DNA Methylation Is Correlated with Oxidative Stress in Myelodysplastic Syndrome—Relevance as Complementary Prognostic Biomarkers
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Sample Preparation
2.3. Uric Acid Determinations
2.4. Vitamin A and E Measurements
2.5. Reduced Glutathione Quantification
2.6. Total Antioxidant Status Evaluation
2.7. Antioxidant Enzyme Activity Determination
2.8. Lipid Peroxidation Measurements
2.9. Plasmatic Nitric Oxide Quantification
2.10. Plasmatic Peroxide Quantification
2.11. Plasmatic 8-Hydroxy-2-Deoxyguanosine Quantification
2.12. Global DNA Methylation Analysis
2.13. Methylation Pattern of Tumor Suppressor Genes
2.14. Statistical Analysis
3. Results
3.1. Biodemographic and Clinical Characteristics of MDS Patients
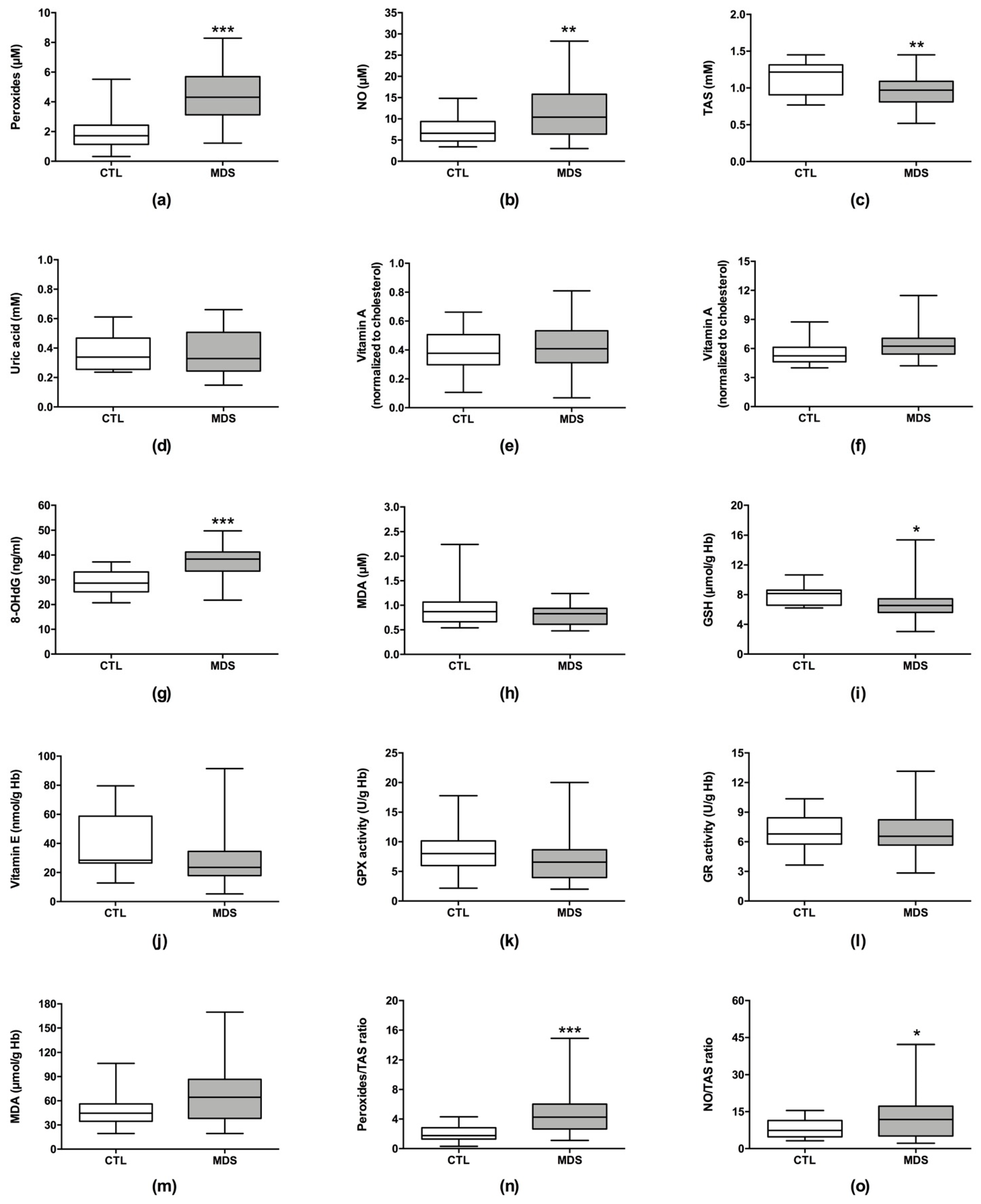
3.2. Oxidative Stress Levels in MDS Patients
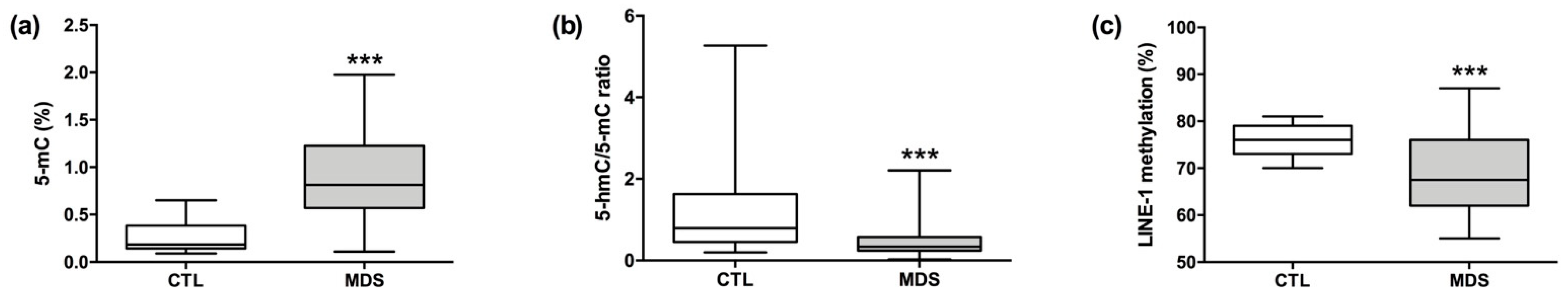
3.3. DNA Methylation Status in MDS Patients
3.4. Oxidative Stress and DNA Methylation as MDS Diagnostic Biomarkers
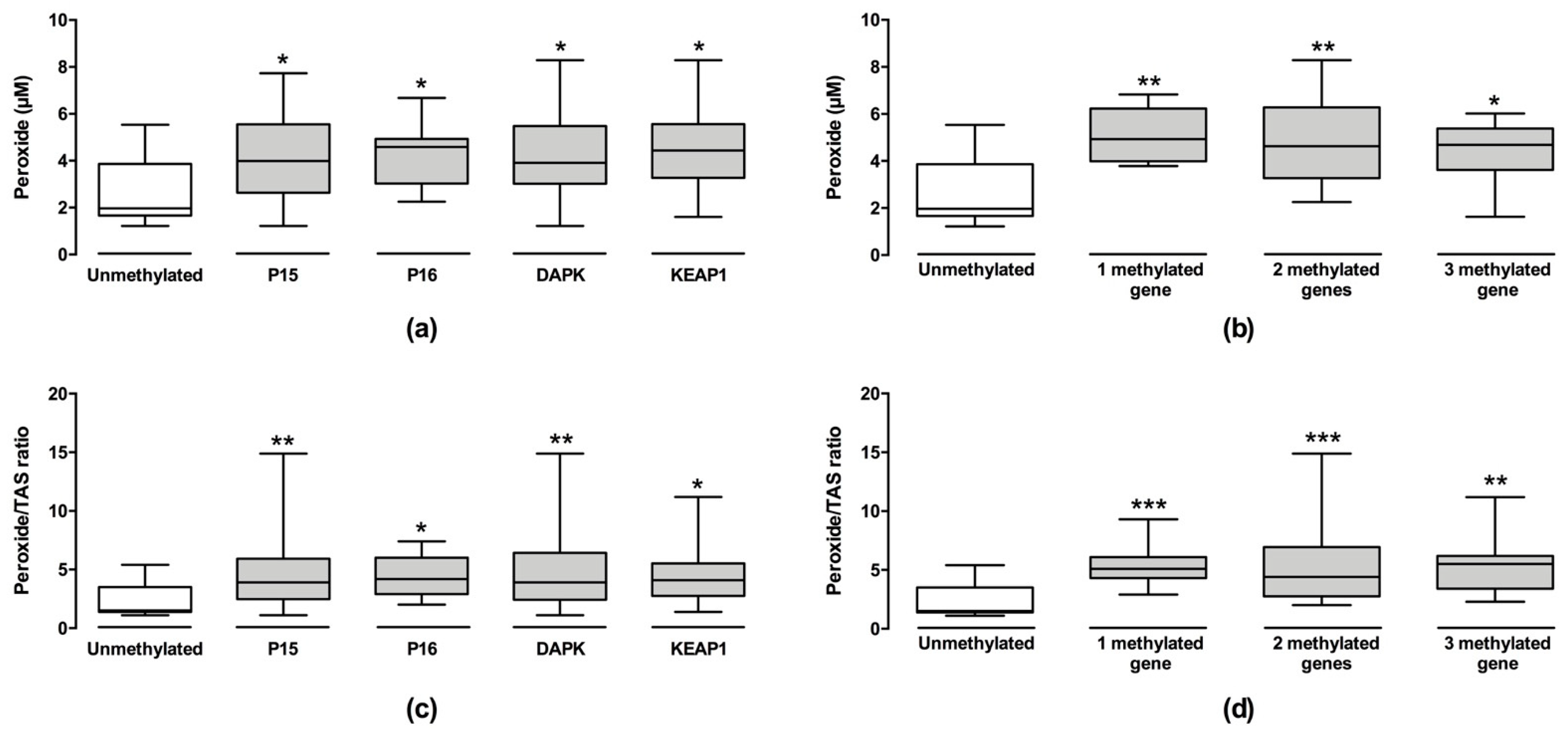
3.5. Correlation between Oxidative Stress and DNA Methylation in MDS Patients
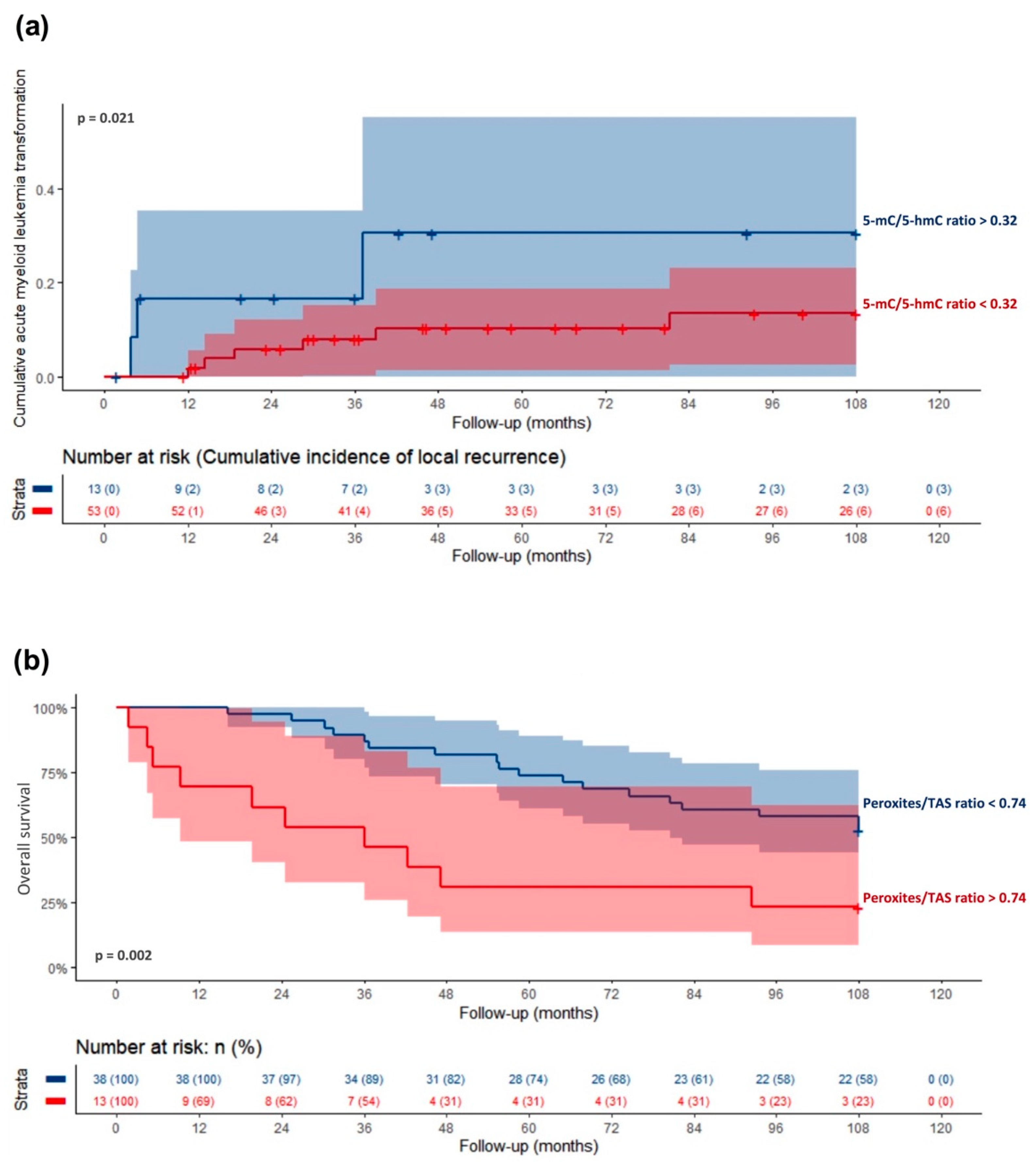
3.6. Association of Oxidative Stress and DNA Methylation with MDS Clinical Outcome
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nimer, S.D. Myelodysplastic syndromes. Blood 2008, 111, 4841–4851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adès, L.; Itzykson, R.; Fenaux, P. Myelodysplastic syndromes. Lancet 2014, 383, 2239–2252. [Google Scholar] [CrossRef]
- Issa, J.-P.J. The myelodysplastic syndrome as a prototypical epigenetic disease. Blood 2013, 121, 3811–3817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cazzola, M. Myelodysplastic Syndromes. N. Engl. J. Med. 2020, 383, 1358–1374. [Google Scholar] [CrossRef]
- Westermann, J.; Bullinger, L. Precision medicine in myeloid malignancies. In Seminars in Cancer Biology; Academic Press: New York, NY, USA, 2021; S1044-579X(21)00084-5. [Google Scholar]
- Palomo, L.; Acha, P.; Solé, F. Genetic Aspects of Myelodysplastic/Myeloproliferative Neoplasms. Cancers 2021, 13, 2120. [Google Scholar]
- Kitamura, T.; Inoue, D.; Okochi-Watanabe, N.; Kato, N.; Komeno, Y.; Lu, Y.; Enomoto, Y.; Doki, N.; Uchida, T.; Kagiyama, Y.; et al. The molecular basis of myeloid malignancies. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2014, 90, 389–404. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.H. Myelodysplastic syndromes and overlap syndromes. Blood Res. 2021, 56, S51–S64. [Google Scholar] [CrossRef]
- Sardina, J.L.; López-Ruano, G.; Sánchez-Sánchez, B.; Llanillo, M.; Hernández-Hernández, A. Reactive oxygen species: Are they important for haematopoiesis? Crit. Rev. Oncol. Hematol. 2012, 81, 257–274. [Google Scholar] [CrossRef]
- Imbesi, S.; Musolino, C.; Allegra, A.; Saija, A.; Morabito, F.; Calapai, G.; Gangemi, S. Oxidative stress in oncohematologic diseases: An update. Expert Rev. Hematol. 2013, 6, 317–325. [Google Scholar] [CrossRef]
