Systematic Review on the Association of Radiomics with Tumor Biological Endpoints
Abstract
Simple Summary
Abstract
1. Introduction
- KRAS and BRAF are the genes responsible for making the proteins K-ras and B-raf, which are, amongst others, involved in important signaling pathways (e.g., Ras-Raf-MAPK, PI3-K-AKT) [7,8]. Mutation and down-/up-regulation of any of those kinases can lead to malignancy and especially cancer formation.
- VEGF is a signaling factor promoting the formation of new blood vessels. To grow and metastasize, solid cancers require blood supply, which they attain by expressing VEGF to form supporting vasculature [9].
- TP-53 is involved in the regulation and progression through the cell cycle; monitors genomic stability and can induce apoptosis. It is one of the most prominent tumor-suppressors [10].
- PD-L1 is involved in suppressing the adaptive arm of the immune system. By upregulating PD-L1 expression, cancer cells may evade the host immune system [11].
- IDH catalyzes the decarboxylation of isocitrate. Through this metabolic deregulation, cancer progression can be initiated or supported [12].
- Ki-67 is a protein that is present during all active phases of the cell cycle but absent in resting (quiescent) cells [13]. Therefore, this cellular proliferation marker is frequently used to distinguish fast growing cell populations, such has cancer cells, from normal cells.
2. Materials and Methods
2.1. Literature Search
2.2. Eligibility Criteria
2.3. Analysis
3. Results
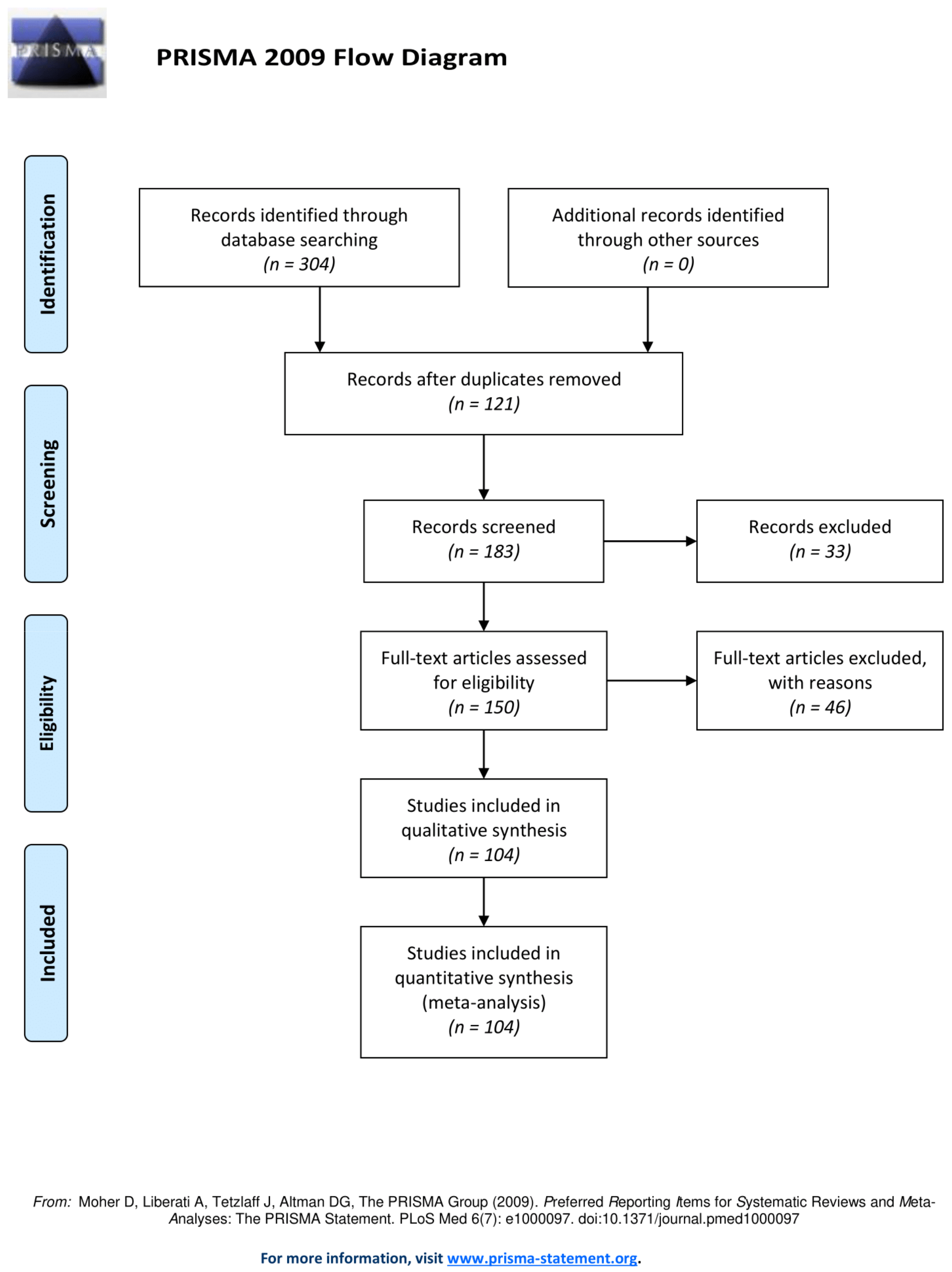
3.1. Literature Search, Eligibility Criteria and Study Selection
3.2. CNS
3.2.1. Summary
3.2.2. IDH
3.2.3. EGFR
3.2.4. Ki-67
3.2.5. TP-53
3.2.6. VEGF
3.3. Breast Cancer
3.3.1. Summary
3.3.2. HER-2
3.3.3. Ki-67
3.3.4. TP-53
3.4. Lung Cancer
3.4.1. Summary
3.4.2. EGFR
3.4.3. KRAS
3.4.4. ALK
3.4.5. PD-L1
3.4.6. Ki-67
3.4.7. TP-53
3.5. Gastrointestinal Cancer
3.5.1. Summary
3.5.2. KRAS
3.5.3. TP-53
3.5.4. HER-2
3.5.5. Ki-67
3.5.6. BRAF
3.6. Liver Cancer
3.6.1. Summary
3.6.2. PD-L1
3.6.3. Ki-67
3.6.4. VEGF
3.7. Other Cancers
3.7.1. Summary
3.7.2. Details
3.8. Feature Interpretation
3.9. Results per Biomarker
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Goossens, N.; Nakagawa, S.; Sun, X.; Hoshida, Y. Cancer biomarker discovery and validation. Transl. Cancer Res. 2015, 4, 256–269. [Google Scholar]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.L.; Siu, L.L. Molecular profiling for precision cancer therapies. Genome Med. 2020, 12, 8. [Google Scholar] [CrossRef]
- Song, J.; Yin, Y.; Wang, H.; Chang, Z.; Liu, Z.; Cui, L. A review of original articles published in the emerging field of radiomics. Eur. J. Radiol. 2020, 127, 108991. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Bublil, E.M.; Yarden, Y. The EGF receptor family: Spearheading a merger of signaling and therapeutics. Curr. Opin. Cell Biol. 2007, 19, 124–134. [Google Scholar] [CrossRef] [PubMed]
- García-Regalado, A.; La Rosa, C.H.G.-D. The Role of Anaplastic Lymphoma Kinase in Human Cancers. Oncol. Hematol. Rev. 2013, 9, 149–153. [Google Scholar] [CrossRef]
- Zaman, A.; Wu, W.; Bivona, T.G. Targeting Oncogenic BRAF: Past, Present, and Future. Cancers 2019, 11, 1197. [Google Scholar] [CrossRef]
- Liu, P.; Wang, Y.; Li, X. Targeting the untargetable KRAS in cancer therapy. Acta Pharm. Sin. B 2019, 9, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Neufeld, G.; Cohen, T.; Gengrinovitch, S.; Poltorak, Z. Vascular endothelial growth factor (VEGF) and its receptors. FASEB J. 1999, 13, 9–22. [Google Scholar] [CrossRef]
- Read, A.; Strachan, T. Chapter 18: Cancer Genetics. Human Molecular Genetics, 2nd ed.; Wiley: New York, NY, USA, 1999. [Google Scholar]
- Salmaninejad, A.; Valilou, S.F.; Shabgah, A.G.; Aslani, S.; Alimardani, M.; Pasdar, A.; Sahebkar, A. PD-1/PD-L1 pathway: Basic biology and role in cancer immunotherapy. J. Cell. Physiol. 2019, 234, 16824–16837. [Google Scholar] [CrossRef]
- Bleeker, F.E.; Molenaar, R.J.; Leenstra, S. Recent advances in the molecular understanding of glioblastoma. J. Neuro-Oncol. 2012, 108, 11–27. [Google Scholar] [CrossRef]
- Scholzen, T.; Gerdes, J. The Ki-67 protein: From the known and the unknown. J. Cell. Physiol. 2000, 182, 311–322. [Google Scholar] [CrossRef]
- Biomarkers in Risk Assessment: Validity and Validation (EHC 222, 2001). Available online: http://www.inchem.org/documents/ehc/ehc/ehc222.htm (accessed on 5 June 2021).
- Moher, D.; Shamseer, L.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A.; PRISMA-P Group. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst. Rev. 2015, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Zotero|Your Personal Research Assistant. Available online: http://www.zotero.org (accessed on 29 September 2020).
- Tripod Statement. Available online: https://www.tripod-statement.org/ (accessed on 6 June 2021).
