Identification and Characterization of Multiple Myeloma Stem Cell-Like Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Markers of MMSCs and Drugs Targeting Them
2.1. Side Population Cell
2.2. ALDH
2.3. CD138
2.4. CD24
2.5. iPS/ES Genes
2.6. BTK
2.7. RARα2
2.8. ROS
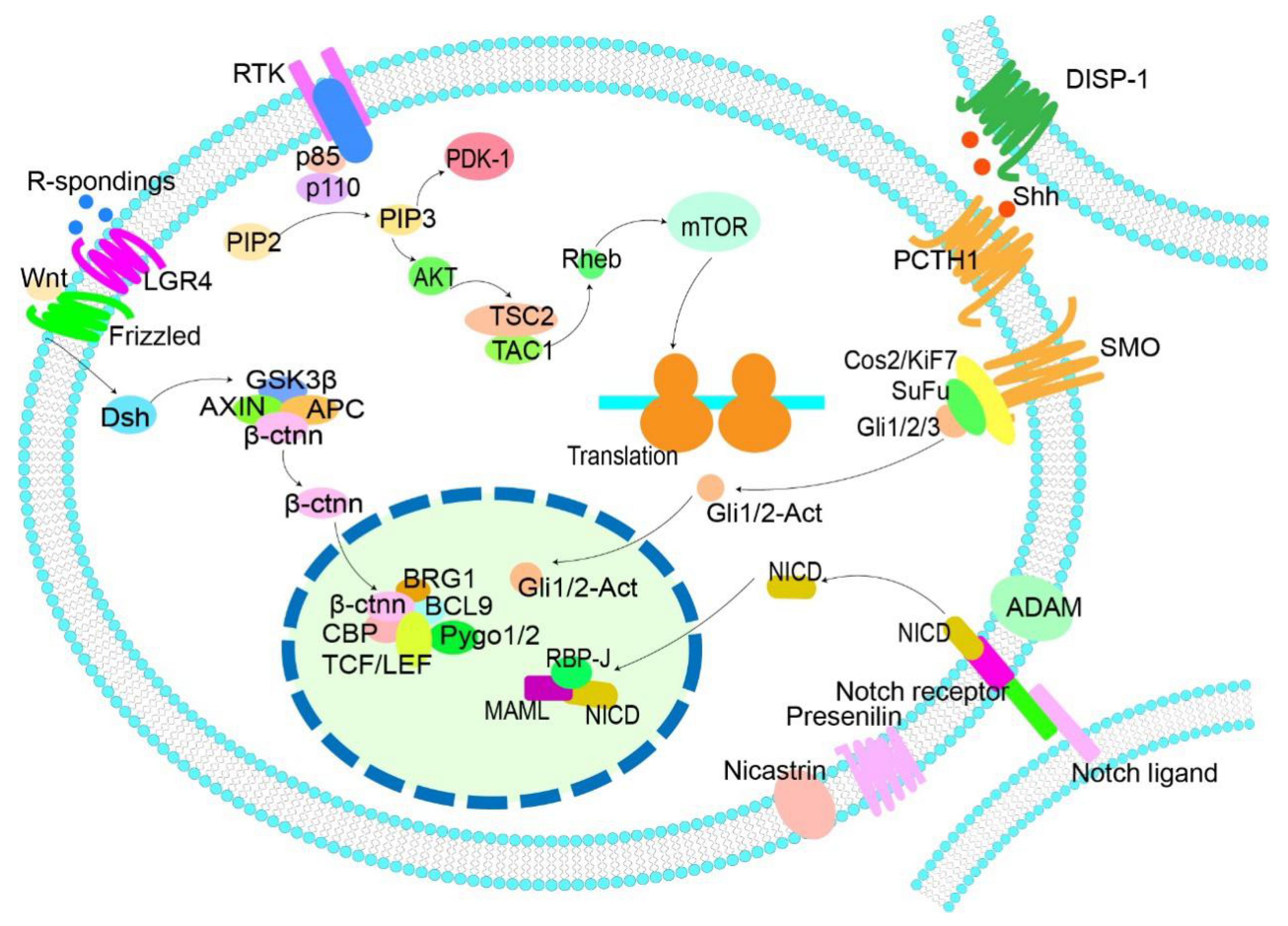
3. Active Signaling Pathways Related to MMSCs and Drugs Targeting Them
3.1. Wnt/β-Catenin
3.2. Hedgehog
3.3. Notch
3.4. PI3K/Akt
4. Epigenetic Regulation of MM Stem Pathway
5. Dispute on MM Stem Cells
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Palumbo, A.; Anderson, K. Multiple myeloma. N. Engl. J. Med. 2011, 364, 1046–1060. [Google Scholar] [CrossRef]
- Kumar, S.K.; Rajkumar, V.; Kyle, R.A.; van Duin, M.; Sonneveld, P.; Mateos, M.V.; Gay, F.; Anderson, K.C. Multiple myeloma. Nat. Rev. Dis. Primers 2017, 3, 17046. [Google Scholar] [CrossRef]
- Li, Y.; Liu, J.; Tang, L.J.; Shi, Y.W.; Ren, W.; Hu, W.X. Apoptosis induced by lycorine in km3 cells is associated with the g0/g1 cell cycle arrest. Oncol. Rep. 2007, 17, 377–384. [Google Scholar] [CrossRef][Green Version]
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.Y.; et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the global burden of disease study 2010. Lancet 2012, 380, 2095–2128. [Google Scholar] [CrossRef]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef] [PubMed]
- Dalerba, P.; Cho, R.W.; Clarke, M.F. Cancer stem cells: Models and concepts. Annu. Rev. Med. 2007, 58, 267–284. [Google Scholar] [CrossRef] [PubMed]
- Huff, C.A.; Matsui, W. Multiple myeloma cancer stem cells. J. Clin. Oncol. 2008, 26, 2895–2900. [Google Scholar] [CrossRef] [PubMed]
- Boutin, L.; Arnautou, P.; Trignol, A.; Segot, A.; Farge, T.; Desterke, C.; Soave, S.; Clay, D.; Raffoux, E.; Sarry, J.E.; et al. Mesenchymal stromal cells confer chemoresistance to myeloid leukemia blasts through side population functionality and abc transporter activation. Haematologica 2020, 105, 987–9998. [Google Scholar] [CrossRef]
- Baeten, J.T.; Waarts, M.R.; Pruitt, M.M.; Chan, W.C.; Andrade, J.; de Jong, J. The side population enriches for leukemia-propagating cell activity and wnt pathway expression in zebrafish acute lymphoblastic leukemia. Haematologica 2019, 104, 1388–1395. [Google Scholar] [CrossRef] [PubMed]
- Grimm, M.; Krimmel, M.; Polligkeit, J.; Alexander, D.; Munz, A.; Kluba, S.; Keutel, C.; Hoffmann, J.; Reinert, S.; Hoefert, S. Abcb5 expression and cancer stem cell hypothesis in oral squamous cell carcinoma. Eur. J. Cancer 2012, 48, 3186–3197. [Google Scholar] [CrossRef] [PubMed]
- Moghbeli, M.; Mosannen, M.H.; Memar, B.; Forghanifard, M.M.; Gholamin, M.; Abbaszadegan, M.R. Role of maml1 in targeted therapy against the esophageal cancer stem cells. J. Transl. Med. 2019, 17, 126. [Google Scholar] [CrossRef] [PubMed]
- Ikegame, A.; Ozaki, S.; Tsuji, D.; Harada, T.; Fujii, S.; Nakamura, S.; Miki, H.; Nakano, A.; Kagawa, K.; Takeuchi, K.; et al. Small molecule antibody targeting hla class i inhibits myeloma cancer stem cells by repressing pluripotency-associated transcription factors. Leukemia 2012, 26, 2124–2134. [Google Scholar] [CrossRef]
- Du, J.; Liu, S.; He, J.; Liu, X.; Qu, Y.; Yan, W.; Fan, J.; Li, R.; Xi, H.; Fu, W.; et al. Microrna-451 regulates stemness of side population cells via pi3k/akt/mtor signaling pathway in multiple myeloma. Oncotarget 2015, 6, 14993–15007. [Google Scholar] [CrossRef] [PubMed]
- Jakubikova, J.; Adamia, S.; Kost-Alimova, M.; Klippel, S.; Cervi, D.; Daley, J.F.; Cholujova, D.; Kong, S.Y.; Leiba, M.; Blotta, S.; et al. Lenalidomide targets clonogenic side population in multiple myeloma: Pathophysiologic and clinical implications. Blood 2011, 117, 4409–4419. [Google Scholar] [CrossRef] [PubMed]
- Nara, M.; Teshima, K.; Watanabe, A.; Ito, M.; Iwamoto, K.; Kitabayashi, A.; Kume, M.; Hatano, Y.; Takahashi, N.; Iida, S.; et al. Bortezomib reduces the tumorigenicity of multiple myeloma via downregulation of upregulated targets in clonogenic side population cells. PLoS ONE 2013, 8, e56954. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Tao, W.; Kuiatse, I.; Lin, P.; Feng, Y.; Jones, R.J.; Orlowski, R.Z.; Zu, Y. Dynamic balance of multiple myeloma clonogenic side population cell percentages controlled by environmental conditions. Int. J. Cancer 2015, 136, 991–1002. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Shi, J.; Gu, Z.; Salama, M.E.; Das, S.; Wendlandt, E.; Xu, H.; Huang, J.; Tao, Y.; Hao, M.; et al. Bruton tyrosine kinase is a therapeutic target in stem-like cells from multiple myeloma. Cancer Res. 2015, 75, 594–604. [Google Scholar] [CrossRef]
