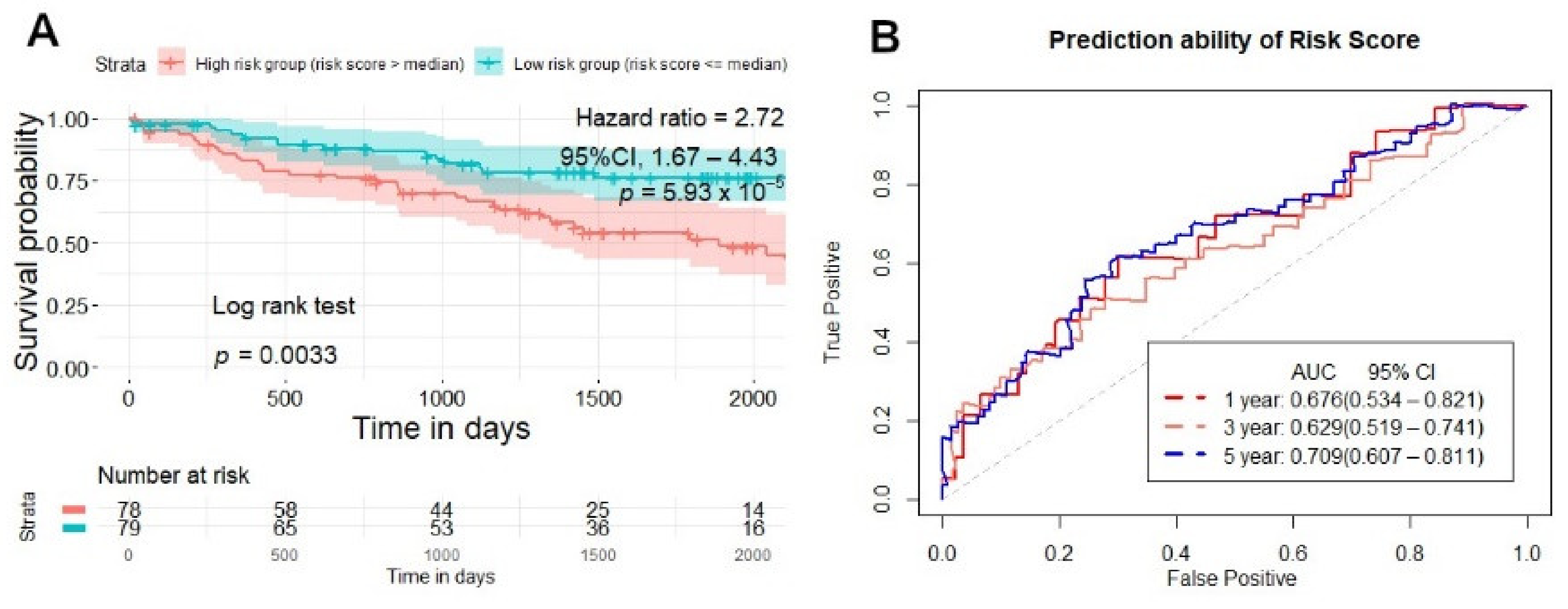
2.5. Testing and Validation of the Risk Score Model for Overall Survival Prediction
3. Discussion
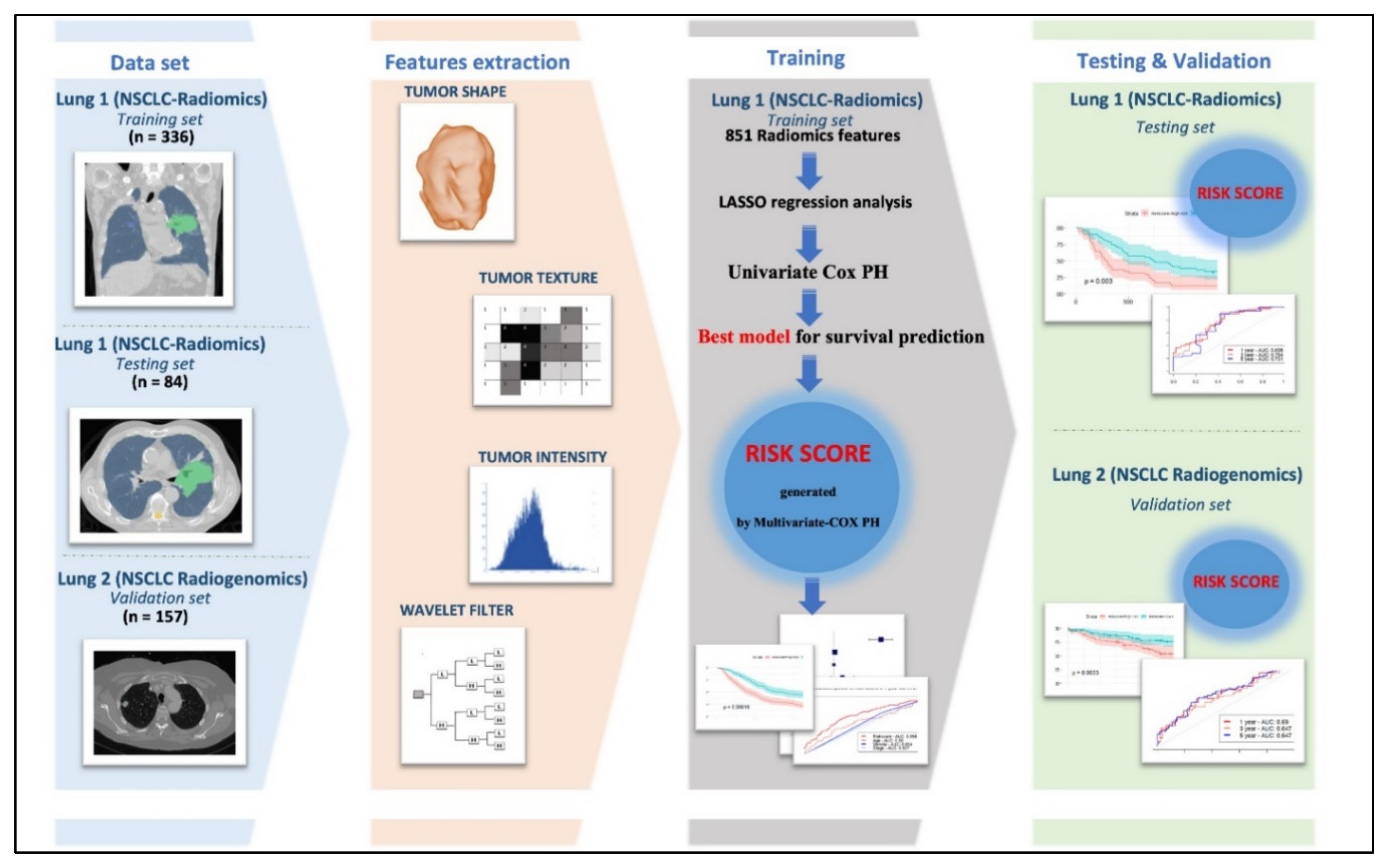
4. Materials and Methods
4.1. Patients Cohort
4.2. Radiomics Features Extraction
4.3. Feature Selection and Construction of the Best Model for Survival Prediction
4.3.1. Removing Highly Correlated Features in the Training Set
4.3.2. Identifying the Best Performing Model and Construction of Risk Score for Survival Prediction
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin. Proc. 2019, 94, 1623–1640. [Google Scholar] [CrossRef] [PubMed]
- Pao, W.; Girard, N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011, 12, 175–180. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H.J.R. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; He, Y.; Yuan, Z.; Benedict, S.; Valicenti, R.; Qiu, J.; Rong, Y. Radiomics for Response and Outcome Assessment for Non-Small Cell Lung Cancer. Technol. Cancer Res. Treat. 2018, 17, 1533033818782788. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, S.M.; Kim, S.; Choi, S.; Kim, W.; Do, K.H.; Seo, J.B. Performance of radiomics models for survival prediction in non-small-cell lung cancer: Influence of CT slice thickness. Eur. Radiol. 2021, 31, 2856–2865. [Google Scholar] [CrossRef]
- Astaraki, M.; Wang, C.; Buizza, G.; Toma-Dasu, I.; Lazzeroni, M.; Smedby, O. Early survival prediction in non-small cell lung cancer from PET/CT images using an intra-tumor partitioning method. Phys. Med. 2019, 60, 58–65. [Google Scholar] [CrossRef] [PubMed]
- van Timmeren, J.E.; van Elmpt, W.; Leijenaar, R.T.H.; Reymen, B.; Monshouwer, R.; Bussink, J.; Paelinck, L.; Bogaert, E.; De Wagter, C.; Elhaseen, E.; et al. Longitudinal radiomics of cone-beam CT images from non-small cell lung cancer patients: Evaluation of the added prognostic value for overall survival and locoregional recurrence. Radiother. Oncol. 2019, 136, 78–85. [Google Scholar] [CrossRef]
- Yang, L.; Yang, J.; Zhou, X.; Huang, L.; Zhao, W.; Wang, T.; Zhuang, J.; Tian, J. Development of a radiomics nomogram based on the 2D and 3D CT features to predict the survival of non-small cell lung cancer patients. Eur. Radiol. 2019, 29, 2196–2206. [Google Scholar] [CrossRef]