- Wu, G.; Chen, Y.; Wang, Y.; Yu, J.; Lv, X.; Ju, X.; Shi, Z.; Chen, L.; Chen, Z. Sparse Representation-Based Radiomics for the Diagnosis of Brain Tumors. IEEE Trans. Med. Imaging 2018, 37, 893–905. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-C.; Bai, H.; Sun, Q.; Zhao, Y.; Lv, Y.; Zhou, J.; Liang, C.; Chen, Y.; Liang, D.; Zheng, H. Multiregional radiomics profiling from multiparametric MRI: Identifying an imaging predictor of IDH1 mutation status in glioblastoma. Cancer Med. 2018, 7, 5999–6009. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Nam, Y.; Lee, Y.S.; Kim, J.; Ahn, K.-J.; Jang, J.; Shin, N.-Y.; Kim, B.-S.; Jeon, S.-S. IDH1 mutation prediction using MR-based radiomics in glioblastoma: Comparison between manual and fully automated deep learning-based approach of tumor segmentation. Eur. J. Radiol. 2020, 128, 109031. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, Y.; Li, S.; Fan, X.; Sun, Z.; Yang, Z.; Wang, K.; Zhang, Z.; Jiang, T.; Liu, Y.; et al. IDH mutation-specific radiomic signature in lower-grade gliomas. Aging 2019, 11, 673–696. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Yu, J.; Guo, Y.; Cao, W. Deep Learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci. Rep. 2017, 7, 5467. [Google Scholar] [CrossRef]
- Yu, J.; Shi, Z.; Lian, Y.; Li, Z.; Liu, T.; Gao, Y.; Wang, Y.; Chen, L.; Mao, Y. Noninvasive IDH1 mutation estimation based on a quantitative radiomics approach for grade II glioma. Eur. Radiol. 2017, 27, 3509–3522. [Google Scholar] [CrossRef]
- Yu, J.; Shi, Z.; Ji, C.; Lian, Y.; Wang, Y.; Chen, L.; Mao, Y. Anatomical location differences between mutated and wild-type isocitrate dehydrogenase 1 in low-grade gliomas. Int. J. Neurosci. 2017, 127, 873–880. [Google Scholar] [CrossRef]
- Liu, T.; Wu, G.; Yu, J.; Guo, Y.; Wang, Y.; Shi, Z.; Chen, L. A mRMRMSRC feature selection method for radiomics approach. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju, Korea, 11–15 July 2017; Volume 2017, pp. 616–619. [Google Scholar] [CrossRef]
- Arita, H.; Kinoshita, M.; Kawaguchi, A.; Takahashi, M.; Narita, Y.; Terakawa, Y.; Tsuyuguchi, N.; Okita, Y.; Nonaka, M.; Moriuchi, S.; et al. Lesion location implemented magnetic resonance imaging radiomics for predicting IDH and TERT promoter mutations in grade II/III gliomas. Sci. Rep. 2018, 8, 11773. [Google Scholar] [CrossRef] [PubMed]
- Fukuma, R.; Yanagisawa, T.; Kinoshita, M.; Shinozaki, T.; Arita, H.; Kawaguchi, A.; Takahashi, M.; Narita, Y.; Terakawa, Y.; Tsuyuguchi, N.; et al. Prediction of IDH and TERT promoter mutations in low-grade glioma from magnetic resonance images using a convolutional neural network. Sci. Rep. 2019, 9, 20311. [Google Scholar] [CrossRef] [PubMed]
- Kuthuru, S.; Deaderick, W.; Bai, H.; Su, C.; Vu, T.; Monga, V.; Rao, A. A Visually Interpretable, Dictionary-Based Approach to Imaging-Genomic Modeling, with Low-Grade Glioma as a Case Study. Cancer Inform. 2018, 17, 1176935118802796. [Google Scholar] [CrossRef]
- Zhou, H.; Vallières, M.; Bai, H.X.; Su, C.; Tang, H.; Oldridge, D.; Zhang, Z.; Xiao, B.; Liao, W.; Tao, Y.; et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro-Oncology 2017, 19, 862–870. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tian, Q.; Wang, L.; Liu, Y.; Li, B.; Liang, Z.; Gao, P.; Zheng, K.; Zhao, B.; Lu, H. Radiomics Strategy for Molecular Subtype Stratification of Lower-Grade Glioma: Detecting IDH andTP53Mutations Based on Multimodal MRI. J. Magn. Reson. Imaging 2018, 48, 916–926. [Google Scholar] [CrossRef]
- Lee, J.; Narang, S.; Martinez, J.J.; Rao, G.; Rao, A. Associating spatial diversity features of radiologically defined tumor habitats with epidermal growth factor receptor driver status and 12-month survival in glioblastoma: Methods and preliminary investigation. J. Med. Imaging 2015, 2, 041006. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Xu, K.; Qian, Z.; Wang, K.; Fan, X.; Li, S.; Wang, Y.; Jiang, T. MRI features can predict EGFR expression in lower grade gliomas: A voxel-based radiomic analysis. Eur. Radiol. 2018, 28, 356–362. [Google Scholar] [CrossRef]
- Li, J.; Liu, S.; Qin, Y.; Zhang, Y.; Wang, N.; Liu, H. High-order radiomics features based on T2 FLAIR MRI predict multiple glioma immunohistochemical features: A more precise and personalized gliomas management. PLoS ONE 2020, 15, e0227703. [Google Scholar] [CrossRef]
- Li, Y.; Qian, Z.; Xu, K.; Wang, K.; Fan, X.; Li, S.; Liu, X.; Wang, Y.; Jiang, T. Radiomic features predict Ki-67 expression level and survival in lower grade gliomas. J. Neuro-Oncol. 2017, 135, 317–324. [Google Scholar] [CrossRef]
- Ugga, L.; Cuocolo, R.; Solari, D.; Guadagno, E.; D’Amico, A.; Somma, T.; Cappabianca, P.; Caro, M.L.D.B.D.; Cavallo, L.M.; Brunetti, A. Prediction of high proliferative index in pituitary macroadenomas using MRI-based radiomics and machine learning. Neuroradiology 2019, 61, 1365–1373. [Google Scholar] [CrossRef]
- Li, Y.; Qian, Z.; Xu, K.; Wang, K.; Fan, X.; Li, S.; Jiang, T.; Liu, X.; Wang, Y. MRI features predict p53 status in lower-grade gliomas via a machine-learning approach. NeuroImage Clin. 2018, 17, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Li, Y.; Wang, Y.; Fan, X.; Xu, K.; Wang, K.; Li, S.; Zhang, Z.; Jiang, T.; Liu, X. Radiogenomic analysis of vascular endothelial growth factor in patients with diffuse gliomas. Cancer Imaging 2019, 19, 68. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Meng, J.; Yu, Q.; Li, P.; Fu, S. Radiomics-based machine learning methods for isocitrate dehydrogenase genotype prediction of diffuse gliomas. J. Cancer Res. Clin. Oncol. 2019, 145, 543–550. [Google Scholar] [CrossRef]
- Binder, Z.A.; Thorne, A.H.; Bakas, S.; Wileyto, E.P.; Bilello, M.; Akbari, H.; Rathore, S.; Ha, S.M.; Zhang, L.; Ferguson, C.J.; et al. Epidermal Growth Factor Receptor Extracellular Domain Mutations in Glioblastoma Present Opportunities for Clinical Imaging and Therapeutic Development. Cancer Cell 2018, 34, 163–177.e7. [Google Scholar] [CrossRef] [PubMed]
- Rathore, S.; Akbari, H.; Rozycki, M.; Abdullah, K.G.; Nasrallah, M.P.; Binder, Z.A.; Davuluri, R.V.; Lustig, R.A.; Dahmane, N.; Bilello, M.; et al. Radiomic MRI signature reveals three distinct subtypes of glioblastoma with different clinical and molecular characteristics, offering prognostic value beyond IDH1. Sci. Rep. 2018, 8, 5087. [Google Scholar] [CrossRef] [PubMed]
- Akbari, H.; Bakas, S.; Pisapia, J.M.; Nasrallah, M.P.; Rozycki, M.; Martinez-Lage, M.; Morrissette, J.J.D.; Dahmane, N.; O’Rourke, D.M.; Davatzikos, C. In vivoevaluation of EGFRvIII mutation in primary glioblastoma patients via complex multiparametric MRI signature. Neuro-Oncology 2018, 20, 1068–1079. [Google Scholar] [CrossRef]
- Tan, Y.; Zhang, S.-T.; Wei, J.-W.; Dong, D.; Wang, X.-C.; Yang, G.-Q.; Tian, J.; Zhang, H. A radiomics nomogram may improve the prediction of IDH genotype for astrocytoma before surgery. Eur. Radiol. 2019, 29, 3325–3337. [Google Scholar] [CrossRef]
- Lu, C.-F.; Hsu, F.-T.; Hsieh, K.L.-C.; Kao, Y.-C.J.; Cheng, S.-J.; Hsu, J.B.-K.; Tsai, P.-H.; Chen, R.-J.; Huang, C.-C.; Yen, Y.; et al. Machine Learning–Based Radiomics for Molecular Subtyping of Gliomas. Clin. Cancer Res. 2018, 24, 4429–4436. [Google Scholar] [CrossRef]
- Su, C.; Jiang, J.; Zhang, S.; Shi, J.; Xu, K.; Shen, N.; Zhang, J.; Li, L.; Zhao, L.; Zhang, J.; et al. Radiomics based on multicontrast MRI can precisely differentiate among glioma subtypes and predict tumour-proliferative behaviour. Eur. Radiol. 2019, 29, 1986–1996. [Google Scholar] [CrossRef]
- Tan, Y.; Mu, W.; Wang, X.-C.; Yang, G.-Q.; Gillies, R.J.; Zhang, H. Whole-tumor radiomics analysis of DKI and DTI may improve the prediction of genotypes for astrocytomas: A preliminary study. Eur. J. Radiol. 2020, 124, 108785. [Google Scholar] [CrossRef]
- Park, C.J.; Choi, Y.S.; Park, Y.W.; Ahn, S.S.; Kang, S.-G.; Chang, J.-H.; Kim, S.H.; Lee, S.-K. Diffusion tensor imaging radiomics in lower-grade glioma: Improving subtyping of isocitrate dehydrogenase mutation status. Neuroradiology 2020, 62, 319–326. [Google Scholar] [CrossRef]
- Kim, M.; Jung, S.Y.; Park, J.E.; Jo, Y.; Park, S.Y.; Nam, S.J.; Kim, J.H.; Kim, H.S. Diffusion- and perfusion-weighted MRI radiomics model may predict isocitrate dehydrogenase (IDH) mutation and tumor aggressiveness in diffuse lower grade glioma. Eur. Radiol. 2020, 30, 2142–2151. [Google Scholar] [CrossRef]
- Lee, M.H.; Kim, J.; Kim, S.-T.; Shin, H.-M.; You, H.-J.; Choi, J.W.; Seol, H.J.; Nam, D.-H.; Lee, J.-I.; Kong, D.-S. Prediction of IDH1 Mutation Status in Glioblastoma Using Machine Learning Technique Based on Quantitative Radiomic Data. World Neurosurg. 2019, 125, e688–e696. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Zhang, X.; Rui, W.; Pang, H.; Qiu, T.; Wang, J.; Xie, Q.; Jin, T.; Zhang, H.; Chen, H.; et al. Noninvasive Prediction of IDH1 Mutation and ATRX Expression Loss in Low-Grade Gliomas Using Multiparametric MR Radiomic Features. J. Magn. Reson. Imaging 2019, 49, 808–817. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Mu, W.; Wang, Y.; Liu, Z.; Liu, Z.; Wang, Y.; Ma, W.; Kong, Z.; Wang, S.; Zhou, X.; et al. A Non-invasive Radiomic Method Using 18F-FDG PET Predicts Isocitrate Dehydrogenase Genotype and Prognosis in Patients with Glioma. Front. Oncol. 2019, 9, 1183. [Google Scholar] [CrossRef]
- Kong, Z.; Li, J.; Liu, Z.; Liu, Z.; Zhao, D.; Cheng, X.; Li, L.; Lin, Y.; Wang, Y.; Tian, J.; et al. Radiomics signature based on FDG-PET predicts proliferative activity in primary glioma. Clin. Radiol. 2019, 74, 815.e15–815.e23. [Google Scholar] [CrossRef] [PubMed]
- Lohmann, P.; Lerche, C.; Bauer, E.K.; Steger, J.; Stoffels, G.; Blau, T.; Dunkl, V.; Kocher, M.; Viswanathan, S.; Filss, C.P.; et al. Predicting IDH genotype in gliomas using FET PET radiomics. Sci. Rep. 2018, 8, 13328. [Google Scholar] [CrossRef]
- Han, Y.; Wang, W.; Yang, Y.; Sun, Y.-Z.; Xiao, G.; Tian, Q.; Zhang, J.; Cui, G.-B.; Yan, L.-F. Amide Proton Transfer Imaging in Predicting Isocitrate Dehydrogenase 1 Mutation Status of Grade II/III Gliomas Based on Support Vector Machine. Front. Neurosci. 2020, 14, 144. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhu, Y.; Burnside, E.S.; Huang, E.; Drukker, K.; Hoadley, K.A.; Fan, C.; Conzen, S.D.; Zuley, M.; Net, J.M.; et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer 2016, 2, 16012. [Google Scholar] [CrossRef]
