Self-Assembling Polypeptide Hydrogels as a Platform to Recapitulate the Tumor Microenvironment
Abstract
:Simple Summary
Abstract
1. Introduction
2. Comprehensive Review
2.1. Collagen
2.2. Alginate
2.3. Hyaluronic Acid
2.4. Polyethylene Glycol
2.5. Polyacrylamide
2.6. Polydimethylsiloxane
2.7. Polypeptides
3. Results
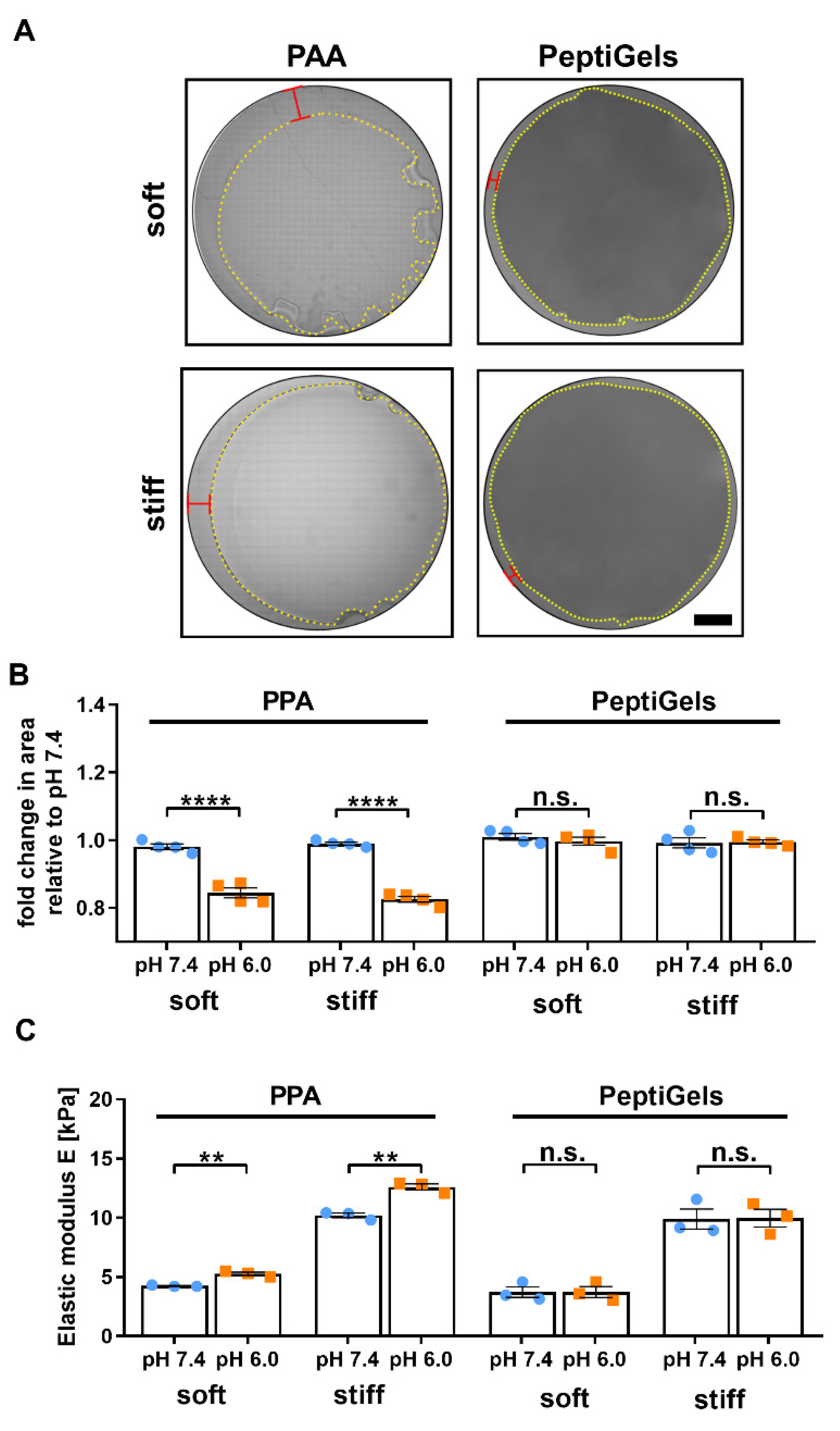
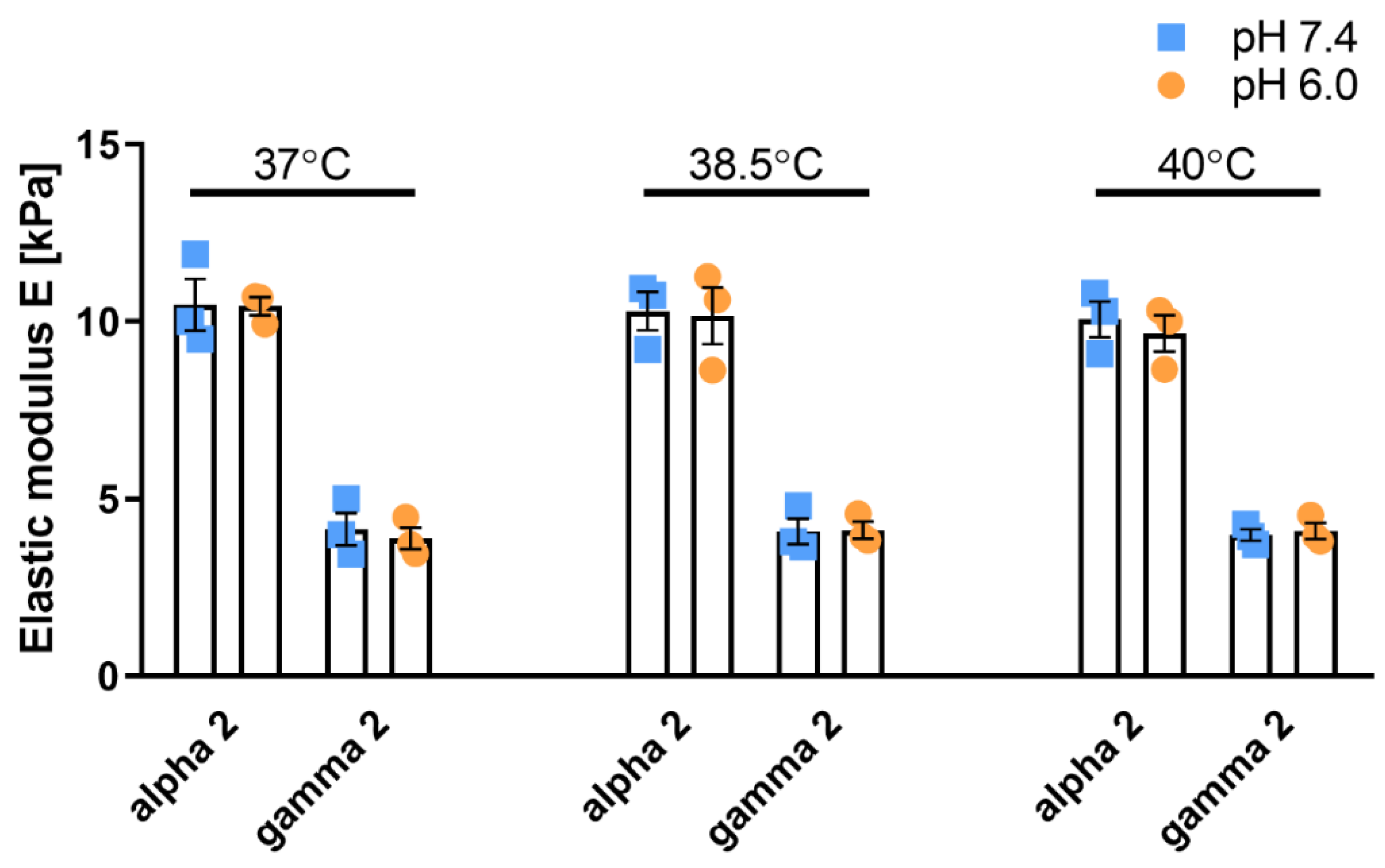
3.1. Self-Assembling Polypeptide Matrices Remain Mechanically Stable at Acidic pH
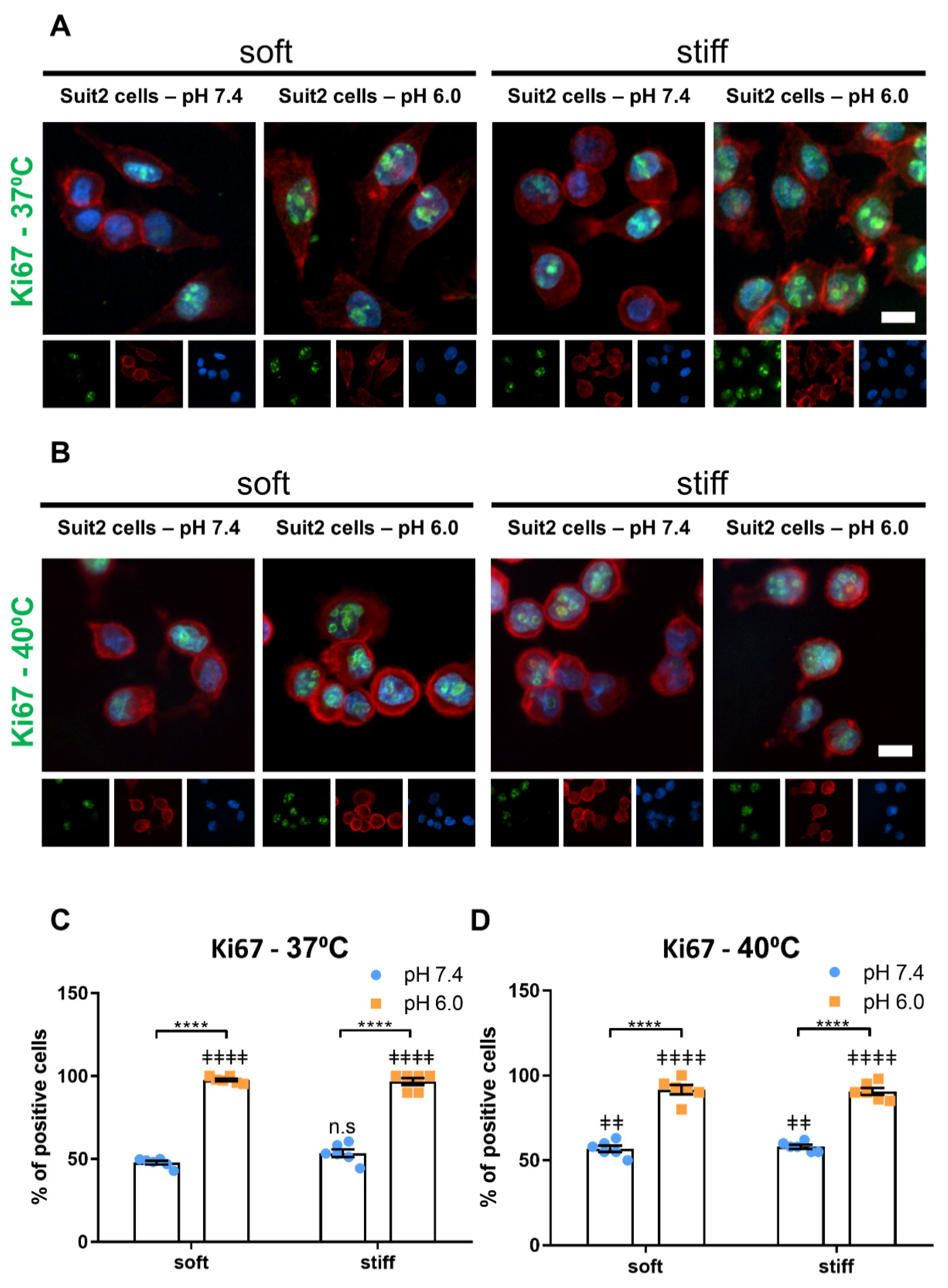
3.2. Extracellular pH and Matrix Stiffness Regulate Cancer Cell Proliferation
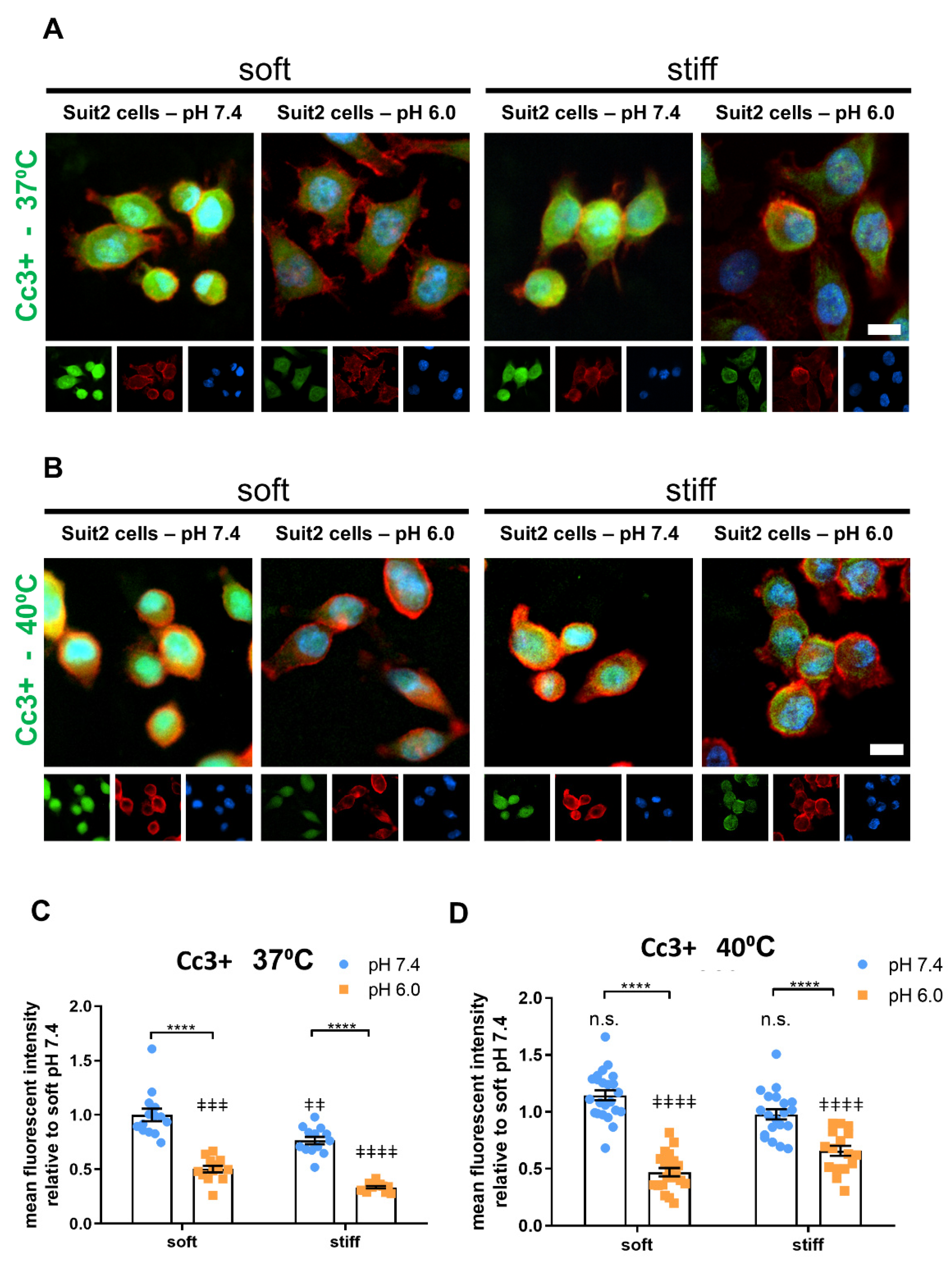
3.3. Extracellular pH and Matrix Stiffness Regulate Cancer Cell Apoptosis
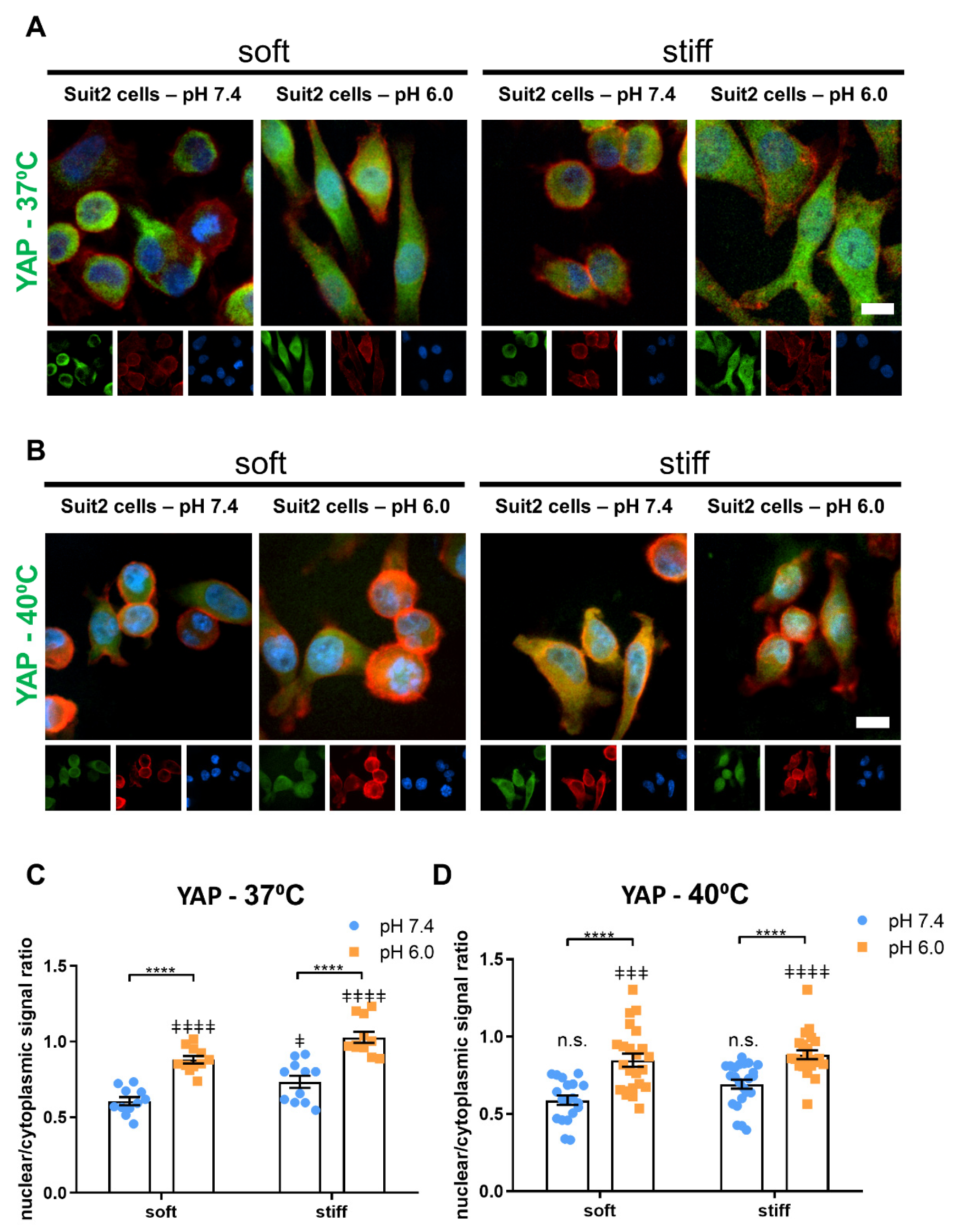
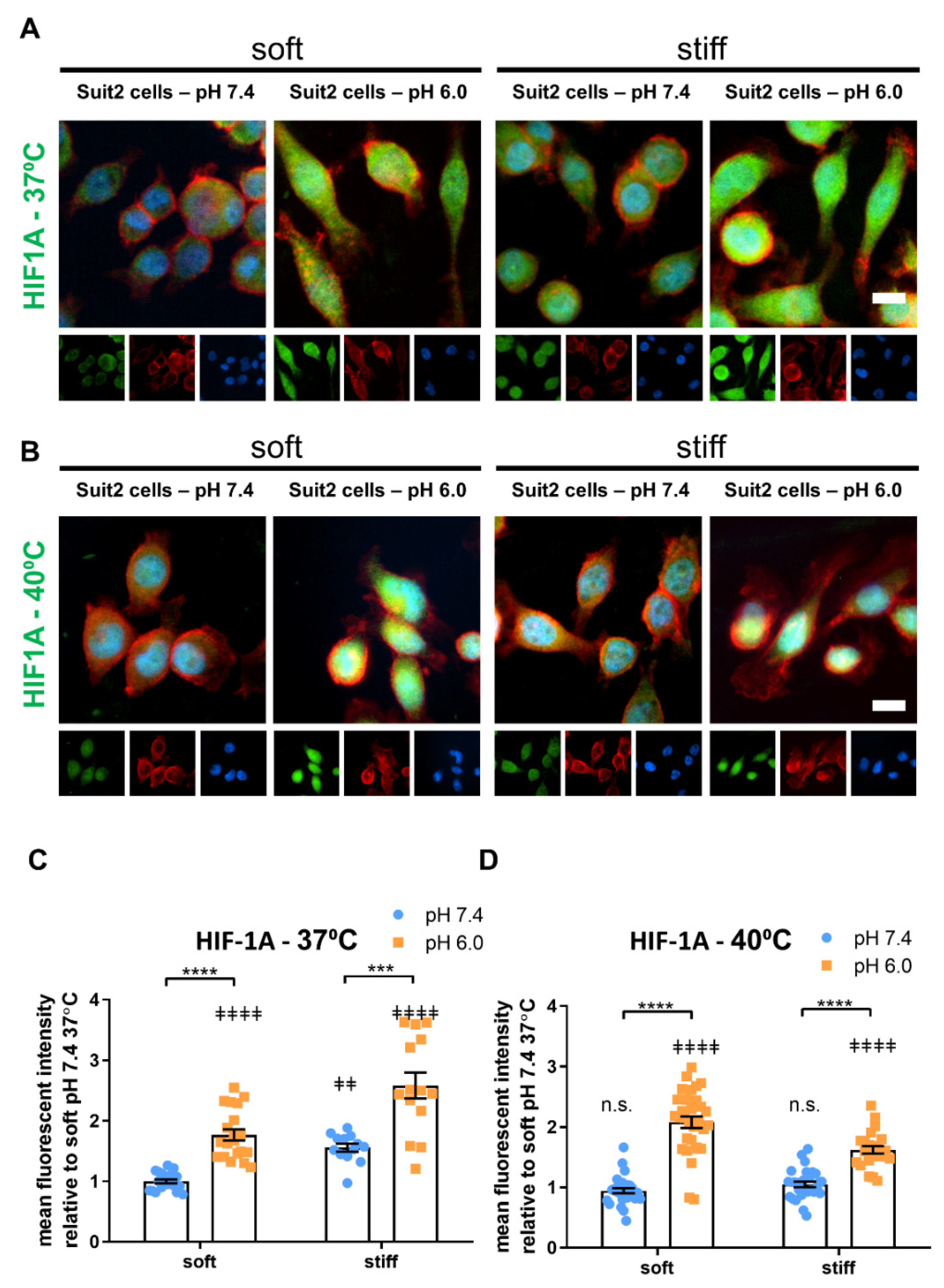
3.4. Extracellular pH, Temperature and Matrix Stiffness Regulate YAP-1 and HIF-1A Signaling
4. Discussion
5. Materials and Methods
5.1. Cell, Reagents, and Antibodies
5.2. Gel Preparation and Gel Contraction Assay
5.3. Rheometry
5.4. Immunofluorescence Staining
5.5. Immunofluorescence Imaging Analysis
5.6. qPCR
5.7. Statistical Analysis
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Matellan, C.; Hernández, A.E.D.R. Engineering the cellular mechanical microenvironment—From bulk mechanics to the nanoscale. J. Cell Sci. 2019, 132, jcs229013. [Google Scholar] [CrossRef] [Green Version]
- Koh, B.; Jeon, H.; Kim, D.; Kang, D.; Kim, K.R. Effect of fibroblast co-culture on the proliferation, viability and drug response of colon cancer cells. Oncol. Lett. 2018, 17, 2409–2417. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.-A.; Lee, E.K.; Kuh, H.-J. Co-culture of 3D tumor spheroids with fibroblasts as a model for epithelial–mesenchymal transition in vitro. Exp. Cell Res. 2015, 335, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Long, L.; Yin, M.; Min, W. 3D Co-culture System of Tumor-associated Macrophages and Ovarian Cancer Cells. Bio-Protocol 2018, 8, e2815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langer, E.M.; Allen-Petersen, B.; King, S.M.; Kendsersky, N.D.; Turnidge, M.A.; Kuziel, G.M.; Riggers, R.; Samatham, R.; Amery, T.S.; Jacques, S.L.; et al. Modeling Tumor Phenotypes In Vitro with Three-Dimensional Bioprinting. Cell Rep. 2019, 26, 608–623.e6. [Google Scholar] [CrossRef] [Green Version]
- Watters, K.M.; Bajwa, P.; Kenny, H.A. Organotypic 3D Models of the Ovarian Cancer Tumor Microenvironment. Cancers 2018, 10, 265. [Google Scholar] [CrossRef] [Green Version]
- Perche, F.; Torchilin, V.P. Cancer cell spheroids as a model to evaluate chemotherapy protocols. Cancer Biol. Ther. 2012, 13, 1205–1213. [Google Scholar] [CrossRef]
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef]
- Shirure, V.S.; Bi, Y.; Curtis, M.B.; Lezia, A.; Goedegebuure, M.M.; Goedegebuure, S.P.; Aft, R.; Fields, R.C.; George, S.C. Tumor-on-a-chip platform to investigate progression and drug sensitivity in cell lines and patient-derived organoids. Lab Chip 2018, 18, 3687–3702. [Google Scholar] [CrossRef] [PubMed]
- Kashaninejad, N.; Nikmaneshi, M.R.; Moghadas, H.; Oskouei, A.K.; Rismanian, M.; Barisam, M.; Saidi, M.S.; Firoozabadi, B. Organ-Tumor-on-a-Chip for Chemosensitivity Assay: A Critical Review. Micromachines 2016, 7, 130. [Google Scholar] [CrossRef]
- Najafi, M.; Farhood, B.; Mortezaee, K. Extracellular matrix (ECM) stiffness and degradation as cancer drivers. J. Cell. Biochem. 2019, 120, 2782–2790. [Google Scholar] [CrossRef]
- Eble, J.A.; Niland, S. The extracellular matrix in tumor progression and metastasis. Clin. Exp. Metastasis 2019, 36, 171–198. [Google Scholar] [CrossRef] [PubMed]
- Chronopoulos, A.; Robinson, B.; Sarper, M.; Cortes, E.; Auernheimer, V.; Lachowski, D.; Attwood, S.; García, R.; Ghassemi, S.; Fabry, B.; et al. ATRA mechanically reprograms pancreatic stellate cells to suppress matrix remodelling and inhibit cancer cell invasion. Nat. Commun. 2016, 7, 12630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, A.J.; Cortes, E.; Lachowski, D.; Cheung, B.C.H.; A Karim, S.; Morton, J.; Hernández, A.D.R. Matrix stiffness induces epithelial–mesenchymal transition and promotes chemoresistance in pancreatic cancer cells. Oncogenesis 2017, 6, e352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeldag, G.; Rice, A.; Hernández, A.D.R. Chemoresistance and the Self-Maintaining Tumor Microenvironment. Cancers 2018, 10, 471. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim-Hashim, A.; Estrella, V. Acidosis and cancer: From mechanism to neutralization. Cancer Metastasis Rev. 2019, 38, 149–155. [Google Scholar] [CrossRef]