- Yang, Y.; Shi, J.; Tolomelli, G.; Xu, H.; Xia, J.; Wang, H.; Zhou, W.; Zhou, Y.; Das, S.; Gu, Z.; et al. Raralpha2 expression confers myeloma stem cell features. Blood 2013, 122, 1437–1447. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Wen, J.; Mike, P.; Choi, D.S.; Eshoa, C.; Shi, Z.Z.; Zu, Y.; Chang, C.C. Bone marrow stromal cells from myeloma patients support the growth of myeloma stem cells. Stem Cells Dev. 2010, 19, 1289–1296. [Google Scholar] [CrossRef]
- Ai, L.; Mu, S.; Sun, C.; Fan, F.; Yan, H.; Qin, Y.; Cui, G.; Wang, Y.; Guo, T.; Mei, H.; et al. Myeloid-derived suppressor cells endow stem-like qualities to multiple myeloma cells by inducing pirna-823 expression and dnmt3b activation. Mol. Cancer 2019, 18, 88. [Google Scholar] [CrossRef]
- Mo, S.L.; Li, J.; Loh, Y.S.; Brown, R.D.; Smith, A.L.; Chen, Y.; Joshua, D.; Roufogalis, B.D.; Li, G.Q.; Fan, K.; et al. Factors influencing the abundance of the side population in a human myeloma cell line. Bone Marrow Res. 2011, 2011, 524845. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Bai, H.; Jethava, Y.; Wu, Y.; Zhu, Y.; Yang, Y.; Xia, J.; Cao, H.; Franqui-Machin, R.; Nadiminti, K.; et al. Identification and characterization of tumor-initiating cells in multiple myeloma. J. Natl. Cancer Inst. 2020, 112, 507–515. [Google Scholar] [CrossRef]
- Yang, Q.; Li, K.; Li, X.; Liu, J. Identification of key genes and pathways in myeloma side population cells by bioinformatics analysis. Int. J. Med. Sci. 2020, 17, 2063–2076. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Chen, B. A 5-gene stemness score for rapid determination of risk in multiple myeloma. Onco Targets Ther. 2020, 13, 4339–4348. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Li, H.; Tao, W.; Savoldo, B.; Foglesong, J.A.; King, L.C.; Zu, Y.; Chang, C.C. High throughput quantitative reverse transcription pcr assays revealing over-expression of cancer testis antigen genes in multiple myeloma stem cell-like side population cells. Br. J. Haematol. 2014, 166, 711–719. [Google Scholar] [CrossRef]
- Loh, Y.S.; Mo, S.; Brown, R.D.; Yamagishi, T.; Yang, S.; Joshua, D.E.; Roufogalis, B.D.; Sze, D.M. Presence of hoechst low side populations in multiple myeloma. Leuk. Lymphoma 2008, 49, 1813–1816. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, M.; Fujita, S.; Katsumoto, T.; Yamagata, K.; Ogawara, Y.; Hattori, A.; Kagiyama, Y.; Honma, D.; Araki, K.; Inoue, T.; et al. Dual inhibition of enhancer of zeste homolog 1/2 overactivateswnt signaling to deplete cancer stem cells in multiple myeloma. Cancer Sci. 2019, 110, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Liu, M.; Pan, J. Blocking ezh2 methylation transferase activity by gsk126 decreases stem cell-like myeloma cells. Oncotarget 2017, 8, 3396–3411. [Google Scholar] [CrossRef]
- Yan, W.; Du, J.; Du, Y.; Pu, H.; Liu, S.; He, J.; Zhang, J.; Hou, J. Fenretinide targets the side population in myeloma cell line nci-h929 and potentiates the efficacy of antimyeloma with bortezomib and dexamethasone regimen. Leuk. Res. 2016, 51, 32–40. [Google Scholar] [CrossRef]
- Ray, A.; Das, D.S.; Song, Y.; Macri, V.; Richardson, P.; Brooks, C.L.; Chauhan, D.; Anderson, K.C. A novel agent sl-401 induces anti-myeloma activity by targeting plasmacytoid dendritic cells, osteoclastogenesis and cancer stem-like cells. Leukemia 2017, 31, 2652–2660. [Google Scholar] [CrossRef] [PubMed]
- Nishida, H.; Hayashi, M.; Morimoto, C.; Sakamoto, M.; Yamada, T. Cd26 is a potential therapeutic target by humanized monoclonal antibody for the treatment of multiple myeloma. Blood Cancer J. 2018, 8, 99. [Google Scholar] [CrossRef]
- Shi, F.; Li, M.; Wang, J.; Wu, D.; Pan, M.; Guo, M.; Dou, J. Induction of multiple myeloma cancer stem cell apoptosis using conjugated anti-abcg2 antibody with epirubicin-loaded microbubbles. Stem Cell Res. Ther. 2018, 9, 144. [Google Scholar] [CrossRef] [PubMed]
- Spradlin, J.N.; Hu, X.; Ward, C.C.; Brittain, S.M.; Jones, M.D.; Ou, L.; To, M.; Proudfoot, A.; Ornelas, E.; Woldegiorgis, M.; et al. Harnessing the anti-cancer natural product nimbolide for targeted protein degradation. Nat. Chem. Biol. 2019, 15, 747–755. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Gao, F.; Zhao, Q.; Zuo, H.; Liu, W.; Li, W. Tanshinoneiia inhibits oral squamous cell carcinoma via reducing akt-c-myc signaling-mediated aerobic glycolysis. Cell Death Dis. 2020, 11, 381. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Fu, Y.; Zheng, Y.; Wang, X.; Liu, B.; Zeng, J. Diallyl thiosulfinate enhanced the anti-cancer activity of dexamethasone in the side population cells of multiple myeloma by promoting mir-127-3p and deactivating the pi3k/akt signaling pathway. BMC Cancer 2021, 21, 125. [Google Scholar] [CrossRef]
- Gu, Y.Y.; Liu, L.P.; Qin, J.; Zhang, M.; Chen, Y.; Wang, D.; Li, Z.; Tang, J.Z.; Mo, S.L. Baicalein decreases side population proportion via inhibition of abcg2 in multiple myeloma cell line rpmi 8226 in vitro. Fitoterapia 2014, 94, 21–28. [Google Scholar] [CrossRef]