- Ghaffari, S. Oxidative stress in the regulation of normal and neoplastic hematopoiesis. Antioxid. Redox Signal. 2008, 10, 1923–1940. [Google Scholar] [CrossRef] [Green Version]
- Picou, F.; Vignon, C.; Debeissat, C.; Lachot, S.; Kosmider, O.; Gallay, N.; Foucault, A.; Estienne, M.H.; Ravalet, N.; Bene, M.C.; et al. Bone marrow oxidative stress and specific antioxidant signatures in myelodysplastic syndromes. Blood Adv. 2019, 3, 4271–4279. [Google Scholar]
- Gonçalves, A.C.; Cortesão, E.; Oliveiros, B.; Alves, V.; Espadana, A.I.; Rito, L.; Magalhães, E.; Lobão, M.J.; Pereira, A.; Nascimento Costa, J.M.; et al. Oxidative stress and mitochondrial dysfunction play a role in myelodysplastic syndrome development, diagnosis, and prognosis: A pilot study. Free Radic. Res. 2015, 49, 1081–1094. [Google Scholar] [CrossRef]
- Ghoti, H.; Amer, J.; Winder, A.; Rachmilewitz, E.; Fibach, E. Oxidative stress in red blood cells, platelets and polymorphonuclear leukocytes from patients with myelodysplastic syndrome. Eur. J. Haematol. 2007, 79, 463–467. [Google Scholar] [CrossRef]
- Wu, Q.; Ni, X. ROS-mediated DNA methylation pattern alterations in carcinogenesis. Curr. Drug Targets 2015, 16, 13–19. [Google Scholar] [CrossRef]
- Weitzman, S.A.; Turk, P.W.; Milkowski, D.H.; Kozlowski, K. Free radical adducts induce alterations in DNA cytosine methylation. Proc. Natl. Acad. Sci. USA 1994, 91, 1261–1264. [Google Scholar] [CrossRef] [Green Version]
- Afanas’ev, I. New nucleophilic mechanisms of ROS-dependent epigenetic modifications: Comparison of aging and cancer. Aging Dis. 2014, 5, 52–62. [Google Scholar] [CrossRef]
- Campos, A.C.E.; Molognoni, F.; Melo, F.H.M.; Galdieri, L.C.; Carneiro, C.R.W.; D’Almeida, V.; Correa, M.; Jasiulionis, M.G. Oxidative stress modulates DNA methylation during melanocyte anchorage blockade associated with malignant transformation. Neoplasia 2007, 9, 1111–1121. [Google Scholar] [CrossRef] [Green Version]
- Santini, V.; Melnick, A.; Maciejewski, J.P.; Duprez, E.; Nervi, C.; Cocco, L.; Ford, K.G.; Mufti, G. Epigenetics in focus: Pathogenesis of myelodysplastic syndromes and the role of hypomethylating agents. Crit. Rev. Oncol. Hematol. 2013, 88, 231–245. [Google Scholar] [CrossRef]
- Maher, M.; Diesch, J.; Le Pannérer, M.M.; Buschbeck, M. Epigenetics in a Spectrum of Myeloid Diseases and Its Exploitation for Therapy. Cancers 2021, 13, 1746. [Google Scholar] [CrossRef]
- Patchsung, M.; Boonla, C.; Amnattrakul, P.; Dissayabutra, T.; Mutirangura, A.; Tosukhowong, P. Long interspersed nuclear element-1 hypomethylation and oxidative stress: Correlation and bladder cancer diagnostic potential. PLoS ONE 2012, 7, e37009. [Google Scholar] [CrossRef] [Green Version]
- Woods, B.A.; Levine, R.L. The role of mutations in epigenetic regulators in myeloid malignancies. Immunol. Rev. 2015, 263, 22–35. [Google Scholar] [CrossRef]
- Zhou, J.D.; Zhang, T.J.; Xu, Z.J.; Deng, Z.Q.; Gu, Y.; Ma, J.C.; Wen, X.M.; Leng, J.Y.; Lin, J.; Chen, S.N.; et al. Genome-wide methylation sequencing identifies progression-related epigenetic drivers in myelodysplastic syndromes. Cell Death Dis. 2020, 11, 997. [Google Scholar] [CrossRef]