- Yu, L.; Chen, X.; Sun, X.; Wang, L.; Chen, S. The Glycolytic Switch in Tumors: How Many Players Are Involved? J. Cancer 2017, 8, 3430–3440. [Google Scholar] [CrossRef]
- Corbet, C.; Feron, O. Tumour acidosis: From the passenger to the driver’s seat. Nat. Rev. Cancer 2017, 17, 577–593. [Google Scholar] [CrossRef]
- Pillai, S.R.; Damaghi, M.; Marunaka, Y.; Spugnini, E.P.; Fais, S.; Gillies, R.J. Causes, consequences, and therapy of tumors acidosis. Cancer Metastasis Rev. 2019, 38, 205–222. [Google Scholar] [CrossRef]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [Green Version]
- Caliari, S.; Burdick, J.A. A practical guide to hydrogels for cell culture. Nat. Methods 2016, 13, 405–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ananthanarayanan, B.; Kim, Y.; Kumar, S. Elucidating the mechanobiology of malignant brain tumors using a brain matrix-mimetic hyaluronic acid hydrogel platform. Biomaterials 2011, 32, 7913–7923. [Google Scholar] [CrossRef] [Green Version]
- Sakai, K.; Kurokawa, T.; Furui, Y.; Kuronuma, Y.; Sekiguchi, M.; Ando, J.; Inagaki, Y.; Tang, W.; Nakata, M.; Fujita-Yamaguchi, Y. Invasion of carcinoma cells into reconstituted type I collagen gels: Visual real-time analysis by time-lapse microscopy. Biosci. Trends 2011, 5, 10–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina, S.H.; Bush, B.; Cam, M.; Sevcik, E.; DelRio, F.W.; Nandy, K.; Schneider, J.P. Identification of a mechanogenetic link between substrate stiffness and chemotherapeutic response in breast cancer. Biomaterials 2019, 202, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zustiak, S.P.; Dadhwal, S.; Medina, C.; Steczina, S.; Chehreghanianzabi, Y.; Ashraf, A.; Asuri, P. Three-dimensional matrix stiffness and adhesive ligands affect cancer cell response to toxins. Biotechnol. Bioeng. 2015, 113, 443–452. [Google Scholar] [CrossRef]
- Willits, R.K.; Skornia, S.L. Effect of collagen gel stiffness on neurite extension. J. Biomater. Sci. Polym. Ed. 2004, 15, 1521–1531. [Google Scholar] [CrossRef] [Green Version]
- Mason, B.N.; Starchenko, A.; Williams, R.M.; Bonassar, L.J.; Reinhart-King, C.A. Tuning three-dimensional collagen matrix stiffness independently of collagen concentration modulates endothelial cell behavior. Acta Biomater. 2013, 9, 4635–4644. [Google Scholar] [CrossRef] [Green Version]
- Mullen, C.A.; Vaughan, T.J.; Billiar, K.L.; McNamara, L.M. The Effect of Substrate Stiffness, Thickness, and Cross-Linking Density on Osteogenic Cell Behavior. Biophys. J. 2015, 108, 1604–1612. [Google Scholar] [CrossRef] [Green Version]
- Mullen, C.; Haugh, M.; Schaffler, M.; Majeska, R.; McNamara, L. Osteocyte differentiation is regulated by extracellular matrix stiffness and intercellular separation. J. Mech. Behav. Biomed. Mater. 2013, 28, 183–194. [Google Scholar] [CrossRef] [Green Version]
- Tirella, A.; Liberto, T.; Ahluwalia, A. Riboflavin and collagen: New crosslinking methods to tailor the stiffness of hydrogels. Mater. Lett. 2012, 74, 58–61. [Google Scholar] [CrossRef]
- Kuo, C.K.; Ma, P.X. Ionically crosslinked alginate hydrogels as scaffolds for tissue engineering: Part Structure, gelation rate and mechanical properties. Biomaterials 2001, 22, 511–521. [Google Scholar] [CrossRef]
- Kaklamani, G.; Cheneler, D.; Grover, L.; Adams, M.J.; Bowen, J. Mechanical properties of alginate hydrogels manufactured using external gelation. J. Mech. Behav. Biomed. Mater. 2014, 36, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Candiello, J.; Singh, S.S.; Task, K.; Kumta, P.N.; Banerjee, I. Early differentiation patterning of mouse embryonic stem cells in response to variations in alginate substrate stiffness. J. Biol. Eng. 2013, 7, 9. [Google Scholar] [CrossRef] [Green Version]
- Burdick, J.A.; Chung, C.; Jia, X.; Randolph, M.A.; Langer, R. Controlled Degradation and Mechanical Behavior of Photopolymerized Hyaluronic Acid Networks. Biomacromolecules 2004, 6, 386–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peyton, S.R.; Raub, C.B.; Keschrumrus, V.P.; Putnam, A.J. The use of poly(ethylene glycol) hydrogels to investigate the impact of ECM chemistry and mechanics on smooth muscle cells. Biomaterials 2006, 27, 4881–4893. [Google Scholar] [CrossRef] [PubMed]