- Lin, M.G.; Liu, L.P.; Li, C.Y.; Zhang, M.; Chen, Y.; Qin, J.; Gu, Y.Y.; Li, Z.; Wu, X.L.; Mo, S.L. Scutellaria extract decreases the proportion of side population cells in a myeloma cell line by down-regulating the expression of abcg2 protein. Asian Pac. J. Cancer Prev. 2013, 14, 7179–7186. [Google Scholar] [CrossRef]
- Xu, X.; Chai, S.; Wang, P.; Zhang, C.; Yang, Y.; Yang, Y.; Wang, K. Aldehyde dehydrogenases and cancer stem cells. Cancer Lett. 2015, 369, 50–57. [Google Scholar] [CrossRef]
- Toledo-Guzmán, M.E.; Hernández, M.I.; Gómez-Gallegos, Á.A.; Ortiz-Sánchez, E. ALDH as a Stem Cell Marker in Solid Tumors. Curr. Stem Cell Res. Ther. 2019, 14, 375–388. [Google Scholar] [CrossRef]
- Zhou, W.; Yang, Y.; Gu, Z.; Wang, H.; Xia, J.; Wu, X.; Zhan, X.; Levasseur, D.; Zhou, Y.; Janz, S.; et al. Aldh1 activity identifies tumor-initiating cells and links to chromosomal instability signatures in multiple myeloma. Leukemia 2014, 28, 1155–1158. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, W.; Xia, J.; Gu, Z.; Wendlandt, E.; Zhan, X.; Janz, S.; Tricot, G.; Zhan, F. Nek2 mediates aldh1a1-dependent drug resistance in multiple myeloma. Oncotarget 2014, 5, 11986–11997. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Gong, Y.; Liang, L.; Xiao, L.; Yi, H.; Ye, M.; Roy, M.; Xia, J.; Zhou, W.; Yang, C.; et al. Lycorine targets multiple myeloma stem cell-like cells by inhibition of wnt/beta-catenin pathway. Br. J. Haematol. 2020, 89, 1151–1164. [Google Scholar] [CrossRef]
- Paino, T.; Ocio, E.M.; Paiva, B.; San-Segundo, L.; Garayoa, M.; Gutierrez, N.C.; Sarasquete, M.E.; Pandiella, A.; Orfao, A.; San, M.J. Cd20 positive cells are undetectable in the majority of multiple myeloma cell lines and are not associated with a cancer stem cell phenotype. Haematologica 2012, 97, 1110–1114. [Google Scholar] [CrossRef]
- Morgenroth, A.; Vogg, A.T.; Zlatopolskiy, B.D.; Siluschek, M.; Oedekoven, C.; Mottaghy, F.M. Breaking the invulnerability of cancer stem cells: Two-step strategy to kill the stem-like cell subpopulation of multiple myeloma. Mol. Cancer Ther. 2014, 13, 144–153. [Google Scholar] [CrossRef]
- Dinavahi, S.S.; Gowda, R.; Bazewicz, C.G.; Battu, M.B.; Lin, J.M.; Chitren, R.J.; Pandey, M.K.; Amin, S.; Robertson, G.P.; Gowda, K. Design, synthesis characterization and biological evaluation of novel multi-isoform ALDH inhibitors as potential anticancer agents. Eur. J. Med. Chem. 2020, 187, 111962. [Google Scholar] [CrossRef] [PubMed]
- Al-Dhfyan, A.; Alhoshani, A.; Korashy, H.M. Aryl hydrocarbon receptor/cytochrome p450 1a1 pathway mediates breast cancer stem cells expansion through pten inhibition and beta-catenin and akt activation. Mol. Cancer 2017, 16, 14. [Google Scholar] [CrossRef]
- Elcheva, I.A.; Wood, T.; Chiarolanzio, K.; Chim, B.; Wong, M.; Singh, V.; Gowda, C.P.; Lu, Q.; Hafner, M.; Dovat, S.; et al. Rna-binding protein igf2bp1 maintains leukemia stem cell properties by regulating hoxb4, myb, and aldh1a1. Leukemia 2020, 34, 1354–1363. [Google Scholar] [CrossRef] [PubMed]
- Annageldiyev, C.; Gowda, K.; Patel, T.; Bhattacharya, P.; Tan, S.F.; Iyer, S.; Desai, D.; Dovat, S.; Feith, D.J.; Loughran, T.J.; et al. The novel isatin analog ks99 targets stemness markers in acute myeloid leukemia. Haematologica 2020, 105, 687–696. [Google Scholar] [CrossRef]
- Jin, N.; Zhu, X.; Cheng, F.; Zhang, L. Disulfiram/copper targets stem cell-like ALDH1+ population of multiple myeloma by inhibition of aldh1a1 and hedgehog pathway. J. Cell. Biochem. 2018, 119, 6882–6893. [Google Scholar] [CrossRef]
- Luna, J.I.; Grossenbacher, S.K.; Sturgill, I.R.; Ames, E.; Judge, S.J.; Bouzid, L.A.; Darrow, M.A.; Murphy, W.J.; Canter, R.J. Bortezomib augments natural killer cell targeting of stem-like tumor cells. Cancers 2019, 11, 85. [Google Scholar] [CrossRef]
- Chilosi, M.; Adami, F.; Lestani, M.; Montagna, L.; Cimarosto, L.; Semenzato, G.; Pizzolo, G.; Menestrina, F. Cd138/syndecan-1: A useful immunohistochemical marker of normal and neoplastic plasma cells on routine trephine bone marrow biopsies. Mod. Pathol. 1999, 12, 1101–1106. [Google Scholar]
- Calame, K.L. Plasma cells: Finding new light at the end of B cell development. Nat. Immunol. 2001, 2, 1103–1108. [Google Scholar] [CrossRef]