- Cabezón, M.; Malinverni, R.; Bargay, J.; Xicoy, B.; Marcé, S.; Garrido, A.; Tormo, M.; Arenillas, L.; Coll, R.; Borras, J.; et al. Different methylation signatures at diagnosis in patients with high-risk myelodysplastic syndromes and secondary acute myeloid leukemia predict azacitidine response and longer survival. Clin. Epigenet. 2021, 13, 9. [Google Scholar] [CrossRef]
- Reilly, B.; Tanaka, T.N.; Diep, D.; Yeerna, H.; Tamayo, P.; Zhang, K.; Bejar, R. DNA methylation identifies genetically and prognostically distinct subtypes of myelodysplastic syndromes. Blood Adv. 2019, 3, 2845–2858. [Google Scholar] [CrossRef]
- Kuan, T.C.; Lin, P.C.; Yang, S.H.; Lin, C.C.; Lan, Y.T.; Lin, H.H.; Liang, W.Y.; Chen, W.S.; Lin, J.K.; Jiang, J.K.; et al. Impact of LINE-1 hypomethylation on the clinicopathological and molecular features of colorectal cancer patients. PLoS ONE 2018, 13, e0197681. [Google Scholar] [CrossRef]
- Gonçalves, A.C.; Cortesão, E.; Oliveiros, B.; Alves, V.; Espadana, A.I.; Rito, L.; Magalhães, E.; Pereira, S.; Pereira, A.; Costa, J.M.; et al. Oxidative stress levels are correlated with P15 and P16 gene promoter methylation in myelodysplastic syndrome patients. Clin. Exp. Med. 2016, 16, 333–343. [Google Scholar] [CrossRef]
- Jorge, J.; Gonçalves, A.C.; Cortesão, E.; Pires, A.C.; Alves, R.; Geraldes, C.; Pereira, A.; Ribeiro, L.; Mascimento Costa, J.M.; Sarmento-Ribeiro, A.B. Can peripheral blood provide good DNA methylation biomarkers in myelodysplastic syndrome? J. Mol. Clin. Med. 2021, 4, 39–45. [Google Scholar] [CrossRef]
- Gonçalves, A.C. Oxidative Stress versus Epigenetic—Role in Susceptibility, Development, and Progression of Myeloid Neoplasms. Ph.D. Thesis, Faculty of Medicine, University of Coimbra, Coimbra, Portugal, 2016. [Google Scholar]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Baldeiras, I.; Santana, I.; Proença, M.T.; Garrucho, M.H.; Pascoal, R.; Rodrigues, A.; Duro, D.; Oliveira, C.R. Oxidative damage and progression to Alzheimer’s disease in patients with mild cognitive impairment. J. Alzheimer’s Dis. 2010, 21, 1165–1177. [Google Scholar] [CrossRef] [PubMed]
- Barham, D.; Trinder, P. An improved colour reagent for the determination of blood glucose by the oxidase system. Analyst 1972, 97, 142–145. [Google Scholar] [CrossRef] [PubMed]
- De Leenheer, A.P.; De Bevere, V.O.; Claeys, A.E. Measurement of alpha-, beta-, and gamma tocopherol in serum by liquid chromatography. Clin. Chem. 1979, 25, 425–428. [Google Scholar] [CrossRef]
- Vatassery, G.T.; Maynard, V.R.; Hagen, D.F. High-performance liquid chromatography of various tocopherols. J. Chromatogr. A 1978, 161, 299–302. [Google Scholar] [CrossRef]
- Paglia, D.E.; Valentine, W.N. Studies on the quantitative and qualitative characterization of erythrocyte glutathione peroxidase. J. Lab. Clin. Med. 1967, 70, 158–169. [Google Scholar]
- Goldberg, D.; Spooner, R. Glutathione reductase. In Methods of Enzymatic Analysis, 3rd ed.; Bergmeyer, H., Bergmeyer, J., Grassl, M., Eds.; Verlag Chemie: Weinheim, Germany, 1983; pp. 258–265. [Google Scholar]