- Kolewe, K.W.; Kalasin, S.; Shave, M.; Schiffman, J.D.; Santore, M.M. Mechanical Properties and Concentrations of Poly(ethylene glycol) in Hydrogels and Brushes Direct the Surface Transport of Staphylococcus aureus. ACS Appl. Mater. Interfaces 2019, 11, 320–330. [Google Scholar] [CrossRef] [Green Version]
- Denisin, A.; Pruitt, B.L. Tuning the Range of Polyacrylamide Gel Stiffness for Mechanobiology Applications. ACS Appl. Mater. Interfaces 2016, 8, 21893–21902. [Google Scholar] [CrossRef]
- Palchesko, R.N.; Zhang, L.; Sun, Y.; Feinberg, A.W. Development of Polydimethylsiloxane Substrates with Tunable Elastic Modulus to Study Cell Mechanobiology in Muscle and Nerve. PLoS ONE 2012, 7, e51499. [Google Scholar] [CrossRef] [Green Version]
- Prager-Khoutorsky, M.; Lichtenstein, A.; Krishnan, R.; Rajendran, K.; Mayo, A.; Kam, Z.; Geiger, B.; Bershadsky, A. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat. Cell Biol. 2011, 13, 1457–1465. [Google Scholar] [CrossRef]
- Yang, B.; Wolfenson, H.; Chung, V.Y.; Nakazawa, N.; Liu, S.; Hu, J.; Huang, R.Y.-J.; Sheetz, M.P. Stopping transformed cancer cell growth by rigidity sensing. Nat. Mater. 2020, 19, 239–250. [Google Scholar] [CrossRef]
- Scelsi, A.; Bochicchio, B.; Smith, A.; Workman, V.L.; Díaz, L.A.C.; Saiani, A.; Pepe, A. Tuning of hydrogel stiffness using a two-component peptide system for mammalian cell culture. J. Biomed. Mater. Res. Part A 2019, 107, 535–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiew, S.H.; Mohanram, H.; Ning, L.; Guo, J.; Sánchez-Ferrer, A.; Shi, X.; Pervushin, K.; Mu, Y.; Mezzenga, R.; Miserez, A. A Short Peptide Hydrogel with High Stiffness Induced by 3 10 -Helices to β-Sheet Transition in Water. Adv. Sci. 2019, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashworth, J.; Thompson, J.; James, J.; Slater, C.; Pijuan-Galitó, S.; Lis-Slimak, K.; Holley, R.; Meade, K.; Thompson, A.; Arkill, K.; et al. Peptide gels of fully-defined composition and mechanics for probing cell-cell and cell-matrix interactions in vitro. Matrix Biol. 2020, 85-86, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Yunoki, S.; Ikoma, T.; Tanaka, J. Development of collagen condensation method to improve mechanical strength of tissue engineering scaffolds. Mater. Charact. 2010, 61, 907–911. [Google Scholar] [CrossRef]
- Doyle, A.D. Generation of 3D Collagen Gels with Controlled Diverse Architectures. Curr. Protoc. Cell Biol. 2016, 72, 10.20.1–10.20.16. [Google Scholar] [CrossRef]
- Sung, K.E.; Su, G.; Pehlke, C.; Trier, S.M.; Eliceiri, K.W.; Keely, P.J.; Friedl, A.; Beebe, D.J. Control of 3-dimensional collagen matrix polymerization for reproducible human mammary fibroblast cell culture in microfluidic devices. Biomaterials 2009, 30, 4833–4841. [Google Scholar] [CrossRef] [Green Version]
- Gobeaux, F.; Mosser, G.; Anglo, A.; Panine, P.; Davidson, P.; Giraud-Guille, M.-M.; Belamie, E. Fibrillogenesis in Dense Collagen Solutions: A Physicochemical Study. J. Mol. Biol. 2008, 376, 1509–1522. [Google Scholar] [CrossRef]
- Antoine, E.E.; Vlachos, P.P.; Rylander, M.N. Review of collagen I hydrogels for bioengineered tissue microenvironments: Characterization of mechanics, structure, and transport. Tissue Eng. Part B Rev. 2014, 20, 683–696. [Google Scholar] [CrossRef] [Green Version]
- Nesrinne, S.; Djamel, A. Synthesis, characterization and rheological behavior of pH sensitive poly(acrylamide-co-acrylic acid) hydrogels. Arab. J. Chem. 2017, 10, 539–547. [Google Scholar] [CrossRef] [Green Version]
- Ranjha, N.M.; Ayub, G.; Naseem, S.; Ansari, M.T. Preparation and characterization of hybrid pH-sensitive hydrogels of chitosan-co-acrylic acid for controlled release of verapamil. J. Mater. Sci. Mater. Electron. 2010, 21, 2805–2816. [Google Scholar] [CrossRef]
- Xu, H.; Matysiak, S. Effect of pH on chitosan hydrogel polymer network structure. Chem. Commun. 2017, 53, 7373–7376. [Google Scholar] [CrossRef]
- Parenteau-Bareil, R.; Gauvin, R.; Berthod, F. Collagen-Based Biomaterials for Tissue Engineering Applications. Materials 2010, 3, 1863–1887. [Google Scholar] [CrossRef] [Green Version]
- Raub, C.B.; Suresh, V.; Krasieva, T.; Lyubovitsky, J.; Mih, J.D.; Putnam, A.; Tromberg, B.J.; George, S.C. Noninvasive Assessment of Collagen Gel Microstructure and Mechanics Using Multiphoton Microscopy. Biophys. J. 2007, 92, 2212–2222. [Google Scholar] [CrossRef] [Green Version]
- Sionkowska, A. Biopolymeric nanocomposites for potential biomedical applications. Polym. Int. 2016, 65, 1123–1131. [Google Scholar] [CrossRef]
