Co-Deregulated miRNA Signatures in Childhood Central Nervous System Tumors: In Search for Common Tumor miRNA-Related Mechanics
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Analysis of Variables
2.2. Microarray Samples
2.3. Microarray Data Pre-Processing
2.4. Microarray Data Post-Processing
2.5. Unsupervised Classification Methods
2.6. Common Expression Patterns in DE miRNAs
2.7. Receiver Operating Characteristic (ROC) Analysis
2.8. Statistical Analysis
2.9. Gene Ontology (GO) Enrichment Analysis
2.10. Pathway Analysis
3. Results
3.1. CNS Sample Cohort
3.2. Deregulated (DE) miRNAs
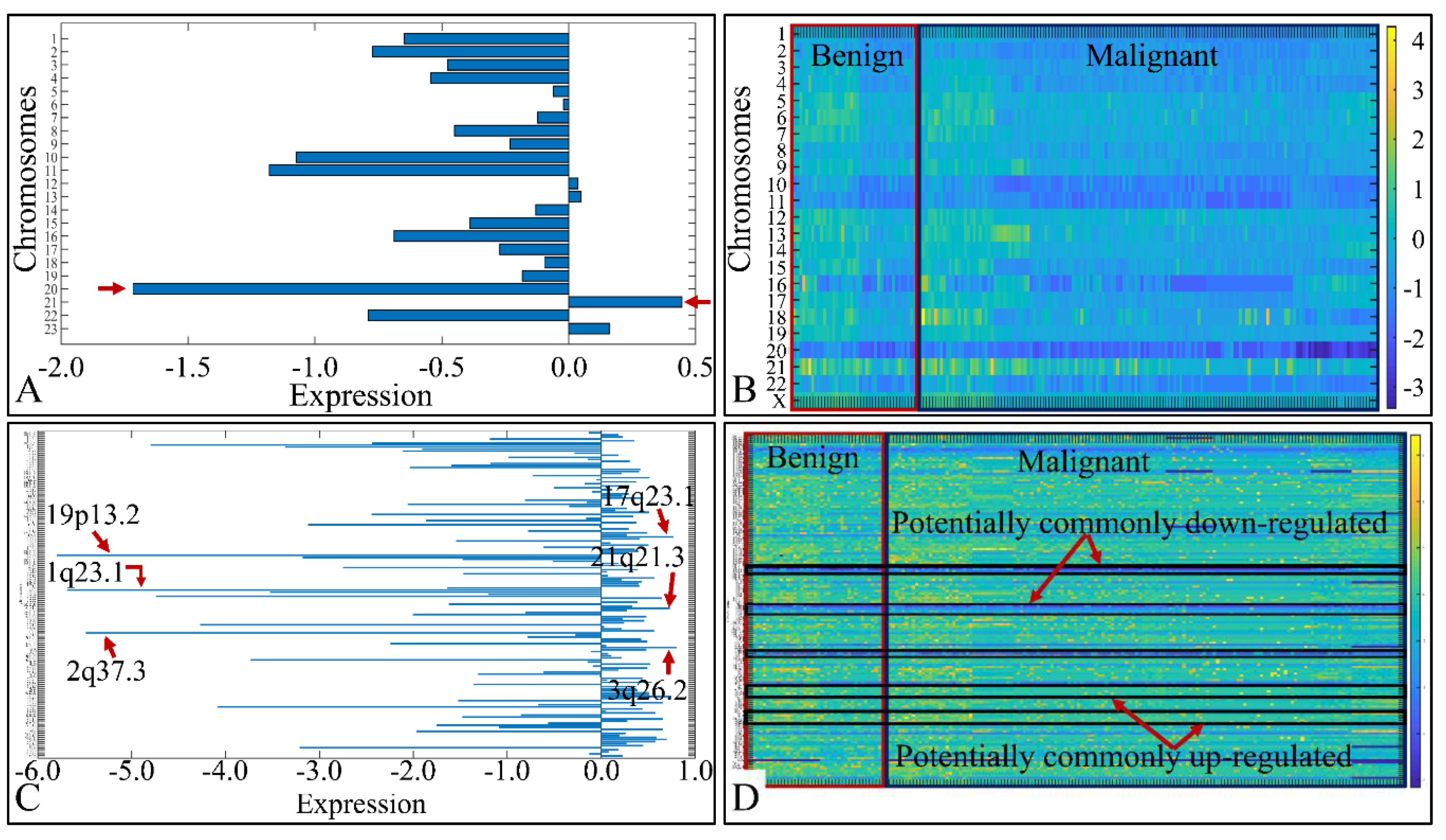
3.3. Chromosomal Distribution of DE miRNAs
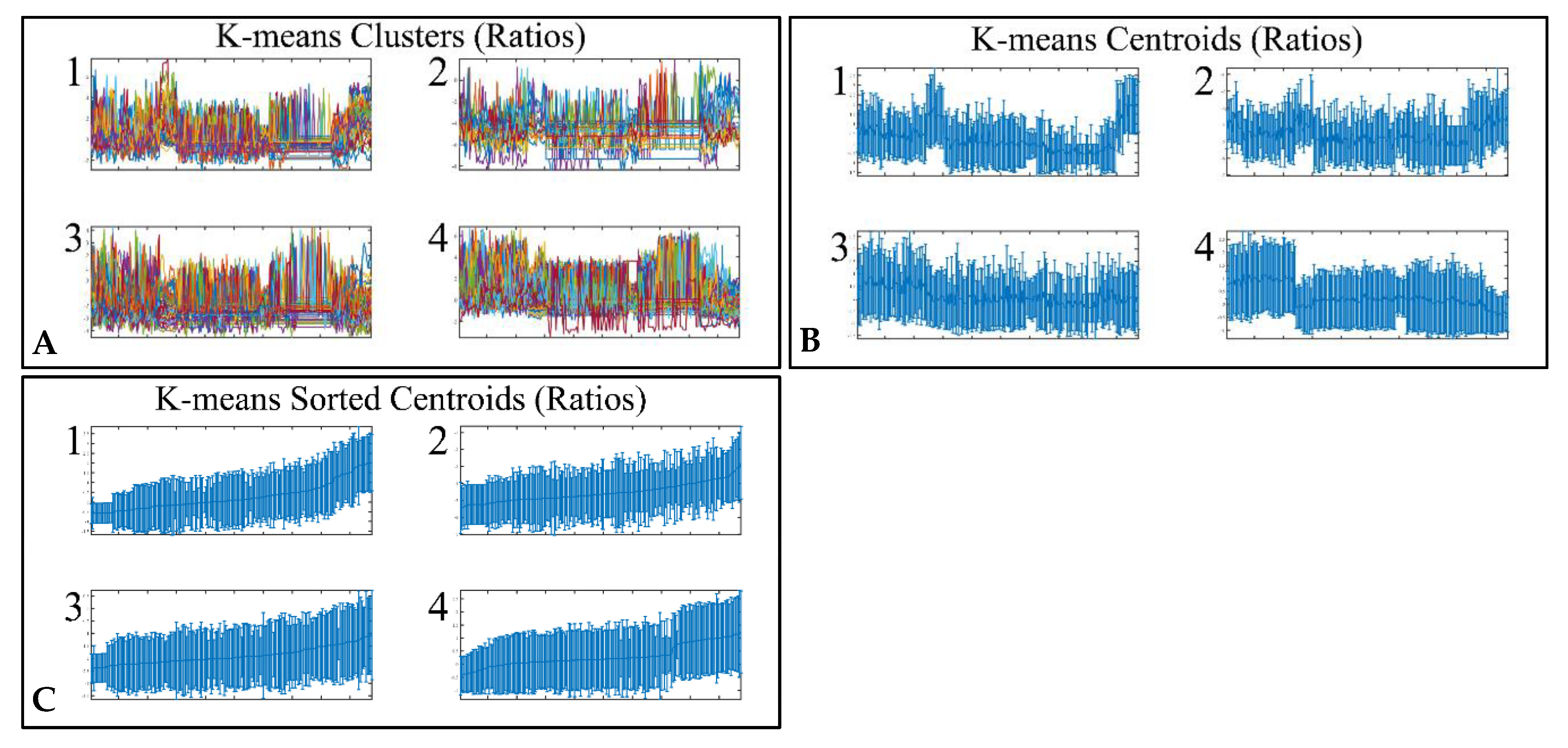
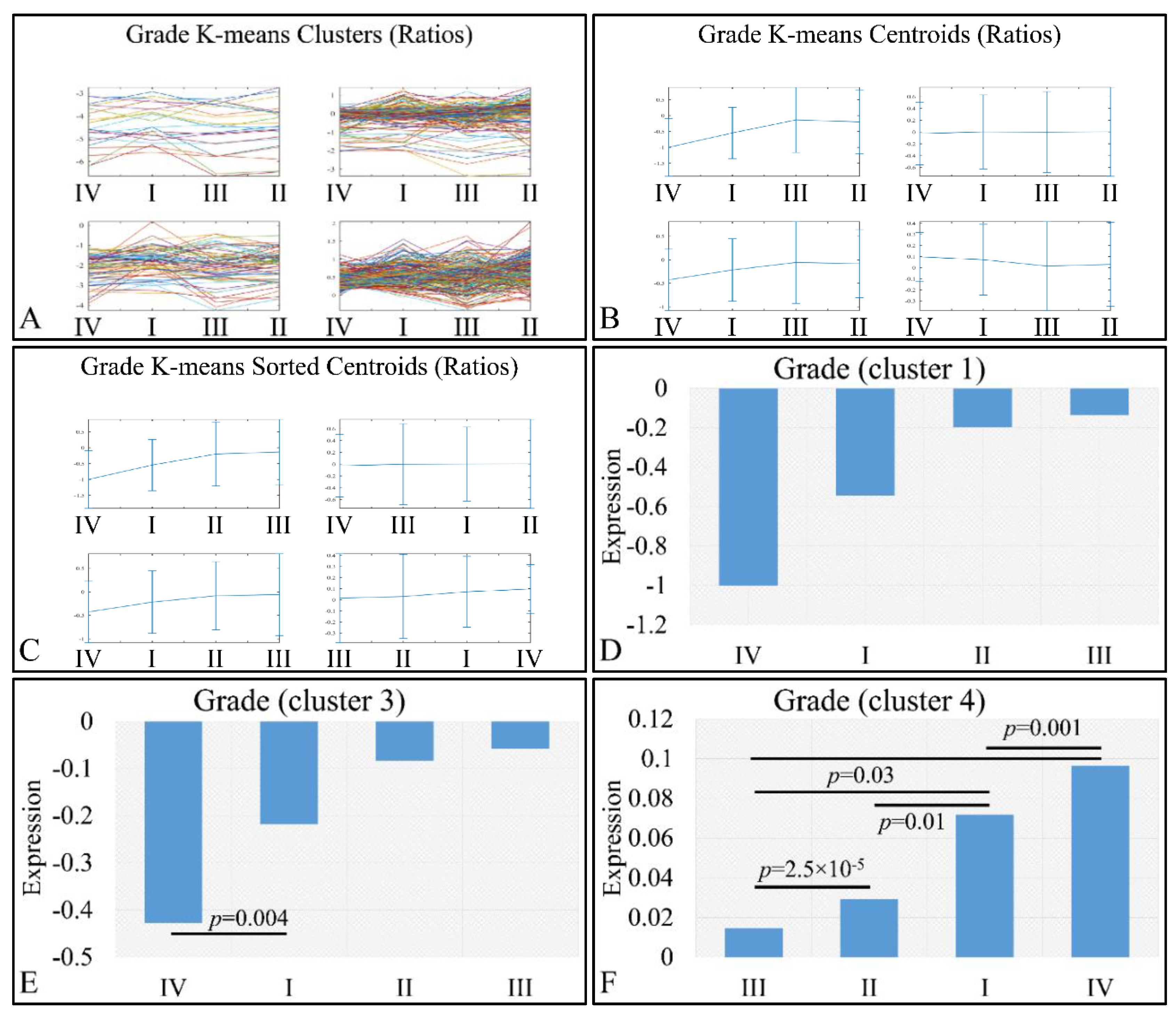
3.4. Unsupervised K-Means Classification
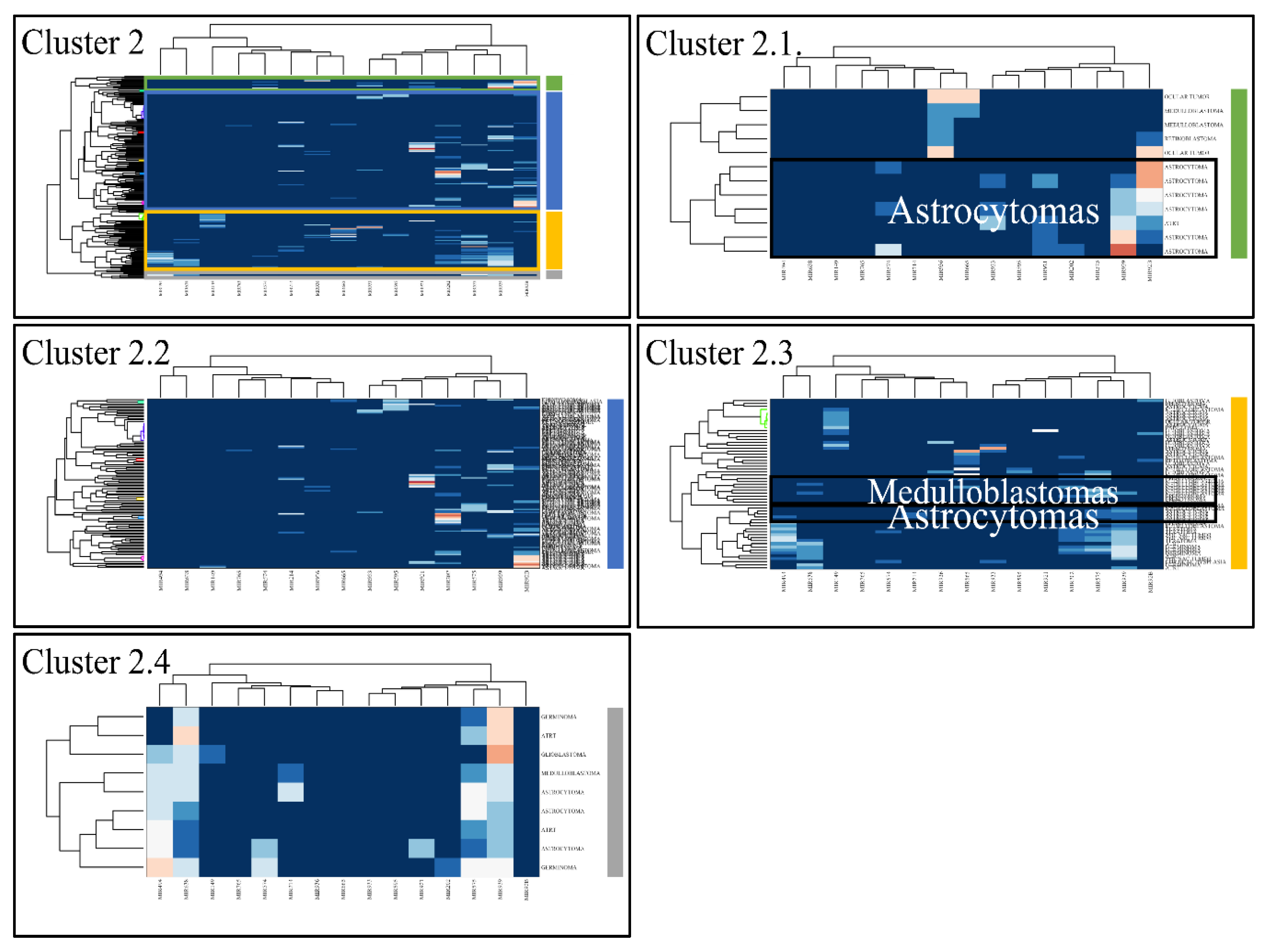
3.5. Common DE miRNAs in Different CNS Tumor Types
3.6. Descriptive K-Means
3.7. Functional Analysis of DE miRNAs
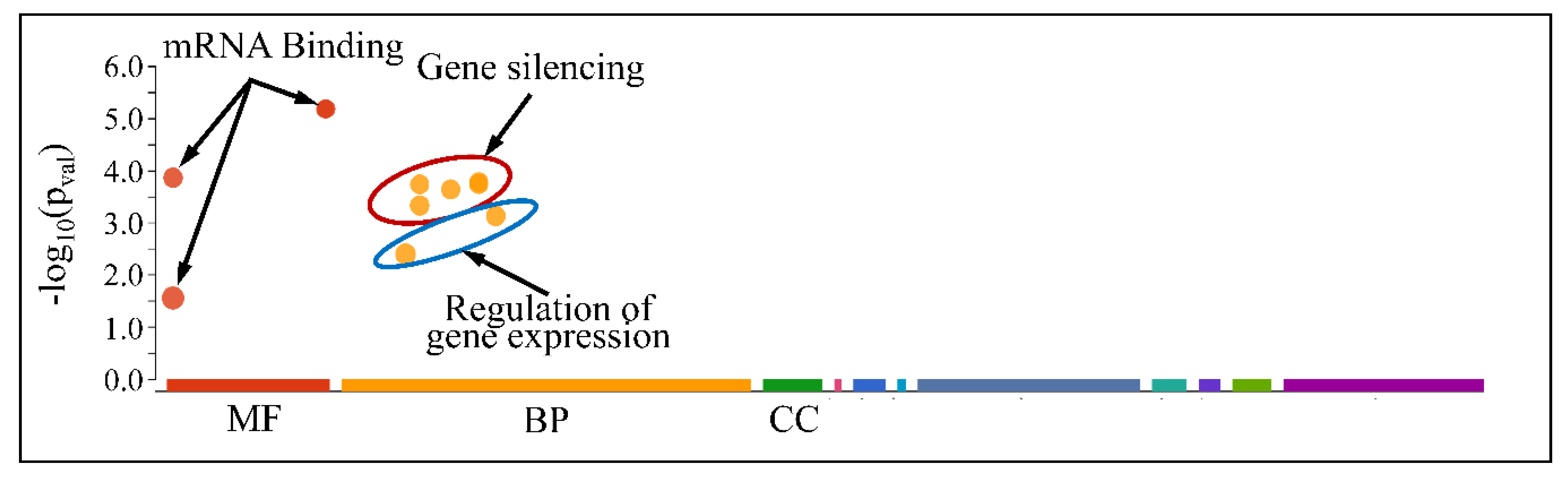
3.7.1. Gene Ontology Enrichment Analysis of Commonly Expressed miRNAs
3.7.2. The Special Case of Down-Regulated miRNAs across All CNS Tumor Samples
3.7.3. Functional Analysis of the DE miRNAs Manifested Expression Patterns
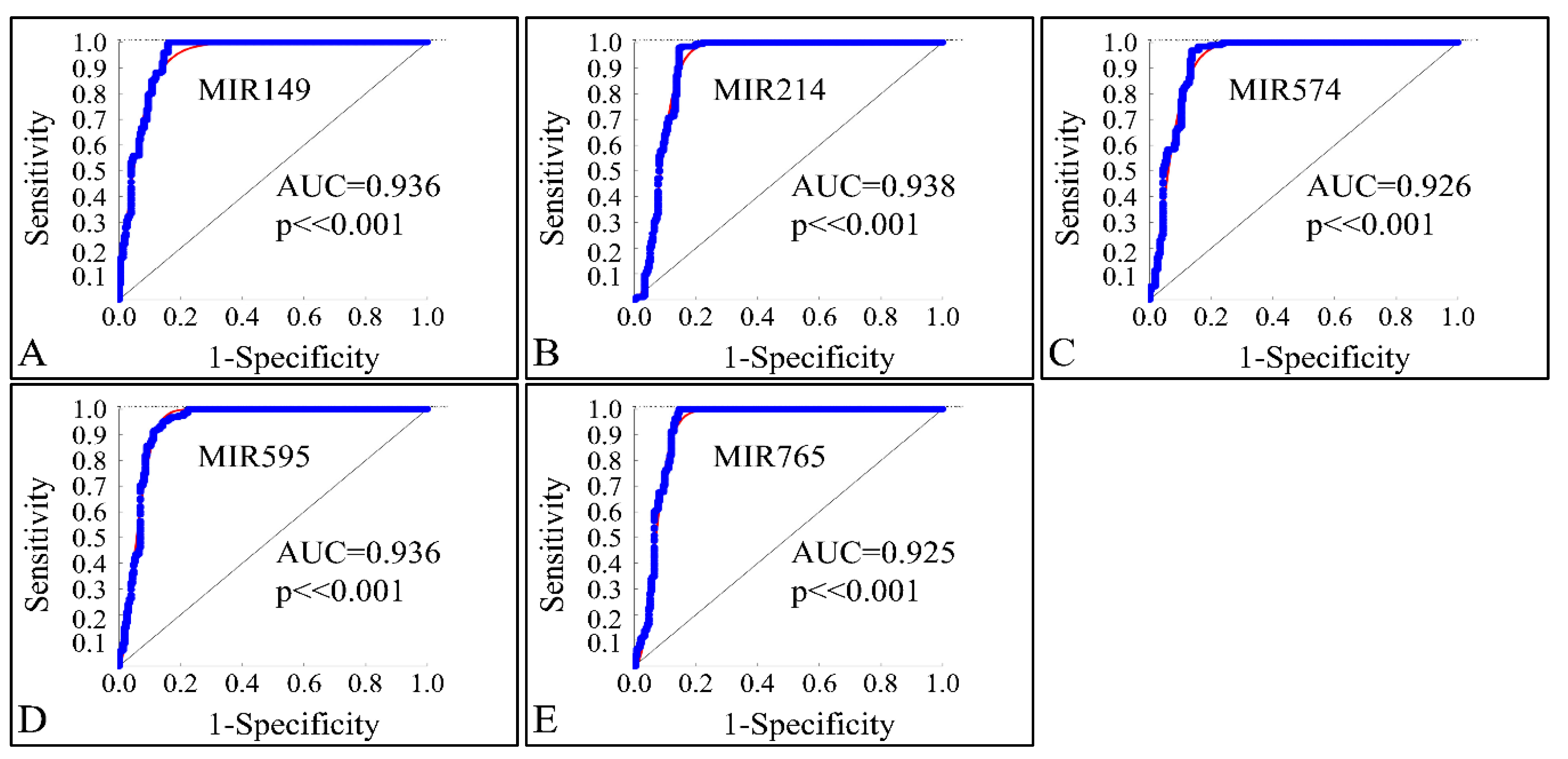
3.8. ROC Analysis of Globally Down-Regulated miRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Braoudaki, M.; Lambrou, G.I. MicroRNAs in pediatric central nervous system embryonal neoplasms: The known unknown. J. Hematol. Oncol. 2015, 8, 6. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA-cancer connection: The beginning of a new tale. Cancer Res. 2006, 66, 7390–7394. [Google Scholar] [CrossRef]
- Georgakopoulos-Soares, I.; Chartoumpekis, D.V.; Kyriazopoulou, V.; Zaravinos, A. Emt factors and metabolic pathways in cancer. Front. Oncol. 2020, 10, 499. [Google Scholar] [CrossRef]
- Garzon, R.; Calin, G.A.; Croce, C.M. MicroRNAs in Cancer. Annu. Rev. Med. 2009, 60, 167–179. [Google Scholar] [CrossRef]
- Zaravinos, A.; Lambrou, G.I.; Mourmouras, N.; Katafygiotis, P.; Papagregoriou, G.; Giannikou, K.; Delakas, D.; Deltas, C. New miRNA profiles accurately distinguish renal cell carcinomas and upper tract urothelial carcinomas from the normal kidney. PLoS ONE 2014, 9, e91646. [Google Scholar] [CrossRef]
- Wang, B.; Li, D.; Kovalchuk, O. p53 Ser15 phosphorylation and histone modifications contribute to IR-induced miR-34a transcription in mammary epithelial cells. Cell Cycle 2013, 12, 2073–2083. [Google Scholar] [CrossRef]
- Migliore, C.; Giordano, S. Resistance to targeted therapies: A role for microRNAs? Trends Mol. Med. 2013, 19, 633–642. [Google Scholar] [CrossRef]
- Petrelli, A.; Perra, A.; Cora, D.; Sulas, P.; Menegon, S.; Manca, C.; Migliore, C.; Kowalik, M.A.; Ledda-Columbano, G.M.; Giordano, S.; et al. MicroRNA/gene profiling unveils early molecular changes and nuclear factor erythroid related factor 2 (NRF2) activation in a rat model recapitulating human hepatocellular carcinoma (HCC). Hepatology 2014, 59, 228–241. [Google Scholar] [CrossRef]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, L.A.; Northcott, P.A.; Taylor, M.D.; Kenney, A.M. Normal and oncogenic roles for microRNAs in the developing brain. Cell Cycle 2009, 8, 4049–4054. [Google Scholar] [CrossRef]
- Northcott, P.A.; Fernandez, L.A.; Hagan, J.P.; Ellison, D.W.; Grajkowska, W.; Gillespie, Y.; Grundy, R.; Van Meter, T.; Rutka, J.T.; Croce, C.M.; et al. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009, 69, 3249–3255. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, G.; Wu, J.H.; Jiang, C.P. Diverse roles of miR-29 in cancer (review). Oncol. Rep. 2014, 31, 1509–1516. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.; Pantel, K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat. Rev. Cancer 2019, 19, 553–567. [Google Scholar] [CrossRef]
- Williams, Z.; Ben-Dov, I.Z.; Elias, R.; Mihailovic, A.; Brown, M.; Rosenwaks, Z.; Tuschl, T. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc. Natl. Acad. Sci. USA 2013, 110, 4255–4260. [Google Scholar] [CrossRef]
- Zampetaki, A.; Willeit, P.; Drozdov, I.; Kiechl, S.; Mayr, M. Profiling of circulating microRNAs: From single biomarkers to re-wired networks. Cardiovasc. Res. 2012, 93, 555–562. [Google Scholar] [CrossRef]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004, 5, R13. [Google Scholar] [CrossRef]