- Titheradge, M.A. The enzymatic measurement of nitrate and nitrite. Methods Mol. Biol. 1998, 100, 83–91. [Google Scholar]
- Bartlett, J.M.S.; White, A. Extraction of DNA from whole blood. Methods Mol. Biol. 2003, 226, 29–32. [Google Scholar]
- Tse, M.Y.; Ashbury, J.E.; Zwingerman, N.; King, W.D.; Taylor, S.A.; Pang, S.C. A refined, rapid and reproducible high resolution melt (HRM)-based method suitable for quantification of global LINE-1 repetitive element methylation. BMC Res. Notes 2011, 4, 565. [Google Scholar] [CrossRef] [Green Version]
- Yeh, K.T.; Chang, J.G.; Lin, T.H.; Wang, Y.F.; Tien, N.; Chang, J.Y.; Chen, J.C.; Shih, M.C. Epigenetic changes of tumor suppressor genes, P15, P16, VHL and TP53 in oral cancer. Oncol. Rep. 2003, 10, 659–663. [Google Scholar] [PubMed]
- Esteller, M.; Garcia-Foncillas, J.; Andion, E.; Goodman, S.N.; Hidalgo, O.F.; Vanaclocha, V.; Baylin, S.B.; Herman, J.G. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N. Engl. J. Med. 2000, 343, 1350–1354. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Gomez, P.; Bello, M.J.; Lomas, J.; Arjona, D.; Alonso, M.E.; Amiñoso, C.; Lopez-Marin, I.; Anselmo, N.P.; Sarasa, J.L.; Gutierrez, M.; et al. Aberrant methylation of multiple genes in neuroblastic tumours. Relationship with MYCN amplification and allelic status at 1p. Eur. J. Cancer 2003, 39, 1478–1485. [Google Scholar] [CrossRef]
- Hanada, N.; Takahata, T.; Zhou, Q.; Ye, X.; Sun, R.; Itoh, J.; Ishiguro, A.; Kijima, H.; Mimura, J.; Itoh, K.; et al. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer 2012, 12, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Souza, G.F.; Ribeiro, H.L.; De Sousa, J.C.; Heredia, F.F.; De Freitas, R.M.; Martins, M.R.; Goncalves, R.P.; Pinheiro, R.F.; Magalhaes, S.M.M. HFE gene mutation and oxidative damage biomarkers in patients with myelodysplastic syndromes and its relation to transfusional iron overload: An observational cross-sectional study. BMJ Open 2015, 5, e006048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saigo, K.; Takenokuchi, M.; Hiramatsu, Y.; Tada, H.; Hishita, T.; Takata, M.; Misawa, M.; Imoto, S.; Imashuku, S. Oxidative stress levels in myelodysplastic syndrome patients: Their relationship to serum ferritin and haemoglobin values. J. Int. Med. Res. 2011, 39, 1941–1945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novotna, B.; Bagryantseva, Y.; Siskova, M.; Neuwirtova, R. Oxidative DNA damage in bone marrow cells of patients with low-risk myelodysplastic syndrome. Leuk. Res. 2009, 33, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Tsamesidis, I.; Pantaleo, A.; Pekou, A.; Gusani, A.; Iliadis, S.; Makedou, K.; Manca, A.; Carruale, A.; Lymperaki, E.; Fozza, C. Correlation of Oxidative Stress Biomarkers and Hematological Parameters in Blood Cancer Patients from Sardinia, Italy. Int. J. Hematol. Oncol. Stem Cell Res. 2019, 13, 49–57. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-Solas, T.; López-Cadenas, F.; Aires-Mejía, I.; Caballero-Berrocal, J.C.; Ortega, R.; Redondo, A.M.; Sánchez-Guijo, F.; Muntión, S.; García-Martín, L.; Albarrán, B.; et al. Deferasirox reduces oxidative DNA damage in bone marrow cells from myelodysplastic patients and improves their differentiation capacity. Br. J. Haematol. 2019, 187, 93–104. [Google Scholar] [CrossRef]
