Detection of Tumor Recurrence via Circulating Tumor DNA Profiling in Patients with Localized Lung Cancer: Clinical Considerations and Challenges
Abstract
:Simple Summary
Abstract
1. Introduction
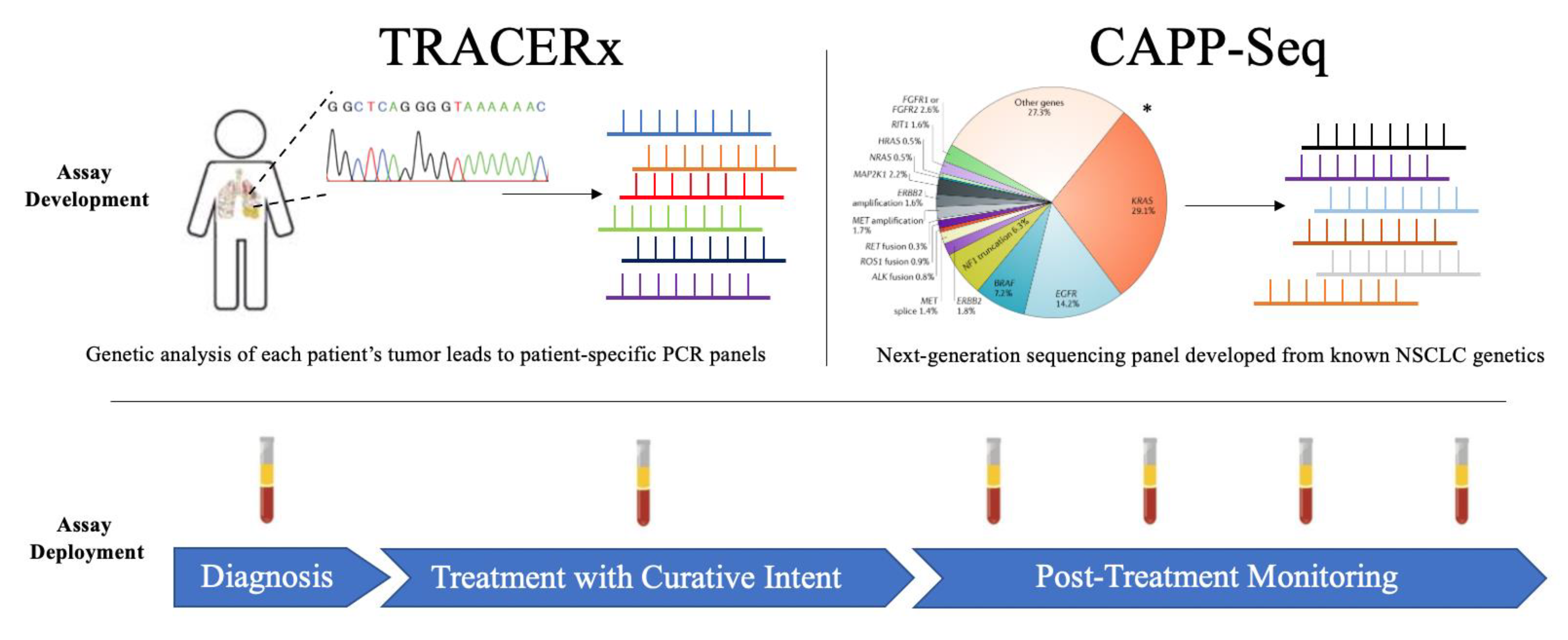
2. TRACERx
3. CAPP-Seq
4. Discussion: Future Opportunities and Challenges
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.; Chirieac, L.R.; D’Amico, T.A.; DeCamp, M.M.; Dilling, T.J.; Dobelbower, M.; et al. Non–Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2017, 15, 504–535. [Google Scholar] [CrossRef]
- Kalemkerian, G.P.; Akerley, W.; Bogner, P.; Borghaei, H.; Chow, L.Q.; Downey, R.J.; Gandhi, L.; Ganti, A.K.P.; Govindan, R.; Grecula, J.C.; et al. Small Cell Lung Cancer. J. Natl. Compr. Cancer Netw. 2013, 11, 78–98. [Google Scholar] [CrossRef] [Green Version]
- Datta, D.; Lahiri, B. Preoperative Evaluation of Patients Undergoing Lung Resection Surgery. Chest 2003, 123, 2096–2103. [Google Scholar] [CrossRef] [Green Version]
- Cagle, P.T.; Allen, T.C.; Olsen, R.J. Lung Cancer Biomarkers: Present Status and Future Developments. Arch. Pathol. Lab. Med. 2013, 137, 1191–1198. [Google Scholar] [CrossRef]
- Le Chevalier, T. Adjuvant chemotherapy for resectable non-small-cell lung cancer: Where is it going? Ann. Oncol. 2010, 21, vii196–vii198. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabieres, C. Tumor microenvironment: Informing on minimal residual disease in solid tumors. Nat. Rev. Clin. Oncol. 2017, 14, 325–326. [Google Scholar] [CrossRef]
- Burgener, J.M.; Rostami, A.; De Carvalho, D.D.; Bratman, S.V. Cell-free DNA as a post-treatment surveillance strategy: Current status. Seimn. Oncol. 2017, 44, 330–346. [Google Scholar] [CrossRef]
- Douillard, J.-Y.; Rosell, R.; De Lena, M.; Carpagnano, F.; Ramlau, R.; Larriba, J.L.G.; Grodzki, T.; Pereira, J.R.; Le Groumellec, A.; Lorusso, V.; et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Asso-ciation [ANITA]): A randomised controlled trial. Lancet Oncol. 2006, 7, 719–727. [Google Scholar] [CrossRef]