- Birks, D.K.; Barton, V.N.; Donson, A.M.; Handler, M.H.; Vibhakar, R.; Foreman, N.K. Survey of MicroRNA expression in pediatric brain tumors. Pediatric Blood Cancer 2011, 56, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Braoudaki, M.; Lambrou, G.I.; Giannikou, K.; Milionis, V.; Stefanaki, K.; Birks, D.K.; Prodromou, N.; Kolialexi, A.; Kattamis, A.; Spiliopoulou, C.A.; et al. Microrna expression signatures predict patient progression and disease outcome in pediatric embryonal central nervous system neoplasms. J. Hematol. Oncol. 2014, 7, 96. [Google Scholar] [CrossRef]
- Wang, H.W.; Wu, Y.H.; Hsieh, J.Y.; Liang, M.L.; Chao, M.E.; Liu, D.J.; Hsu, M.T.; Wong, T.T. Pediatric primary central nervous system germ cell tumors of different prognosis groups show characteristic miRNome traits and chromosome copy number variations. BMC Genom. 2010, 11, 132. [Google Scholar] [CrossRef]
- Jones, T.A.; Jeyapalan, J.N.; Forshew, T.; Tatevossian, R.G.; Lawson, A.R.; Patel, S.N.; Doctor, G.T.; Mumin, M.A.; Picker, S.R.; Phipps, K.P.; et al. Molecular analysis of pediatric brain tumors identifies microRNAs in pilocytic astrocytomas that target the MAPK and NF-κB pathways. Acta Neuropathol. Commun. 2015, 3, 86. [Google Scholar] [CrossRef]
- Moreau, M.P.; Bruse, S.E.; Jornsten, R.; Liu, Y.; Brzustowicz, L.M. Chronological changes in microRNA expression in the developing human brain. PLoS ONE 2013, 8, e60480. [Google Scholar] [CrossRef] [PubMed]
- Edward, D.P.; Alkatan, H.; Rafiq, Q.; Eberhart, C.; Al Mesfer, S.; Ghazi, N.; Al Safieh, L.; Kondkar, A.A.; Abu Amero, K.K. MicroRNA profiling in intraocular medulloepitheliomas. PLoS ONE 2015, 10, e0121706. [Google Scholar] [CrossRef]
- Castro-Magdonel, B.E.; Orjuela, M.; Camacho, J.; García-Chéquer, A.J.; Cabrera-Muñoz, L.; Sadowinski-Pine, S.; Durán-Figueroa, N.; Orozco-Romero, M.J.; Velázquez-Wong, A.C.; Hernández-Ángeles, A.; et al. miRNome landscape analysis reveals a 30 miRNA core in retinoblastoma. BMC Cancer 2017, 17, 458. [Google Scholar] [CrossRef] [PubMed]
- Darrigo Júnior, L.G.; Lira, R.C.P.; Fedatto, P.F.; Marco Antonio, D.S.; Valera, E.T.; Aguiar, S.; Yunes, J.A.; Brandalise, S.R.; Neder, L.; Saggioro, F.P.; et al. MicroRNA profile of pediatric pilocytic astrocytomas identifies two tumor-specific signatures when compared to non-neoplastic white matter. J. Neuro-Oncol. 2019, 141, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Braoudaki, M.; Lambrou, G.I.; Giannikou, K.; Papadodima, S.A.; Lykoudi, A.; Stefanaki, K.; Sfakianos, G.; Kolialexi, A.; Tzortzatou-Stathopoulou, F.; Tzetis, M.; et al. miR-15a and miR-24-1 as putative prognostic microRNA signatures for pediatric pilocytic astrocytomas and ependymomas. Tumour Biol. 2016, 37, 9887–9897. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, M.; Wells, M.T. Multiplicative background correction for spotted microarrays to improve reproducibility. Genet. Res. 2006, 87, 195–206. [Google Scholar] [CrossRef]
- Cleveland, W.S. Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 1979, 74, 829–836. [Google Scholar] [CrossRef]
- Uzman, B.G.; Foley, G.E.; Farber, S.; Lazarus, H. Morphologic variations in human leukemic lymphoblasts (CCRF-CEM cells) after long-term culture and exposure to chemotherapeutic agents. A study with the electron microscope. Cancer 1966, 19, 1725–1742. [Google Scholar] [CrossRef]
- Yang, I.V.; Chen, E.; Hasseman, J.P.; Liang, W.; Frank, B.C.; Wang, S.; Sharov, V.; Saeed, A.I.; White, J.; Li, J.; et al. Within the fold: Assessing differential expression measures and reproducibility in microarray assays. Genome Biol. 2002, 3, research0062. [Google Scholar]
- Klipper-Aurbach, Y.; Wasserman, M.; Braunspiegel-Weintrob, N.; Borstein, D.; Peleg, S.; Assa, S.; Karp, M.; Benjamini, Y.; Hochberg, Y.; Laron, Z. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med. Hypotheses 1995, 45, 486–490. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical methods for identifying differentially expressed genes in DNA microarrays. In Functional Genomics; Humana Press: Totowa, NJ, USA, 2003; Volume 224, pp. 149–157. [Google Scholar]
- Forgy, E.W. Cluster analysis of multivariate data: Efficiency vs. interpretability of classifications, 1965. Biometrics 1965, 21, 768769. [Google Scholar]
- Lloyd, S. Least squares quantization in PCM. IEEE Trans. Inf. Theory 1982, 28, 129–137. [Google Scholar] [CrossRef]
- Freyhult, E.; Landfors, M.; Onskog, J.; Hvidsten, T.R.; Ryden, P. Challenges in microarray class discovery: A comprehensive examination of normalization, gene selection and clustering. BMC Bioinform. 2010, 11, 503. [Google Scholar] [CrossRef]
- Gibbons, F.D.; Roth, F.P. Judging the quality of gene expression-based clustering methods using gene annotation. Genome Res. 2002, 12, 1574–1581. [Google Scholar] [CrossRef]