- Honda, M.; Yamada, Y.; Tomonaga, M.; Ichinose, H.; Kamihira, S. Correlation of urinary 8-hydroxy-2′-deoxyguanosine (8-OHdG), a biomarker of oxidative DNA damage, and clinical features of hematological disorders: A pilot study. Leuk. Res. 2000, 24, 461–468. [Google Scholar] [CrossRef]
- Jankowska, A.M.; Gondek, L.P.; Szpurka, H.; Nearman, Z.P.; Tiu, R.V.; Maciejewski, J.P. Base excision repair dysfunction in a subgroup of patients with myelodysplastic syndrome. Leukemia 2008, 22, 551–558. [Google Scholar] [CrossRef] [Green Version]
- Circu, M.L.; Aw, T.Y. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic. Biol. Med. 2010, 48, 749–762. [Google Scholar] [CrossRef] [Green Version]
- Solomon, P.R.; Munirajan, A.K.; Tsuchida, N.; Muthukumarasamy, K.; Rathinavel, A.; Selvam, G.S.; Shanmugam, G. Promoter hypermethylation analysis in myelodysplastic syndromes: Diagnostic prognostic implication. Indian J. Med. Res. 2008, 127, 52–57. [Google Scholar]
- Qian, J.; Yao, D.-M.; Lin, J.; Wang, Y.-L.; Han, L.-X.; Xu, W.-R.; Wu, C.-Y. Methylation of DAPK1 promoter: Frequent but not an adverse prognostic factor in myelodysplastic syndrome. Int. J. Lab. Hematol. 2010, 32, 74–81. [Google Scholar] [CrossRef]
- Baird, L.; Yamamoto, M. The Molecular Mechanisms Regulating the KEAP1-NRF2 Pathway. Mol. Cell. Biol. 2020, 40, e00099-20. [Google Scholar] [CrossRef]
- Zhou, J.D.; Lin, J.; Zhang, T.J.; Ma, J.C.; Yang, L.; Wen, X.M.; Guo, H.; Yang, J.; Deng, Z.Q.; Qian, J. GPX3 methylation in bone marrow predicts adverse prognosis and leukemia transformation in myelodysplastic syndrome. Cancer Med. 2017, 6, 267–274. [Google Scholar] [CrossRef]
- Zhang, T.J.; Zhou, J.D.; Yang, D.Q.; Wang, Y.X.; Yao, D.M.; Ma, J.C.; Wen, X.M.; Guo, H.; Lin, J.; Qian, J. Hypermethylation of DLX4 predicts poor clinical outcome in patients with myelodysplastic syndrome. Clin. Chem. Lab. Med. 2016, 54, 865–871. [Google Scholar] [CrossRef] [PubMed]
- Ko, M.; Huang, Y.; Jankowska, A.M.; Pape, U.J.; Tahiliani, M.; Bandukwala, H.S.; An, J.; Lamperti, E.D.; Koh, K.P.; Ganetzky, R.; et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 2010, 468, 839–843. [Google Scholar] [CrossRef] [Green Version]
- Figueroa, M.E.; Abdel-Wahab, O.; Lu, C.; Ward, P.S.; Patel, J.; Shih, A.; Li, Y.; Bhagwat, N.; Vasanthakumar, A.; Fernandez, H.F.; et al. Leukemic IDH1 and IDH2 Mutations Result in a Hypermethylation Phenotype, Disrupt TET2 Function, and Impair Hematopoietic Differentiation. Cancer Cell 2010, 18, 553–567. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki, J.; Taby, R.; Vasanthakumar, A.; Macrae, T.; Ostler, K.R.; Shen, L.; Kantarjian, H.M.; Estecio, M.R.; Jelinek, J.; Godley, L.; et al. Effects of TET2 mutations on DNA methylation in chronic myelomonocytic leukemia. Epigenetics 2012, 7, 201–207. [Google Scholar] [CrossRef] [Green Version]