- Wijdenes, J.; Vooijs, W.C.; Clement, C.; Post, J.; Morard, F.; Vita, N.; Laurent, P.; Sun, R.X.; Klein, B.; Dore, J.M. A plasmocyte selective monoclonal antibody (b-b4) recognizes syndecan-1. Br. J. Haematol. 1996, 94, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, M.M.; Fojo, T.; Bates, S.E. Multidrug resistance in cancer: Role of atp-dependent transporters. Nat. Rev. Cancer 2002, 2, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Matsui, W.; Wang, Q.; Barber, J.P.; Brennan, S.; Smith, B.D.; Borrello, I.; Mcniece, I.; Lin, L.; Ambinder, R.F.; Peacock, C.; et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res. 2008, 68, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Matsui, W.; Huff, C.A.; Wang, Q.; Malehorn, M.T.; Barber, J.; Tanhehco, Y.; Smith, B.D.; Civin, C.I.; Jones, R.J. Characterization of clonogenic multiple myeloma cells. Blood 2004, 103, 2332–2336. [Google Scholar] [CrossRef] [PubMed]
- Atanackovic, D.; Panse, J.; Hildebrandt, Y.; Jadczak, A.; Kobold, S.; Cao, Y.; Templin, J.; Meyer, S.; Reinhard, H.; Bartels, K.; et al. Surface molecule cd229 as a novel target for the diagnosis and treatment of multiple myeloma. Haematologica 2011, 96, 1512–1520. [Google Scholar] [CrossRef]
- Reghunathan, R.; Bi, C.; Liu, S.C.; Loong, K.T.; Chung, T.H.; Huang, G.; Chng, W.J. Clonogenic multiple myeloma cells have shared stemness signature associated with patient survival. Oncotarget 2013, 4, 1230–1240. [Google Scholar] [CrossRef]
- Chen, R.; Zhao, H.; Wu, D.; Zhao, C.; Zhao, W.; Zhou, X. The role of sh3gl3 in myeloma cell migration/invasion, stemness and chemo-resistance. Oncotarget 2016, 7, 73101–73113. [Google Scholar] [CrossRef]
- Mondala, P.K.; Vora, A.A.; Zhou, T.; Lazzari, E.; Ladel, L.; Luo, X.; Kim, Y.; Costello, C.; Macleod, A.R.; Jamieson, C.; et al. Selective antisense oligonucleotide inhibition of human irf4 prevents malignant myeloma regeneration via cell cycle disruption. Cell Stem Cell 2021, 28, 623–636. [Google Scholar] [CrossRef] [PubMed]
- Tarhriz, V.; Bandehpour, M.; Dastmalchi, S.; Ouladsahebmadarek, E.; Zarredar, H.; Eyvazi, S. Overview of cd24 as a new molecular marker in ovarian cancer. J. Cell. Physiol. 2019, 234, 2134–2142. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, Y.; Tsung, A.; Huang, H.; Du, Q.; Yang, M.; Deng, M.; Xiong, S.; Wang, X.; Zhang, L.; et al. Inos promotes cd24(+) cd133(+) liver cancer stem cell phenotype through a tace/adam17-dependent notch signaling pathway. Proc. Natl. Acad. Sci. USA 2018, 115, E10127–E10136. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Qiu, T.; Jiang, D.; Xu, H.; Zou, L.; Yang, Q.; Chen, C.; Jiao, B. Sgce promotes breast cancer stem cells by stabilizing egfr. Adv. Sci. 2020, 7, 1903700. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, T.; Lengerke, C. Sox2 protein biochemistry in stemness, reprogramming, and cancer: The pi3k/akt/sox2 axis and beyond. Oncogene 2020, 39, 278–292. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Kang, L.; Zhang, H.; Huang, Y.; Fang, L.; Li, M.; Brown, P.J.; Arrowsmith, C.H.; Li, J.; Wong, J. Akt drives sox2 overexpression and cancer cell stemness in esophageal cancer by protecting sox2 from ubr5-mediated degradation. Oncogene 2019, 38, 5250–5264. [Google Scholar] [CrossRef] [PubMed]
- Bak, M.J.; Furmanski, P.; Shan, N.L.; Lee, H.J.; Bao, C.; Lin, Y.; Shih, W.J.; Yang, C.S.; Suh, N. Tocopherols inhibit estrogen-induced cancer stemness and oct4 signaling in breast cancer. Carcinogenesis 2018, 39, 1045–1055. [Google Scholar] [CrossRef]
- Song, B.; Kim, D.K.; Shin, J.; Bae, S.H.; Kim, H.Y.; Won, B.; Kim, J.K.; Youn, H.D.; Kim, S.T.; Kang, S.W.; et al. Oct4 directly regulates stemness and extracellular matrix-related genes in human germ cell tumours. Biochem. Biophys. Res. Commun. 2018, 503, 1980–1986. [Google Scholar] [CrossRef]
- Takahashi, T.; Honma, Y.; Miyake, T.; Adachi, K.; Takami, S.; Okada, M.; Kumanomidou, S.; Ikejiri, F.; Jo, Y.; Onishi, C.; et al. Synergistic combination therapy with cotylenin a and vincristine in multiple myeloma models. Int. J. Oncol. 2015, 46, 1801–1809. [Google Scholar] [CrossRef]
- Zhao, P.; Chen, Y.; Yue, Z.; Yuan, Y.; Wang, X. Bone marrow mesenchymal stem cells regulate stemness of multiple myeloma cell lines via btk signaling pathway. Leuk. Res. 2017, 57, 20–26. [Google Scholar] [CrossRef]
- Wang, S.; Tricot, G.; Shi, L.; Xiong, W.; Zeng, Z.; Xu, H.; Zangari, M.; Barlogie, B.; Shaughnessy, J.J.; Zhan, F. Raralpha2 expression is associated with disease progression and plays a crucial role in efficacy of atra treatment in myeloma. Blood 2009, 114, 600–607. [Google Scholar] [CrossRef]
- Pei, S.; Minhajuddin, M.; Adane, B.; Khan, N.; Stevens, B.M.; Mack, S.C.; Lai, S.; Rich, J.N.; Inguva, A.; Shannon, K.M.; et al. Ampk/fis1-mediated mitophagy is required for self-renewal of human aml stem cells. Cell Stem Cell 2018, 23, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Lagadinou, E.D.; Sach, A.; Callahan, K.; Rossi, R.M.; Neering, S.J.; Minhajuddin, M.; Ashton, J.M.; Pei, S.; Grose, V.; O’Dwyer, K.M.; et al. Bcl-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 2013, 12, 329–341. [Google Scholar] [CrossRef]