- Yan, L.; Wang, Y.-J.; Ren, L.; Wu, G.; Caridade, S.G.; Fan, J.-B.; Wang, L.-Y.; Ji, P.-H.; Oliveira, J.M.; Oliveira, J.T.; et al. Genipin-cross-linked collagen/chitosan biomimetic scaffolds for articular cartilage tissue engineering applications. J. Biomed. Mater. Res. Part A 2010, 95, 465–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sionkowska, A.; Skrzyński, S.; Śmiechowski, K.; Kołodziejczak, A. The review of versatile application of collagen. Polym. Adv. Technol. 2017, 28, 4–9. [Google Scholar] [CrossRef]
- Davidenko, N.; Schuster, C.F.; Bax, D.V.; Farndale, R.W.; Hamaia, S.; Best, S.M.; Cameron, R.E. Evaluation of cell binding to collagen and gelatin: A study of the effect of 2D and 3D architecture and surface chemistry. J. Mater. Sci. Mater. Med. 2016, 27, 148. [Google Scholar] [CrossRef] [Green Version]
- Plant, A.L.; Bhadriraju, K.; Spurlin, T.A.; Elliott, J.T. Cell response to matrix mechanics: Focus on collagen. Biochim. Biophys. Acta Mol. Cell Res. 2009, 1793, 893–902. [Google Scholar] [CrossRef] [Green Version]
- Robinson, B.K.; Cortes, E.; Rice, A.J.; Sarper, M.; Hernández, A.D.R. Quantitative analysis of 3D extracellular matrix remodelling by pancreatic stellate cells. Biol. Open 2016, 5, 875–882. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.Y.; Mooney, D.J. Alginate: Properties and biomedical applications. Prog. Polym. Sci. 2012, 37, 106–126. [Google Scholar] [CrossRef] [Green Version]
- Augst, A.D.; Kong, H.J.; Mooney, D.J. Alginate Hydrogels as Biomaterials. Macromol. Biosci. 2006, 6, 623–633. [Google Scholar] [CrossRef]
- Genes, N.G.; Rowley, J.A.; Mooney, D.; Bonassar, L.J. Effect of substrate mechanics on chondrocyte adhesion to modified alginate surfaces. Arch. Biochem. Biophys. 2004, 422, 161–167. [Google Scholar] [CrossRef]
- Kong, H.-J.; Lee, K.Y.; Mooney, D.J. Decoupling the dependence of rheological/mechanical properties of hydrogels from solids con-centration. Polymer 2002, 43, 6239–6246. [Google Scholar] [CrossRef]
- Freeman, F.; Kelly, D.J. Tuning Alginate Bioink Stiffness and Composition for Controlled Growth Factor Delivery and to Spatially Direct MSC Fate within Bioprinted Tissues. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eiselt, P.; Lee, K.Y.; Mooney, D. Rigidity of Two-Component Hydrogels Prepared from Alginate and Poly(ethylene glycol)−Diamines. Macromolecules 1999, 32, 5561–5566. [Google Scholar] [CrossRef]
- Hashemnejad, S.M.; Kundu, S. Rheological properties and failure of alginate hydrogels with ionic and covalent crosslinks. Soft Matter 2019, 15, 7852–7862. [Google Scholar] [CrossRef] [PubMed]
- Somo, S.I.; Langert, K.; Yang, C.-Y.; Vaicik, M.K.; Ibarra, V.; Appel, A.A.; Akar, B.; Cheng, M.-H.; Brey, E.M. Synthesis and evaluation of dual crosslinked alginate microbeads. Acta Biomater. 2018, 65, 53–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, A.; Gu, L.; Kwee, B.; Li, W.A.; Dellacherie, M.; Celiz, A.D.; Mooney, D.J. Hydrogel substrate stress-relaxation regulates the spreading and proliferation of mouse myoblasts. Acta Biomater. 2017, 62, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Huebsch, N.; Arany, P.; Mao, A.S.; Shvartsman, D.; Ali, O.A.; Bencherif, S.; Rivera-Feliciano, J.; Mooney, D. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat. Mater. 2010, 9, 518–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burdick, J.A.; Prestwich, G.D. Hyaluronic acid hydrogels for biomedical applications. Adv. Mater. 2011, 23, H41–H56. [Google Scholar] [CrossRef] [PubMed]
- Khunmanee, S.; Jeong, Y.; Park, H. Crosslinking method of hyaluronic-based hydrogel for biomedical applications. J. Tissue Eng. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, J.L.; Engler, A.J. Hydrogels with time-dependent material properties enhance cardiomyocyte differentiation in vitro. Biomaterials 2011, 32, 1002–1009. [Google Scholar] [CrossRef] [Green Version]
- Jin, R.; Teixeira, L.M.; Krouwels, A.; Dijkstra, P.; van Blitterswijk, C.; Karperien, M.; Feijen, J. Synthesis and characterization of hyaluronic acid–poly(ethylene glycol) hydrogels via Michael addition: An injectable biomaterial for cartilage repair. Acta Biomater. 2010, 6, 1968–1977. [Google Scholar] [CrossRef] [PubMed]
- Caliari, S.; Perepelyuk, M.; Soulas, E.M.; Lee, G.Y.; Wells, R.G.; Burdick, J.A. Gradually softening hydrogels for modeling hepatic stellate cell behavior during fibrosis regression. Integr. Biol. 2016, 8, 720–728. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.J.; Fernandes-Cunha, G.M.; Myung, D. In situ-forming hyaluronic acid hydrogel through visible light-induced thiol-ene reaction. React. Funct. Polym. 2018, 131, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Guvendiren, M.; Burdick, J.A. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat. Commun. 2012, 3, 792. [Google Scholar] [CrossRef] [Green Version]