- van Timmeren, J.E.; Leijenaar, R.T.H.; van Elmpt, W.; Reymen, B.; Oberije, C.; Monshouwer, R.; Bussink, J.; Brink, C.; Hansen, O.; Lambin, P. Survival prediction of non-small cell lung cancer patients using radiomics analyses of cone-beam CT images. Radiother. Oncol. 2017, 123, 363–369. [Google Scholar] [CrossRef] [Green Version]
- Afshar, P.; Mohammadi, A.; Tyrrell, P.N.; Cheung, P.; Sigiuk, A.; Plataniotis, K.N.; Nguyen, E.T.; Oikonomou, A. DRTOP: Deep learning-based radiomics for the time-to-event outcome prediction in lung cancer. Sci. Rep. 2020, 10, 12366. [Google Scholar] [CrossRef] [PubMed]
- Botta, F.; Raimondi, S.; Rinaldi, L.; Bellerba, F.; Corso, F.; Bagnardi, V.; Origgi, D.; Minelli, R.; Pitoni, G.; Petrella, F.; et al. Association of a CT-Based Clinical and Radiomics Score of Non-Small Cell Lung Cancer (NSCLC) with Lymph Node Status and Overall Survival. Cancers 2020, 12, 1432. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, R.; Wang, S.; Fang, M.; Zhu, Y.; Hu, Z.; Dong, D.; Shi, J.; Tian, J. CT-Based Radiomic Signature as a Prognostic Factor in Stage IV ALK-Positive Non-small-cell Lung Cancer Treated With TKI Crizotinib: A Proof-of-Concept Study. Front. Oncol. 2020, 10, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugai, Y.; Kadoya, N.; Tanaka, S.; Tanabe, S.; Umeda, M.; Yamamoto, T.; Takeda, K.; Dobashi, S.; Ohashi, H.; Takeda, K.; et al. Impact of feature selection methods and subgroup factors on prognostic analysis with CT-based radiomics in non-small cell lung cancer patients. Radiat. Oncol. 2021, 16, 80. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Chen, Y.; Chen, X.; Sun, X.; Xing, L. CT-based radiomics for predicting brain metastases as the first failure in patients with curatively resected locally advanced non-small cell lung cancer. Eur. J. Radiol. 2021, 134, 109411. [Google Scholar] [CrossRef]
- Lathwal, A.; Arora, C.; Raghava, G.P.S. Prediction of risk scores for colorectal cancer patients from the concentration of proteins involved in mitochondrial apoptotic pathway. PLoS ONE 2019, 14, e0217527. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Long, Q.; Zhang, D.; Li, J.; Zhang, X. Identification of a four-gene panel predicting overall survival for lung adenocarcinoma. BMC Cancer 2020, 20, 1198. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Qin, X.; Li, L.; Zhou, J.; Zhou, M.; Li, X.; Xu, Y.; Yuan, L.; Liu, Q.N.; Xing, H. A 15-long non-coding RNA signature to improve prognosis prediction of cervical squamous cell carcinoma. Gynecol. Oncol. 2018, 149, 181–187. [Google Scholar] [CrossRef]
- Zengin, T.; Onal-Suzek, T. Comprehensive Profiling of Genomic and Transcriptomic Differences between Risk Groups of Lung Adenocarcinoma and Lung Squamous Cell Carcinoma. J. Pers. Med. 2021, 11, 154. [Google Scholar] [CrossRef]
- Sana, J.; Radova, L.; Lakomy, R.; Kren, L.; Fadrus, P.; Smrcka, M.; Besse, A.; Nekvindova, J.; Hermanova, M.; Jancalek, R.; et al. Risk Score based on microRNA expression signature is independent prognostic classifier of glioblastoma patients. Carcinogenesis 2014, 35, 2756–2762. [Google Scholar] [CrossRef] [Green Version]
- Leger, S.; Zwanenburg, A.; Pilz, K.; Lohaus, F.; Linge, A.; Zophel, K.; Kotzerke, J.; Schreiber, A.; Tinhofer, I.; Budach, V.; et al. A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci. Rep. 2017, 7, 13206. [Google Scholar] [CrossRef]
- Saeys, Y.; Inza, I.; Larranaga, P. A review of feature selection techniques in bioinformatics. Bioinformatics 2007, 23, 2507–2517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meinshausen, N.; Bühlmann, P. High-dimensional graphs and variable selection with the Lasso. Ann. Stat. 2006, 34, 1436–1462. [Google Scholar] [CrossRef] [Green Version]
- Mattonen, S.A.; Davidzon, G.A.; Benson, J.; Leung, A.N.C.; Vasanawala, M.; Horng, G.; Shrager, J.B.; Napel, S.; Nair, V.S. Bone Marrow and Tumor Radiomics at (18)F-FDG PET/CT: Impact on Outcome Prediction in Non-Small Cell Lung Cancer. Radiology 2019, 293, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Parmar, C.; Grossmann, P.; Bussink, J.; Lambin, P.; Aerts, H. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci. Rep. 2015, 5, 13087. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, S.H.; Korecki, J.N.; Balagurunathan, Y.; Yuhua, G.; Kumar, V.; Basu, S.; Hall, L.O.; Goldgof, D.B.; Gatenby, R.A.; Gillies, R.J. Predicting Outcomes of Nonsmall Cell Lung Cancer Using CT Image Features. IEEE Access 2014, 2, 1418–1426. [Google Scholar] [CrossRef]