- Lambrou, G.; Braoudaki, M. A Novel Method for the Analysis of Gene Expression Microarray Data with K-Means Clustering: Sorted K-Means. Int. J. Eng. Res. Sci. 2016, 2, 99–105. [Google Scholar]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, w191–w198. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Schmoyer, D.; Kirov, S.; Snoddy, J. GOTree Machine (GOTM): A web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinform. 2004, 5, 16. [Google Scholar]
- Braoudaki, M.; Koutsouris, D.D.; Kouris, I.; Paidi, A.; Koutsouri, G.; George Lambrou, I. Bioinformatics and Regression Analyses Manifest Tumor-Specific miRNA Expression Dynamics in Pediatric Embryonal Malignancies. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 5834–5837. [Google Scholar] [CrossRef]
- Braoudaki, M.; Sarafidis, M.; Koutsouris, D.D.; Koutsouri, G.; Lambrou, G.I. Bioinformatics Analysis Reveals Ki-67 Specific microRNA Functions in Pediatric Embryonal Tumors. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 1346–1349. [Google Scholar] [CrossRef]
- Hennchen, M.; Stubbusch, J.; Abarchan-El Makhfi, I.; Kramer, M.; Deller, T.; Pierre-Eugene, C.; Janoueix-Lerosey, I.; Delattre, O.; Ernsberger, U.; Schulte, J.B.; et al. Lin28B and Let-7 in the Control of Sympathetic Neurogenesis and Neuroblastoma Development. J. Neurosci. 2015, 35, 16531–16544. [Google Scholar] [CrossRef]
- Kusakabe, K.; Kohno, S.; Inoue, A.; Seno, T.; Yonezawa, S.; Moritani, K.; Mizuno, Y.; Kurata, M.; Kitazawa, R.; Tauchi, H.; et al. Combined morphological, immunohistochemical and genetic analyses of medulloepithelioma in the posterior cranial fossa. Neuropathology 2018, 38, 179–184. [Google Scholar] [CrossRef]
- Lee, Y.Y.; Yang, Y.P.; Huang, M.C.; Wang, M.L.; Yen, S.H.; Huang, P.I.; Chen, Y.W.; Chiou, S.H.; Lan, Y.T.; Ma, H.I.; et al. MicroRNA142-3p promotes tumor-initiating and radioresistant properties in malignant pediatric brain tumors. Cell Transplant. 2014, 23, 669–690. [Google Scholar] [CrossRef]
- Roussel, M.F.; Hatten, M.E. Cerebellum development and medulloblastoma. Curr. Top. Dev. Biol. 2011, 94, 235–282. [Google Scholar] [CrossRef] [PubMed]
- Shalaby, T.; Fiaschetti, G.; Baumgartner, M.; Grotzer, M.A. MicroRNA signatures as biomarkers and therapeutic target for CNS embryonal tumors: The pros and the cons. Int. J. Mol. Sci. 2014, 15, 21554–21586. [Google Scholar] [CrossRef] [PubMed]
- Conti, A.; Romeo, S.G.; Cama, A.; La Torre, D.; Barresi, V.; Pezzino, G.; Tomasello, C.; Cardali, S.; Angileri, F.F.; Polito, F.; et al. MiRNA expression profiling in human gliomas: Upregulated miR-363 increases cell survival and proliferation. Tumour Biol. 2016, 37, 14035–14048. [Google Scholar] [CrossRef] [PubMed]
- Pezuk, J.A.; Salomão, K.B.; Baroni, M.; Pereira, C.A.; Geron, L.; Brassesco, M.S. Aberrantly expressed microRNAs and their implications in childhood central nervous system tumors. Cancer Metastasis Rev. 2019, 38, 813–828. [Google Scholar] [CrossRef]
- Conti, L.; Crisafulli, L.; Caldera, V.; Tortoreto, M.; Brilli, E.; Conforti, P.; Zunino, F.; Magrassi, L.; Schiffer, D.; Cattaneo, E. REST controls self-renewal and tumorigenic competence of human glioblastoma cells. PLoS ONE 2012, 7, e38486. [Google Scholar] [CrossRef]
- Fox, J.L.; Dews, M.; Minn, A.J.; Thomas-Tikhonenko, A. Targeting of TGFβ signature and its essential component CTGF by miR-18 correlates with improved survival in glioblastoma. RNA 2013, 19, 177–190. [Google Scholar] [CrossRef]
- Mazzacurati, L.; Marzulli, M.; Reinhart, B.; Miyagawa, Y.; Uchida, H.; Goins, W.F.; Li, A.; Kaur, B.; Caligiuri, M.; Cripe, T.; et al. Use of miRNA response sequences to block off-target replication and increase the safety of an unattenuated, glioblastoma-targeted oncolytic HSV. Mol. Ther. 2015, 23, 99–107. [Google Scholar] [CrossRef]
- Zakrzewska, M.; Fendler, W.; Zakrzewski, K.; Sikorska, B.; Grajkowska, W.; Dembowska-Bagińska, B.; Filipek, I.; Stefańczyk, Ł.; Liberski, P.P. Altered MicroRNA Expression Is Associated with Tumor Grade, Molecular Background and Outcome in Childhood Infratentorial Ependymoma. PLoS ONE 2016, 11, e0158464. [Google Scholar] [CrossRef]
- Hao, Y.; Zhang, S.; Sun, S.; Zhu, J.; Xiao, Y. MiR-595 targeting regulation of SOX7 expression promoted cell proliferation of human glioblastoma. Biomed. Pharmacother. 2016, 80, 121–126. [Google Scholar] [CrossRef]
- Zaravinos, A. The Regulatory Role of MicroRNAs in EMT and Cancer. J. Oncol. 2015, 2015, 865816. [Google Scholar] [CrossRef]
- Koutsaki, M.; Spandidos, D.A.; Zaravinos, A. Epithelial-mesenchymal transition-associated miRNAs in ovarian carcinoma, with highlight on the miR-200 family: Prognostic value and prospective role in ovarian cancer therapeutics. Cancer Lett. 2014, 351, 173–181. [Google Scholar] [CrossRef]
