Expression of Retroelements in Cervical Cancer and Their Interplay with HPV Infection and Host Gene Expression
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. RNA-Seq
2.3. Host Gene and Retrotransposon Expression Analyses
3. Results
3.1. Retrotransposon Expression in Cervical Cancer
3.2. Retrotransposons Are Differentially Expressed between Cervical Cancer Histological Types and HPV Genotype Infections
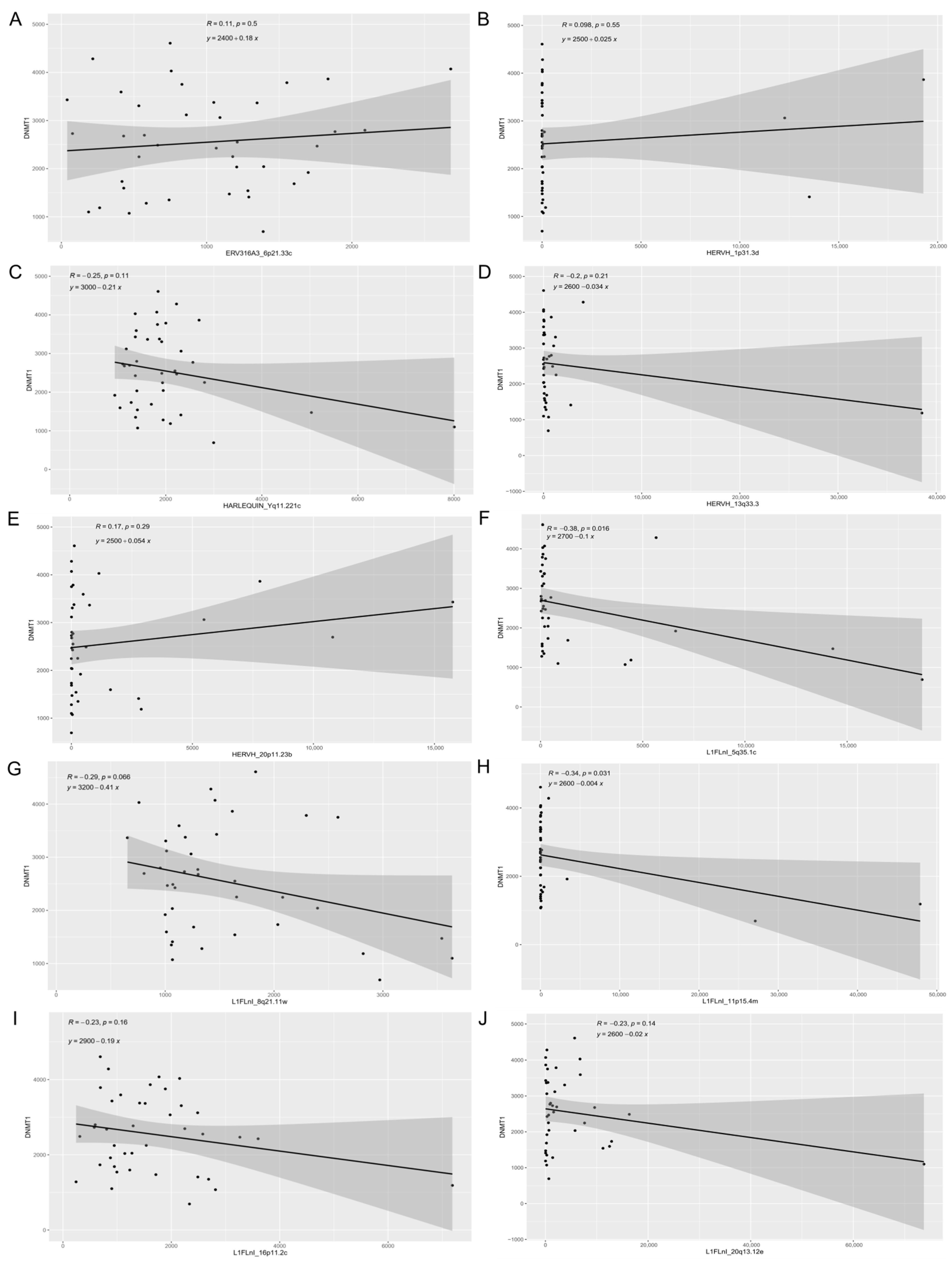
3.3. Expression of Retroelements and Nearby Host Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arbyn, M.; Weiderpass, E.; Bruni, L.; de Sanjosé, S.; Saraiya, M.; Ferlay, J.; Bray, F. Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob. Health 2020, 8, e191–e203. [Google Scholar] [CrossRef]
- Kocjan, B.J.; Bzhalava, D.; Forslund, O.; Dillner, J.; Poljak, M. Molecular methods for identification and characterization of novel papillomaviruses. Clin. Microbiol. Infect. 2015, 21, 808–816. [Google Scholar] [CrossRef]
- Muñoz, N.; Bosch, F.X.; de Sanjosé, S.; Herrero, R.; Castellsagué, X.; Shah, K.V.; Snijders, P.J.F.; Meijer, C.J.L.M. Epidemiologic Classification of Human Papillomavirus Types Associated with Cervical Cancer. N. Engl. J. Med. 2003, 348, 518–527. [Google Scholar] [CrossRef]
- Clifford, G.M.; Smith, J.S.; Plummer, M.; Muñoz, N.; Franceschi, S. Human papillomavirus types in invasive cervical cancer worldwide: A meta-analysis. Br. J. Cancer 2003, 88, 63–73. [Google Scholar] [CrossRef]
- Bosch, F.X.; Manos, M.M.; Munoz, N.; Sherman, M.; Jansen, A.M.; Peto, J.; Schiffman, M.H.; Moreno, V.; Kurman, R.; Shan, K.V. Prevalence of Human Papillomavirus in Cervical Cancer: A Worldwide Perspective. J. Natl. Cancer Inst. 1995, 87, 796–802. [Google Scholar] [CrossRef]
- De Sanjose, S.; Quint, W.G.; Alemany, L.; Geraets, D.T.; Klaustermeier, J.E.; Lloveras, B.; Tous, S.; Felix, A.; Bravo, L.E.; Shin, H.-R.; et al. Human papillomavirus genotype attribution in invasive cervical cancer: A retrospective cross-sectional worldwide study. Lancet Oncol. 2010, 11, 1048–1056. [Google Scholar] [CrossRef]
- Roden, R.B.S.; Stern, P.L. Opportunities and challenges for human papillomavirus vaccination in cancer. Nat. Rev. Cancer 2018, 18, 240–254. [Google Scholar] [CrossRef]