- Bujko, M.; Musialik, E.; Olbromski, R.; Przestrzelska, M.; Libura, M.; Pastwińska, A.; Juszczyński, P.; Zwierzchowski, L.; Baranowski, P.; Siedlecki, J.A. Repetitive genomic elements and overall DNA methylation changes in acute myeloid and childhood B-cell lymphoblastic leukemia patients. Int. J. Hematol. 2014, 100, 79–87. [Google Scholar] [CrossRef]
- Sucic, M.; Ljubic, N.; Krmek, D.Z.; Perkovic, L.; Ivanovic, D.; Magazin, M.; Solomun, G.F. Immunoexpression of 5-methylcytosine (5mc) in Bone Marrow Haematopoietic Cells in Patients with Myelodysplastic Syndromes. Ann. Clin. Lab. Sci. 2019, 49, 810–817. [Google Scholar]
- Scopim-Ribeiro, R.; Machado-Neto, J.A.; Campos, P.D.M.; Silva, C.A.M.; Favaro, P.; Lorand-Metze, I.; Costa, F.F.; Saad, S.T.O.; Traina, F. Ten-Eleven-Translocation 2 (TET2) is downregulated in myelodysplastic syndromes. Eur. J. Haematol. 2015, 94, 413–418. [Google Scholar] [CrossRef]
- Rasmussen, K.D.; Helin, K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016, 30, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.P.; Rand, K.N.; Molloy, P.L. Hypomethylation of repeated DNA sequences in cancer. Epigenomics 2010, 2, 245–269. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.A.; Wardell, C.P.; Chiecchio, L.; Smith, E.M.; Boyd, K.D.; Neri, A.; Davies, F.E.; Ross, F.M.; Morgan, G.J. Aberrant global methylation patterns affect the molecular pathogenesis and prognosis of multiple myeloma. Blood 2011, 117, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Römermann, D.; Hasemeier, B.; Metzig, K.; Göhring, G.; Schlegelberger, B.; Länger, F.; Kreipe, H.; Lehmann, U. Global increase in DNA methylation in patients with myelodysplastic syndrome. Leukemia 2008, 22, 1954–1956. [Google Scholar] [CrossRef]
- Bollati, V.; Fabris, S.; Pegoraro, V.; Ronchetti, D.; Mosca, L.; Deliliers, G.L.; Motta, V.; Bertazzi, P.A.; Baccarelli, A.; Neri, A. Differential repetitive DNA methylation in multiple myeloma molecular subgroups. Carcinogenesis 2009, 30, 1330–1335. [Google Scholar] [CrossRef] [Green Version]
- Roman-Gomez, J.; Jimenez-Velasco, A.; Agirre, X.; Castillejo, J.A.; Navarro, G.; San Jose-Eneriz, E.; Garate, L.; Cordeu, L.; Cervantes, F.; Prosper, F.; et al. Repetitive DNA hypomethylation in the advanced phase of chronic myeloid leukemia. Leuk. Res. 2008, 32, 487–490. [Google Scholar] [CrossRef]
- Sarmento-Ribeiro, A.B.; Scorilas, A.; Gonçalves, A.C.; Efferth, T.; Trougakos, I.P. The emergence of drug resistance to targeted cancer therapies: Clinical evidence. Dug Resist. Updates 2019, 47, 100646. [Google Scholar] [CrossRef]
- Stomper, J.; Rotondo, J.C.; Greve, G.; Lübbert, M. Hypomethylating agents (HMA) for the treatment of acute myeloid leukemia and myelodysplastic syndromes: Mechanisms of resistance and novel HMA-based therapies. Leukemia 2021. [Google Scholar] [CrossRef]
- Hitchler, M.J.; Domann, F.E. An epigenetic perspective on the free radical theory of development. Free Radic. Biol. Med. 2007, 43, 1023–1036. [Google Scholar] [CrossRef] [Green Version]