- Lin, P.; Liu, W.; Li, X.; Wan, D.; Qin, H.; Li, Q.; Chen, G.; He, Y.; Yang, H. MRI-based radiogenomics analysis for predicting genetic alterations in oncogenic signalling pathways in invasive breast carcinoma. Clin. Radiol. 2020, 75, 561.e1–561.e11. [Google Scholar] [CrossRef]
- Ma, W.; Ji, Y.; Qi, L.; Guo, X.; Jian, X.; Liu, P. Breast cancer Ki67 expression prediction by DCE-MRI radiomics features. Clin. Radiol. 2018, 73, 909.e1–909.e5. [Google Scholar] [CrossRef]
- Monti, S.; Aiello, M.; Incoronato, M.; Grimaldi, A.M.; Moscarino, M.; Mirabelli, P.; Ferbo, U.; Cavaliere, C.; Salvatore, M. DCE-MRI Pharmacokinetic-Based Phenotyping of Invasive Ductal Carcinoma: A Radiomic Study for Prediction of Histological Outcomes. Contrast Media Mol. Imaging 2018, 2018, 5076269. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yu, K.; Feng, C.; Zhao, D. Molecular Subtypes Recognition of Breast Cancer in Dynamic Contrast-Enhanced Breast Magnetic Resonance Imaging Phenotypes from Radiomics Data. Comput. Math. Methods Med. 2019, 2019, 6978650. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Li, H.; Wang, S.; Zheng, B.; Zhang, J.; Li, L. Radiomic analysis reveals DCE-MRI features for prediction of molecular subtypes of breast cancer. PLoS ONE 2017, 12, e0171683. [Google Scholar] [CrossRef] [PubMed]
- Castaldo, R.; Pane, K.; Nicolai, E.; Salvatore, M.; Franzese, M. The Impact of Normalization Approaches to Automatically Detect Radiogenomic Phenotypes Characterizing Breast Cancer Receptors Status. Cancers 2020, 12, 518. [Google Scholar] [CrossRef]
- Braman, N.; Prasanna, P.; Whitney, J.; Singh, S.; Beig, N.; Etesami, M.; Bates, D.D.B.; Gallagher, K.; Bloch, B.N.; Vulchi, M.; et al. Association of Peritumoral Radiomics with Tumor Biology and Pathologic Response to Preoperative Targeted Therapy for HER2 (ERBB2)–Positive Breast Cancer. JAMA Netw. Open 2019, 2, e192561. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Gao, F.; Duan, S.; Zhang, L.; Liu, Y.; Zhou, J.; Bai, G.; Tao, W. Radiomic features of Pk-DCE MRI parameters based on the extensive Tofts model in application of breast cancer. Phys. Eng. Sci. Med. 2020, 43, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Cheng, Z.; Huang, Y.; He, L.; Chen, X.; Ma, Z.; Huang, X.; Liang, C.; Liu, Z. An MRI-based Radiomics Classifier for Preoperative Prediction of Ki-67 Status in Breast Cancer. Acad. Radiol. 2018, 25, 1111–1117. [Google Scholar] [CrossRef]
- Leithner, D.; Horvat, J.V.; Marino, M.A.; Bernard-Davila, B.; Jochelson, M.S.; Ochoa-Albiztegui, R.E.; Martinez, D.; Morris, E.A.; Thakur, S.; Pinker, K. Radiomic signatures with contrast-enhanced magnetic resonance imaging for the assessment of breast cancer receptor status and molecular subtypes: Initial results. Breast Cancer Res. 2019, 21, 106. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, Y.; Zhang, K.; Liu, Y.; Cui, J.; Tao, J.; Wang, Y.; Wang, S. Invasive ductal breast cancer: Preoperative predict Ki-67 index based on radiomics of ADC maps. Radiol. Med. 2020, 125, 109–116. [Google Scholar] [CrossRef]
- Fan, M.; Yuan, W.; Zhao, W.; Xu, M.; Wang, S.; Gao, X.; Li, L. Joint Prediction of Breast Cancer Histological Grade and Ki-67 Expression Level Based on DCE-MRI and DWI Radiomics. IEEE J. Biomed. Health Inform. 2020, 24, 1632–1642. [Google Scholar] [CrossRef]
- Tagliafico, A.S.; Bignotti, B.; Rossi, F.; Matos, J.; Calabrese, M.; Valdora, F.; Houssami, N. Breast cancer Ki-67 expression prediction by digital breast tomosynthesis radiomics features. Eur. Radiol. Exp. 2019, 3, 36. [Google Scholar] [CrossRef]
- Antunovic, L.; Gallivanone, F.; Sollini, M.; Sagona, A.; Invento, A.; Manfrinato, G.; Kirienko, M.; Tinterri, C.; Chiti, A.; Castiglioni, I. [18F]FDG PET/CT features for the molecular characterization of primary breast tumors. Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 1945–1954. [Google Scholar] [CrossRef]
- Zhou, J.; Tan, H.; Bai, Y.; Li, J.; Lu, Q.; Chen, R.; Zhang, M.; Feng, Q.; Wang, M. Evaluating the HER-2 status of breast cancer using mammography radiomics features. Eur. J. Radiol. 2019, 121, 108718. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Grossmann, P.; Tan, Y.; Oxnard, G.R.; Rizvi, N.; Schwartz, L.H.; Zhao, B. Defining a Radiomic Response Phenotype: A Pilot Study using targeted therapy in NSCLC. Sci. Rep. 2016, 6, 33860. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Lu, L.; Dercle, L.; Lichtenstein, P.; Li, Y.; Yin, Q.; Zong, M.; Schwartz, L.; Zhao, B. Interobserver variability in tumor contouring affects the use of radiomics to predict mutational status. J. Med. Imaging 2018, 5, 011005. [Google Scholar] [CrossRef] [PubMed]
- Jia, T.-Y.; Xiong, J.-F.; Li, X.-Y.; Yu, W.; Xu, Z.-Y.; Cai, X.-W.; Ma, J.-C.; Ren, Y.-C.; Larsson, R.; Zhang, J.; et al. Identifying EGFR mutations in lung adenocarcinoma by noninvasive imaging using radiomics features and random forest modeling. Eur. Radiol. 2019, 29, 4742–4750. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Li, M.; Zhang, H.-M.; Hua, S.; Meng, F.; Yang, H.; Li, X.; Cao, D. A novel radiomic nomogram for predicting epidermal growth factor receptor mutation in peripheral lung adenocarcinoma. Phys. Med. Biol. 2020, 65, 055012. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lu, L.; Xiao, M.; Dercle, L.; Huang, Y.; Zhang, Z.; Schwartz, L.H.; Li, D.; Zhao, B. CT Slice Thickness and Convolution Kernel Affect Performance of a Radiomic Model for Predicting EGFR Status in Non-Small Cell Lung Cancer: A Preliminary Study. Sci. Rep. 2018, 8, 17913. [Google Scholar] [CrossRef]
- Li, X.-Y.; Xiong, J.-F.; Jia, T.-Y.; Shen, T.-L.; Hou, R.-P.; Zhao, J.; Fu, X.-L. Detection of epithelial growth factor receptor (EGFR) mutations on CT images of patients with lung adenocarcinoma using radiomics and/or multi-level residual convolutionary neural networks. J. Thorac. Dis. 2018, 10, 6624–6635. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Ding, C.; Zhang, H.; Song, J.; Wu, L. Radiomics for the prediction of EGFR mutation subtypes in non-small cell lung cancer. Med. Phys. 2019, 46, 4545–4552. [Google Scholar] [CrossRef]
- Mei, D.; Luo, Y.; Wang, Y.; Gong, J. CT texture analysis of lung adenocarcinoma: Can Radiomic features be surrogate biomarkers for EGFR mutation statuses. Cancer Imaging 2018, 18, 52. [Google Scholar] [CrossRef]
- Tu, W.; Sun, G.; Fan, L.; Wang, Y.; Xia, Y.; Guan, Y.; Li, Q.; Zhang, D.; Liu, S.; Li, Z. Radiomics signature: A potential and incremental predictor for EGFR mutation status in NSCLC patients, comparison with CT morphology. Lung Cancer 2019, 132, 28–35. [Google Scholar] [CrossRef]
- Yang, X.; Dong, X.; Wang, J.; Li, W.; Gu, Z.; Gao, D.; Zhong, N.; Guan, Y. Computed Tomography-Based Radiomics Signature: A Potential Indicator of Epidermal Growth Factor Receptor Mutation in Pulmonary Adenocarcinoma Appearing as a Subsolid Nodule. Oncologist 2019, 24, e1156–e1164. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, B.; Liu, X.; Song, J.; Fang, M.; Hu, C.; Dong, D.; Li, W.; Tian, J. Quantitative Biomarkers for Prediction of Epidermal Growth Factor Receptor Mutation in Non-Small Cell Lung Cancer. Transl. Oncol. 2018, 11, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wu, Y.; Xu, Y.; Sun, Y.; Gao, P.; Tan, M.; Ma, W.; Li, C.; Jin, L.; Hua, Y.; et al. The Potential of Radiomics Nomogram in Non-invasively Prediction of Epidermal Growth Factor Receptor Mutation Status and Subtypes in Lung Adenocarcinoma. Front. Oncol. 2019, 9, 1485. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Yang, J.; Ni, B.; Bi, D.; Sun, Y.; Xu, M.; Zhu, X.; Li, C.; Jin, L.; Gao, P.; et al. Toward automatic prediction of EGFR mutation status in pulmonary adenocarcinoma with 3D deep learning. Cancer Med. 2019, 8, 3532–3543. [Google Scholar] [CrossRef] [PubMed]
- Velazquez, E.R.; Parmar, C.; Liu, Y.; Coroller, T.P.; Cruz, G.; Stringfield, O.; Ye, Z.; Makrigiorgos, M.; Fennessy, F.; Mak, R.H.; et al. Somatic Mutations Drive Distinct Imaging Phenotypes in Lung Cancer. Cancer Res. 2017, 77, 3922–3930. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kong, C.; Xu, W.; Yang, S.; Shi, D.; Zhang, J.; Du, M.; Wang, S.; Bai, Y.; Zhang, T.; et al. Decoding tumor mutation burden and driver mutations in early stage lung adenocarcinoma using CT-based radiomics signature. Thorac. Cancer 2019, 10, 1904–1912. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.; Xu, K.; Zhang, L.; Wan, X.; Guo, Y. Radiomics Signature as a Predictive Factor for EGFR Mutations in Advanced Lung Adenocarcinoma. Front. Oncol. 2020, 10, 28. [Google Scholar] [CrossRef]
- Liu, Y.; Kim, J.; Balagurunathan, Y.; Li, Q.; Garcia, A.L.; Stringfield, O.; Ye, Z.; Gillies, R.J. Radiomic Features Are Associated with EGFR Mutation Status in Lung Adenocarcinomas. Clin. Lung Cancer 2016, 17, 441–448.e6. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Zhu, Z.; Mao, L.; Li, X.; Han, W.; Du, H.; Wu, H.; Song, W.; Jin, Z. Clinical, Conventional CT and Radiomic Feature-Based Machine Learning Models for Predicting ALK Rearrangement Status in Lung Adenocarcinoma Patients. Front. Oncol. 2020, 10, 369. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Hu, S.; Ge, Y.; Wang, J.; Duan, S.; Song, J.; Hu, C.; Li, Y. Radiomics study for predicting the expression of PD-L1 in non-small cell lung cancer based on CT images and clinicopathologic features. J. X-Ray Sci. Technol. 2020, 28, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Suh, Y.J.; Han, K.; Cho, H.; Lee, H.-J.; Hur, J.; Choi, B.W. Utility of CT radiomics for prediction of PD-L1 expression in advanced lung adenocarcinomas. Thorac. Cancer 2020, 11, 993–1004. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Xu, J.; Tian, Y.; Yuan, S.; Li, X. Correlation between radiomic features based on contrast-enhanced computed tomography images and Ki-67 proliferation index in lung cancer: A preliminary study. Thorac. Cancer 2018, 9, 1235–1240. [Google Scholar] [CrossRef]