- Thomas, J.T.; Laimins, L.A. Human Papillomavirus Oncoproteins E6 and E7 Independently Abrogate the Mitotic Spindle Checkpoint. J. Virol. 1998, 72, 1131–1137. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Kundu, R. Human Papillomavirus E6 and E7: The Cervical Cancer Hallmarks and Targets for Therapy. Front. Microbiol. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Dizanzo, M.P.; Marziali, F.; Brunet Avalos, C.; Bugnon Valdano, M.; Leiva, S.; Cavatorta, A.L.; Gardiol, D. HPV E6 and E7 oncoproteins cooperatively alter the expression of Disc Large 1 polarity protein in epithelial cells. BMC Cancer 2020, 20, 293. [Google Scholar] [CrossRef] [PubMed]
- Arias-Pulido, H.; Peyton, C.L.; Joste, N.E.; Vargas, H.; Wheeler, C.M. Human Papillomavirus Type 16 Integration in Cervical Carcinoma In Situ and in Invasive Cervical Cancer. J. Clin. Microbiol. 2006, 44, 1755–1762. [Google Scholar] [CrossRef]
- Das, B.C.; Sharma, J.K.; Gopalakrishna, V.; Luthra, U.K. Analysis by polymerase chain reaction of the physical state of human papillomavirus type 16 DNA in cervical preneoplastic and neoplastic lesions. J. Gen. Virol. 1992, 73, 2327–2336. [Google Scholar] [CrossRef] [PubMed]
- McBride, A.A.; Warburton, A. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog. 2017, 13, e1006211. [Google Scholar] [CrossRef]
- Curty, G.; Marston, J.L.; de Mulder Rougvie, M.; Leal, F.E.; Nixon, D.F.; Soares, M.A. Human Endogenous Retrovirus K in Cancer: A Potential Biomarker and Immunotherapeutic Target. Viruses 2020, 12, 726. [Google Scholar] [CrossRef]
- Bannert, N.; Hofmann, H.; Block, A.; Hohn, O. HERVs New Role in Cancer: From Accused Perpetrators to Cheerful Protectors. Front. Microbiol. 2018, 9, 178. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liang, J.Q.; Zheng, S. Expressional activation and functional roles of human endogenous retroviruses in cancers. Rev. Med. Virol. 2019, 29, e2025. [Google Scholar] [CrossRef]
- Wang, Z.; Yin, J.; Zhou, W.; Bai, J.; Xie, Y.; Xu, K.; Zheng, X.; Xiao, J.; Zhou, L.; Qi, X.; et al. Complex impact of DNA methylation on transcriptional dysregulation across 22 human cancer types. Nucleic Acids Res. 2020, 48, 2287–2302. [Google Scholar] [CrossRef]
- Ando, M.; Saito, Y.; Xu, G.; Bui, N.Q.; Medetgul-Ernar, K.; Pu, M.; Fisch, K.; Ren, S.; Sakai, A.; Fukusumi, T.; et al. Chromatin dysregulation and DNA methylation at transcription start sites associated with transcriptional repression in cancers. Nat. Commun. 2019, 10, 2188. [Google Scholar] [CrossRef] [PubMed]
- Ma, J. Retrotransposons. In Brenner’s Encyclopedia of Genetics; Elsevier: Amsterdam, The Netherlands, 2013; pp. 208–210. [Google Scholar]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef]