- Khetan, S.; Guvendiren, M.; Legant, W.R.; Cohen, D.M.; Chen, C.; Burdick, J.A. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat. Mater. 2013, 12, 458–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caliari, S.; Perepelyuk, M.; Cosgrove, B.D.; Tsai, S.J.; Lee, G.Y.; Mauck, R.; Wells, R.G.; Burdick, J.A. Stiffening hydrogels for investigating the dynamics of hepatic stellate cell mechanotransduction during myofibroblast activation. Sci. Rep. 2016, 6, 21387. [Google Scholar] [CrossRef] [Green Version]
- Dicker, K.T.; Gurski, L.A.; Pradhan-Bhatt, S.; Witt, R.L.; Farach-Carson, M.; Jia, X. Hyaluronan: A simple polysaccharide with diverse biological functions. Acta Biomater. 2014, 10, 1558–1570. [Google Scholar] [CrossRef] [Green Version]
- Guvendiren, M.; Perepelyuk, M.; Wells, R.G.; Burdick, J.A. Hydrogels with differential and patterned mechanics to study stiffness-mediated myofibroblastic differentiation of hepatic stellate cells. J. Mech. Behav. Biomed. Mater. 2014, 38, 198–208. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J. Bioactive modification of poly(ethylene glycol) hydrogels for tissue engineering. Biomaterials 2010, 31, 4639–4656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yesildag, C.; Ouyang, Z.; Zhang, Z.; Lensen, M.C. Micro-Patterning of PEG-Based Hydrogels with Gold Nanoparticles Using a Reactive Micro-Contact-Printing Approach. Front. Chem. 2019, 6, 667. [Google Scholar] [CrossRef] [PubMed]
- Peter, M.; Tayalia, P. An alternative technique for patterning cells on poly(ethylene glycol) diacrylate hydrogels. RSC Adv. 2016, 6, 40878–40885. [Google Scholar] [CrossRef]
- Hansen, T.D.; Koepsel, J.T.; Le, N.N.; Nguyen, E.H.; Zorn, S.; Parlato, M.; Loveland, S.G.; Schwartz, M.P.; Murphy, W.L. Biomaterial arrays with defined adhesion ligand densities and matrix stiffness identify distinct phenotypes for tumorigenic and non-tumorigenic human mesenchymal cell types. Biomater. Sci. 2014, 2, 745–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metters, A.; Hubbell, J. Network Formation and Degradation Behavior of Hydrogels Formed by Michael-Type Addition Reactions. Biomacromolecules 2005, 6, 290–301. [Google Scholar] [CrossRef]
- Fairbanks, B.D.; Schwartz, M.; Halevi, A.E.; Nuttelman, C.R.; Bowman, C.N.; Anseth, K.S. A Versatile Synthetic Extracellular Matrix Mimic via Thiol-Norbornene Photopolymerization. Adv. Mater. 2009, 21, 5005–5010. [Google Scholar] [CrossRef] [Green Version]
- Toepke, M.W.; Impellitteri, N.A.; Theisen, J.M.; Murphy, W.L. Characterization of Thiol-Ene Crosslinked PEG Hydrogels. Macromol. Mater. Eng. 2013, 298, 699–703. [Google Scholar] [CrossRef]
- Nguyen, K.T.; West, J.L. Photopolymerizable hydrogels for tissue engineering applications. Biomaterials 2002, 23, 4307–4314. [Google Scholar] [CrossRef]
- Kloxin, A.M.; Kasko, A.M.; Salinas, C.N.; Anseth, K.S. Photodegradable Hydrogels for Dynamic Tuning of Physical and Chemical Properties. Science 2009, 324, 59–63. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.-Y.; Greene, T.; Lin, T.-Y.; Dawes, C.S.; Korc, M.; Lin, C.-C. Enzyme-mediated stiffening hydrogels for probing activation of pancreatic stellate cells. Acta Biomater. 2017, 48, 258–269. [Google Scholar] [CrossRef] [Green Version]
- Günay, K.A.; Ceccato, T.L.; Silver, J.; Bannister, K.L.; Bednarski, O.J.; Leinwand, L.A.; Anseth, K.S. PEG–Anthracene Hydrogels as an On-Demand Stiffening Matrix to Study Mechanobiology. Angew. Chem. Int. Ed. 2019, 58, 9912–9916. [Google Scholar] [CrossRef] [PubMed]
- Rosales, A.M.; Mabry, K.M.; Nehls, E.; Anseth, K.S. Photoresponsive Elastic Properties of Azobenzene-Containing Poly(ethylene-glycol)-Based Hydrogels. Biomacromolecules 2015, 16, 798–806. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, S.; Benoit, D.S.W. Degradable poly(ethylene glycol) (PEG)-based hydrogels for spatiotemporal control of siR-NA/nanoparticle delivery. J. Control. Release 2018, 287, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Metters, T.A.; Anseth, K.S.; Bowman, C.N. Fundamental studies of a novel, biodegradable PEG-b-PLA hydrogel. Polymer 2000, 41, 3993–4004. [Google Scholar] [CrossRef]