- Costa, F.F.; Bischof, J.M.; Vanin, E.F.; Lulla, R.R.; Wang, M.; Sredni, S.T.; Rajaram, V.; Bonaldo Mde, F.; Wang, D.; Goldman, S.; et al. Identification of microRNAs as potential prognostic markers in ependymoma. PLoS ONE 2011, 6, e25114. [Google Scholar] [CrossRef]
- Ruiz Esparza-Garrido, R.; Velazquez-Flores, M.A.; Diegoperez-Ramirez, J.; Lopez-Aguilar, E.; Siordia-Reyes, G.; Hernandez-Ortiz, M.; Martinez-Batallar, A.G.; Encarnacion-Guevara, S.; Salamanca-Gomez, F.; Arenas-Aranda, D.J. A proteomic approach of pediatric astrocytomas: MiRNAs and network insight. J. Proteom. 2013, 94, 162–175. [Google Scholar] [CrossRef]
- Zhang, L.; Liao, Y.; Tang, L. MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 2019, 38, 53. [Google Scholar] [CrossRef]
- Aran, D.; Camarda, R.; Odegaard, J.; Paik, H.; Oskotsky, B.; Krings, G.; Goga, A.; Sirota, M.; Butte, A.J. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun. 2017, 8, 1077. [Google Scholar] [CrossRef]
- Slaughter, D.P.; Southwick, H.W.; Smejkal, W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer 1953, 6, 963–968. [Google Scholar] [CrossRef]
- Heaphy, C.M.; Griffith, J.K.; Bisoffi, M. Mammary field cancerization: Molecular evidence and clinical importance. Breast Cancer Res. Treat. 2009, 118, 229–239. [Google Scholar] [CrossRef]
- Bakhshinyan, D.; Savage, N.; Salim, S.K.; Venugopal, C.; Singh, S.K. The Strange Case of Jekyll and Hyde: Parallels Between Neural Stem Cells and Glioblastoma-Initiating Cells. Front. Oncol. 2020, 10, 603738. [Google Scholar] [CrossRef]
- Wechsler-Reya, R.; Scott, M.P. The developmental biology of brain tumors. Annu. Rev. Neurosci. 2001, 24, 385–428. [Google Scholar] [CrossRef]
- Grimmer, M.R.; Weiss, W.A. Childhood tumors of the nervous system as disorders of normal development. Curr. Opin. Pediatrics 2006, 18, 634–638. [Google Scholar] [CrossRef]
- Lu, Q.R.; Qian, L.; Zhou, X. Developmental origins and oncogenic pathways in malignant brain tumors. Wiley Interdiscip. Rev. Dev. Biol. 2019, 8, e342. [Google Scholar] [CrossRef]
- Liu, C.; Zong, H. Developmental origins of brain tumors. Curr. Opin. Neurobiol. 2012, 22, 844–849. [Google Scholar] [CrossRef]
- Madhusoodanan, J. Elusive cancer cells dissected using developmental-biology toolkit. Nature 2021, 592, 647–649. [Google Scholar] [CrossRef]
- Wu, W.; Yu, T.; Wu, Y.; Tian, W.; Zhang, J.; Wang, Y. The miR155HG/miR-185/ANXA2 loop contributes to glioblastoma growth and progression. J. Exp. Clin. Cancer Res. 2019, 38, 133. [Google Scholar] [CrossRef]
- Dong, H.; Cao, W.; Xue, J. Long noncoding FOXD2-AS1 is activated by CREB1 and promotes cell proliferation and metastasis in glioma by sponging miR-185 through targeting AKT1. Biochem. Biophys. Res. Commun. 2019, 508, 1074–1081. [Google Scholar] [CrossRef]
- Tang, H.; Wang, Z.; Liu, X.; Liu, Q.; Xu, G.; Li, G.; Wu, M. LRRC4 inhibits glioma cell growth and invasion through a miR-185-dependent pathway. Curr. Cancer Drug Targets 2012, 12, 1032–1042. [Google Scholar] [CrossRef]
- Zhang, Z.; Tang, H.; Wang, Z.; Zhang, B.; Liu, W.; Lu, H.; Xiao, L.; Liu, X.; Wang, R.; Li, X.; et al. MiR-185 targets the DNA methyltransferases 1 and regulates global DNA methylation in human glioma. Mol. Cancer 2011, 10, 124. [Google Scholar] [CrossRef]
- Shahar, T.; Granit, A.; Zrihan, D.; Canello, T.; Charbit, H.; Einstein, O.; Rozovski, U.; Elgavish, S.; Ram, Z.; Siegal, T.; et al. Expression level of miRNAs on chromosome 14q32.31 region correlates with tumor aggressiveness and survival of glioblastoma patients. J. Neuro-Oncol. 2016, 130, 413–422. [Google Scholar] [CrossRef]
- Sun, S.; Wang, X.; Xu, X.; Di, H.; Du, J.; Xu, B.; Wang, Q.; Wang, J. MiR-433-3p suppresses cell growth and enhances chemosensitivity by targeting CREB in human glioma. Oncotarget 2017, 8, 5057–5068. [Google Scholar] [CrossRef]
- Visani, M.; de Biase, D.; Marucci, G.; Cerasoli, S.; Nigrisoli, E.; Bacchi Reggiani, M.L.; Albani, F.; Baruzzi, A.; Pession, A. Expression of 19 microRNAs in glioblastoma and comparison with other brain neoplasia of grades I-III. Mol. Oncol. 2014, 8, 417–430. [Google Scholar] [CrossRef]
- Xu, G.; Li, J.Y. Differential expression of PDGFRB and EGFR in microvascular proliferation in glioblastoma. Tumour Biol. 2016, 37, 10577–10586. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, F.; Chen, X.; Ying, Q. MicroRNA-518b functions as a tumor suppressor in glioblastoma by targeting PDGFRB. Mol. Med. Rep. 2017, 16, 5326–5332. [Google Scholar] [CrossRef]