- Flavell, A.J. Long terminal repeat retrotransposons jump between species. Proc. Natl. Acad. Sci. USA 1999, 96, 12211–12212. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.; Devine, S. The Role of Somatic L1 Retrotransposition in Human Cancers. Viruses 2017, 9, 131. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, R.; Yu, J. New Understanding of the Relevant Role of LINE-1 Retrotransposition in Human Disease and Immune Modulation. Front. Cell Dev. Biol. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Brant, A.C.; Menezes, A.N.; Felix, S.P.; Almeida, L.M.; Moreira, M.A.M. Preferential expression of a HPV genotype in invasive cervical carcinomas infected by multiple genotypes. Genomics 2020, 112, 2942–2948. [Google Scholar] [CrossRef]
- Brant, A.C.; Menezes, A.N.; Felix, S.P.; de Almeida, L.M.; Sammeth, M.; Moreira, M.A.M. Characterization of HPV integration, viral gene expression and E6E7 alternative transcripts by RNA-Seq: A descriptive study in invasive cervical cancer. Genomics 2019, 111, 1853–1861. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Bendall, M.L.; de Mulder, M.; Iñiguez, L.P.; Lecanda-Sánchez, A.; Pérez-Losada, M.; Ostrowski, M.A.; Jones, R.B.; Mulder, L.C.F.; Reyes-Terán, G.; Crandall, K.A.; et al. Telescope: Characterization of the retrotranscriptome by accurate estimation of transposable element expression. PLoS Comput. Biol. 2019, 15, e1006453. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Vasaikar, S.; Shi, Z.; Greer, M.; Zhang, B. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017, 45, W130–W137. [Google Scholar] [CrossRef]
- Xiao-Jie, L.; Hui-Ying, X.; Qi, X.; Jiang, X.; Shi-Jie, M. LINE-1 in cancer: Multifaceted functions and potential clinical implications. Genet. Med. 2016, 18, 431–439. [Google Scholar] [CrossRef]
- Kolbe, A.R.; Bendall, M.L.; Pearson, A.T.; Paul, D.; Nixon, D.F.; Pérez-Losada, M.; Crandall, K.A. Human Endogenous Retrovirus Expression Is Associated with Head and Neck Cancer and Differential Survival. Viruses 2020, 12, 956. [Google Scholar] [CrossRef]
- Hurst, T.; Magiorkinis, G. Epigenetic Control of Human Endogenous Retrovirus Expression: Focus on Regulation of Long-Terminal Repeats (LTRs). Viruses 2017, 9, 130. [Google Scholar] [CrossRef] [PubMed]
- Curty, G.; Beckerle, G.A.; Iñiguez, L.P.; Furler, R.L.; de Carvalho, P.S.; Marston, J.L.; Champiat, S.; Heymann, J.J.; Ormsby, C.E.; Reyes-Terán, G.; et al. Human Endogenous Retrovirus Expression Is Upregulated in the Breast Cancer Microenvironment of HIV Infected Women: A Pilot Study. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef]