- Chen, D.; She, Y.; Wang, T.; Xie, H.; Li, J.; Jiang, G.; Chen, Y.; Zhang, L.; Xie, D.; Chen, C. Radiomics-based prediction for tumour spread through air spaces in stage I lung adenocarcinoma using machine learning. Eur. J. Cardio-Thorac. Surg. 2020, 58, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Paul, R.; Hawkins, S.H.; Balagurunathan, Y.; Schabath, M.B.; Gillies, R.J.; Hall, L.O.; Goldgof, D.B. Deep Feature Transfer Learning in Combination with Traditional Features Predicts Survival Among Patients with Lung Adenocarcinoma. Tomography 2016, 2, 388–395. [Google Scholar] [CrossRef]
- Vial, A.; Stirling, D.; Field, M.; Ros, M.; Ritz, C.; Carolan, M.; Holloway, L.; Miller, A.A. The role of deep learning and radiomic feature extraction in cancer-specific predictive modelling: A review. Transl. Cancer Res. 2018, 7, 803–816. [Google Scholar] [CrossRef]
- Liu, J.; Xu, H.; Qing, H.; Li, Y.; Yang, X.; He, C.; Ren, J.; Zhou, P. Comparison of Radiomic Models Based on Low-Dose and Standard-Dose CT for Prediction of Adenocarcinomas and Benign Lesions in Solid Pulmonary Nodules. Front. Oncol. 2020, 10, 634298. [Google Scholar] [CrossRef]
- Choi, W.; Oh, J.H.; Riyahi, S.; Liu, C.J.; Jiang, F.; Chen, W.; White, C.; Rimner, A.; Mechalakos, J.G.; Deasy, J.O.; et al. Radiomics analysis of pulmonary nodules in low-dose CT for early detection of lung cancer. Med. Phys. 2018, 45, 1537–1549. [Google Scholar] [CrossRef]
- Bakr, S.; Gevaert, O.; Echegaray, S.; Ayers, K.; Zhou, M.; Shafiq, M.; Zheng, H.; Benson, J.A.; Zhang, W.; Leung, A.N.C.; et al. A radiogenomic dataset of non-small cell lung cancer. Sci. Data 2018, 5, 180202. [Google Scholar] [CrossRef] [Green Version]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.J.R. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Zhao, X.; Li, Q.; Xia, W.; Peng, Z.; Zhang, R.; Li, Q.; Jian, J.; Wang, W.; Tang, Y.; et al. Can peritumoral radiomics increase the efficiency of the prediction for lymph node metastasis in clinical stage T1 lung adenocarcinoma on CT? Eur. Radiol. 2019, 29, 6049–6058. [Google Scholar] [CrossRef] [PubMed]