- Kaid, C.; Jordan, D.; Bueno, H.M.S.; Araujo, B.H.S.; Assoni, A.; Okamoto, O.K. miR-367 as a therapeutic target in stem-like cells from embryonal central nervous system tumors. Mol. Oncol. 2019, 13, 2574–2587. [Google Scholar] [CrossRef]
- Kaid, C.; Silva, P.B.; Cortez, B.A.; Rodini, C.O.; Semedo-Kuriki, P.; Okamoto, O.K. miR-367 promotes proliferation and stem-like traits in medulloblastoma cells. Cancer Sci. 2015, 106, 1188–1195. [Google Scholar] [CrossRef]
- Lavon, I.; Zrihan, D.; Granit, A.; Einstein, O.; Fainstein, N.; Cohen, M.A.; Cohen, M.A.; Zelikovitch, B.; Shoshan, Y.; Spektor, S.; et al. Gliomas display a microRNA expression profile reminiscent of neural precursor cells. Neuro-Oncol. 2010, 12, 422–433. [Google Scholar] [CrossRef]
- Yu, X.; Wang, W. Tumor suppressor microRNA-613 inhibits glioma cell proliferation, invasion and angiogenesis by targeting vascular endothelial growth factor A. Mol. Med. Rep. 2017, 16, 6729–6735. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, K.; Shi, L.; Zhang, L.; Zhao, Z.; Xu, H.; Liang, F.; Li, H.; Zhao, Y.; Xu, X.; et al. Overexpression of MicroRNA-216a Suppresses Proliferation, Migration, and Invasion of Glioma Cells by Targeting Leucine-Rich Repeat-Containing G Protein-Coupled Receptor 5. Oncol. Res. 2017, 25, 1317–1327. [Google Scholar] [CrossRef]
- Zhang, T.; Ma, G.; Zhang, Y.; Huo, H.; Zhao, Y. miR-599 inhibits proliferation and invasion of glioma by targeting periostin. Biotechnol. Lett. 2017, 39, 1325–1333. [Google Scholar] [CrossRef]
- Zhu, X.Y.; Li, G.X.; Liu, Z.L. MiR-599 as a potential biomarker for prognosis of glioma. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 294–298. [Google Scholar] [CrossRef]
- Wei, N.; Wei, H.; Zhang, H. Long non-coding RNA ZEB1-AS1 promotes glioma cell proliferation, migration and invasion through regulating miR-577. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3085–3093. [Google Scholar] [CrossRef]
- Wu, P.; Gao, Y.; Shen, S.; Xue, Y.; Liu, X.; Ruan, X.; Shao, L.; Liu, Y.; Wang, P. KHDRBS3 regulates the permeability of blood-tumor barrier via cDENND4C/miR-577 axis. Cell Death Dis. 2019, 10, 536. [Google Scholar] [CrossRef]
- Zhang, W.; Shen, C.; Li, C.; Yang, G.; Liu, H.; Chen, X.; Zhu, D.; Zou, H.; Zhen, Y.; Zhang, D.; et al. miR-577 inhibits glioblastoma tumor growth via the Wnt signaling pathway. Mol. Carcinog. 2016, 55, 575–585. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, B.; Guo, W.; Gao, L.; Shi, L.; Li, H.; Lu, S.; Liu, Y.; Li, X. miR-429 inhibits glioma invasion through BMK1 suppression. J. Neuro-Oncol. 2015, 125, 43–54. [Google Scholar] [CrossRef]
- Sun, X.; Li, Z.; Chen, Y. The Potential Prognostic Value of MicroRNA-429 for Human Gliomas. Ann. Clin. Lab. Sci. 2016, 46, 44–48. [Google Scholar]
- Zaravinos, A.; Lambrou, G.I.; Boulalas, I.; Delakas, D.; Spandidos, D.A. Identification of common differentially expressed genes in urinary bladder cancer. PLoS ONE 2011, 6, e18135. [Google Scholar] [CrossRef]
- Gao, X.; Chen, Y.; Chen, M.; Wang, S.; Wen, X.; Zhang, S. Identification of key candidate genes and biological pathways in bladder cancer. PeerJ 2018, 6, e6036. [Google Scholar] [CrossRef]
- Shen, P.; He, X.; Lan, L.; Hong, Y.; Lin, M. Identification of cell division cycle 20 as a candidate biomarker and potential therapeutic target in bladder cancer using bioinformatics analysis. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, J.; Wan, L.; Zhou, X.; Wang, Z.; Wei, W. Targeting Cdc20 as a novel cancer therapeutic strategy. Pharmacol. Ther. 2015, 151, 141–151. [Google Scholar] [CrossRef]











| Series | Platform | Diagnosis | Sample Number | Publication |
|---|---|---|---|---|
| GSE19347 | GPL8227 | GCTs-Germinoma | 6 | Wang et al. (2010) [19] |
| GSE19347 | GPL8227 | GCTs-Teratoma | 3 | Wang et al. (2010) [19] |
| GSE19347 | GPL8227 | GCTs-Yoc sac tumor | 3 | Wang et al. (2010) [19] |
| GSE34016 | GPL8786 | Control (Neural progenitor cells) | 6 | N/A |
| GSE42657 | GPL8179 | Pilocytic Astrocytoma | 15 | Jones et al. (2015) [20] |
| GSE42657 | GPL8179 | Papillary Neuroglial Tumor | 1 | Jones et al. (2015) [20] |
| GSE42657 | GPL8179 | Diffuse Astrocytoma | 3 | Jones et al. (2015) [20] |
| GSE42657 | GPL8179 | Anaplastic Astrocytoma | 2 | Jones et al. (2015) [20] |
| GSE42657 | GPL8179 | Glioblastoma | 5 | Jones et al. (2015) [20] |
| GSE42657 | GPL8179 | Ependymoma | 14 | Jones et al. (2015) [20] |
| GSE42657 | GPL8179 | Medulloblastoma | 9 | Jones et al. (2015) [20] |
| GSE42657 | GPL8179 | Atypical teratoid rhabdoid tumor (ATRT) | 5 | Jones et al. (2015) [20] |