- Gu, Q.; Feng, Z.; Liang, Q.; Li, M.; Deng, J.; Ma, M.; Wang, W.; Liu, J.; Liu, P.; Rong, P. Machine learning-based radiomics strategy for prediction of cell proliferation in non-small cell lung cancer. Eur. J. Radiol. 2019, 118, 32–37. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, Y.; Xu, J.; Ji, M.; Guo, Y.; Guo, Y.; Xiao, J.; Yao, X.; Shi, H.; Zeng, M. Assessing EGFR gene mutation status in non-small cell lung cancer with imaging features from PET/CT. Nucl. Med. Commun. 2019, 40, 842–849. [Google Scholar] [CrossRef]
- Koyasu, S.; Nishio, M.; Isoda, H.; Nakamoto, Y.; Togashi, K. Usefulness of gradient tree boosting for predicting histological subtype and EGFR mutation status of non-small cell lung cancer on 18F FDG-PET/CT. Ann. Nucl. Med. 2020, 34, 49–57. [Google Scholar] [CrossRef]
- Li, X.; Yin, G.; Zhang, Y.; Dai, D.; Liu, J.; Chen, P.; Zhu, L.; Ma, W.; Xu, W. Predictive Power of a Radiomic Signature Based on 18F-FDG PET/CT Images for EGFR Mutational Status in NSCLC. Front. Oncol. 2019, 9, 1062. [Google Scholar] [CrossRef]
- Nair, J.K.R.; Saeed, U.A.; McDougall, C.C.; Sabri, A.; Kovacina, B.; Raidu, B.V.S.; Khokhar, R.A.; Probst, S.; Hirsh, V.; Chankowsky, J.; et al. Radiogenomic Models Using Machine Learning Techniques to Predict EGFR Mutations in Non-Small Cell Lung Cancer. Can. Assoc. Radiol. J. 2020, 72, 109–119. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, X.; Zhao, Y.; Zhang, J.; Zhang, Z.; Wang, J.; Wang, Y.; Dai, M.; Han, J. Value of pre-therapy 18F-FDG PET/CT radiomics in predicting EGFR mutation status in patients with non-small cell lung cancer. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 1137–1146. [Google Scholar] [CrossRef]
- Shiri, I.; Maleki, H.; Hajianfar, G.; Abdollahi, H.; Ashrafinia, S.; Hatt, M.; Zaidi, H.; Oveisi, M.; Rahmim, A. Next-Generation Radiogenomics Sequencing for Prediction of EGFR and KRAS Mutation Status in NSCLC Patients Using Multimodal Imaging and Machine Learning Algorithms. Mol. Imaging Biol. 2020, 22, 1132–1148. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.J.; Sohn, I.; Cho, J.H.; Lee, H.Y.; Kim, J.-H.; Choi, Y.-L.; Kim, H.; Lee, G.; Lee, K.S.; Kim, J. Decoding Tumor Phenotypes for ALK, ROS1, and RET Fusions in Lung Adenocarcinoma Using a Radiomics Approach. Medicine 2015, 94, e1753. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Sun, D.; Guo, Y.; Guo, Y.; Xiao, J.; Wang, L.; Yao, X. Assessing PD-L1 Expression Level by Radiomic Features from PET/CT in Nonsmall Cell Lung Cancer Patients: An Initial Result. Acad. Radiol. 2020, 27, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.S.; Kim, J.; Coroller, T.P.; Parmar, C.; Velazquez, E.R.; Huynh, E.; Mak, R.H.; Aerts, H.J. Associations Between Somatic Mutations and Metabolic Imaging Phenotypes in Non–Small Cell Lung Cancer. J. Nucl. Med. 2017, 58, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.S.; Parmar, C.; Kim, J.; Huynh, E.; Mak, R.H.; Aerts, H.J. Impact of experimental design on PET radiomics in predicting somatic mutation status. Eur. J. Radiol. 2017, 97, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.T.; Jin, T.; Ye, N.; Mambetsariev, I.; Daniel, E.; Wang, T.; Wong, C.W.; Rockne, R.C.; Colen, R.; Holodny, A.I.; et al. Radiomic prediction of mutation status based on MR imaging of lung cancer brain metastases. Magn. Reson. Imaging 2020, 69, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cheng, Z.; Gevaert, O.; He, L.; Huang, Y.; Chen, X.; Huang, X.; Wu, X.; Zhang, W.; Dong, M.; et al. A CT-based radiomics nomogram for prediction of human epidermal growth factor receptor 2 status in patients with gastric cancer. Chin. J. Cancer Res. 2020, 32, 62–71. [Google Scholar] [CrossRef]
- Zhang, Q.; Gao, Y.; Zhang, R.; Zhou, X.; Chen, S.; Zhang, Y.; Liu, Q.; Xu, J.; Ge, Z. Personalized CT-based radiomics nomogram preoperative predicting Ki-67 expression in gastrointestinal stromal tumors: A multicenter development and validation cohort. Clin. Transl. Med. 2020, 9, 12. [Google Scholar] [CrossRef]
- Liang, W.; Yang, P.; Huang, R.; Xu, L.; Wang, J.; Liu, W.; Zhang, L.; Wan, D.; Huang, Q.; Lu, Y.; et al. A Combined Nomogram Model to Preoperatively Predict Histologic Grade in Pancreatic Neuroendocrine Tumors. Clin. Cancer Res. 2019, 25, 584–594. [Google Scholar] [CrossRef]
- Yang, L.; Dong, D.; Fang, M.; Zhu, Y.; Zang, Y.; Liu, Z.; Zhang, H.; Ying, J.; Zhao, X.; Tian, J. Can CT-based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer? Eur. Radiol. 2018, 28, 2058–2067. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, Y.; Chen, X.; Huang, Y.; He, L.; Zhao, K.; Huang, X.; Zhang, W.; Huang, Y.; Li, Y.; et al. Deep Learning Features Improve the Performance of a Radiomics Signature for Predicting KRAS Status in Patients with Colorectal Cancer. Acad. Radiol. 2020, 27, e254–e262. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.H.; Cho, Y.S.; Choi, J.Y.; Lee, K.-H.; Lee, J.K.; Min, J.H.; Hyun, S.H. Imaging phenotype using 18F-fluorodeoxyglucose positron emission tomography–based radiomics and genetic alterations of pancreatic ductal adenocarcinoma. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 2113–2122. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-W.; Shen, W.-C.; Chen, W.T.-L.; Hsieh, T.-C.; Yen, K.-Y.; Chang, J.-G.; Kao, C.-H. Metabolic Imaging Phenotype Using Radiomics of [18F]FDG PET/CT Associated with Genetic Alterations of Colorectal Cancer. Mol. Imaging Biol. 2019, 21, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Liu, H.; Ren, J.; Du, X.; Xin, L.; Li, D.; Yang, X.; Wang, D. Development and validation of a MRI-based radiomics signature for prediction of KRAS mutation in rectal cancer. Eur. Radiol. 2020, 30, 1948–1958. [Google Scholar] [CrossRef]
- Oh, J.E.; Kim, M.J.; Lee, J.; Hur, B.Y.; Kim, B.; Kim, D.Y.; Baek, J.Y.; Chang, H.J.; Park, S.C.; Oh, J.H.; et al. Magnetic Resonance-Based Texture Analysis Differentiating KRAS Mutation Status in Rectal Cancer. Cancer Res. Treat. 2020, 52, 51–59. [Google Scholar] [CrossRef]
- Meng, X.; Xia, W.; Xie, P.; Zhang, R.; Li, W.; Wang, M.; Xiong, F.; Liu, Y.; Fan, X.; Xie, Y.; et al. Preoperative radiomic signature based on multiparametric magnetic resonance imaging for noninvasive evaluation of biological characteristics in rectal cancer. Eur. Radiol. 2019, 29, 3200–3209. [Google Scholar] [CrossRef]
- Hectors, S.J.; Lewis, S.; Besa, C.; King, M.J.; Said, D.; Putra, J.; Ward, S.; Higashi, T.; Thung, S.; Yao, S.; et al. MRI radiomics features predict immuno-oncological characteristics of hepatocellular carcinoma. Eur. Radiol. 2020, 30, 3759–3769. [Google Scholar] [CrossRef]
- Ye, Z.; Jiang, H.; Chen, J.; Liu, X.; Wei, Y.; Xia, C.; Duan, T.; Cao, L.; Zhang, Z.; Song, B. Texture analysis on gadoxetic acid enhanced-MRI for predicting Ki-67 status in hepatocellular carcinoma: A prospective study. Chin. J. Cancer Res. 2019, 31, 806–817. [Google Scholar] [CrossRef]
- Yao, Z.; Dong, Y.; Wu, G.; Zhang, Q.; Yang, D.; Yu, J.-H.; Wang, W.-P. Preoperative diagnosis and prediction of hepatocellular carcinoma: Radiomics analysis based on multi-modal ultrasound images. BMC Cancer 2018, 18, 1089. [Google Scholar] [CrossRef]
- Peng, Y.-T.; Zhou, C.-Y.; Lin, P.; Wen, D.-Y.; Wang, X.-D.; Zhong, X.-Z.; Pan, D.-H.; Que, Q.; Li, X.; Chen, L.; et al. Preoperative Ultrasound Radiomics Signatures for Noninvasive Evaluation of Biological Characteristics of Intrahepatic Cholangiocarcinoma. Acad. Radiol. 2019, 27, 785–797. [Google Scholar] [CrossRef]
- Saadani, H.; Van Der Hiel, B.; Aalbersberg, E.A.; Zavrakidis, I.; Haanen, J.B.; Hoekstra, O.S.; Boellaard, R.; Stokkel, M.P. Metabolic Biomarker–Based BRAFV600 Mutation Association and Prediction in Melanoma. J. Nucl. Med. 2019, 60, 1545–1552. [Google Scholar] [CrossRef]
- Yoon, J.H.; Han, K.; Lee, E.; Lee, J.; Kim, E.-K.; Moon, H.J.; Park, V.; Nam, K.-H.; Kwak, J.Y. Radiomics in predicting mutation status for thyroid cancer: A preliminary study using radiomics features for predicting BRAFV600E mutations in papillary thyroid carcinoma. PLoS ONE 2020, 15, e0228968. [Google Scholar] [CrossRef]
- Zhu, Y.; Mohamed, A.S.; Lai, S.; Yang, S.; Kanwar, A.; Wei, L.; Kamal, M.; Sengupta, S.; ElHalawani, H.; Skinner, H.; et al. Imaging-Genomic Study of Head and Neck Squamous Cell Carcinoma: Associations Between Radiomic Phenotypes and Genomic Mechanisms via Integration of The Cancer Genome Atlas and The Cancer Imaging Archive. JCO Clin. Cancer Inform. 2019, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.-Y.; Lin, Y.-C.; Shen, W.-C.; Hsieh, T.-C.; Yen, K.-Y.; Chen, S.-W.; Kao, C.-H. Associations of Tumor PD-1 Ligands, Immunohistochemical Studies, and Textural Features in 18F-FDG PET in Squamous Cell Carcinoma of the Head and Neck. Sci. Rep. 2018, 8, 105. [Google Scholar] [CrossRef]
- Ahmed, A.; Elmohr, M.; Fuentes, D.; Habra, M.; Fisher, S.; Perrier, N.; Zhang, M.; Elsayes, K. Radiomic mapping model for prediction of Ki-67 expression in adrenocortical carcinoma. Clin. Radiol. 2020, 75, 479.e17–479.e22. [Google Scholar] [CrossRef] [PubMed]
- Bogowicz, M.; Jochems, A.; Deist, T.M.; Tanadini-Lang, S.; Huang, S.H.; Chan, B.; Waldron, J.N.; Bratman, S.; O’Sullivan, B.; Riesterer, O.; et al. Privacy-preserving distributed learning of radiomics to predict overall survival and HPV status in head and neck cancer. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Zhovannik, I.; Traverso, A.; Dankers, F.J.W.M.; Deist, T.M.; Kalendralis, P.; Monshouwer, R.; Bussink, J.; Fijten, R.; Aerts, H.J.W.L.; et al. Distributed radiomics as a signature validation study using the Personal Health Train infrastructure. Sci. Data 2019, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bogowicz, M.; Vuong, D.; Huellner, M.W.; Pavic, M.; Andratschke, N.; Gabrys, H.S.; Guckenberger, M.; Tanadini-Lang, S. CT radiomics and PET radiomics: Ready for clinical implementation? Q. J. Nucl. Med. Mol. Imaging 2019, 63, 355–370. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Steinmann, A.; Ding, Y.; Lee, H.; Owens, C.; Wang, J.; Yang, J.; Followill, D.; Ger, R.; MacKin, D.; et al. Radiomics feature robustness as measured using an MRI phantom. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef]
- Van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in medical imaging—“How-to” guide and critical reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- The Image Biomarker Standardisation Initiative—IBSI 0.0.1dev Documentation. Available online: https://ibsi.readthedocs.io/en/latest/ (accessed on 17 November 2020).