- Kuchino, Y.; Mori, F.; Kasai, H.; Inoue, H.; Iwai, S.; Miura, K.; Ohtsuka, E.; Nishimura, S. Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature 1987, 327, 77–79. [Google Scholar] [CrossRef]
- Franco, R.; Schoneveld, O.; Georgakilas, A.G.; Panayiotidis, M.I. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. 2008, 266, 6–11. [Google Scholar] [CrossRef]
- Christman, J.K.; Sheikhnejad, G.; Marasco, C.J.; Sufrin, J.R. 5-Methyl-2′-deoxycytidine in single-stranded DNA can act in cis to signal de novo DNA methylation. Proc. Natl. Acad. Sci. USA 1995, 92, 7347–7351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, K.A.; Zhang, R.; Kim, G.Y.; Bae, S.C.; Hyun, J.W. Epigenetic changes induced by oxidative stress in colorectal cancer cells: Methylation of tumor suppressor RUNX3. Tumor Biol. 2012, 33, 403–412. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, H.M.; Wang, W.; Sen, S.; DeStefano Shields, C.; Lee, S.S.; Zhang, Y.W.; Clements, E.G.; Cai, Y.; Van Neste, L.; Easwaran, H.; et al. Oxidative Damage Targets Complexes Containing DNA Methyltransferases, SIRT1, and Polycomb Members to Promoter CpG Islands. Cancer Cell 2011, 20, 606–619. [Google Scholar] [CrossRef] [Green Version]

| Characteristics | MDS (n = 66) | Controls (n = 26) | ||
|---|---|---|---|---|
| Demographic Features | ||||
| Gender (%) | ||||
| Male | 26 | (39.9%) | 12 | (46.2%) |
| Female | 40 | (60.1%) | 14 | (53.8%) |
| Age (years) | ||||
| Median | 74 | 67 | ||
| Range | 22–89 | 32–79 | ||
| Clinical features | ||||
| Hematological parameters (median, range) | ||||
| WBC (×109/L) | 3.5 | (1.3–13.0) | ||
| Hb (g/L) | 10.6 | (5.4–16.0) | ||
| Platelets (×109/L) | 98 | (12–324) | ||
| WHO 2016 classification | ||||
| MDS-SLD (%) | 9 | (13.6) | ||
| MDS-RS (%) | 10 | (15.2) | ||
| MDS-MD (%) | 40 | (60.6) | ||
| MDS-EB (%) | 7 | (10.6) | ||
| IPSS-R risk groups | ||||
| Lower-risk (IPSS-R score ≤3.5) | 38 | |||
| Higher-risk (IPSS-R score >3.5) | 13 | |||
| Not recorded | 15 | |||
| Cytogenetics | ||||
| Good | 37 | |||
| Intermediate | 11 | |||
| Poor | 2 | |||
| Not recorded | 15 | |||
| Parameter | MDS-SLD Median (Q1–Q3) | MDS-RS Median (Q1–Q3) | MDS-MD Median (Q1–Q3) | MDS-EB Median (Q1–Q3) | p-Value |
|---|---|---|---|---|---|
| Peroxides (µM) | 2.27 (1.70–4.02) | 4.59 (3.03–5.85) | 4.54 (3.48–6.07) | 4.34 (2.62–5.92) | 0.048 |
| Peroxide/TAS ratio | 2.30 (1.40–4.15) | 4.40 (2.65–5.78) | 4.75 (3.50–6.68) | 4.50 (2.40–6.50) | 0.036 |
| Condition | Methylation Frequency | |||||||
|---|---|---|---|---|---|---|---|---|
| P15 | P16 | DAPK | KEAP1 | |||||
| n | % | n | % | n | % | n | % | |
| MDS | 32/66 | 48.5 ** | 13/66 | 19.7 | 28/66 | 42.4 *** | 26/66 | 33.3 * |
| MDS-SLD | 3/9 | 33.3 | 3/9 | 11.1 | 3/9 | 33.3 | 1/9 | 11.1 |
| MDS-RS | 4/10 | 40.0 | 4/10 | 10.0 | 7/10 | 70.0 | 2/10 | 20.0 |
| MDS-MD | 20/40 | 50.0 | 20/40 | 20.0 | 14/40 | 35.0 | 17/40 | 42.5 |
| MDS-EB | 5/7 | 71.4 | 3/7 | 42.9 | 4/7 | 57.1 | 2/7 | 28.6 |
| Controls | 3/26 | 11.5 | 1/26 | 3.8 | 0/26 | 0.0 | 3/26 | 11.5 |
| TP53 | MGMT | 1 ≥ methylated genes | 2 ≥ methylated genes | |||||