- Kenmotsu, H.; Yamamoto, N.; Yamanaka, T.; Yoshiya, K.; Takahashi, T.; Ueno, T.; Goto, K.; Daga, H.; Ikeda, N.; Sugio, K.; et al. Randomized Phase III Study of Pemetrexed Plus Cisplatin Versus Vinorelbine Plus Cisplatin for Completely Resected Stage II to IIIA Nonsquamous Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2020, 38, 2187–2196. [Google Scholar] [CrossRef]
- Zhong, W.-Z.; Wang, Q.; Mao, W.-M.; Xu, S.-T.; Wu, L.; Shen, Y.; Liu, Y.-Y.; Chen, C.; Cheng, Y.; Xu, L.; et al. Gefitinib versus vinorelbine plus cisplatin as adjuvant treatment for stage II–IIIA (N1–N2) EGFR-mutant NSCLC (ADJUVANT/CTONG1104): A randomised, open-label, phase 3 study. Lancet Oncol. 2018, 19, 139–148. [Google Scholar] [CrossRef]
- Yue, D.; Xu, S.; Wang, Q.; Li, X.; Shen, Y.; Zhao, H.; Chen, C.; Mao, W.; Liu, W.; Liu, J.; et al. Erlotinib versus vinorelbine plus cisplatin as adjuvant therapy in Chinese patients with stage IIIA EGFR mutation-positive non-small-cell lung cancer (EVAN): A randomised, open-label, phase 2 trial. Lancet Respir. Med. 2018, 6, 863–873. [Google Scholar] [CrossRef]
- Li, N.; Ou, W.; Ye, X.; Sun, H.-B.; Zhang, L.; Fang, Q.; Zhang, S.-L.; Wang, B.-X.; Wang, S.-Y. Pemetrexed-Carboplatin Adjuvant Chemotherapy With or Without Gefitinib in Resected Stage IIIA-N2 Non-Small Cell Lung Cancer Harbouring EGFR Mutations: A Randomized, Phase II Study. Ann. Surg. Oncol. 2014, 21, 2091–2096. [Google Scholar] [CrossRef] [PubMed]
- Winton, T.; Livingston, R.; Johnson, D.; Rigas, J.; Johnston, M.; Butts, C.; Cormier, Y.; Goss, G.; Inculet, R.; Vallieres, E.; et al. Vinorelbine plus Cisplatin vs. Observation in Resected Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2005, 352, 2589–2597. [Google Scholar] [CrossRef] [Green Version]
- Lou, F.; Sima, C.S.; Rusch, V.; Jones, D.R.; Huang, J. Differences in Patterns of Recurrence in Early-Stage Versus Locally Advanced Non-Small Cell Lung Cancer. Ann. Thorac. Surg. 2014, 98, 1755–1761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelsey, C.R.; Marks, L.B.; Hollis, D.; Hubbs, J.L.; Ready, N.E.; D’Amico, T.A.; Boyd, J.A. Local recurrence after surgery for early stage lung cancer: An 11-year ex-perience with 975 patients. Cancer 2009, 115, 5218–5227. [Google Scholar] [CrossRef]
