A Novel Screen for Expression Regulators of the Telomeric Protein TRF2 Identified Small Molecules That Impair TRF2 Dependent Immunosuppression and Tumor Growth
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells
2.2. SDS-PAGE and Western Blotting
2.3. Cloning Strategy
2.4. Lentivirus Production
2.5. Flow Cytometry Screening
2.6. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
2.7. AlamarBlue
2.8. Animals
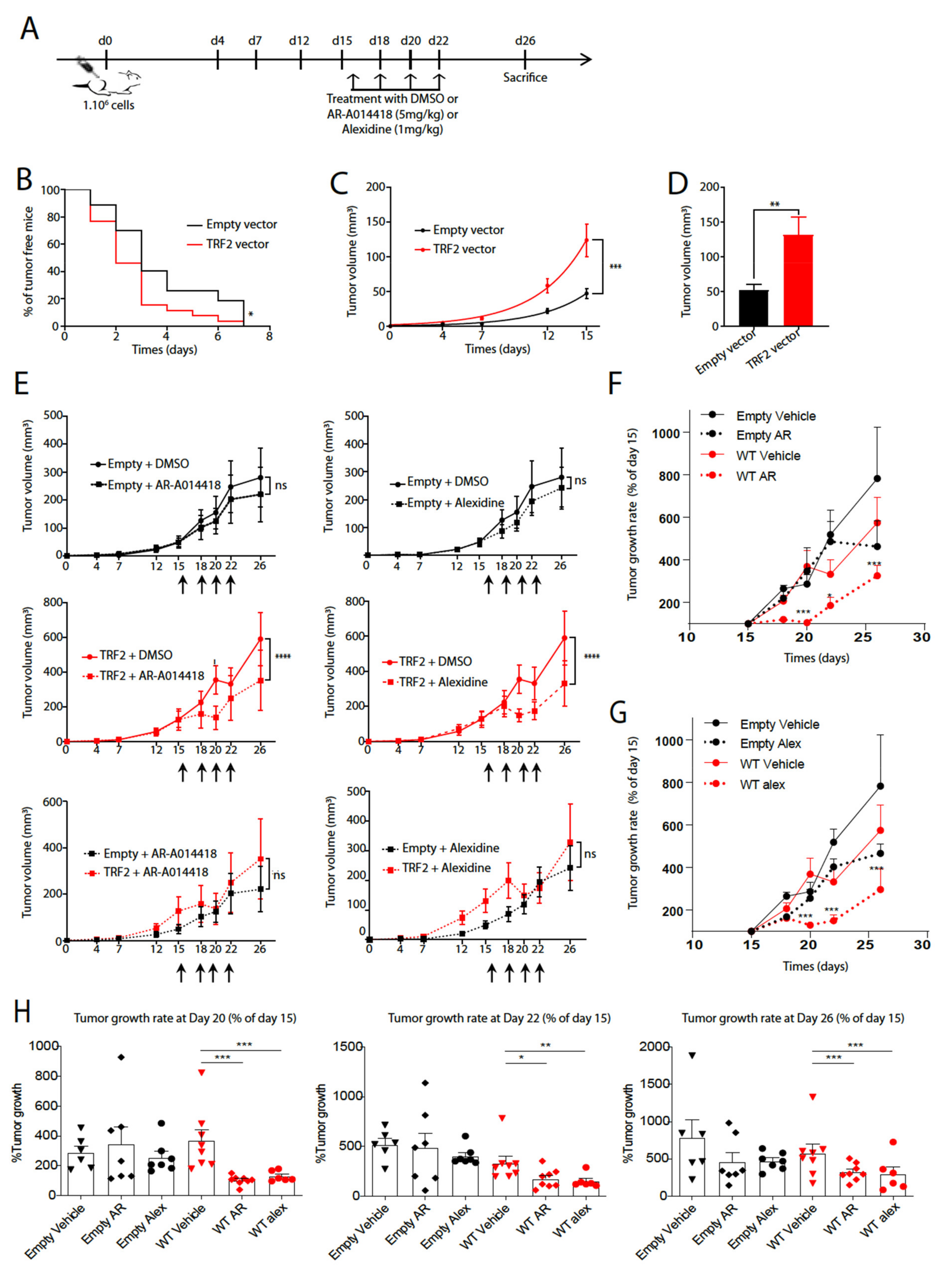
2.9. Tumor Growth Experiments
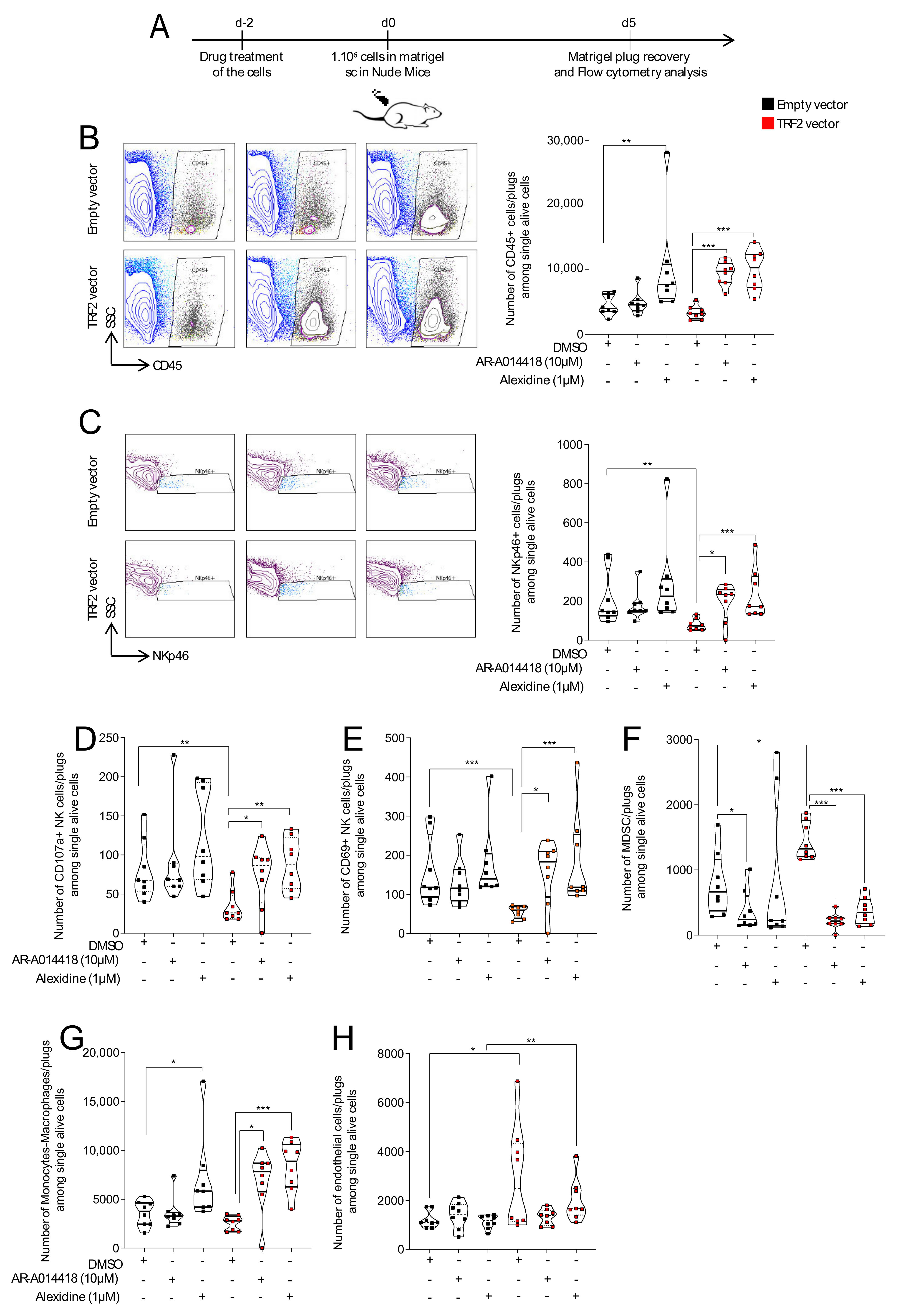
2.10. Matrigel Plug Assays
2.11. Statistics
3. Results
3.1. Identification of TRF2 Inhibitory Molecules
3.2. AR-A014418 and Alexidine·2HCl Reversed the Tumorigenicity Conferred by High TRF2 Expression Levels
3.3. AR-A014418 and Alexidine·2HCl Counteracted TRF2-Dependent Immunosuppression and Neo-Angiogenesis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giraud-Panis, M.-J.; Pisano, S.; Benarroch-Popivker, D.; Pei, B.; Le Du, M.-H.; Gilson, E. One Identity or More for Telomeres? Front. Oncol. 2013, 3, 48. [Google Scholar] [CrossRef]
- Gilson, E.; Géli, V. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 2007, 8, 825–838. [Google Scholar] [CrossRef]
- Armanios, M.; Blackburn, E.H. The telomere syndromes. Nat. Rev. Genet. 2012, 13, 693–704. [Google Scholar] [CrossRef]
- Kordinas, V.; Ioannidis, A.; Chatzipanagiotou, S. The Telomere/Telomerase System in Chronic Inflammatory Diseases. Cause or Effect? Genes 2016, 7, 60. [Google Scholar] [CrossRef]
- Kim, N.; Piatyszek, M.; Prowse, K.; Harley, C.; West, M.; Ho, P.; Coviello, G.; Wright, W.; Weinrich, S.; Shay, J. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef]