- Iramaneerat, K.; Rattanatunyong, P.; Khemapech, N.; Triratanachat, S.; Mutirangura, A. HERV-K Hypomethylation in Ovarian Clear Cell Carcinoma Is Associated with a Poor Prognosis and Platinum Resistance. Int. J. Gynecol. Cancer 2011, 21, 51–57. [Google Scholar] [CrossRef]
- Shuangshoti, S.; Hourpai, N.; Pumsuk, U.; Mutirangura, A. Line-1 hypomethylation in multistage carcinogenesis of the uterine cervix. Asian Pac. J. Cancer Prev. 2007, 8, 307–309. [Google Scholar] [PubMed]
- Zhang, J.; Yang, C.; Wu, C.; Cui, W.; Wang, L. DNA Methyltransferases in Cancer: Biology, Paradox, Aberrations, and Targeted Therapy. Cancers 2020, 12, 2123. [Google Scholar] [CrossRef]
- Jönsson, M.E.; Ludvik Brattås, P.; Gustafsson, C.; Petri, R.; Yudovich, D.; Pircs, K.; Verschuere, S.; Madsen, S.; Hansson, J.; Larsson, J.; et al. Activation of neuronal genes via LINE-1 elements upon global DNA demethylation in human neural progenitors. Nat. Commun. 2019, 10, 3182. [Google Scholar] [CrossRef] [PubMed]
- Koito, A.; Ikeda, T. Intrinsic immunity against retrotransposons by APOBEC cytidine deaminases. Front. Microbiol. 2013, 4, 1–9. [Google Scholar] [CrossRef]
- Jo, J.-O.; Kang, Y.-J.; Ock, M.S.; Song, K.S.; Jeong, M.-J.; Jeong, S.-J.; Choi, Y.H.; Ko, E.-J.; Leem, S.-H.; Kim, S.; et al. Expression profiles of HERV-K Env protein in normal and cancerous tissues. Genes Genom. 2016, 38, 91–107. [Google Scholar] [CrossRef]
- Zhang, R.; Shen, C.; Zhao, L.; Wang, J.; McCrae, M.; Chen, X.; Lu, F. Dysregulation of host cellular genes targeted by human papillomavirus (HPV) integration contributes to HPV-related cervical carcinogenesis. Int. J. Cancer 2016, 138, 1163–1174. [Google Scholar] [CrossRef]
- Matovina, M.; Sabol, I.; Grubišić, G.; Gašperov, N.M.; Grce, M. Identification of human papillomavirus type 16 integration sites in high-grade precancerous cervical lesions. Gynecol. Oncol. 2009, 113, 120–127. [Google Scholar] [CrossRef]
- Cohen, C.J.; Lock, W.M.; Mager, D.L. Endogenous retroviral LTRs as promoters for human genes: A critical assessment. Gene 2009, 448, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, D.R.; Swigut, T.; Wysocka, J. Systematic perturbation of retroviral LTRs reveals widespread long-range effects on human gene regulation. Elife 2018, 7, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Babaian, A.; Mager, D.L. Endogenous retroviral promoter exaptation in human cancer. Mob. DNA 2016, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Lamprecht, B.; Walter, K.; Kreher, S.; Kumar, R.; Hummel, M.; Lenze, D.; Köchert, K.; Bouhlel, M.A.; Richter, J.; Soler, E.; et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat. Med. 2010, 16, 571–579. [Google Scholar] [CrossRef]