| GSE42657 | GPL8179 | Choroid Plexus Papilloma | 4 | Jones et al. (2015) [20] |
| GSE42657 | GPL8179 | Controls | 7 | Jones et al. (2015) [20] |
| GSE45126 | GPL16783 | Controls (Mixture of all RNA samples) | 98 | Moreau et al. (2013) [21] |
| GSE45126 | GPL16783 | Controls (Fetal brain) | 98 | Moreau et al. (2013) [21] |
| GSE62367 | GPL16384 | Ocular Medulloepithelioma | 5 | Edward et al. (2015) [22] |
| GSE62367 | GPL16384 | Controls | 8 | Edward et al. (2015) [22] |
| GSE63319 | GPL16384 | Glioblastoma | 11 | N/A |
| GSE63319 | GPL16384 | Anaplastic Astrocytoma | 3 | N/A |
| GSE63319 | GPL16384 | Controls | 4 | N/A |
| GSE66968 | GPL8227 | Medulloblastoma | 29 | N/A |
| GSE84747 | GPL21572 | Retinoblastoma | 12 | Castro-Magdonel et al. (2017) [23] |
| GSE135189 | GPL20906 | Pilocytic Astrocytoma | 16 | Darrigo et al. (2019) [24] |
| GSE135189 | GPL20906 | Ocular Medulloepithelioma | 1 | Darrigo et al. (2019) [24] |
| GSE135189 | GPL20906 | Controls | 11 | Darrigo et al. (2019) [24] |
| In-house | miRLink (https://appliedmicroarrays.com/, Last Accessed on 5 September 2020) | Pilocytic Astrocytoma | 19 | Braoudaki et al. (2016) [25] |
| In-house | miRLink (https://appliedmicroarrays.com/, Last Accessed on 5 September 2020) | Ependymoma | 7 | Braoudaki et al. (2016) [25] |
| In-house | miRLink (https://appliedmicroarrays.com/, Last Accessed on 5 September 2020) | Medulloblastoma | 15 | Braoudaki et al. (2014) [18] |
| In-house | miRLink (https://appliedmicroarrays.com/, Last Accessed on 5 September 2020) | ATRT | 4 | Braoudaki et al. (2014) [18] |
| In-house | miRLink (https://appliedmicroarrays.com/, Last Accessed on 5 September 2020) | Cortical Dysplasia | 2 | Braoudaki et al. (2014) [18] |
| In-house | miRLink (https://appliedmicroarrays.com/, Last Accessed on 5 September 2020) | Controls | 14 | Braoudaki et al. (2014, 2016) [18,25] |
| Primary Nominal Variables | Secondary Nominal Variables | N | Descriptive Variables | Age (Years) ¥ | Gest Age (Years) α |
|---|---|---|---|---|---|
| Total Population | 439 | Mean ± SD | 4.20 ± 5.24 | 4.84 ± 5.31 | |
| Median (range) | 1.73 (0.00–27.00) | 2.47 (0.19–27.74) | |||
| Gender 1 | FEMALES | 97 | Mean ± SD | 3.99 ± 4.76 | 4.60 ± 4.88 |
| Median | 2.00 (0.00–16.00) | 2.74 (0.27–16.74) | |||
| MALES | 165 | Mean ± SD | 5.55 ± 6.32 | 6.19 ± 6.41 | |
| Median | 3.00 (0.00–27.00) | 3.74 (0.19–27.74) | |||
| FETUS | 6 | Mean ± SD | 0.00 ± 0.00 | 0.23 ± 0.00 | |
| Median | 0.00 (0.00–0.00) | 0.23 (0.23–0.23) | |||
| Not Available | 171 | NaN | NaN | NaN | |
| Sampling 2 | VIVUS | 181 | Mean ± SD | 7.86 ± 5.62 | 8.60 ± 5.62 |
| Median | 7.00 (0.03–27.00) | 7.74 (0.77–27.74) | |||
| POST-MORTEM | 200 | Mean ± SD | 0.89 ± 0.84 | 1.46 ± 1.02 | |
| Median | 0.73 (0.00–1.73) | 1.45 (0.19–2.47) | |||
| First Diagnosis 3 | NEOPLASM | 195 | Mean ± SD | 7.29 ± 4.93 | 8.03 ± 4.93 |
| Median | 7.00 (0.03–19.00) | 7.74 (0.77–19.74) | |||
| CONTROL | 244 | Mean ± SD | 1.69 ± 4.01 | 2.26 ± 4.07 | |
| Median | 1.73 (0.00–27.00) | 2.47 (0.19–27.74) | |||
| Second Diagnosis 4 | MALIGNANCY | 154 | Mean ± SD | 7.55 ± 5.05 | 8.29 ± 5.05 |
| Median | 7.00 (0.03–19.00) | 7.74 (0.77–19.74) | |||
| BENIGN | 41 | Mean ± SD | 6.35 ± 4.41 | 7.09 ± 4.41 | |
| Median | 6.00 (0.83–16.00) | 6.74 (1.57–16.74) | |||
| CONTROL | 244 | Mean ± SD | 1.69 ± 4.01 | 2.26 ± 4.07 | |
| Median | 1.73 (0.00–27.00) | 2.47 (0.19–27.74) | |||
| Third Diagnosis 5 | MEDULLOBLASTOMA | 53 | Mean ± SD | 6.38 ± 4.06 | 7.12 ± 4.06 |
| Median | 6.00 (0.50–16.06) | 6.74 (1.24–16.80) | |||
| ASTROCYTOMA | 58 | Mean ± SD | 8.08 ± 5.14 | 8.82 ± 5.14 | |
| Median | 7.00 (0.92–19.00) | 7.74 (1.66–19.74) | |||
| EPENDYMOMA | 21 | Mean ± SD | 5.19 ± 4.34 | 5.93 ± 4.34 | |
| Median | 4.00 (1.00–16.01) | 4.74 (1.74–16.75) | |||
| ATRT | 9 | Mean ± SD | 1.59 ± 2.37 | 2.33 ± 2.37 | |
| Median | 0.75 (0.03–7.61) | 1.49 (0.77–8.35) | |||
| CONTROL | 244 | Mean ± SD | 1.69 ± 4.01 | 2.26 ± 4.07 | |
| Median | 1.73 (0.00–27.00) | 2.47 (0.19–27.74) | |||
| CORTICAL DYSPLASIA | 2 | Mean ± SD | 10.78 ± 3.87 | 11.52 ± 3.87 | |
| Median | 10.78 (8.04–13.52) | 11.52 (8.78–14.25) | |||
| GLIOBLASTOMA | 16 | Mean ± SD | 12.73 ± 2.29 | 13.47 ± 2.29 | |
| Median | 12.75 (10.40–15.90) | 13.49 (11.14–16.64) | |||
| GERMINOMA | 6 | Mean ± SD | 4.86 ± 5.13 | 5.60 ± 5.13 | |
| Median | 4.30 (0.03–10.25) | 5.04 (0.77–10.99) | |||
| TERATOMA | 3 | Mean ± SD | 10.93 ± 3.41 | 11.67 ± 3.41 | |
| Median | 10.60 (7.70–14.50) | 11.34 (8.44–15.24) | |||
| YOC SAC TUMOR | 3 | Mean ± SD | 14.00 ± 0.00 | 14.74 ± 0.00 | |
| Median | 14.00 (14.00–14.00) | 14.74 (14.74–14.74) | |||