- Jones, C.L.; Stevens, B.M.; D’Alessandro, A.; Reisz, J.A.; Culp-Hill, R.; Nemkov, T.; Pei, S.; Khan, N.; Adane, B.; Ye, H.; et al. Inhibition of amino acid metabolism selectively targets human leukemia stem cells. Cancer Cell 2018, 34, 724–740. [Google Scholar] [CrossRef] [PubMed]
- Das, D.S.; Das, A.; Ray, A.; Song, Y.; Samur, M.K.; Munshi, N.C.; Chauhan, D.; Anderson, K.C. Blockade of deubiquitylating enzyme usp1 inhibits dna repair and triggers apoptosis in multiple myeloma cells. Clin. Cancer Res. 2017, 23, 4280–4289. [Google Scholar] [CrossRef]
- Liang, L.; He, Y.; Wang, H.; Zhou, H.; Xiao, L.; Ye, M.; Kuang, Y.; Luo, S.; Zuo, Y.; Feng, P.; et al. The wee1 kinase inhibitor mk 1775 suppresses cell growth, attenuates stemness and synergises with bortezomib in multiple myeloma. Br. J. Haematol. 2020, 191, 62–76. [Google Scholar] [CrossRef]
- Yan, Y.; Zuo, X.; Wei, D. Concise review: Emerging role of cd44 in cancer stem cells: A promising biomarker and therapeutic target. Stem Cells Transl. Med. 2015, 4, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Katz, B.Z. Adhesion molecules--the lifelines of multiple myeloma cells. Semin. Cancer Biol. 2010, 20, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Bjorklund, C.C.; Baladandayuthapani, V.; Lin, H.Y.; Jones, R.J.; Kuiatse, I.; Wang, H.; Yang, J.; Shah, J.J.; Thomas, S.K.; Wang, M.; et al. Evidence of a role for cd44 and cell adhesion in mediating resistance to lenalidomide in multiple myeloma: Therapeutic implications. Leukemia 2014, 28, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Zou, C.; Zhu, Y.; Luo, Y.; Chen, L.; Lei, Y.; Tang, K.; Sun, Y.; Zhang, W.; Li, S.; et al. Hif-1a-regulated mir- 1275 maintains stem cell-like phenotypes and promotes the progression of luad by simultaneously activating wnt/beta-catenin and notch signaling. Theranostics 2020, 10, 2553–2570. [Google Scholar] [CrossRef]
- Yin, Y.; Liu, L.; Zhao, Z.; Yin, L.; Bauer, N.; Nwaeburu, C.C.; Gladkich, J.; Gross, W.; Hackert, T.; Sticht, C.; et al. Simvastatin inhibits sonic hedgehog signaling and stemness features of pancreatic cancer. Cancer Lett. 2018, 426, 14–24. [Google Scholar] [CrossRef]
- Kashyap, T.; Pramanik, K.K.; Nath, N.; Mishra, P.; Singh, A.K.; Nagini, S.; Rana, A.; Mishra, R. Crosstalk between raf-mek-erk and pi3k-akt-gsk3beta signaling networks promotes chemoresistance, invasion/migration and stemness via expression of cd44 variants (v4 and v6) in oral cancer. Oral Oncol. 2018, 86, 234–243. [Google Scholar] [CrossRef]
- Asati, V.; Mahapatra, D.K.; Bharti, S.K. Pi3k/akt/mtor and ras/raf/mek/erk signaling pathways inhibitors as anticancer agents: Structural and pharmacological perspectives. Eur. J. Med. Chem. 2016, 109, 314–341. [Google Scholar] [CrossRef]
- Holland, J.D.; Klaus, A.; Garratt, A.N.; Birchmeier, W. Wnt signaling in stem and cancer stem cells. Curr. Opin. Cell Biol. 2013, 25, 254–264. [Google Scholar] [CrossRef]
- Takebe, N.; Miele, L.; Harris, P.J.; Jeong, W.; Bando, H.; Kahn, M.; Yang, S.X.; Ivy, S.P. Targeting notch, hedgehog, and wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 2015, 12, 445–464. [Google Scholar] [CrossRef]
- Spaan, I.; Raymakers, R.A.; van de Stolpe, A.; Peperzak, V. Wnt signaling in multiple myeloma: A central player in disease with therapeutic potential. J. Hematol. Oncol. 2018, 11, 67. [Google Scholar] [CrossRef]
- Geng, W.; Guo, X.; Zhang, L.; Ma, Y.; Wang, L.; Liu, Z.; Ji, H.; Xiong, Y. Resveratrol inhibits proliferation, migration and invasion of multiple myeloma cells via neat1-mediated wnt/beta-catenin signaling pathway. Biomed. Pharmacother. 2018, 107, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Savvidou, I.; Khong, T.; Cuddihy, A.; Mclean, C.; Horrigan, S.; Spencer, A. Beta-catenin inhibitor bc 2059 is efficacious as monotherapy or in combination with proteasome inhibitor bortezomib in multiple myeloma. Mol. Cancer Ther. 2017, 16, 1765–1778. [Google Scholar] [CrossRef] [PubMed]
- Alonso, S.; Hernandez, D.; Chang, Y.T.; Gocke, C.B.; Mccray, M.; Varadhan, R.; Matsui, W.H.; Jones, R.J.; Ghiaur, G. Hedgehog and retinoid signaling alters multiple myeloma microenvironment and generates bortezomib resistance. J. Clin. Investig. 2016, 126, 4460–4468. [Google Scholar] [CrossRef] [PubMed]
- Martello, M.; Remondini, D.; Borsi, E.; Santacroce, B.; Procacci, M.; Pezzi, A.; Dico, F.A.; Martinelli, G.; Zamagni, E.; Tacchetti, P.; et al. Opposite activation of the hedgehog pathway in cd138+ plasma cells and cd138-cd19+ b cells identifies two subgroups of patients with multiple myeloma and different prognosis. Leukemia 2016, 30, 1869–1876. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Xu, A.; Xu, J.; Huang, H.; Chen, L.; Su, Y.; Zhang, L.; Li, J.; Fan, F.; Deng, J.; et al. Microrna-324-5p regulates stemness, pathogenesis and sensitivity to bortezomib in multiple myeloma cells by targeting hedgehog signaling. Int. J. Cancer 2018, 142, 109–120. [Google Scholar] [CrossRef]