- Sugimura, H.; Nichols, F.C.; Yang, P.; Allen, M.S.; Cassivi, S.D.; Deschamps, C.; Williams, B.A.; Pairolero, P.C. Survival after recurrent non small cell lung cancer after complete pul-monary resection. Ann. Thorac. Surg. 2007, 83, 409–417. [Google Scholar] [CrossRef]
- Dziedzic, D.A.; Rudzinski, P.; Langfort, R.; Orlowski, T.; Polish Lung Cancer Study Group. Risk factors for local and distant recurrence after surgical treat-ment in patients with non-small-cell lung cancer. Clin. Lung Cancer 2016, 17, e157–e167. [Google Scholar] [CrossRef]
- Hung, J.-J.; Hsu, W.-H.; Hsieh, C.-C.; Huang, B.-S.; Huang, M.-H.; Liu, J.-S.; Wu, Y.-C. Post-recurrence survival in completely resected stage I non-small cell lung cancer with local recurrence. Thorax 2009, 64, 192–196. [Google Scholar] [CrossRef] [Green Version]
- Pignon, J.-P.; Tribodet, H.; Scagliotti, G.V.; Douillard, J.-Y.; Shepherd, F.A.; Stephens, R.J.; Dunant, A.; Torri, V.; Rosell, R.; Seymour, L.; et al. Lung adjuvant cisplatin evaluation: A pooled analysis by the LACE Collaborative Group. J. Clin. Oncol. 2008, 26, 3552–3559. [Google Scholar] [CrossRef]
- Butts, C.A.; Ding, K.; Seymour, L.; Twumasi-Ankrah, P.; Graham, B.; Gandara, D.; Johnson, D.H.; Kesler, K.A.; Green, M.; Vincent, M.; et al. Randomized Phase III Trial of Vinorelbine Plus Cisplatin Compared With Observation in Completely Resected Stage IB and II Non–Small-Cell Lung Cancer: Updated Survival Analysis of JBR-10. J. Clin. Oncol. 2010, 28, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Kris, M.G.; Gaspar, L.E.; Chaft, J.E.; Kennedy, E.B.; Azzoli, C.G.; Ellis, P.M.; Lin, S.H.; Pass, H.; Seth, R.; Shepherd, F.A.; et al. Adjuvant systemic therapy and adjuvant radiation therapy for stage I to IIIA completely resected non-small-cell lung cancers: American Society of Clinical Oncology/Cancer Care On-tario clinical practice guideline update. J. Clin. Oncol. 2017, 35, 2960–2974. [Google Scholar] [CrossRef] [Green Version]
- National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology. Non-Small Cell LUNG Cancer. Available online: https://www.nccn.org/store/login/login.aspx?ReturnURL=https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf (accessed on 2 March 2021).