- Lao, J.; Chen, Y.; Li, Z.C.; Li, Q.; Zhang, J.; Liu, J.; Zhai, G. A Deep Learning-Based Radiomics Model for Prediction of Survival in Glioblastoma Multiforme. Sci. Rep. 2017, 7, 10353. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Evans, J.R.; Malik, R.; Feng, F.Y.; Dhanasekaran, S.M.; Cao, X.; Chen, G.; Beer, D.G.; Jiang, H.; Chinnaiyan, A.M. Development of a RNA-Seq Based Prognostic Signature in Lung Adenocarcinoma. J. Natl. Cancer Inst. 2017, 109, djw200. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Shang, J.; Chen, B.; Yang, Z.; Song, Q.; Sun, X.; Chen, J.; Yang, J. Identification of Prognostic Signatures for Predicting the Overall Survival of Uveal Melanoma Patients. J. Cancer 2019, 10, 4921–4931. [Google Scholar] [CrossRef]
- Shang, J.; Song, Q.; Yang, Z.; Li, D.; Chen, W.; Luo, L.; Wang, Y.; Yang, J.; Li, S.J.O. Identification of lung adenocarcinoma specific dysregulated genes with diagnostic and prognostic value across 27 TCGA cancer types. Oncotarget 2017, 8, 87292–87306. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.-Y.; Yu, S.-L.; Chen, C.-H.; Chang, G.-C.; Chen, C.-Y.; Yuan, A.; Cheng, C.-L.; Wang, C.-H.; Terng, H.-J.; Kao, S.-F.; et al. A five-gene signature and clinical outcome in non–small-cell lung cancer. N. Engl. J. Med. 2007, 356, 11–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aerts, H.J.; Grossmann, P.; Tan, Y.; Oxnard, G.R.; Rizvi, N.; Schwartz, L.H.; Zhao, B. Defining a Radiomic Response Phenotype: A Pilot Study using targeted therapy in NSCLC. Sci. Rep. 2016, 6, 33860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fave, X.; Zhang, L.; Yang, J.; Mackin, D.; Balter, P.; Gomez, D.; Followill, D.; Jones, A.K.; Stingo, F.; Court, L.E. Impact of image preprocessing on the volume dependence and prognostic potential of radiomics features in non-small cell lung cancer. Transl. Cancer Res. 2016, 5, 349–363. [Google Scholar] [CrossRef] [Green Version]
- Shafiq-Ul-Hassan, M.; Latifi, K.; Zhang, G.; Ullah, G.; Gillies, R.; Moros, E. Voxel size and gray level normalization of CT radiomic features in lung cancer. Sci. Rep. 2018, 8, 10545. [Google Scholar] [CrossRef] [PubMed]
- Welch, M.L.; McIntosh, C.; Haibe-Kains, B.; Milosevic, M.F.; Wee, L.; Dekker, A.; Huang, S.H.; Purdie, T.G.; O’Sullivan, B.; Aerts, H.; et al. Vulnerabilities of radiomic signature development: The need for safeguards. Radiother. Oncol. 2019, 130, 2–9. [Google Scholar] [CrossRef] [Green Version]