- Clara, J.A.; Monge, C.; Yang, Y.; Takebe, N. Targeting signalling pathways and the immune microenvironment of cancer stem cells—A clinical update. Nat. Rev. Clin. Oncol. 2020, 17, 204–232. [Google Scholar] [CrossRef]
- Fendler, A.; Bauer, D.; Busch, J.; Jung, K.; Wulf-Goldenberg, A.; Kunz, S.; Song, K.; Myszczyszyn, A.; Elezkurtaj, S.; Erguen, B.; et al. Inhibiting wnt and notch in renal cancer stem cells and the implications for human patients. Nat. Commun. 2020, 11, 929. [Google Scholar] [CrossRef] [PubMed]
- Nefedova, Y.; Gabrilovich, D. Mechanisms and clinical prospects of notch inhibitors in the therapy of hematological malignancies. Drug Resist. Updat. 2008, 11, 210–218. [Google Scholar] [CrossRef][Green Version]
- Muguruma, Y.; Yahata, T.; Warita, T.; Hozumi, K.; Nakamura, Y.; Suzuki, R.; Ito, M.; Ando, K. Jagged1-induced notch activation contributes to the acquisition of bortezomib resistance in myeloma cells. Blood Cancer J. 2017, 7, 650. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Garavelli, S.; Mazzola, M.; Platonova, N.; Giannandrea, D.; Colella, R.; Apicella, L.; Lancellotti, M.; Lesma, E.; Ancona, S.; et al. Multiple myeloma exploits jagged1 and jagged2 to promote intrinsic and bone marrow-dependent drug resistance. Haematologica 2020, 105, 1925–1936. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Bai, X.; Feng, X.; Ni, J.; Beretov, J.; Graham, P.; Li, Y. Inhibition of pi3k/akt/mtor signaling pathway alleviates ovarian cancer chemoresistance through reversing epithelial-mesenchymal transition and decreasing cancer stem cell marker expression. BMC Cancer 2019, 19, 618. [Google Scholar] [CrossRef]
- Kahraman, D.C.; Kahraman, T.; Cetin-Atalay, R. Targeting pi3k/akt/mtor pathway identifies differential expression and functional role of il8 in liver cancer stem cell enrichment. Mol. Cancer Ther. 2019, 18, 2146–2157. [Google Scholar]
- Liu, H.; Liu, Z.; Du, J.; He, J.; Lin, P.; Amini, B.; Starbuck, M.W.; Novane, N.; Shah, J.J.; Davis, R.E.; et al. Thymidine phosphorylase exerts complex effects on bone resorption and formation in myeloma. Sci. Transl. Med. 2016, 8, 113r–353r. [Google Scholar] [CrossRef]
- Wang, L.; Lin, N.; Li, Y. The pi3k/akt signaling pathway regulates abcg2 expression and confers resistance to chemotherapy in human multiple myeloma. Oncol. Rep. 2019, 41, 1678–1690. [Google Scholar] [CrossRef]
- Schmeel, F.C.; Schmeel, L.C.; Kim, Y.; Schmidt-Wolf, I.G. Piceatannol exhibits selective toxicity to multiple myeloma cells and influences thewnt/beta-catenin pathway. Hematol. Oncol. 2014, 32, 197–204. [Google Scholar] [CrossRef]
- Xu, Y.; Yang, Z.; Yuan, H.; Li, Z.; Li, Y.; Liu, Q.; Chen, J. Pcdh10 inhibits cell proliferation of multiple myeloma via the negative regulation of the wnt/beta-catenin/bcl-9 signaling pathway. Oncol. Rep. 2015, 34, 747–754. [Google Scholar] [CrossRef]
- Park, S.; Yun, E.; Hwang, I.H.; Yoon, S.; Kim, D.E.; Kim, J.S.; Na, M.; Song, G.Y.; Oh, S. Ilimaquinone and ethylsmenoquinone, marine sponge metabolites, suppress the proliferation of multiple myeloma cells by down-regulating the level of beta-catenin. Mar. Drugs 2014, 12, 3231–3244. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Sun, X.; Xie, Y.; Liu, L.; Han, D.; Yao, Y.; Li, H.; Li, Z.; Xu, K. Lithium chloride inhibits cell survival, overcomes drug resistance, and triggers apoptosis in multiple myeloma via activation of the wnt/beta-catenin pathway. Am. J. Transl. Res. 2018, 10, 2610–2618. [Google Scholar]
- Liu, Z.; Xu, J.; He, J.; Zheng, Y.; Li, H.; Lu, Y.; Qian, J.; Lin, P.; Weber, D.M.; Yang, J.; et al. A critical role of autocrine sonic hedgehog signaling in human cd138+ myeloma cell survival and drug resistance. Blood 2014, 124, 2061–2071. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Xiao, Z.; Li, H.P.; Han, D.H.; Zhang, Y.P. The mechanism study of mir-125b in occurrence and progression of multiple myeloma. Cancer Med. 2018, 7, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, Y.; Huang, C.; Lin, S. Mir-215-5p is an anticancer gene in multiple myeloma by targeting runx1 and deactivating the pi3k/akt/mtor pathway. J. Cell. Biochem. 2020, 121, 1475–1490. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Chen, H.; Zhang, X.; Pan, Y.; Jing, R.; Shen, L.; Wang, X.; Ju, S.; Jin, C.; Cong, H. Serum mir-30d as a novel biomarker for multiple myeloma and its antitumor role in u266 cells through the targeting of the mtdh/pi3k/akt signaling pathway. Int. J. Oncol. 2018, 53, 2131–2144. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Jin, Z.; Yu, K.; Liu, Q. Nvp-bez235-induced autophagy as a potential therapeutic approach for multiple myeloma. Am. J. Transl. Res. 2019, 11, 87–105. [Google Scholar]