- Sugarman, E.T.; Zhang, G.; Shay, J.W. In perspective: An update on telomere targeting in cancer. Mol. Carcinog. 2019, 58, 1581–1588. [Google Scholar] [CrossRef]
- Artandi, S.E.; Chang, S.; Lee, S.-L.; Alson, S.; Gottlieb, G.J.; Chin, L.; DePinho, R.A. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nat. Cell Biol. 2000, 406, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Perera, S.A.; Maser, R.S.; Xia, H.; McNamara, K.; Protopopov, A.; Chen, L.; Hezel, A.F.; Kim, C.F.; Bronson, R.T.; Castrillon, D.H.; et al. Telomere dysfunction promotes genome instability and metastatic potential in a K-ras p53 mouse model of lung cancer. Carcinogenesis 2008, 29, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Englezou, A.; Dalla-Pozza, L.; Dunham, M.A.; Reddel, R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997, 3, 1271–1274. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Hwang, S.S.; Liesa, M.; Gan, B.; Sahin, E.; Jaskelioff, M.; Ding, Z.; Ying, H.; Boutin, A.T.; Zhang, H.; et al. Antitelomerase Therapy Provokes ALT and Mitochondrial Adaptive Mechanisms in Cancer. Cell 2012, 148, 651–663. [Google Scholar] [CrossRef]
- Fernandes, S.G.; Dsouza, R.; Pandya, G.; Kirtonia, A.; Tergaonkar, V.; Lee, S.Y.; Garg, M.; Khattar, E. Role of Telomeres and Telomeric Proteins in Human Malignancies and Their Therapeutic Potential. Cancers 2020, 12, 1901. [Google Scholar] [CrossRef]
- Augereau, A.; De Roodenbeke, C.T.; Simonet, T.; Bauwens, S.; Horard, B.; Callanan, M.; Leroux, M.; Jallades, L.; Salles, G.; Gilson, E.; et al. Telomeric damage in early stage of chronic lymphocytic leukemia correlates with shelterin dysregulation. Blood 2011, 118, 1316–1322. [Google Scholar] [CrossRef]