- Chouaid, C.; Danson, S.; Andreas, S.; Siakpere, O.; Benjamin, L.; Ehness, R.; Dramard-Goasdoue, M.-H.; Barth, J.; Hoffmann, H.; Potter, V.; et al. Adjuvant treatment patterns and outcomes in patients with stage IB-IIIA non-small cell lung cancer in France, Germany, and the United Kingdom based on the LuCaBIS burden of illness study. Lung Cancer 2018, 124, 310–316. [Google Scholar] [CrossRef] [Green Version]
- Buck, P.O.; Saverno, K.R.; Miller, P.J.; Arondekar, B.; Walker, M.S. Treatment Patterns and Health Resource Utilization Among Patients Diagnosed With Early Stage Resected Non–Small Cell Lung Cancer at US Community Oncology Practices. Clin. Lung Cancer 2015, 16, 486–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.-L.; Tsuboi, M.; He, J.; John, T.; Grohe, C.; Majem, M.; Goldman, J.W.; Laktionov, K.; Kim, S.-W.; Kato, T.; et al. Osimertinib in Resected EGFR-Mutated Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Sands, J.; Mandrekar, S.J.; Oxnard, G.R.; Kozono, D.E.; Hillman, S.L.; Dahlberg, S.E.; Sun, Z.; Chaft, J.E.; Govindan, R.; Gerber, D.E.; et al. ALCHEMIST: Adjuvant targeted therapy or immunotherapy for high-risk resected NSCLC. J. Clin. Oncol. 2020, 38, TPS9077. [Google Scholar] [CrossRef]
- Sands, J.M.; Mandrekar, S.J.; Kozono, D.; Oxnard, G.R.; Hillman, S.L.; Wigle, D.A.; Govindan, R.; Carlisle, J.; Gray, J.; Salama, J.K.; et al. Integration of immunotherapy into adjuvant therapy for resected non-small-cell lung cancer: ALCHEMIST chemo-IO (ACCIO). Immunotherapy 2021, 13, 727–734. [Google Scholar] [CrossRef]
- The National Lung Screening Trial Research Team. Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [CrossRef] [Green Version]
- De Koning, H.J.; van der Aalst, C.M.; de Jong, P.A. Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N. Engl. J. Med. 2020, 382, 503–513. [Google Scholar] [CrossRef]
- Jonas, D.E.; Reuland, D.S.; Reddy, S.M.; Nagle, M.; Clark, S.D.; Weber, R.P.; Enyioha, C.; Malo, T.L.; Brenner, A.T.; Armstrong, C.; et al. Screening for Lung Cancer With Low-Dose Computed Tomography. JAMA 2021, 325, 971–987. [Google Scholar] [CrossRef]
- Ulrich, B.C.; Paweletz, C.P. Cell-Free DNA in Oncology: Gearing up for Clinic. Ann. Lab. Med. 2018, 38, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Ulrich, B.C.; Guibert, N. Toward a Comprehensive Framework for cell-free DNA analysis: Lessons from TRAC-ERx. Ann. Transl. Med. 2017, 5, 428. [Google Scholar] [CrossRef]
- Durin, L.; Pradines, A.; Basset, C.; Ulrich, B.; Keller, L.; Dongay, V.; Favre, G.; Mazieres, J.; Guibert, N. Liquid Biopsy of Non-Plasma Body Fluids in Non-Small Cell Lung Cancer: Look Closer to the Tumor! Cells 2020, 9, 2486. [Google Scholar] [CrossRef]
- Corcoran, R.B.; Chabner, B.A. Application of Cell-free DNA Analysis to Cancer Treatment. N. Engl. J. Med. 2018, 379, 1754–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Said, R.; Guibert, N.; Oxnard, G.R.; Tsimberidou, A.M. Circulating tumor DNA analysis in the era of precision oncology. Oncotarget 2020, 11, 188–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aggarwal, C.; Rolfo, C.D.; Oxnard, G.R.; Gray, J.E.; Sholl, L.M.; Gandara, D.R. Strategies for the successful implementation of plasma-based NSCLC genotyping in clinical practice. Nat. Rev. Clin. Oncol. 2021, 18, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar]