- Aronson, J.K.; Ferner, R. Biomarkers—A General Review. Curr. Protoc. Pharmacol. 2017, 76, 9.23.1–9.23.17. [Google Scholar] [CrossRef] [PubMed]

| Breast | CNS | GI | Liver | Lung | Others | Total | |
|---|---|---|---|---|---|---|---|
| ALK | 0 | 0 | 0 | 0 | 3 | 0 | 3 |
| BRAF | 0 | 0 | 1 | 0 | 0 | 2 | 3 |
| EGFR | 0 | 5 | 0 | 0 | 26 | 1 | 32 |
| HER-2 | 10 | 0 | 2 | 0 | 0 | 0 | 12 |
| IDH | 0 | 24 | 0 | 0 | 0 | 0 | 24 |
| Ki-67 | 8 | 5 | 3 | 3 | 2 | 2 | 23 |
| KRAS | 0 | 0 | 7 | 0 | 5 | 0 | 12 |
| PD-L1 | 0 | 0 | 0 | 2 | 3 | 1 | 6 |
| TP-53 | 1 | 2 | 2 | 0 | 1 | 1 | 7 |
| VEGF | 0 | 1 | 0 | 1 | 0 | 1 | 3 |
| TOTAL | 19 | 37 | 15 | 6 | 40 | 8 | 125 |
| Study | Biomarker | Alteration | Modality | Dataset Origin | Training | Validation | Feature Reduction | Feature Robustness | # Radiomic Features | Additional Features | Predictive Power Measure = Mean (95% Confidence Interval) | Open Source |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Akbari et al. [41] | EGFR | Variant III mutation (deletion of exons 2–7) | MRI, DWI, PWI | Hospital of the University of Pennsylvania, Philadelphia, US | 75 | 54 * | no | no | 421 | 16 tumor spatial location features; peritumoral heterogeneity index | AUC = 0.92 Accuracy = 88.9% | Code |
| Arita et al. [26] | IDH | Isoforms 1 (codon 132) and 2 (codon 172) mutations | MRI | Osaka International Cancer Institute, Osaka, Japan; National Cancer Center Research Institute, Tokyo, Japan | 111 | 58 * | yes | no | 50 | 59 tumor spatial location features | Accuracy = 87% | Code Features |
| Binder et al. [39] | EGFR | Extracellular A289D/T/V, R108G/K and G598V mutations | MRI, PWI, DWI | Hospital of the University of Pennsylvania, Philadelphia, US | 260 | - | yes | no | 2088 | 11 tumor spatial location features; 5 glioma diffusion properties from tumor biophysical models | Significant correlation (p < 0.0444) | Code |
| Choi et al. [20] | IDH | Isoforms 1 (codon 132) mutation | MRI | TCIA/TCGA-GBM; St. Mary’s Hospital, Seoul, South Korea | 45 | 91 ** | yes | no | 107 | - | AUC = 0.904 (0.805, 1.0) Accuracy = 86.8% (63.7, 97.8) | Images and ROI partially |
| Fukuma et al. [27] | IDH | Isoforms 1 (codon 132) and 2 (codon 172) mutations | MRI | Osaka International Cancer Institute, Osaka, Japan; National Cancer Center Research Institute, Tokyo, Japan | 127 | 10-CV | yes | no | 61 | 3 tumor spatial location features; 4000 DL features; age | Accuracy = 73.1% | - |
| Han et al. [53] | IDH | Isoforms 1 (codon 132) mutation | APTw | Tangdu Hospital, Xian, China | 49 | 10 * | yes | yes | 1044 | - |
AUC = 0.952 Accuracy = 0.892 | Images on request |
| Kim et al. [47] | IDH | Isoforms 1 (codon 132) mutation | MRI, DWI, PWI | Asan Medical Center, Seoul, South Korea | 127 | 28 *** | yes | yes | 6472 | - | AUC = 0.747 (0.66–0.83) Accuracy = 65.3% | - |
| Kong et al. [51] | Ki-67 | High Ki-67 expression as > 10% | FDG-PET | Peking Union Medical College Hospital, Beijing, China | 82 | 41 * | yes | no | 1561 | Age; sex; metabolic pattern; SUVmax; SUVmean | AUC = 0.73 Accuracy = 78% | - |
| Kuthuru et al. [28] | IDH | Isoforms 1 (codon 132) mutation | MRI | TCGA/TCIA-LGG | 108 | 10-CV | no | no | No | > 35,000 histogram of oriented gradients, scale-invariant feature transform and voxel intensities | AUC = 0.8224 (0.7856–0.8575) | Images and ROI |
| Lee et al. [31] | EGFR | mutation | MRI | TCGA/TCIA-GBM | 44 | 3-CV | no | no | - | 36 spatial diversity features | AUC = 0.845 Accuracy = 0.79 | Images and ROI |
| Lee et al. [48] | IDH | Isoforms 1 (codon 132) mutation | MRI, DWI, PWI | Samsung Medical Center, Seoul, South Korea | 88 | 35 *** | yes | no | 82 | - | Accuracy = 83.4% | - |
| Li et al. [50] | IDH | Isoforms 1 (codon 132) and 2 (codon 172) mutations | FDG-PET | Peking Union Medical College Hospital, Beijing, China | 84 | 43 * | yes | no | 1561 | Age; sex; metabolic pattern; SUVmax; SUVmean | AUC = 0.900 (0.877–0.923) | Code |
| Li et al. [19] | IDH | Isoforms 1 (codon 132) and 2 (codon 172) mutations | MRI | TCGA/TCIA-GBM; Sun Yat-sen University Cancer Center, Guangzhou, China; The 3rd Affiliated Hospital of Sun Yat-sen University, Guangzhou, China; Guangzhou General Hospital of Guangzhou Military Command, Guangzhou, China | 118 | 107 ** | yes | no | 1614 | Sex; age; KPS | AUC = 0.96 Accuracy = 97% | Images and ROI (partially) |
| Li et al. [33] | Ki-67 | High Ki-67 expression as > 25% | MRI | The Second Hospital of Hebei Medical University, Tangshan, Hebei, China | 50 | 3-CV, 5-CV, bootstrap | yes | no | 396 | - | AUC = 0.713 (0.568–0.832) Accuracy = 66.0% | - |
| Li et al. [32] | EGFR | High EGFR expression as > 30% | MRI | Beijing Tiantan Hospital, Beijing, China | 200 | 70 * | yes | no | 431 | - | AUC = 0.95 Accuracy = 90.0% | |
| Li et al. [36] | TP-53 | mutation | MRI | Chinese Glioma Genome Atlas, Beijing Tiantan Hospital, Beijing, China | 180 | 92 * | yes | no | 431 | - | AUC = 0.763 Accuracy = 70.7% | Images and ROI |
| Li et al. [34] | Ki-67 | High Ki-67 expression as > 10% | MRI | Beijing Tiantan Hospital, Beijing, China; Chinese Glioma Genome Atlas | 78 | 39 * | yes | no | 431 | - | AUC = 0.90 Accuracy = 88.6% | - |
| Li et al. [22] | IDH | Isoforms 1 (codon 132) mutation | MRI | Huashan Hospital, Shangai, China | 229 | LOOCV | yes | no | 671 | 16,384 DLR features | AUC = 0.9521 Accuracy = 92.44% | - |
| Liu et al. [21] | IDH | Isoforms 1 (codon 132) mutation | MRI | Beijing Tiantan Hospital, Beijing, China; | 158 | 102 *** | yes | yes | 431 | - | AUC = 0.99 | - |
| Lohmann et al. [52] | IDH | Isoforms 1 (codon 132) mutation | FET-PET | University Hospital RWTH Aachen | 84 | 5-CV, 10-CV | no | yes | 33 | Slope; TTP; mean tumor-to-brain ratio; maximum tumor-to-brain ratio | AUC = 0.79 Accuracy = 80.0% | - |
| Lu et al. [43] | IDH | mutation | MRI, DWI | TCGA/TCIA-LGG; TCGA/TCIA-GBM; TCIA-REMBRANDT; Taipei Medical University, Taipei, Taiwan | 214 | 70 ** | yes | no | 39,212 | - | Accuracy = 88.9–91.7% | Images and ROI (partially) |
| Park et al. [46] | IDH | Isoforms 1 (codon 132) mutation | MRI, DWI | Yonsei University, Seoul, South Korea | 168 | 10-CV | yes | no | 411 | - | AUC = 0.900 (0.855–0.945) | - |
| Rathore et al. [40] | EGFR | Variant III mutation (deletion of exons 2–7) | MRI, DWI, PWI | Hospital of the University of Pennsylvania, Philadelphia, US | 107 | 10-CV | yes | no | 255 | 9 tumor spatial location features; 3 biophysical growth model-based features | Accuracy = 80.19% | - |
| Ren et al. [49] | IDH | Isoforms 1 (codon 132) mutation | MRI, DWI, PWI | Huashan Hospital, Shangai, China | 57 | 10-CV | yes | no | 260 | 10 VASARI features; age; sex; Ki-67 | AUC = 0.931 Accuracy = 94.74% | - |
| Su et al. [44] | Ki-67 | High Ki-67 expression as > 25% | MRI, DWI, PWI | Tongji Hospital, Wuhan, Hubei, China | 220 | bootstrap | yes | no | 431 | - | AUC = 0.936 | - |
| Sun et al. [37] | VEGF | VEGF expression at < 5%, 6–25%, 26–50% and > 50% | MRI | Beijing Tiantan Hospital, Beijing, China; | 160 | 79 * | yes | no | 431 | - | AUC = 0.702 Accuracy = 72.3% | Images on request |
| Tan et al. [45] | IDH | mutation | DKI, DWI | Shanxi Medical University Shanxi, China | 62 | bootstrap | yes | no | 728 | Age; sex; grade; tumor size; tumor border; hemorrhage; cystic and necrosis; edema degree; enhancement style; enhancement degree; signal characteristics; 6 tumor location features; mean diffusivity value; mean kurtosis value | AUC = 0.885 (0.802–0.955) Accuracy = 80.6% (71.0–90.3) | - |
| Tan et al. [42] | IDH | Isoforms 1 (codon 132) and 2 (codon 172) mutations | MRI, DWI | Shanxi Medical University Shanxi, China | 74 | 31 * | yes | yes | 3882 | Age; sex; grade; tumor size; tumor border; hemorrhage; cystic and necrosis; edema degree; enhancement style; enhancement degree; signal characteristics; 6 tumor location features | AUC = 0.900 (0.859–0.941) Accuracy = 87.1% | Features, ROI |
| Tontong Liu et al. [25] | IDH | Isoforms 1 (codon 132) mutation | MRI | Huashan Hospital, Shangai, China | 110 | LOOCV | yes | no | 671 | - | AUC = 0.90 Accuracy = 0.85 | - |
| Ugga et al. [35] | Ki-67 | High Ki-67 expression as > 3% | MRI | University of Naples “Federico II” Neurosurgery, Naples, Italy | 53 | 36 * | yes | yes | 1128 | - | AUC = 0.87 Accuracy = 91.67% | - |