- Vanderhooft, J.L.; Mann, B.K.; Prestwich, G.D. Synthesis and Characterization of Novel Thiol-Reactive Poly(ethylene glycol) Cross-Linkers for Extracellular-Matrix-Mimetic Biomaterials. Biomacromolecules 2007, 8, 2883–2889. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, P.; Kirkland, B.; Liu, Y.; Guan, J. Microcontact printing of polyelectrolytes on PEG using an unmodified PDMS stamp for micropatterning nanoparticles, DNA, proteins and cells. Soft Matter 2012, 8, 7630. [Google Scholar] [CrossRef]
- Jo, S.; Park, K. Surface modification using silanated poly(ethylene glycol)s. Biomaterials 2000, 21, 605–616. [Google Scholar] [CrossRef]
- Ren, D.; Xia, Y.; Wang, J.; You, Z. Micropatterning of single cell arrays using the PEG-Silane and Biotin–(Strept)Avidin System with photolithography and chemical vapor deposition. Sens. Actuators B Chem. 2013, 188, 340–346. [Google Scholar] [CrossRef]
- Kumai, J.; Sasagawa, S.; Horie, M.; Yui, Y. A Novel Method for Polyacrylamide Gel Preparation Using N-hydroxysuccinimide-acrylamide Ester to Study Cell-Extracellular Matrix Mechanical Interactions. Front. Mater. 2021, 8. [Google Scholar] [CrossRef]
- Damljanović, V.; Lagerholm, C.; Jacobson, K. Bulk and micropatterned conjugation of extracellular matrix proteins to characterized polyacrylamide substrates for cell mechanotransduction assays. Biotechniques 2005, 39, 847–851. [Google Scholar] [CrossRef] [Green Version]
- Kandow, C.E.; Georges, P.; Janmey, P.A.; Beningo, K.A. Polyacrylamide Hydrogels for Cell Mechanics: Steps Toward Optimization and Alternative Uses. Methods Cell Biol. 2007, 83, 29–46. [Google Scholar] [PubMed]
- Rowlands, A.S.; George, P.A.; Cooper-White, J. Directing osteogenic and myogenic differentiation of MSCs: Interplay of stiffness and adhesive ligand presentation. Am. J. Physiol. Physiol. 2008, 295, C1037–C1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuh, E.; Kramer, J.; Rohwedel, J.; Notbohm, H.; Müller, R.; Gutsmann, T.; Rotter, N. Effect of Matrix Elasticity on the Maintenance of the Chondrogenic Phenotype. Tissue Eng. Part A 2010, 16, 1281–1290. [Google Scholar] [CrossRef] [PubMed]
- Schrader, J.; Gordon-Walker, T.T.; Aucott, R.L.; Van Deemter, M.; Quaas, A.; Walsh, S.; Benten, D.; Forbes, S.J.; Wells, R.G.; Iredale, J.P. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology 2011, 53, 1192–1205. [Google Scholar] [CrossRef] [Green Version]
- Bajaj, P.; Tang, X.; Saif, T.A.; Bashir, R. Stiffness of the substrate influences the phenotype of embryonic chicken cardiac myocytes. J. Biomed. Mater. Res. Part A 2010, 95, 1261–1269. [Google Scholar] [CrossRef] [Green Version]
- Engler, A.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [Green Version]
- Lachowski, D.; Cortes, E.; Pink, D.; Chronopoulos, A.; Karim, S.A.; Morton, J.P.; Hernández, A.E.D.R. Substrate Rigidity Controls Activation and Durotaxis in Pancreatic Stellate Cells. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Hartman, C.D.; Isenberg, B.C.; Chua, S.G.; Wong, J.Y. Vascular smooth muscle cell durotaxis depends on extracellular matrix composition. Proc. Natl. Acad. Sci. USA 2016, 113, 11190–11195. [Google Scholar] [CrossRef] [Green Version]
- Masterton, S.; Ahearne, M. Influence of polydimethylsiloxane substrate stiffness on corneal epithelial cells. R. Soc. Open Sci. 2019, 6, 191796. [Google Scholar] [CrossRef] [Green Version]
- Prauzner-Bechcicki, S.; Raczkowska, J.; Madej, E.; Pabijan, J.; Lukes, J.; Sepitka, J.; Rysz, J.; Awsiuk, K.; Bernasik, A.; Budkowski, A.; et al. PDMS substrate stiffness affects the morphology and growth profiles of cancerous prostate and melanoma cells. J. Mech. Behav. Biomed. Mater. 2015, 41, 13–22. [Google Scholar] [CrossRef]
- Chuah, Y.J.; Heng, Z.T.; Tan, J.S.; Tay, L.M.; Lim, C.S.; Kang, Y.; Wang, D.-A. Surface modifications to polydimethylsiloxane substrate for stabilizing prolonged bone marrow stromal cell culture. Colloids Surf. B Biointerfaces 2020, 191, 110995. [Google Scholar] [CrossRef]
- Chuah, Y.J.; Zhang, Y.; Wu, Y.; Menon, N.V.; Goh, G.H.; Lee, A.C.; Chan, V.; Zhang, Y.; Kang, Y. Combinatorial effect of substratum properties on mesenchymal stem cell sheet engineering and subsequent multi-lineage differentiation. Acta Biomater. 2015, 23, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Lantoine, J.; Grevesse, T.; Villers, A.; Delhaye, G.; Mestdagh, C.; Versaevel, M.; Mohammed, D.; Bruyère, C.; Alaimo, L.; Lacour, S.P.; et al. Matrix stiffness modulates formation and activity of neuronal networks of controlled architectures. Biomaterials 2016, 89, 14–24. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.-Y.; Zhang, Y.-Y.; Xie, J.; Li, C.-X.; Chen, W.-Y.; Liu, B.-L.; Wu, X.-A.; Li, S.-N.; Huo, B.; Jiang, L.-H.; et al. Stiff substrates enhance cultured neuronal network activity. Sci. Rep. 2015, 4, srep06215. [Google Scholar] [CrossRef] [Green Version]