| Lung 1 (NSCLC-Radiomics) | Lung 2 (NSCLC Radiogenomics) | |||||
|---|---|---|---|---|---|---|
| Training Set | Testing Set | Validation Set | ** p-Value | |||
| Parameter | (n = 336) | (n = 84) | * p-value | Parameter | (n = 157) | |
| Age (mean SD) | 67.67 (10.04) | 69.36 (10.19) | 0.183 | Age (mean SD) | 68.97 (9.52) | 0.302 |
| Gender | Gender | |||||
| Men (%) | 229 (68.2) | 60 (71.4) | 0.654 | Men (%) | 109 (69.4) | 0.967 |
| Type of NSCLC (%) | 0.545 | Type of NSCLC (%) | <0.001 | |||
| Adenocarcinoma | 40 (13.3) | 11 (14.1) | Adenocarcinoma | 126 (80.3) | ||
| Large cell | 95 (31.7) | 19 (24.4) | Squamous cell | 28 (17.8) | ||
| Squamous cell | 115 (38.3) | 36 (46.2) | Nos | 3 (1.9) | ||
| Nos | 50 (16.7) | 12 (15.4) | Stage (%) | <0.001 | ||
| Stage (%) | 0.126 | Tis | 6 (3.8) | |||
| I | 77 (23.0) | 15 (17.9) | I | 86 (54.8) | ||
| II | 30 (9.0) | 10 (11.9) | II | 23 (14.6) | ||
| IIIa | 95 (28.4) | 16 (19.0) | IIIa | 15 (9.6) | ||
| IIIb | 133 (39.7) | 43 (51.2) | IIIb | 5 (3.2) | ||
| IV | 0 | 0 | IV | 4 (2.5) | ||
| Survival time (median(IQR))(days) | 579.00 (286.25, 1510.25) | 456.50 (229.00, 1038.50) | 0.073 | Survival time (median (IQR))(days) | 1315.00 (630.00, 1921.00) | <0.001 |
| Feature | Hazard Ratio | p-Value | Concordance |
|---|---|---|---|
| original.shape.Elongation | 0.49 (0.25–0.95) | 0.04 * | 0.54 (se = 0.019) |
| original.gldm.DependenceVariance | 1.01 (1.00–1.02) | 0.02 * | 0.55 (se = 0.017) |
| wavelet.LHL.glcm.MCC | 2.71 (0.75–9.88) | 0.13 | 0.517 (se = 0.018) |
| wavelet.LHL.firstorder.Skewness | 0.70 (0.56–0.90) | <0.01 * | 0.57 (se = 0.018) |
| wavelet.LHH.gldm.LargeDependenceHighGrayLevelEmphasis | 1.00 (1.00–1.01) | <0.001 * | 0.57 (se = 0.017) |
| wavelet.LHH.gldm.LowGrayLevelEmphasis | 18.82 (0.00–1662852) | 0.61 | 0.48 (se = 0.017) |
| wavelet.LHH.firstorder.Mean | 0.85 (0.74–0.98) | 0.02 * | 0.54 (se = 0.017) |
| wavelet.LLH.glcm.ClusterShade | 1.0 (0.99–1.00) | 0.04 * | 0.54 (se = 0.018) |
| wavelet.LLH.firstorder.Maximum | 1.00 (1.00–1.01) | <0.01 * | 0.57 (se = 0.018) |
| wavelet.HLH.firstorder.Energy | 1.00 (1.00–1.01) | <0.001 * | 0.58 (se = 0.018) |