| Wu et al. [38] | IDH | Isoforms 1 (codon 132) mutation | MRI | TCGA/TCIA-LGG; TCGA/TCIA-GBM | 126 | bootstrap | yes | no | 698 | 6 tumor growth model parameters |
AUC = 0.931 Accuracy = 0.885 | Images, ROI |
| Wu et al. [18] | IDH | Isoforms 1 (codon 132) mutation | MRI | Huashan Hospital, Shangai, China | 80 | 25 * | yes | no | - | 968 dictionary features | Accuracy = 88.0% | - |
| Yu et al. [24] | IDH | Isoforms 1 (codon 132) mutation | MRI | Huashan Hospital, Shangai, China | 92 | LOOCV | no | no | - | 116 tumor spatial location features | AUC = 0.71 Accuracy = 72.0% | - |
| Yu et al. [23] | IDH | Isoforms 1 (codon 132) mutation | MRI | Huashan Hospital, Shangai, China | 110 | LOOCV | yes | no | 671 | - | AUC = 0.86 Accuracy = 80.0% | - |
| Zhang et al. [30] | IDH, TP-53 | mutation | MRI | TCGA/TCIA-LGG | 73 | 30 * | yes | yes | 260 | 16 VASARI features | IDH: AUC = 0.792 Accuracy = 80.0% TP-53: AUC = 0.869 Accuracy = 85.0% | Images, ROI |
| Zhou et al. [29] | IDH | mutation | MRI | TCGA/TCIA-LGG | 84 | bootstrap | yes | no | 3360 | 30 VASARI features; age; sex; KPS; histological type; grade; laterality; location | AUC = 0.86 | Images, ROI, code |
| Study | Biomarker | Alteration | Modality | Dataset Origin | Training | Validation | Feature Reduction | Feature Robustness | # Radiomic Features | Additional Features | Predictive Power Measure = Mean (95% Confidence Interval) | Open Source |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antunovic et al. [68] | HER-2, Ki-67 | HER-2: positive (IHC 3+) vs. negative (IHC 0 or 1) Ki-67: High expression at >20% | FDG-PET/CT | Humanitas Hospital, Milan, Italy | 43 | - | yes | no | 20 | MTV, SUVmean and TLG | HER-2: Significant correlation (p = 0.021–0.046) Ki-67: No significant correlation | - |
| Braman et al. [61] | HER-2 | mutation | PWI | Cleveland Medical Center, Cleveland, Ohio, US; City of Hope Comprehensive Cancer Center, Duarte, California, US; Yale Cancer Center, New Haven, Connecticut, US; Brown University Oncology Research Group, Providence, Rhode Island, US; TCIA/TCGA-BRCA | 117 | 3-CV | yes | no | 495 | - | AUC = 0.71 (0.63–0.79) | images |
| Castaldo et al. [60] | HER-2 | mutation | PWI | TCIA/TCGA-BRCA | 55 | 36 * | no | no | 36 | - | AUC = 0.91 Accuracy = 81–88% | images |
| Fan et al. [59] | HER-2 | positive (IHC 3+) vs. negative (IHC 0 or 1) | PWI | Zhejiang Cancer Hospital, Hangzhou, China | 60 | 36 * | yes | no | 65 | Age, menopausal status; 29 dynamic features from BPE and the lesion; 9 bilateral differences in BPE | AUC = 0.947 | - |
| Fan et al. [66] | Ki-67 | mutation | PWI, DWI | First Affiliated Hospital of Zhejiang Chinese Medical University, Hangzhou, China | 144 | LOOCV | yes | no | 97 | - | AUC = 0.811 | Code |
| Leithner et al. [64] | HER-2 | mutation | MRI | Memorial Sloan Kettering Cancer Center, New York, USA; Medical University Vienna, Vienna, Austria | 91 | - | yes | no | 352 | - | Accuracy = 73.6% | Code |
| Li et al. [54] | HER-2 | mutation | PWI | TCGA/TCIA-BRCA | 91 | LOOCV | yes | no | 24 | 10 kinetic features (maximum contrast enhancement, TTP, uptake rate, washout rate, curve shape index, enhancement at first post-contrast, SER, volume of most enhancing voxels, total rate variation, normalized total rate variation) and 4 enhancement-variance kinetic features (maximum variance of enhancement, TTP, variance increase rate, and variance decrease rate) | AUC = 0.65 | images |
| Li et al. [58] | HER-2 | mutation | PWI | Cancer Hospital of Liaoning, China | 637 | LOOCV | yes | no | 137 | 5 kinetic features (standard deviation, mean, maximum value, enhancement rate, absorption rate) | AUC = 0.83 Accuracy = 87.0% | - |
| Liang et al. [63] | Ki-67 | High expression at >14% | MRI | Guangdong General Hospital, Guangdong Academy of Medical Sciences, Guangzhou, China & Southern Medical University, Guangzhou, Guangdong, China | 200 | 118 *** | yes | yes | 10,207 | - | AUC = 0.740 (0.645,0.836) Accuracy = 0.729 | - |
| Lin et al. [55] | TP-53 | mutation | PWI | TCGA/TCIA-BRCA | 88 | LOOCV | yes | no | 5234 | - | AUC = 0.886 (0.817–0.955) | images |
| Ma et al. [56] | Ki-67 | High expression at >14% | PWI | Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center for Cancer, Tianjin, China | 159 | 10-CV | yes | no | 56 | - | AUC = 0.773 Accuracy = 0.757 | - |
| Monti et al. [57] | HER-2, Ki-67 | mutation | PWI | Hospital of Moscati, Avellino, Italy; Institute for Hospitalization and Healthcare SDN, Naples, Italy | HER-2: 48 Ki-67: 49 | bootstrap | yes | no | 163 | Pharmacokinetic maps | HER-2: AUC = 0.838 Accuracy = 0.785 Ki-67: AUC = 0.811 Accuracy = 0.677 | |
| Tagliafico et al. [67] | Ki-67 | High expression at >14% | DBT | Emergency Radiology, IRCCS Policlinico San Martino, Genova, Italy | 70 | bootstrap | yes | no | 106 | - | AUC = 0.698 | Code, features |
| Zhang et al. [65] | Ki-67 | High expression at >14% | DWI | The Second Hospital, Dalian Medical University, Dalian, China | 101 | 27 * | yes | no | 1029 | - | AUC = 0.72 (0.495–0.857) Accuracy = 0.70 | - |
| Zhou et al. [69] | HER-2 | positive (IHC 3+) vs. negative (IHC 0 or 1) | DMG | Henan Provincial People’s Hospital, Henan, China | 244 | 62 * | yes | no | 186 | - | AUC = 0.787 (0.673–0.885) Accuracy = 77.00% | |
| Zhou et al. [62] | HER-2; Ki-67 | HER-2: positive (IHC 3+) vs. negative (IHC 0 or 1) Ki-67: High expression at >20% | PWI | The Affiliated Huaian No. 1 People’s Hospital of Nanjing Medical University, China | 126 | 5-CV | yes | yes | 386 | - | HER-2: AUC = 0.68 Accuracy = 0.60 Ki-67: AUC = 0.74 Accuracy = 0.69 |
| Study | Biomarker | Alteration | Modality | Dataset Origin | Training | Validation | Feature Reduction | Feature Robustness | # Radiomic Features | Additional Features | Predictive Power Measure = Mean (95% Confidence Interval) | Open Source |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aerts et al. [70] | EGFR | Exons 19 and 21 mutations | CT | Memorial Sloan-Kettering Cancer Center, New York City, New York, US | 47 | - | yes | yes | 183 | - | AUC = 0.91 | Images |
| Chen et al. [102] | EGFR; KRAS; ALK | mutation | MR | City of Hope Medical Center, Duarte, California, US | 110 | LOOCV | yes | yes | 2786 | Age; sex; ethnicity; history of smoking; histology type; other metastatic sites | EGFR: AUC = 0.912 Accuracy = 77.7% ALK: AUC = 0.915 Accuracy = 86.7% KRAS: AUC = 0.985 Accuracy = 96.7% | - |
| Gu et al. [91] | Ki-67 | High Ki-67 expression as >50% | CT | The Third Xiangya Hospital of Central South University, Hunan, China | 245 | 10-CV | yes | no | 103 | Lobulation sign; spicule sign; cavitation; cystic necrosis; pleural indentation; pleural effusion | AUC = 0.782 | - |
| Hong et al. [85] | EGFR | Exons 18, 19, 20, and 21 mutations | CT | The First Hospital of China Medical University, Shenyang, China | 140 | 61 * | yes | no | 396 | Age; sex; history of smoking | AUC = 0.851 (0.750–0.951) c-index = 0.835 (0.825–0.845) | - |
| Huang et al. [71] | EGFR | mutation | CT | The University of Texas MD Anderson Cancer Center, Houston, Texas | 46 | - | yes | yes | 89 | - | AUC = 0.88 | Images |
| Jia et al. [72] | EGFR | Exons 19 and 21 mutations | CT | Shanghai Chest Hospital, Shanghai, China | 345 | 158 * | no | no | 440 | Age; sex; smoking history; TNM stage | AUC = 0.828 (0.764–0.892) | - |
| Jiang et al. [99] | PD-L1 | PD-L1 cutoff value of 1% and 50% | PET/CT | Shanghai Institute of Medical Imaging, Zhongshan Hospital of Fudan University, Shanghai, China | 266 | 133 * | yes | no | 1744 | SUVmax; age; sex; smoking status; TNM stage; histology type | AUC = 0.97 | - |
| Jiang et al. [92] | EGFR | mutation | PET/CT | Shanghai Institute of Medical Imaging, Zhongshan Hospital of Fudan University, Shanghai, China | 80 | 10-CV | yes | no | 512 | 12 semantic features | AUC = 0.953 | - |
| Koyasu et al. [93] | EGFR | mutation | PET/CT | TCIA- NSCLC Radiogenomics | 138 | 10-CV | yes | no | Not disclosed | SUVmax; SUVmean; TLG; MTV | AUC = 0.659 Accuracy = 81.2% | Images, ROI |
| Li et al. [94] | EGFR | Exons 18–24 mutations | PET/CT | Tianjin Medical University Cancer Hospital, Tianjin, China | 115 | 10-CV | yes | no | 38 | SUVmax; SUVmean; SUVpeak; TLG; MTV; age; sex; smoking status; TNM stage; lesion location | AUC = 0.822 Accuracy = 82.65% | - |