- Hopp, L.; Löffler-Wirth, H.; Binder, H. Epigenetic Heterogeneity of B-Cell Lymphoma: DNA Methylation, Gene Expression and Chromatin States. Genes 2015, 6, 812–840. [Google Scholar] [CrossRef] [PubMed]
- Vogl, D.T.; Raje, N.; Jagannath, S.; Richardson, P.; Hari, P.; Orlowski, R.; Supko, J.G.; Tamang, D.; Yang, M.; Jones, S.S.; et al. Ricolinostat, the First Selective Histone Deacetylase 6 Inhibitor, in Combination with Bortezomib and Dexamethasone for Relapsed or Refractory Multiple Myeloma. Clin. Cancer Res. 2017, 23, 3307–3315. [Google Scholar] [CrossRef]
- Chaidos, A.; Barnes, C.P.; Cowan, G.; May, P.C.; Melo, V.; Hatjiharissi, E.; Papaioannou, M.; Harrington, H.; Doolittle, H.; Terpos, E.; et al. Clinical drug resistance linked to interconvertible phenotypic and functional states of tumor-propagating cells in multiple myeloma. Blood 2013, 121, 318–328. [Google Scholar] [CrossRef]
- Adamia, S.; Abiatari, I.; Amin, S.B.; Fulciniti, M.; Minvielle, S.; Li, C.; Moreau, P.; Avet-Loiseau, H.; Munshi, N.C.; Anderson, K.C. The effects of MicroRNA deregulation on pre-RNA processing network in multiple myeloma. Leukemia 2020, 34, 167–179. [Google Scholar] [CrossRef]
- Valent, P.; Bonnet, D.; De Maria, R.; Lapidot, T.; Copland, M.; Melo, J.V.; Chomienne, C.; Ishikawa, F.; Schuringa, J.J.; Stassi, G.; et al. Cancer stem cell definitions and terminology: The devil is in the details. Nat. Rev. Cancer 2012, 12, 767–775. [Google Scholar] [CrossRef]
- Rycaj, K.; Tang, D.G. Cell-of-Origin of Cancer versus Cancer Stem Cells: Assays and Interpretations. Cancer Res. 2015, 75, 4003–4011. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, P.; Shuai, L.; Chen, K.; Li, Z.; Zhang, Y.; Jiang, Y.; Li, X. Mir-589-5p inhibits map3k8 and suppresses cd90(+) cancer stem cells in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2016, 35, 176. [Google Scholar] [CrossRef]
- Chaudhary, K.; Poirion, O.B.; Lu, L.; Garmire, L.X. Deep learning-based multi-omics integration robustly predicts survival in liver cancer. Clin. Cancer Res. 2018, 24, 1248–1259. [Google Scholar] [CrossRef] [PubMed]
- Alison, M.R.; Lin, W.R.; Lim, S.M.; Nicholson, L.J. Cancer stem cells: In the line of fire. Cancer Treat. Rev. 2012, 38, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Najafi, M.; Farhood, B.; Mortezaee, K. Cancer stem cells (CSCs) in cancer progression and therapy. J. Cell. Physiol. 2019, 234, 8381–8395. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, S.; Abdel-Malek, M.A.; Malek, E.; Vad, N.; Latif, T.; Anderson, K.C.; Driscoll, J.J. Pharmacologic screens reveal metformin that suppresses grp78-dependent autophagy to enhance the anti-myeloma effect of bortezomib. Leukemia 2015, 29, 2184–2191. [Google Scholar] [CrossRef] [PubMed]
- Saltarella, I.; Desantis, V.; Melaccio, A.; Solimando, A.G.; Lamanuzzi, A.; Ria, R.; Storlazzi, C.T.; Mariggiò, M.A.; Vacca, A.; Frassanito, M.A. Mechanisms of Resistance to Anti-CD38 Daratumumab in Multiple Myeloma. Cells 2020, 9, 167. [Google Scholar] [CrossRef]
- Desantis, V.; Saltarella, I.; Lamanuzzi, A.; Melaccio, A.; Solimando, A.G.; Mariggiò, M.A.; Racanelli, V.; Paradiso, A.; Vacca, A.; Frassanito, M.A. MicroRNAs-Based Nano-Strategies as New Therapeutic Approach in Multiple Myeloma to Overcome Disease Progression and Drug Resistance. Int. J. Mol. Sci. 2020, 21, 3084. [Google Scholar] [CrossRef] [PubMed]
| Surface Marker | Function | Drug | Mechanism | Reference Study |
|---|---|---|---|---|
| SP cells/ABC transporter | Trans-membrane transportation; related to drug resistance of CSCs; regulating oxidation reduction status; regulating membrane lipid composition; regulating the release of nutrients and metabolites, and regulating the tumor microenvironment | HLA class I small molecule antibody Lenalidomide Ibrutinib | Targeting β-catenin and suppressing stem genes such as SOX2, OCT3/4, and Nanog. Affecting phosphorylation of AKT, GSK-3-α/β, MEK1, c-JUN, P53, and P70S6K Targeting Bruton’s tyrosine kinase | [10,14,17] |
| EZH1/2 double inhibitor (or-s1) | Targeting EZH1 and EZH2, causing Wnt signaling repression | [27] | ||
| HuCD26mAb | Specifically inhibiting SP cells | [29] | ||
| Fenretinide | Targeting IL-3, blocking pDC-induced MM cell proliferation | [30] | ||
| Targeting CD26 in SP cells | [31] | |||
| ALDH | Promoting the dehydrogenation of acetaldehyde, participating in cell detoxification through oxidation, regulating the differentiation, apoptosis, and growth of CSC through the Ra signal pathway, and reducing the ROS in CSCs | GSK126 | Abrogating the methylated histone 3 level, blocking the Wnt/β-catenin pathway, and inhibiting of EZH2 methyltransferase activity | [28] |