| GLIONEURONAL | 1 | Mean ± SD | 11.31 ± 3.75 | 12.05 ± 3.75 | |
| Median | 12.00 (4.00–18.00) | 12.74 (4.74–18.74) | |||
| PAPILLOMA | 4 | Mean ± SD | 1.61 ± 1.21 | 2.35 ± 1.21 | |
| Median | 1.00 (0.83–3.00) | 1.74 (1.57–3.74) | |||
| OCULAR TUMOR 5a | 6 | Mean ± SD | NaN | NaN | |
| Median | NaN | NaN | |||
| RETINOBLASTOMA 5b | 12 | Mean ± SD | NaN | NaN | |
| Median | NaN | NaN | |||
| Grade 6 | I | 53 | Mean ± SD | 7.53 ± 4.96 | 8.27 ± 4.96 |
| Median | 6.69 (0.83–19.00) | 7.43 (1.57–19.74) | |||
| II | 18 | Mean ± SD | 6.85 ± 5.26 | 7.59 ± 5.26 | |
| Median | 4.47 (0.26–16.01) | 5.21 (1.00–16.75) | |||
| III | 12 | Mean ± SD | 4.19 ± 3.88 | 4.93 ± 3.88 | |
| Median | 2.55 (1.00–15.00) | 3.29 (1.74–15.74) | |||
| IV | 92 | Mean ± SD | 7.64 ± 4.90 | 8.38 ± 4.90 | |
| Median | 7.82 (0.03–18.00) | 8.56 (0.77–18.74) | |||
| CONTROL | 244 | Mean ± SD | 1.69 ± 4.01 | 2.26 ± 4.07 | |
| Median | 1.73 (0.00–27.00) | 2.47 (0.19–27.74) | |||
| Not Available | 20 | Mean ± SD | NaN | NaN | |
| Median | NaN | NaN | |||
| Developmental Status 7 | CHILD | 125 | Mean ± SD | 6.13 ± 3.29 | 6.87 ± 3.29 |
| Median | 6.00 (0.47–12.00) | 6.74 (1.21–12.74) | |||
| INFANT | 37 | Mean ± SD | 0.81 ± 0.65 | 1.55 ± 0.65 | |
| Median | 0.74 (0.16–2.02) | 1.48 (0.90–2.76) | |||
| NEONATE | 4 | Mean ± SD | 0.02 ± 0.01 | 0.76 ± 0.01 | |
| Median | 0.02 (0.01–0.03) | 0.76 (0.75–0.77) | |||
| ADOLESCENT | 43 | Mean ± SD | 13.38 ± 3.24 | 14.12 ± 3.24 | |
| Median | 14.00 (1.90–18.00) | 14.74 (2.64–18.74) | |||
| ADULT | 6 | Mean ± SD | 0.01 ± 0.08 | 0.35 ± 0.13 | |
| Median | 0.00 (0.00–0.71) | 0.35 (0.19–1.45) | |||
| FETUS | 81 | Mean ± SD | 23.33 ± 3.14 | 24.07 ± 3.14 | |
| Median | 23.50 (19.00–27.00) | 24.24 (19.74–27.74) | |||
| ALL STAGES | 100 | Mean ± SD | 1.73 ± 0.00 | 2.45 ± 0.21 | |
| Median | 1.73 (1.73–1.73) | 2.47 (0.38–2.47) |
| Inv. | miRNA | Pattern | f | f (%) | K-Means Cluster | Mean Expression |
|---|---|---|---|---|---|---|
| 1 | MIR376B | Up-regulated | 165 | 84.61 | 1 | 0.722 |
| 2 | MIR372 | Up-regulated | 148 | 75.90 | 1 | 0.624 |
| 3 | MIR149 | Down-regulated | 195 | 100.00 | 2 | −5.491 |
| 4 | MIR214 | Down-regulated | 195 | 100.00 | 2 | −4.742 |
| 5 | MIR574 | Down-regulated | 195 | 100.00 | 2 | −4.975 |
| 6 | MIR595 | Down-regulated | 195 | 100.00 | 2 | −4.083 |
| 7 | MIR765 | Down-regulated | 195 | 100.00 | 2 | −5.690 |
| 8 | MIR92B | Down-regulated | 182 | 93.33 | 2 | −3.622 |
| 9 | MIR939 | Down-regulated | 189 | 96.92 | 2 | −3.225 |
| 10 | MIR202 | Down-regulated | 190 | 97.43 | 2 | −3.369 |
| 11 | MIR921 | Down-regulated | 191 | 97.95 | 2 | −3.527 |
| 12 | MIR494 | Down-regulated | 193 | 98.97 | 2 | −6.053 |
| 13 | MIR665 | Down-regulated | 193 | 98.97 | 2 | −4.835 |
| 14 | MIR936 | Down-regulated | 193 | 98.97 | 2 | −4.799 |
| 15 | MIR575 | Down-regulated | 194 | 99.49 | 2 | −3.731 |
| 16 | MIR638 | Down-regulated | 194 | 99.49 | 2 | −5.799 |
| 17 | MIR933 | Down-regulated | 194 | 99.49 | 2 | −4.271 |
| 18 | MIR32 | Down-regulated | 176 | 90.25 | 3 | −1.7540219 |
| 19 | MIR557 | Down-regulated | 176 | 90.25 | 3 | −1.2031172 |
| 20 | MIR583 | Down-regulated | 177 | 90.77 | 3 | −1.3094348 |
| 21 | MIR675 | Down-regulated | 177 | 90.77 | 3 | −1.8494352 |
| 22 | MIR370 | Down-regulated | 178 | 91.28 | 3 | −1.7722773 |
| 23 | MIR760 | Down-regulated | 178 | 91.28 | 3 | −1.4657573 |
| 24 | MIR198 | Down-regulated | 179 | 91.80 | 3 | −2.2433618 |
| 25 | MIR210 | Down-regulated | 179 | 91.80 | 3 | −2.3769263 |
| 26 | MIR627 | Down-regulated | 179 | 91.80 | 3 | −1.0966398 |
| 27 | MIR650 | Down-regulated | 179 | 91.80 | 3 | −2.0081156 |
| 28 | MIR647 | Down-regulated | 180 | 92.31 | 3 | −1.6223603 |
| 29 | MIR632 | Down-regulated | 181 | 92.82 | 3 | −1.5932408 |
| 30 | MIR498 | Down-regulated | 183 | 93.85 | 3 | −1.7145673 |
| 31 | MIR608 | Down-regulated | 184 | 94.36 | 3 | −2.4389162 |
| 32 | MIR610 | Down-regulated | 184 | 94.36 | 3 | −1.9065001 |
| 33 | MIR564 | Down-regulated | 185 | 94.87 | 3 | −2.1568429 |
| 34 | MIR206 | Down-regulated | 186 | 95.38 | 3 | −2.9338987 |
| 35 | MIR671 | Down-regulated | 186 | 95.38 | 3 | −1.6370801 |
| 36 | MIR297 | Down-regulated | 188 | 96.41 | 3 | −2.8914463 |
| 37 | MIR637 | Down-regulated | 188 | 96.41 | 3 | −3.1802687 |
| 38 | MIR891A | Down-regulated | 188 | 96.41 | 3 | −2.5938886 |
| 39 | MIR185 | Down-regulated | 193 | 98.97 | 3 | −2.8633022 |
| 40 | MIR183 | Up-regulated | 176 | 90.26 | 4 | −1.7540219 |
| 41 | MIR889 | Up-regulated | 176 | 90.26 | 4 | −1.2031172 |
| 42 | MIR520H | Up-regulated | 177 | 90.77 | 4 | −1.3094348 |
| 43 | MIR563 | Up-regulated | 177 | 90.77 | 4 | −1.8494352 |