- Kuddannaya, S.; Bao, J.; Zhang, Y. EnhancedIn VitroBiocompatibility of Chemically Modified Poly(dimethylsiloxane) Surfaces for Stable Adhesion and Long-term Investigation of Brain Cerebral Cortex Cells. ACS Appl. Mater. Interfaces 2015, 7, 25529–25538. [Google Scholar] [CrossRef]
- Kuddannaya, S.; Chuah, Y.J.; Lee, M.H.A.; Menon, N.V.; Kang, Y.; Zhang, Y. Surface Chemical Modification of Poly(dimethylsiloxane) for the Enhanced Adhesion and Proliferation of Mesenchymal Stem Cells. ACS Appl. Mater. Interfaces 2013, 5, 9777–9784. [Google Scholar] [CrossRef] [PubMed]
- Chuah, Y.J.; Kuddannaya, S.; Lee, M.H.A.; Zhang, Y.; Kang, Y. The effects of poly(dimethylsiloxane) surface silanization on the mesenchymal stem cell fate. Biomater. Sci. 2014, 3, 383–390. [Google Scholar] [CrossRef]
- Chen, J.; Zou, X. Self-assemble peptide biomaterials and their biomedical applications. Bioact. Mater. 2019, 4, 120–131. [Google Scholar] [CrossRef]
- Li, R.; Horgan, C.; Long, B.; Rodriguez, A.L.; Mather, L.; Barrow, C.J.; Nisbet, D.R.; Williams, R.J. Tuning the mechanical and morphological properties of self-assembled peptide hydrogels via control over the gelation mechanism through regulation of ionic strength and the rate of pH change. RSC Adv. 2014, 5, 301–307. [Google Scholar] [CrossRef]
- Gao, J.; Tang, C.; Elsawy, M.; Smith, A.M.; Miller, A.F.; Saiani, A. Controlling Self-Assembling Peptide Hydrogel Properties through Network Topology. Biomacromolecules 2017, 18, 826–834. [Google Scholar] [CrossRef]
- Burgess, K.; Frati, C.; Meade, K.; Gao, J.; Diaz, L.C.; Madeddu, D.; Graiani, G.; Cavalli, S.; Miller, A.; Oceandy, D.; et al. Functionalised peptide hydrogel for the delivery of cardiac progenitor cells. Mater. Sci. Eng. C 2021, 119, 111539. [Google Scholar] [CrossRef]
- Yang, S.; Wang, C.; Zhu, J.; Lu, C.; Li, H.; Chen, F.; Lu, J.; Zhang, Z.; Yan, X.; Zhao, H.; et al. Self-assembling peptide hydrogels functionalized with LN- and BDNF- mimicking epitopes synergistically enhance peripheral nerve regeneration. Theranostics 2020, 10, 8227–8249. [Google Scholar] [CrossRef]
- Tao, H.; Wu, Y.; Li, H.; Wang, C.; Zhang, Y.; Li, C.; Wen, T.; Wang, X.; He, Q.; Wang, D.; et al. BMP7-Based Functionalized Self-Assembling Peptides for Nucleus Pulposus Tissue Engineering. ACS Appl. Mater. Interfaces 2015, 7, 17076–17087. [Google Scholar] [CrossRef]
- Zhang, S.; Holmes, T.; Lockshin, C.; Rich, A. Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane. Proc. Natl. Acad. Sci. USA 1993, 90, 3334–3338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clough, H.C.; O’Brien, M.; Zhu, X.; Miller, A.F.; Saiani, A.; Tsigkou, O. Neutrally charged self-assembling peptide hydrogel recapitulates in vitro mechanisms of breast cancer progression. Mater. Sci. Eng. C 2021, 127, 112200. [Google Scholar] [CrossRef]
- Wychowaniec, J.K.; Smith, A.M.; Ligorio, C.; Mykhaylyk, O.O.; Miller, A.F.; Saiani, A. Role of Sheet-Edge Interactions in β-sheet Self-Assembling Peptide Hydrogels. Biomacromolecules 2020, 21, 2285–2297. [Google Scholar] [CrossRef]
- Kalli, M.; Stylianopoulos, T. Defining the Role of Solid Stress and Matrix Stiffness in Cancer Cell Proliferation and Metastasis. Front. Oncol. 2018, 8, 55. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, N.A.; Liu, T.-C.; Cavatiao, A.; Mawad, K.; Chen, L.; Strasberg, S.S.; Linehan, D.C.; Cao, D.; Hawkins, W.G. Ki-67 predicts disease recurrence and poor prognosis in pancreatic neuroendocrine neoplasms. Surgery 2012, 152, 107–113. [Google Scholar] [CrossRef] [Green Version]
- Melling, N.; Kowitz, C.M.; Simon, R.; Bokemeyer, C.; Terracciano, L.; Sauter, G.; Izbicki, J.R.; Marx, A.H. High Ki67 expression is an independent good prognostic marker in colorectal cancer. J. Clin. Pathol. 2016, 69, 209–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimura, R.; Osako, T.; Okumura, Y.; Hayashi, M.; Toyozumi, Y.; Arima, N. Ki-67 as a prognostic marker according to breast cancer subtype and a predictor of recurrence time in primary breast cancer. Exp. Ther. Med. 2010, 1, 747–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soliman, A.N.; Yussif, S.M. Ki-67 as a prognostic marker according to breast cancer molecular subtype. Cancer Biol. Med. 2016, 13, 496–504. [Google Scholar] [PubMed] [Green Version]