- Biroccio, A.; Cherfils-Vicini, J.; Augereau, A.; Pinte, S.; Bauwens, S.; Ye, J.; Simonet, T.; Horard, B.; Jamet, K.; Cervera, L.; et al. TRF2 inhibits a cell-extrinsic pathway through which natural killer cells eliminate cancer cells. Nat. Cell Biol. 2013, 15, 818–828. [Google Scholar] [CrossRef] [PubMed]
- El Maï, M.; Wagner, K.-D.; Michiels, J.-F.; Ambrosetti, D.; Borderie, A.; Destree, S.; Renault, V.; Djerbi, N.; Giraud-Panis, M.-J.; Gilson, E.; et al. The Telomeric Protein TRF2 Regulates Angiogenesis by Binding and Activating the PDGFRβ Promoter. Cell Rep. 2014, 9, 1047–1060. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, A.J.; Quesada, V.; Foronda, M.; Conde, L.; Martínez-Trillos, A.; Villamor, N.; Rodríguez, D.; Kwarciak, A.; Garabaya, C.; Gallardo, M.; et al. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat. Genet. 2013, 45, 526–530. [Google Scholar] [CrossRef]
- Bejarano, L.; Schuhmacher, A.J.; Méndez, M.; Megías, D.; Blanco-Aparicio, C.; Martínez, S.; Pastor, J.; Squatrito, M.; Blasco, M.A. Inhibition of TRF1 Telomere Protein Impairs Tumor Initiation and Progression in Glioblastoma Mouse Models and Patient-Derived Xenografts. Cancer Cell 2017, 32, 590–607. [Google Scholar] [CrossRef]
- Benhamou, Y.; Picco, V.; Raybaud, H.; Sudaka, A.; Chamorey, E.; Brolih, S.; Monteverde, M.; Merlano, M.; Nigro, C.L.; Ambrosetti, D.; et al. Telomeric repeat-binding factor 2: A marker for survival and anti-EGFR efficacy in oral carcinoma. Oncotarget 2016, 7, 44236–44251. [Google Scholar] [CrossRef] [PubMed]
- Cherfils-Vicini, J.; Zizza, P.; Gilson, E.; Biroccio, A. A novel pathway links telomeres to NK-cell activity. OncoImmunology 2014, 3, e27358. [Google Scholar] [CrossRef]
- Cherfils-Vicini, J.; Gilson, E. Inhibiting TRF 1 upstream signaling pathways to target telomeres in cancer cells. EMBO Mol. Med. 2019, 11, 11. [Google Scholar] [CrossRef]