| Lycorine | Inhibiting ALDH1+ cells through the Wnt/βcatenin pathway | [42] | ||
| I-5-iodo-4′-thio-2′-deoxyuridine | Decreasing the activity of ALDH | [44] | ||
| KS99 | Targeting leukemia stem cells with high aldehyde dehydrogenase activity and inhibiting STAT3 phosphorylation and inhibiting the activation of Bruton’s tyrosine kinase. | [48] | ||
| Disulfiram/Cu | Targeting ALDH1A1, inhibiting the expression of NANOG and OCT, and suppressing the Hh pathway by inhibiting transcription factors Gli1 and Gli2. | [49] | ||
| MK1775 | Inhibiting ALDH1+ cells through Wee1 kinase | [75] | ||
| CD24 | Cell adhesion protein, mediating B cell antigen-dependent activation, distinguishing pre-B cells from Mature B cells, and overexpressed in SP cells | SWA11(CD24 antibody) | Targeting CD24 | [22] |
| SOX2 | Related to tumor invasion, metastasis, and EMT; an important index in clinical trials at present | cotylenin A and vincristine | Inhibiting SOX2 mRNA expression in myeloma cells | [68] |
| CD44 | Necessary medium for the bone-marrow adhesion of MM cells; participating in cell-adhesion-mediated drug resistance | All-trans retinoic acid (ATRA) | Downregulating the expression of total β-catenin, cell surface, and total CD44 in a mice xenotransplantation model; decreasing lenalidomide-resistant MM cells’ adhesion; and enhancing the effect of lenalidomide. | [78] |
| Pathway | Function | Drug/Target Gene | Mechanism | References |
|---|---|---|---|---|
| Wnt/β-catenin | Mediating the proliferation, migration, and drug resistance of MM cells; promoting the differentiation of osteoblasts | Resveratrol | Downregulating the expression of lncrna-neat1 in MM cells by suppressing the Wnt signaling pathway and UPR | [86] |
| BC2059 | Downregulating β-catenin protein | [87] | ||
| Piceatannol | Decreasing the level of β-catenin, the transcriptional activity of Tcf4/lef complex, and the level of its target gene Axin 2 | [100] | ||
| Tumor suppressor gene PCDH10 | Inhibiting the nuclear localization, lef/TCF activity, bcl-9, and Akt expression of β-catenin | [101] | ||
| ilimaquinone and ethylsmenoquinone | Decreasing the level of β-catenin in the cell | [102] | ||
| LiCl | Inducing G2/M phase arrest of the MM cell cycle; activating the Wnt/β-catenin signaling pathway to induce MM cell apoptosis | [103] | ||
| Hedgehog | Changing the tumor microenvironment, participating in BORTEZOMIB resistance, inhibiting the apoptosis of MM cells | CYP26 | Forming a low retinoic acid environment and producing resistance to BTZ | [88] |
| SHH (sonic hedgehog) | Inhibiting the apoptosis of myeloma cells | [104] | ||
| Notch | Participating in the development of MM, related to strom-mediated drug resistance. | GSI (γ-secretase inhibitor) | Inhibiting the second cleavage of Notch receptor | [93] |
| miR-125b | Targeting MALAT1 and regulating the proliferation of MM cells | [105] | ||
| PI3K/Akt | Tumor-suppressor gene inactivation, related to bone lysis | miR-215-5p | Targeting RUNX1 and inhibiting the PI3K/AKT/mTOR pathway | [106] |
| miR-30d | Targeting metaherherin and inhibiting the PI3K/Akt signal pathway | [107] | ||
| NVP-BEZ235 | Binding to the ATP binding gap of PI3K and mTOR kinases | [108] |
| MM with Specific Markers | Biological Function Characteristics | References |
|---|---|---|
| Side population cell | High clonogenicity, tumorigenicity, and self-renewal ability | [14,15] |
| ALDH1+ MM | High colony-forming ability, resistance to bortezomib and adriamycin | [40,41] |
| CD138− MM | Strong colony-forming and tumor initiation ability | [58] |
| CD24+ MM | Strong colony-forming ability and tumorigenicity, high iPS/ES genes expression, and strong resistance to MM clinical drugs | [22] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, W.; Wang, H.; Chen, P.; Shen, X.; Zhang, B.; Liu, J.; Peng, H.; Xiao, X. Identification and Characterization of Multiple Myeloma Stem Cell-Like Cells. Cancers 2021, 13, 3523. https://doi.org/10.3390/cancers13143523
Guo W, Wang H, Chen P, Shen X, Zhang B, Liu J, Peng H, Xiao X. Identification and Characterization of Multiple Myeloma Stem Cell-Like Cells. Cancers. 2021; 13(14):3523. https://doi.org/10.3390/cancers13143523
Chicago/Turabian StyleGuo, Wancheng, Haiqin Wang, Peng Chen, Xiaokai Shen, Boxin Zhang, Jing Liu, Hongling Peng, and Xiaojuan Xiao. 2021. "Identification and Characterization of Multiple Myeloma Stem Cell-Like Cells" Cancers 13, no. 14: 3523. https://doi.org/10.3390/cancers13143523
APA StyleGuo, W., Wang, H., Chen, P., Shen, X., Zhang, B., Liu, J., Peng, H., & Xiao, X. (2021). Identification and Characterization of Multiple Myeloma Stem Cell-Like Cells. Cancers, 13(14), 3523. https://doi.org/10.3390/cancers13143523