- Estrella, V.; Chen, T.; Lloyd, M.; Wojtkowiak, J.; Cornnell, H.; Ibrahim-Hashim, A.; Bailey, K.; Balagurunathan, Y.; Rothberg, J.M.; Sloane, B.F.; et al. Acidity Generated by the Tumor Microenvironment Drives Local Invasion. Cancer Res. 2013, 73, 1524–1535. [Google Scholar] [CrossRef] [Green Version]
- Busco, G.; Cardone, R.A.; Greco, M.R.; Bellizzi, A.; Colella, M.; Antelmi, E.; Mancini, M.T.; Dell’Aquila, M.E.; Casavola, V.; Paradiso, A.; et al. NHE1 promotes invadopodial ECM proteolysis through acidification of the peri-invadopodial space. FASEB J. 2010, 24, 3903–3915. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Zaguilan, R.; Seftor, E.A.; Seftor, R.E.B.; Chu, Y.-W.; Gillies, R.J.; Hendrix, M.J.C. Acidic pH enhances the invasive behavior of human melanoma cells. Clin. Exp. Metastasis 1996, 14, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Cortes, E.; Lachowski, D.; Robinson, B.; Sarper, M.; Teppo, J.S.; Thorpe, S.D.; Lieberthal, T.; Iwamoto, K.; A Lee, D.; Okada-Hatakeyama, M.; et al. Tamoxifen mechanically reprograms the tumor microenvironment via HIF-1A and reduces cancer cell survival. EMBO Rep. 2019, 20, 46557. [Google Scholar] [CrossRef]
- Feng, S.; Bowden, N.; Fragiadaki, M.; Souilhol, C.; Hsiao, S.; Mahmoud, M.; Allen, S.; Pirri, D.; Ayllon, B.T.; Akhtar, S.; et al. Mechanical Activation of Hypoxia-Inducible Factor 1α Drives Endothelial Dysfunction at Atheroprone Sites. Arter. Thromb. Vasc. Biol. 2017, 37, 2087–2101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.-H.; Cho, Y.-S.; Chun, Y.-S.; Park, J.-W.; Kim, M.-S. Early Expression of Myocardial HIF-1α in Response to Mechanical Stresses. Circ. Res. 2002, 90, e25–e33. [Google Scholar] [CrossRef] [Green Version]
- Wen, J.H.; Vincent, L.G.; Fuhrmann, A.; Choi, Y.S.; Hribar, K.C.; Taylor-Weiner, H.; Chen, S.; Engler, A.J. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat. Mater. 2014, 13, 979–987. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
| Material | Stiffness Range | Advantages | Disadvantages | References |
|---|---|---|---|---|
| Collagen | 10–100 Pa ~1 kPa * | Mimics the physiological ECM Suitable for 3D fibrous matrices | Low stiffness range Stiffness and ligands cannot be controlled independently | [26,27,28,29,30] |
| Alginate | 0.2–550 kPa | Excellent bioprinting properties Ionic and/or covalent crosslinking | Stiffness difficult to tune independently | [31,32,33] |
| Hyaluronic Acid | 2–100 kPa | Multiple functional groups and crosslinking methods Dynamic hydrogels | Confounding biological signaling Requires chemical expertise | [34] |
| PEG | 2–1000 kPa ** 10–400 kPa | High degree of design flexibility | Requires expertise to synthesize and functionalize | [35,36] |
| PAA | 1–1000 kPa | Easy to tune over a large range of rigidity. Independent control of stiffness and ligand presentation | Not suitable for 3D culture | [37] |
| PDMS | 5–2000 kPa | Easy to tune over a large range of rigidity. Chemically inert | Difficult to functionalize for long term culture Not suitable for 3D culture | [38,39,40] |
| Peptides | 0.1–120 kPa ** | High design flexibility Direct functionalization Highly reproducible | Early development | [41,42,43] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lachowski, D.; Matellan, C.; Cortes, E.; Saiani, A.; Miller, A.F.; del Río Hernández, A.E. Self-Assembling Polypeptide Hydrogels as a Platform to Recapitulate the Tumor Microenvironment. Cancers 2021, 13, 3286. https://doi.org/10.3390/cancers13133286
Lachowski D, Matellan C, Cortes E, Saiani A, Miller AF, del Río Hernández AE. Self-Assembling Polypeptide Hydrogels as a Platform to Recapitulate the Tumor Microenvironment. Cancers. 2021; 13(13):3286. https://doi.org/10.3390/cancers13133286
Chicago/Turabian StyleLachowski, Dariusz, Carlos Matellan, Ernesto Cortes, Alberto Saiani, Aline F. Miller, and Armando E. del Río Hernández. 2021. "Self-Assembling Polypeptide Hydrogels as a Platform to Recapitulate the Tumor Microenvironment" Cancers 13, no. 13: 3286. https://doi.org/10.3390/cancers13133286
APA StyleLachowski, D., Matellan, C., Cortes, E., Saiani, A., Miller, A. F., & del Río Hernández, A. E. (2021). Self-Assembling Polypeptide Hydrogels as a Platform to Recapitulate the Tumor Microenvironment. Cancers, 13(13), 3286. https://doi.org/10.3390/cancers13133286