| Li et al. [74] | EGFR | mutation | CT | Second Xiangya Hospital of Central South University, Hunan, China | 51 | 10-CV | yes | yes | 1695 | - | AUC = 0.83 (0.68–0.92) | Images, ROI |
| Li et al. [75] | EGFR | Exon 19 and 21 mutations | CT | Shanghai Chest Hospital, Shanghai, China | 810 | 200 * | yes | no | 440 | DL prediction; age; sex; smoking history; pathological stage | AUC = 0.834 (0.776–0.892) | - |
| Li et al. [76] | EGFR | Exon 19 and 21 mutations | CT | Shengjing Hospital of China Medical University, Liaoning, China | 236 | 76 *** | yes | yes | 580 | Age; sex; tumor grade; lobe; smoking history; intrapulmonary metastasis | AUC = 0.7750–0.7925 | - |
| Liu et al. [86] | EGFR | Exons 18–21 mutations | CT | Tianjin Medical University Cancer Institute and Hospital, Tianjin, China | 298 | bootstrap | yes | no | 209 | 10 tumor spatial location features; age; sex; histological subtype; pathological stage; smoking history | AUC = 0.709 (0.654–0.766) | - |
| Lu et al. [73] | EGFR | mutation | CT | The First Hospital of Jilin University, China | 83 | 21 * | yes | yes | 1025 | 45 categorical variables including: age, sex, smoking status, CEA level, vascular infiltration, visceral pleural infiltration, lymph node metastasis, histological subtype, pathological stage, type of lesion, tumor location, tumor size, tumor necrosis, lobulation, spiculation, vacuolization, etc. | AUC = 0.894 | code |
| Mei et al. [77] | EGFR | Exon 18–21 mutations | CT | Shenzhen People’s Hospital, Guangdong, China | 296 | - | yes | no | 94 | Age; sex; smoking status | AUC = 0.75 | code |
| Nair et al. [95] | EGFR | Exons 19 and 21 mutations | PET/CT | McGill University Health Centre, 2011 and 2015 | 50 | LOOCV | yes | no | 326 | - | AUC = 0.8713 | - |
| Rios Velazquez et al. [83] | EGFR; KRAS | mutation | CT | Profile and Harvard-RT (Dana-Farber/Harvard Cancer Center IRB, Boston, MA), Tianjin (Tianjin Medical University IRB, Tianjin, China), Moffitt (IRB Moffitt Cancer Center, Tampa, FL) | 353 | 352 *** | yes | yes | 635 | Age; sex; smoking status; ethnicity; clinical stage | EGFR: AUC = 0.75 (0.69–0.81) Accuracy = 65.0% KRAS: AUC = 0.75 (0.69–080) Accuracy = 66.0% EGFR+ vs. KRAS+: 0.86 (0.80–0.91) Accuracy = 79.0% | Images |
| Shiri et al. [97] | EGFR; KRAS | EGFR: Exons 18–21 mutations KRAS: Exon 2 codons 12 and 13 mutations | PET/CT | TCIA | 82 | 68 * | yes | no | 109 | MTV, SUVmax, SUVpeak, SULmax, SULpeak | EGFR: AUC = 0.82 KRAS: AUC = 0.83 | Images, ROI, code |
| Song et al. [87] | ALK | mutation | CT | Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, November 2015 to October 2018 | 268 | 67 * | yes | no | 1218 | Age; sex; smoking history; smoking index; clinical stage; distal metastasis; pathological invasiveness of tumor; maximum diameter; mean CT attenuation; lesion location; involved lobe; density; margin; cavity; calcification; pleural retraction sign; pleural effusion; pericardial effusion; local lymphadenopathy | AUC = 0.88 (0.77–0.94) Accuracy = 79.0% | Images (partially), code |
| Sun et al. [88] | PD-L1 | High PD-L1 expression as ≥ 50% | CT | The First Affiliated Hospital of Soochow University, Suzhou City, China | 260 | 130 * | yes | yes | 200 | Age; sex; tumor location; CEA level; TNM stage; smoking status; histologic type; histologic grade | AUC = 0.848 | - |
| Tu et al. [78] | EGFR | Exons 18–21 mutations | CT | Changzheng Hospital, Second Military Medical University, Shanghai, China | 243 | 130 * | yes | yes | 234 | Age; sex; smoking status; CEA level; clinical stage; maximum diameter; density; tumor location; interface; shape; lobulation; pleural indentation; spiculation; cusp angle; spine-like process; vacuole sign; cavity sign; air bronchograms; vascular convergence; pleura thickening; pleural effusion; lymphadenopathy | AUC = 0.818 (0.751–0885) Accuracy = 75.8% | - |
| Wang et al. [84] | EGFR; TP-53 | mutation | CT | Nanjing Medical University Affiliated Cancer Hospital, Nanjing, China | 41 | 20 * | yes | no | 718 | 78 clinical and pathological features (age, sex, smoking status, histological subtypes, pathological stages, etc.) |
EGFR: AUC = 0.697 TP-53: AUC = 0.656 | code |
| Yang et al. [79] | EGFR | Exons 18–21 mutations | CT | The First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China | 306 | 161 *** | yes | no | 1063 | Age; sex; smoking history; CT pattern; histopathological subtype | AUC = 0.779 (0.702–0.856) | code |
| Yip et al. [100] | EGFR; KRAS | EGFR: Exons 18–24 mutations KRAS: Exons 2–3 mutations | PET | Dana-Farber Cancer Institute, Brigham and Women’s Hospital, and Harvard Medical School, Boston, Massachusetts | 348 | bootstrap | yes | no | 68 | MTV, SUVmax, SUVpeak, SUVmean, and SUVtot | EGFR: AUC = 0.67 KRAS:- EGFR+ vs. KRAS+: AUC = 0.65 | - |
| Yip et al. [101] | EGFR; KRAS | mutation | PET | Dana-Farber Cancer Institute, Brigham and Women’s Hospital, and Harvard Medical School, Boston, Massachusetts | 348 | - | yes | yes | 66 | - | EGFR: AUC = 0.66 KRAS:- | - |
| Yoon et al. [89] | PD-L1 | High PD-L1 expression as ≥50% | CT | Severance Hospital, Yonsei University College of Medicine, Seoul, South Korea | 153 | bootstrap | yes | yes | 58 | Age; sex; smoking history; stage; tumor size; tumor location; tumor type; tumor margin; internal characteristics of tumor; external characteristics of tumor; lung metastasis; pleural effusion; pleural nodularity; pericardial effusion; lymphadenopathy | c-index = 0.646 | - |
| Yoon et al. [98] | ALK/ROS1/RET | mutation | PET/CT | Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, South Korea | 128 | 10-CV | yes | yes | 50 | Age; sex; smoking history; stage; SUVmax; tumor solidity; tumor size; tumor location; lymphangitic metastasis; pleural effusion | Sensitivity = 0.73 Specificity = 0.70 | - |
| Zhang et al. [96] | EGFR | Exons 18–21 mutations | PET/CT | The Fourth Hospital of Hebei Medical University, Hebei, China | 175 | 73 * | yes | no | 92 | Age; sex; smoking history; pathological stage; CEA level | AUC = 0.87 (0.79–0.95) | - |
| Zhang et al. [80] | EGFR | Exons 18–21 mutations | CT | West China Hospital, Sichuan, China | 140 | 40 * | yes | no | 485 | Age; sex; smoking status | AUC = 0.8725 Accuracy = 72.5% | - |
| Zhao et al. [82] | EGFR | Exons 18–21 mutations | CT | Huadong Hospital Affiliated to Fudan University, Shanghai, China; TCIA | 464 nodules | 115 nodules * 37 nodules ** | yes | yes | 475 | DL prediction | AUC = 0.76 | Images, ROI |
| Zhao et al. [81] | EGFR | Exon 19 and 21 mutations | CT | Second Xiangya Hospital, Central South University, Changsha, China; Huadong Hospital Affiliated to Fudan University, Shanghai, China | 322 | 315 * | yes | yes | 475 | Age; sex; smoking status; tumor size; tumor location; histological subtype; TNM stage; tumor solidity; tumor margin; tumor type; pleural retraction; bubble lucency; vascular change; bronchiole change; lobulation; spiculation; peripheral emphysema; peripheral fibrosis; pleural effusion | AUC = 0.734 | - |
| Zhou et al. [90] | Ki-67 | High Ki-67 expression as > 40% | CT | Tianjin Medical University Cancer Institute and Hospital, Tianjin, China | 110 | - | yes | no | 105 | Age; sex; smoking history; histological subtype; TNM stage | AUC = 0.77 | code |
| Study | Biomarker | Alteration | Modality | Dataset Origin | Training | Validation | Feature Reduction | Feature Robustness | # Radiomic Features | Additional Features | Predictive Power Measure = Mean (95% Confidence Interval) | Open Source |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chen et al. [109] | KRAS; TP-53 | KRAS: Exon 2 codons 12 and 13 mutation TP-53: Exons 2–11 mutations | FDG-PET/CT | China Medical University Hospital, Taichung, Taiwan | 74 | - | yes | no | 56 | SUVmax, SUVpeak, SUVtot, MTV, TLGmax, TLGpeak, and TLGmean | KRAS: AUC = 0.79 Accuracy = 77% TP-53: AUC = 0.71 Accuracy = 62% | - |
| Cui et al. [110] | KRAS | KRAS: Exons 2–4 mutations | MRI | Shanxi Province Cancer Hospital, Taiyuan, China; Xinhua Hospital, Shanghai, China | 213 | 91 * 86 ** | yes | no | 960 | - |
AUC * = 0.682 (0.569–0.794) AUC ** = 0.714 (0.602–0.827) | - |
| Li et al. [103] | HER-2 | positive (HER-2/CEP17 ≥ 2) vs. negative (HER-2/CEP17 < 2) | CT | Guangdong Provincial People’s Hospital, Guangzhou, China | 94 | 40 * | yes | yes | 12,410 | CEA level | AUC = 0.771 (0.607−0.934) | - |