| 44 | MIR433 | Up-regulated | 178 | 91.28 | 4 | −1.7722773 |
| 45 | MIR519D | Up-regulated | 178 | 91.28 | 4 | −1.4657573 |
| 46 | MIR891B | Up-regulated | 179 | 91.79 | 4 | −2.2433618 |
| 47 | MIR631 | Up-regulated | 179 | 91.79 | 4 | −2.3769263 |
| 48 | MIR518B | Up-regulated | 179 | 91.79 | 4 | −1.0966398 |
| 49 | MIR367 | Up-regulated | 179 | 91.79 | 4 | −2.0081156 |
| 50 | MIR613 | Up-regulated | 180 | 92.31 | 4 | −1.6223603 |
| 51 | MIR651 | Up-regulated | 181 | 92.82 | 4 | −1.5932408 |
| 52 | MIR216A | Up-regulated | 183 | 93.85 | 4 | −1.7145673 |
| 53 | MIR374A | Up-regulated | 184 | 94.36 | 4 | −2.4389162 |
| 54 | MIR599 | Up-regulated | 184 | 94.36 | 4 | −1.9065001 |
| 55 | MIR577 | Up-regulated | 185 | 94.87 | 4 | −2.1568429 |
| 56 | MIR190B | Up-regulated | 186 | 95.38 | 4 | −2.9338987 |
| 57 | MIR429 | Up-regulated | 186 | 95.38 | 4 | −1.6370801 |
| 58 | MIR567 | Up-regulated | 188 | 96.41 | 4 | −2.8914463 |
| 59 | MIR618 | Up-regulated | 188 | 96.41 | 4 | −3.1802687 |
| 60 | MIR581 | Up-regulated | 188 | 96.41 | 4 | −2.5938886 |
| 61 | MIR183 | Up-regulated | 193 | 98.97 | 4 | −2.8633022 |
| Source | Term Name | −log10(p) |
|---|---|---|
| GO:MF | SH3 domain binding | 2.936 |
| GO:MF | Ras GTPase binding | 1.731 |
| GO:BP | nervous system development | 19.815 |
| GO:BP | generation of neurons | 13.643 |
| GO:BP | neuron projection development | 11.23 |
| GO:BP | neuron projection morphogenesis | 9.512 |
| GO:BP | regulation of nervous system development | 8.701 |
| GO:BP | central nervous system development | 6.965 |
| GO:BP | cell morphogenesis involved in neuron differentiation | 6.74 |
| GO:BP | regulation of neuron projection development | 6.543 |
| GO:BP | axonogenesis | 5.644 |
| GO:BP | synaptic vesicle cycle | 4.855 |
| GO:BP | axon development | 4.648 |
| GO:BP | neurotransmitter transport | 4.407 |
| GO:BP | regulation of synaptic plasticity | 2.796 |
| GO:BP | neurotransmitter secretion | 2.601 |
| GO:BP | regulation of axonogenesis | 2.352 |
| GO:BP | neuron projection guidance | 2.344 |
| GO:BP | dendritic spine development | 2.197 |
| GO:BP | central nervous system neuron axonogenesis | 2.195 |
| GO:BP | synaptic vesicle exocytosis | 2.078 |
| GO:BP | central nervous system neuron differentiation | 1.375 |
| GO:BP | central nervous system projection neuron axonogenesis | 1.345 |
| GO:BP | camera-type eye morphogenesis | 1.332 |
| GO:CC | synapse | 18.837 |
| GO:CC | neuron projection | 11.917 |
| GO:CC | somatodendritic compartment | 11.311 |
| GO:CC | dendrite | 10.215 |
| GO:CC | dendritic tree | 10.092 |
| GO:CC | neuron spine | 3.125 |
| GO:CC | postsynaptic membrane | 2.872 |
| GO:CC | neuron projection terminus | 2.794 |
| GO:CC | main axon | 2.58 |
| KEGG | Pathways in cancer | 7.647 |
| KEGG | Axon guidance | 4.293 |
| KEGG | MicroRNAs in cancer | 3.479 |
| REAC | Neuronal System | 3.262 |
| REAC | Axon guidance | 2.046 |
| REAC | Nervous system development | 2.046 |
| REAC | Transmission across Chemical Synapses | 1.421 |
| REAC | Vesicle-mediated transport | 1.391 |
| HPA | cerebral cortex; neuropil [Approved, Medium] | 4.339 |
| HPA | cerebral cortex; neuropil [Approved, Low] | 3.99 |
| HPA | cerebellum; cells in granular layer [Approved, Medium] | 3.958 |
| HPA | cerebellum; cells in granular layer [Approved, Low] | 3.439 |
| HPA | hippocampus; neuronal cells [Approved Low] | 1.66 |
| HPA | cerebral cortex; neuropil [Approved, High] | 1.507 |
| HPA | cerebral cortex; neuronal cells [Approved, Low] | 1.47 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lambrou, G.I.; Zaravinos, A.; Braoudaki, M. Co-Deregulated miRNA Signatures in Childhood Central Nervous System Tumors: In Search for Common Tumor miRNA-Related Mechanics. Cancers 2021, 13, 3028. https://doi.org/10.3390/cancers13123028
Lambrou GI, Zaravinos A, Braoudaki M. Co-Deregulated miRNA Signatures in Childhood Central Nervous System Tumors: In Search for Common Tumor miRNA-Related Mechanics. Cancers. 2021; 13(12):3028. https://doi.org/10.3390/cancers13123028
Chicago/Turabian StyleLambrou, George I., Apostolos Zaravinos, and Maria Braoudaki. 2021. "Co-Deregulated miRNA Signatures in Childhood Central Nervous System Tumors: In Search for Common Tumor miRNA-Related Mechanics" Cancers 13, no. 12: 3028. https://doi.org/10.3390/cancers13123028
APA StyleLambrou, G. I., Zaravinos, A., & Braoudaki, M. (2021). Co-Deregulated miRNA Signatures in Childhood Central Nervous System Tumors: In Search for Common Tumor miRNA-Related Mechanics. Cancers, 13(12), 3028. https://doi.org/10.3390/cancers13123028