| n | % | n | % | n | % | n | % | |
| MDS | 0/66 | 0.0 | 0/66 | 0.0 | 43/66 | 65.2 *** | 32/66 | 48.5 *** |
| MDS-SLD | 0/9 | 0.0 | 0/9 | 0.0 | 4/9 | 44.4 | 1/9 | 11.1 |
| MDS-RS | 0/10 | 0.0 | 0/10 | 0.0 | 5/10 | 70.0 | 3/10 | 30.0 |
| MDS-MD | 0/40 | 0.0 | 0/40 | 0.0 | 29/40 | 72.5 | 23/40 | 57.5 |
| MDS-EB | 0/7 | 0.0 | 0/7 | 0.0 | 5/7 | 71.4 | 5/7 | 71.4 |
| Controls | 0/26 | 0.0 | 0/26 | 0.0 | 4/26 | 15.4 | 0/26 | 0.0 |
| Biomarkers | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | p-Value | OR | 95% CI | p-Value | |
| Peroxide | 3.36 | 1.96–5.76 | <0.001 | 1.31 | 1.23–1.45 | 0.008 |
| 8-OHdG | 1.27 | 1.43–1.41 | <0.001 | 1.23 | 1.01–1.59 | 0.041 |
| Peroxide/TAS | 2.54 | 1.61–3.99 | <0.001 | – | ||
| NO | 1.18 | 1.05–1.31 | 0.004 | – | ||
| NO/TAS | 1.11 | 1.02–1.21 | 0.017 | – | ||
| 5-mC | 2.43 | 1.62–3.64 | <0.001 | 2.13 | 1.30–3.48 | 0.003 |
| 5-hmC | 1.54 | 1.14–2.09 | 0.005 | – | ||
| LINE-1 | 0.83 | 0.75–0.91 | <0.001 | – | ||
| Methylated p15 | 8.42 | 3.85–9.40 | <0.001 | – | ||
| Methylated KEAP1 | 3.83 | 1.04–7.18 | 0.044 | – | ||
| 2 ≥ methylated genes | 7.51 | 4.59–9.34 | <0.001 | 4.52 | 3.39–8.80 | <0.001 |
| Biomarkers | AUC | Cut-Off | |||||
|---|---|---|---|---|---|---|---|
| Value (95% CI) | p-Value | Value | SEN (%) | SPE (%) | PPV (%) | NPV (%) | |
| Peroxide (µM) | 0.877 (0.800–0.955) | <0.001 | 3.28 | 73 | 92 | 96 | 77 |
| 8-OHdG (ng/mL) | 0.863 (0.790–0.937) | <0.001 | 34.7 | 72 | 92 | 96 | 80 |
| 5-mC (%) | 0.936 (0.887–0.984) | <0.001 | 0.48 | 82 | 96 | 93 | 78 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonçalves, A.C.; Alves, R.; Baldeiras, I.; Marques, B.; Oliveiros, B.; Pereira, A.; Nascimento Costa, J.M.; Cortesão, E.; Mota Vieira, L.; Sarmento Ribeiro, A.B. DNA Methylation Is Correlated with Oxidative Stress in Myelodysplastic Syndrome—Relevance as Complementary Prognostic Biomarkers. Cancers 2021, 13, 3138. https://doi.org/10.3390/cancers13133138
Gonçalves AC, Alves R, Baldeiras I, Marques B, Oliveiros B, Pereira A, Nascimento Costa JM, Cortesão E, Mota Vieira L, Sarmento Ribeiro AB. DNA Methylation Is Correlated with Oxidative Stress in Myelodysplastic Syndrome—Relevance as Complementary Prognostic Biomarkers. Cancers. 2021; 13(13):3138. https://doi.org/10.3390/cancers13133138
Chicago/Turabian StyleGonçalves, Ana Cristina, Raquel Alves, Inês Baldeiras, Bárbara Marques, Bárbara Oliveiros, Amélia Pereira, José Manuel Nascimento Costa, Emília Cortesão, Luisa Mota Vieira, and Ana Bela Sarmento Ribeiro. 2021. "DNA Methylation Is Correlated with Oxidative Stress in Myelodysplastic Syndrome—Relevance as Complementary Prognostic Biomarkers" Cancers 13, no. 13: 3138. https://doi.org/10.3390/cancers13133138
APA StyleGonçalves, A. C., Alves, R., Baldeiras, I., Marques, B., Oliveiros, B., Pereira, A., Nascimento Costa, J. M., Cortesão, E., Mota Vieira, L., & Sarmento Ribeiro, A. B. (2021). DNA Methylation Is Correlated with Oxidative Stress in Myelodysplastic Syndrome—Relevance as Complementary Prognostic Biomarkers. Cancers, 13(13), 3138. https://doi.org/10.3390/cancers13133138