| Liang et al. [105] | Ki-67 | mutation | CT | The First Affiliated Hospital, Hangzhou, Zhejiang, China; Second Affiliated Hospital, Hangzhou, Zhejiang, China | 86 | 51 ** | yes | no | 467 | Clinical stage | Significant correlation (p < 0.0001) | - |
| Lim et al. [108] | KRAS; TP-53 | mutation | FDG-PET/CT | Samsung Medical Center, Sungkyunkwan University School of Medicine, Gangnam-gu, Seoul, South Korea | 48 | - | no | yes | 27 | SUVmax, SUVmean, SUVstd, SUVkurt, SUVskew, SUVent, MTV, TLG | KRAS: AUC = 0.829 TP-53:- | Code (partially) |
| Meng et al. [112] | Ki-67, KRAS, HER-2 | KRAS: exon 2 codons 12 and 13 mutation Ki-67: High expression at >40% HER-2: | MRI, DWI, PWI | Sixth Affiliated Hospital of Sun Yat-sen University. Guangzhou, China | 197 | 148 *** | yes | yes | 2534 | - | HER-2: AUC = 0.696 (0.610–0.782) Accuracy = 0.621 Ki-67: AUC = 0.699 (0.611–0.788) Accuracy = 0.582 KRAS: AUC = 0.651 (0.539–0.763) Accuracy = 0.616 | - |
| Oh et al. [111] | KRAS | A59T, G12A, G12C, G12D, G12F, G12R, G12S, G12V, G13D, G61H, and Q61 mutation | MRI | Research Institute and Hospital, National Cancer Center, Goyang, Korea | 60 | - | no | no | 44 | - |
AUC = 0.884 Accuracy = 81.7% | - |
| Wu et al. [107] | KRAS | Exons 2–4 mutations | CT | South China University of Technology, Guangzhou, Guangdong Province, China | 279 | 119 *** | yes | yes | 2634 | 2208 DL features | c-index = 0.832 (0.762–0.905) | - |
| Yang et al. [106] | KRAS; BRAF | KRAS: Exons 2–4 mutations BRAF: v600E mutation | CT | National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China | 61 | 57 *** | yes | yes | 346 | - | AUC = 0.829 (0.718–0.939) Accuracy = 0.750 (0.623–0.845) | - |
| Zhang et al. [104] | Ki-67 | High expression as ≥ 10% | CT | Renji Hospital, Huangpu, Shanghai, China; Zhongshan Hospital, Shanghai, China; Sir Run Shaw Hospital, Hangzhou, Zhejiang, China and First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China | 148 | 41 * 150 ** | yes | yes | 833 | Tumor size | AUC * = 0.828 (0.681–0.974) AUC ** = 0.784 (0.701–0.868) Accuracy * = 68.29% Accuracy ** = 73.33% | Images/data on request |
| Study | Biomarker | Alteration | Modality | Dataset Origin | Training | Validation | Feature Reduction | Feature Robustness | # Radiomic Features | Additional Features | Predictive Power Measure = Mean (95% Confidence Interval) | Open Source |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hectors et al. [113] | PD-L1 | expression | MRI, DWI | Icahn School of Medicine at Mount Sinai, New York, USA | 48 | - | no | no | 196 | Infiltrative pattern; presence of multiple lesions; extra-nodular growth; macrovascular invasion; tumor necrosis; tumor hemorrhage; tumor fat content; mosaic appearance; internal arteries; capsule; T2 hyper-intensity; ADC hypo-intensity; wash-in/wash-out; hepatobiliary phase hypo-intensity; ADCmin; ADCmean; ER in EA, LA, PV, LV and hepatobiliary phases; tumor size | Significant correlation (p < 0.029) | - |
| Peng et al. [116] | Ki-67; VEGF | Ki-67: High expression at ≥10% VEGF: expression | US | First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi, China | Ki-67: 63 VEGF: 39 | Ki-67: 27 * VEGF: 18 * | yes | no | 1,076 | - | Ki-67: AUC = 0.848 Accuracy = 0.889 VEGF: AUC = 0.864 Accuracy = 0.833 | - |
| Yao et al. [115] | Ki-67; PD-L1 | Ki-67: High expression at ≥25% PD-L1: expression | US | Zhongshan Hospital, Fudan University, Shanghai, China | 47 | LOOCV | yes | no | - | 2560 dictionary-based image features |
PD-L1: AUC = 0.97 (0.89–0.98) Accuracy = 92% Ki-67: AUC = 0.94 (0.87–0.97) Accuracy = 93% | Images on request |
| Ye et al. [114] | Ki-67 | High expression at ≥15% | MRI | West China Hospital, Sichuan, China | 89 | 10-CV | yes | no | 396 | Serum level of alpha-fetoprotein; hepatitis B surface antigen; hepatitis C antibody; Barcelona-Clinic Liver Cancer classification; cirrhosis; multifocality; arterial phase hyper-enhancement; washout, capsule integrity, internal arteries, tumor margin, enhancing capsule, hepato-biliary phase hypo-intensity | c-index: 0.936 (0.863–0.977) | - |
| Study | Biomarker | Alteration | Modality | Dataset Origin | Training | Validation | Feature Reduction | Feature Robustness | # Radiomic Features | Additional Features | Predictive Power Measure = Mean (95% Confidence Interval) | Open Source |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ahmed et al. [121] | Ki-67 | High expression at ≥10% | CT | MD Anderson Cancer Center, Texas, US | 53 | - | no | no | 106 | - | AUC = 0.78 | - |
| Chen et al. [120] | PD-L1; EGFR; VEGF; Ki-67 | PD-L1: High expression at ≥5% and ≥1% EGFR: expression VEGF: expression Ki-67: expression | FDG-PET | China Medical University, Taichung City, Taiwan | 53 | - | no | no | 41 | SUVmax, MTV, TLGmean; smoking history; tumor origin; TNM stage | PD-L1: AUC = 0.24 1; EGFR: no correlation. VEGF: Correlation (p < 0.05); Ki-67: Correlation (p < 0.05) | - |
| Saadani et al. [117] | BRAF | v600E mutation | FDG-PET/CT | Netherlands Cancer Institute, Amsterdam, The Netherlands | 70 | 10-CV | yes | no | 480 | SUVmax; SUVmean; SUVpeak; MTV; TLG; longest diameter | AUC = 0.62 | - |
| Yoon et al. [118] | BRAF | v600E mutation | US | Severance Hospital, Yonsei University College of Medicine, Seoul, South Korea | 387 | 140 *** | yes | no | 730 | Age; tumor size; sex; | AUC = 0.629 (0.516–0.742) | - |
| Zhu et al. [119] | TP-53 | mutation | CT | TCIA/TCGA-HNSCC | 126 | 5-CV | yes | yes | 187 | - | AUC = 0.641 | Images, ROI |
| MRI | EGFR | Ki-67 | KRAS | TP-53 | VEGF | IDH |
|---|---|---|---|---|---|---|
| CNS | EGFR+ more heterogeneous, less spherical [32] | Ki-67 high expression more heterogeneous [34] | TP-53+ higher intensity [36] | VEGF+ more heterogeneous [37] | IDH+ more homogeneous, more regularly shaped [21] | |
| GI | KRAS+ more heterogeneous [110] | |||||
| Liver | Ki-67 high expression more heterogeneous [114] |
| CT | EGFR | Ki-67 | KRAS/BRAF | TP-53 | HER-2 | ALK | PD-L1 |
|---|---|---|---|---|---|---|---|
| HNC | Ki-67 high expression more heterogeneous [120] | TP-53+ more heterogeneous [119] | |||||
| Lung | EGFR+ more heterogeneous, smaller [83] | Ki-67 high expression more homogeneous, more elongated [90] | KRAS+ more homogeneous [83] | ALK+ higher density [87] | PD-L1+ more homogeneous [89] | ||
| GI | Ki-67 high expression more heterogeneous [104] | KRAS/BRAF+ more heterogeneous [106] | HER-2+ more heterogeneous [103] |
| PET | EGFR | Ki-67 | KRAS | TP-53 | VEGF | IDH | PD-L1 |
|---|---|---|---|---|---|---|---|
| CNS | IDH+ more homogeneous, less spherical [50] | ||||||
| HNC | VEGF+ more heterogeneous [120] | PD-L1+ more heterogeneous [120] | |||||
| Lung | EGFR+ more heterogeneous, more compact [100] | ||||||
| GI | KRAS+ lower intensity [108] | TP-53+ more heterogeneous [109] | |||||
| Adrenal gland carcinoma | Ki-67 high expression more elongated and flatter [121] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
La Greca Saint-Esteven, A.; Vuong, D.; Tschanz, F.; van Timmeren, J.E.; Dal Bello, R.; Waller, V.; Pruschy, M.; Guckenberger, M.; Tanadini-Lang, S. Systematic Review on the Association of Radiomics with Tumor Biological Endpoints. Cancers 2021, 13, 3015. https://doi.org/10.3390/cancers13123015
La Greca Saint-Esteven A, Vuong D, Tschanz F, van Timmeren JE, Dal Bello R, Waller V, Pruschy M, Guckenberger M, Tanadini-Lang S. Systematic Review on the Association of Radiomics with Tumor Biological Endpoints. Cancers. 2021; 13(12):3015. https://doi.org/10.3390/cancers13123015
Chicago/Turabian StyleLa Greca Saint-Esteven, Agustina, Diem Vuong, Fabienne Tschanz, Janita E. van Timmeren, Riccardo Dal Bello, Verena Waller, Martin Pruschy, Matthias Guckenberger, and Stephanie Tanadini-Lang. 2021. "Systematic Review on the Association of Radiomics with Tumor Biological Endpoints" Cancers 13, no. 12: 3015. https://doi.org/10.3390/cancers13123015
APA StyleLa Greca Saint-Esteven, A., Vuong, D., Tschanz, F., van Timmeren, J. E., Dal Bello, R., Waller, V., Pruschy, M., Guckenberger, M., & Tanadini-Lang, S. (2021). Systematic Review on the Association of Radiomics with Tumor Biological Endpoints. Cancers, 13(12), 3015. https://doi.org/10.3390/cancers13123015