- Stroun, M.; Maurice, P.; Vasioukhin, V.; Lyautey, J.; Lederrey, C.; Lefort, F.; Rossier, A.; Chen, X.Q.; Anker, P. The origin and mechanism of circu-lating DNA. Ann. N. Y. Acad. Sci. 2000, 906, 161–168. [Google Scholar] [CrossRef]
- Stroun, M.; Lyautey, J.; Lederrey, C.; Olson-Sand, A.; Anker, P. About the possible origin and mechanism of circulating DNA. Clin. Chim. Acta 2001, 313, 139–142. [Google Scholar] [CrossRef]
- Liu, M.; Oxnard, G.; Klein, E.; Swanton, C.; Seiden, M.; Smith, D.; Richards, D.; Yeatman, T.J.; Cohn, A.L.; Lapham, R.; et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [Green Version]
- Diaz, L.A., Jr.; Williams, R.T.; Wu, J.; Kinde, I.; Hecht, J.R.; Berlin, J.; Allen, B.; Bozic, I.; Reiter, J.; Nowak, M.A.; et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nat. Cell Biol. 2012, 486, 537–540. [Google Scholar] [CrossRef] [Green Version]
- Oxnard, G.R.; Thress, K.S.; Alden, R.S.; Lawrance, R.; Paweletz, C.P.; Cantarini, M.; Yang, J.C.-H.; Barrett, J.C.; Jänne, P.A. Association between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2016, 34, 3375–3382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guibert, N.; Mazieres, J.; Delaunay, M.; Casanova, A.; Farella, M.; Keller, L.; Favre, G.; Pradines, A. Monitoring of KRAS-mutated ctDNA to discriminate pseudo-progression from true progression during anti-PD-1 treatment of lung adenocarcinoma. Oncotarget 2017, 8, 38056–38060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, A.; Bratman, S.; To, J.; Wynne, J.F.; Eclov, N.C.W.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Tie, J.; Wang, Y.; Tomasetti, C.; Li, L.; Springer, S.; Kinde, I.; Silliman, N.; Tacey, M.; Wong, H.-L.; Christie, M.; et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016, 8, 346ra92. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Murillas, I.; Schiavon, G.; Weigelt, B.; Ng, C.K.Y.; Hrebien, S.; Cutts, R.J.; Cheang, M.; Osin, P.; Nerurkar, A.; Kozarewa, I.; et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 2015, 7, 302ra133. [Google Scholar] [CrossRef]
- Beaver, J.A.; Jelovac, D.; Balukrishna, S.; Cochran, R.L.; Croessmann, S.; Zabransky, D.J.; Wong, H.Y.; Toro, P.V.; Cidado, J.; Blair, B.G.; et al. Detection of Cancer DNA in Plasma of Patients with Early-Stage Breast Cancer. Clin. Cancer Res. 2014, 20, 2643–2650. [Google Scholar] [CrossRef] [Green Version]
- Olsson, E.; Winter, C.; George, A.; Chen, Y.; Howlin, J.; Tang, M.-H.E.; Dahlgren, M.; Schulz, R.; Grabau, D.; van Westen, D.; et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol. Med. 2015, 7, 1034–1047. [Google Scholar] [CrossRef]
- Reinert, T.; Schøler, L.V.; Thomsen, R.; Tobiasen, H.; Vang, S.; Nordentoft, I.K.; Lamy, P.-J.; Kannerup, A.-S.; Mortensen, F.V.; Stribolt, K.; et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016, 65, 625–634. [Google Scholar] [CrossRef] [Green Version]
- Jamal-Hanjani, M.; Hackshaw, A.; Ngai, Y.; Shaw, J.; Dive, C.; Quezada, S.; Middleton, G.; De Bruin, E.; Le Quesne, J.; Shafi, S.; et al. Tracking Genomic Cancer Evolution for Precision Medicine: The Lung TRACERx Study. PLoS Biol. 2014, 12, e1001906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamal-Hanjani, M. TRACERx—Tracking Non-Small Cell Lung Cancer Evolution. N. Engl. J. Med. 2017, 376, 2109–2121. [Google Scholar] [CrossRef] [Green Version]
- Jamal-Hanjani, M.; Wilson, G.A.; Horswell, S.; Mitter, R.; Sakarya, O.; Constantin, T.; Salari, R.; Kirkizlar, E.; Sigurjonsson, S.; Pelham, R.; et al. Detection of ubiquitous and heterogeneous mutations in cell-free DNA from patients with early-stage non-small-cell lung cancer. Ann. Oncol. 2016, 27, 862–867. [Google Scholar] [CrossRef]