- Rutz, S.; Wang, X.; Ouyang, W. The IL-20 subfamily of cytokines—From host defence to tissue homeostasis. Nat. Rev. Immunol. 2014, 14, 783–795. [Google Scholar] [CrossRef]
- Cui, X.-F.; Cui, X.-G.; Leng, N. Overexpression of interleukin-20 receptor subunit beta (IL20RB) correlates with cell proliferation, invasion and migration enhancement and poor prognosis in papillary renal cell carcinoma. J. Toxicol. Pathol. 2019, 32, 245–251. [Google Scholar] [CrossRef] [PubMed]




| Gene Set | Description | p-Value | q-Value |
|---|---|---|---|
| R-HSA-854691 | Interleukin-20 family signaling | 7.0383 × 10−7 | 0.0012162 |
| R-HSA-419408 | Lysosphingolipid and LPA receptors | 0.00011395 | 0.098454 |
| R-HSA-799990 | Metal sequestration by antimicrobial proteins | 0.0007144 | 0.35011 |
| R-HSA-162588 | Budding and maturation of HIV virion | 0.0010607 | 0.35011 |
| R-HSA-420029 | Tight junction interactions | 0.0010607 | 0.35011 |
| R-HSA-941332 | RUNX2 regulates genes involved in cell migration | 0.0013215 | 0.35011 |
| R-HSA-917729 | Endosomal Sorting Complex Required For Transport (ESCRT) | 0.0014183 | 0.35011 |
| R-HSA-803205 | TP53 signaling | 0.0041793 | 0.75655 |
| R-HSA-803157 | Antimicrobial peptides | 0.0047203 | 0.75655 |
| R-HSA-162123 | Synthesis of Prostaglandins (PG) and Thromboxanes (TX) | 0.0048005 | 0.75655 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Curty, G.; Menezes, A.N.; Brant, A.C.; de Mulder Rougvie, M.; Moreira, M.Â.M.; Soares, M.A. Expression of Retroelements in Cervical Cancer and Their Interplay with HPV Infection and Host Gene Expression. Cancers 2021, 13, 3513. https://doi.org/10.3390/cancers13143513
Curty G, Menezes AN, Brant AC, de Mulder Rougvie M, Moreira MÂM, Soares MA. Expression of Retroelements in Cervical Cancer and Their Interplay with HPV Infection and Host Gene Expression. Cancers. 2021; 13(14):3513. https://doi.org/10.3390/cancers13143513
Chicago/Turabian StyleCurty, Gislaine, Albert N. Menezes, Ayslan C. Brant, Miguel de Mulder Rougvie, Miguel Ângelo M. Moreira, and Marcelo A. Soares. 2021. "Expression of Retroelements in Cervical Cancer and Their Interplay with HPV Infection and Host Gene Expression" Cancers 13, no. 14: 3513. https://doi.org/10.3390/cancers13143513
APA StyleCurty, G., Menezes, A. N., Brant, A. C., de Mulder Rougvie, M., Moreira, M. Â. M., & Soares, M. A. (2021). Expression of Retroelements in Cervical Cancer and Their Interplay with HPV Infection and Host Gene Expression. Cancers, 13(14), 3513. https://doi.org/10.3390/cancers13143513