| wavelet.HLH.firstorder.Skewness | 1.48 (1.10–1.99) | <0.01 * | 0.513 (se = 0.019) |
| wavelet.LHH.glcm.Correlation | 0.02 (0.00–1.40) | 0.07 | 0.54 (se = 0.017) |
| wavelet.HLL.glszm.GrayLevelNonUniformityNormalized | 130.61 (3.30–5169) | <0.01 * | 0.53 (se = 0.018) |
| Feature | β Coefficient | Hazard Ratio | p-Value |
|---|---|---|---|
| original.shape.Elongation | −9.936 × 10−1 | 0.37 (0.18–0.75) | 0.006 |
| original.gldm.DependenceVariance | 5.230 × 10−3 | 1.01 (0.99–1.02) | 0.349 |
| wavelet.LHL.firstorder.Skewness | −1.693 × 10−1 | 0.84 (0.66–1.08) | 0.184 |
| wavelet.LHH.gldm.LargeDependenceHighGrayLevelEmphasis | 4.764 × 10−6 | 1.00 (1.00–1.00) | 0.627 |
| wavelet.LHH.firstorder.Mean | −1.489 × 10−1 | 0.86 (0.73–1.01) | 0.073 |
| wavelet.LLH.glcm.ClusterShade | −6.460 × 10−6 | 1.00 (1.00–1.00) | 0.654 |
| wavelet.LLH.firstorder.Maximum | 1.074 × 10−4 | 1.00 (1.00–1.00) | 0.734 |
| wavelet.HLH.firstorder.Energy | 8.738 × 10−9 | 1.00 (1.00–1.00) | <0.001 |
| wavelet.HLH.firstorder.Skewness | 3.567 × 10−1 | 1.43 (1.06–1.93) | 0.02 |
| wavelet.HLL.glszm.GrayLevelNonUniformityNormalized | 2.988 | 19.85 (0.36–1108.62) | 0.145 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, V.-H.; Kha, Q.-H.; Hung, T.N.K.; Le, N.Q.K. Risk Score Generated from CT-Based Radiomics Signatures for Overall Survival Prediction in Non-Small Cell Lung Cancer. Cancers 2021, 13, 3616. https://doi.org/10.3390/cancers13143616
Le V-H, Kha Q-H, Hung TNK, Le NQK. Risk Score Generated from CT-Based Radiomics Signatures for Overall Survival Prediction in Non-Small Cell Lung Cancer. Cancers. 2021; 13(14):3616. https://doi.org/10.3390/cancers13143616
Chicago/Turabian StyleLe, Viet-Huan, Quang-Hien Kha, Truong Nguyen Khanh Hung, and Nguyen Quoc Khanh Le. 2021. "Risk Score Generated from CT-Based Radiomics Signatures for Overall Survival Prediction in Non-Small Cell Lung Cancer" Cancers 13, no. 14: 3616. https://doi.org/10.3390/cancers13143616
APA StyleLe, V.-H., Kha, Q.-H., Hung, T. N. K., & Le, N. Q. K. (2021). Risk Score Generated from CT-Based Radiomics Signatures for Overall Survival Prediction in Non-Small Cell Lung Cancer. Cancers, 13(14), 3616. https://doi.org/10.3390/cancers13143616