- Abbosh, C.; The TRACERx Consortium; Birkbak, N.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nat. Cell Biol. 2017, 545, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.; Lovejoy, A.F.; Klass, D.M.; Kurtz, D.; Chabon, J.J.; Scherer, F.; Stehr, H.; Liu, C.L.; Bratman, S.; Say, C.; et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 2016, 34, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.A.; Chabon, J.J.; Lovejoy, A.F.; Newman, A.; Stehr, H.; Azad, T.; Khodadoust, M.S.; Esfahani, M.S.; Liu, C.L.; Zhou, L.; et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017, 7, 1394–1403. [Google Scholar] [CrossRef] [Green Version]
- Skoulidis, F.; Heymach, J.V. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer 2019, 19, 495–509. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012, 489, 519–525. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network; Cancer Genome Atlas Research Network; Li, M.O. Author Correction: Comprehensive molecular profiling of lung adenocarcinoma. Nat. Cell Biol. 2018, 559, E12. [Google Scholar] [CrossRef] [Green Version]
- George, J.; Lim, J.S.; Jang, S.J.; Cun, Y.; Ozretić, L.; Kong, G.; Leenders, F.; Lu, X.; Fernandez-Cuesta, L.; Bosco, G.; et al. Comprehensive genomic profiles of small cell lung cancer. Nat. Cell Biol. 2015, 524, 47–53. [Google Scholar] [CrossRef]
- Kocak, Z.; Evans, E.S.; Zhou, S.-M.; Miller, K.L.; Folz, R.J.; Shafman, T.D.; Marks, L.B. Challenges in defining radiation pneumonitis in patients with lung cancer. Int. J. Radiat. Oncol. 2005, 62, 635–638. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Dahele, M.; Senan, S.; Guckenberger, M.; Rodrigues, G.B.; Ward, A.; Boldt, R.G.; Palma, D.A. Radiographic changes after lung stereotactic ablative radiotherapy (SABR)—Can we distinguish recurrence from fibrosis? A systematic review of the literature. Radiother. Oncol. 2012, 102, 335–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larici, A.R.; del Ciello, A.; Maggi, F.; Santoro, S.I.; Meduri, B.; Valentini, V.; Giordano, A.; Bonomo, L. Lung Abnormalities at Multimodality Imaging after Radiation Therapy for Non–Small Cell Lung Cancer. Radiographic 2011, 31, 771–789. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ulrich, B.; Pradines, A.; Mazières, J.; Guibert, N. Detection of Tumor Recurrence via Circulating Tumor DNA Profiling in Patients with Localized Lung Cancer: Clinical Considerations and Challenges. Cancers 2021, 13, 3759. https://doi.org/10.3390/cancers13153759
Ulrich B, Pradines A, Mazières J, Guibert N. Detection of Tumor Recurrence via Circulating Tumor DNA Profiling in Patients with Localized Lung Cancer: Clinical Considerations and Challenges. Cancers. 2021; 13(15):3759. https://doi.org/10.3390/cancers13153759
Chicago/Turabian StyleUlrich, Bryan, Anne Pradines, Julien Mazières, and Nicolas Guibert. 2021. "Detection of Tumor Recurrence via Circulating Tumor DNA Profiling in Patients with Localized Lung Cancer: Clinical Considerations and Challenges" Cancers 13, no. 15: 3759. https://doi.org/10.3390/cancers13153759
APA StyleUlrich, B., Pradines, A., Mazières, J., & Guibert, N. (2021). Detection of Tumor Recurrence via Circulating Tumor DNA Profiling in Patients with Localized Lung Cancer: Clinical Considerations and Challenges. Cancers, 13(15), 3759. https://doi.org/10.3390/cancers13153759