- Zizza, P.; Dinami, R.; Porru, M.; Cingolani, C.; Salvati, E.; Rizzo, A.; D’Angelo, C.; Petti, E.; Amoreo, C.A.; Mottolese, M.; et al. TRF2 positively regulates SULF2 expression increasing VEGF-A release and activity in tumor microenvironment. Nucleic Acids Res. 2019, 47, 3365–3382. [Google Scholar] [CrossRef] [PubMed]
- Cherfils-Vicini, J.; Iltis, C.; Cervera, L.; Pisano, S.; Croce, O.; Sadouni, N.; Győrffy, B.; Collet, R.; Renault, V.M.; Rey-Millet, M.; et al. Cancer cells induce immune escape via glycocalyx changes controlled by the telomeric protein TRF 2. EMBO J. 2019, 38. [Google Scholar] [CrossRef] [PubMed]
- Di Maro, S.; Zizza, P.; Salvati, E.; DE Luca, V.; Capasso, C.; Fotticchia, I.; Pagano, B.; Marinelli, L.; Gilson, E.; Novellino, E.; et al. Shading the TRF2 Recruiting Function: A New Horizon in Drug Development. J. Am. Chem. Soc. 2014, 136, 16708–16711. [Google Scholar] [CrossRef]
- Roisin, A.; Buchsbaum, S.; Mocquet, V.; Jalinot, P. The fluorescent protein stability assay: An efficient method for monitoring intracellular protein stability. Biotechniques 2021, 70, btn-2021-0032. [Google Scholar] [CrossRef]
- Su, C.-H.; Chu, W.-C.; Lan, K.-H.; Li, C.-P.; Chao, Y.; Lin, H.-C.; Lee, S.-D.; Tsai, Y.-C.; Lee, W.-P. Gemcitabine causes telomere attrition by stabilizing TRF2. Eur. J. Cancer 2012, 48, 3465–3474. [Google Scholar] [CrossRef]
- Fujita, K.; Horikawa, I.; Mondal, A.M.; Jenkins, L.M.M.; Appella, E.; Vojtesek, B.; Bourdon, J.-C.; Lane, D.P.; Harris, C.C. Positive feedback between p53 and TRF2 during telomere-damage signalling and cellular senescence. Nat. Cell Biol. 2010, 12, 1205–1212. [Google Scholar] [CrossRef] [PubMed]
- Shakoori, A.; Mai, W.; Miyashita, K.; Yasumoto, K.; Takahashi, Y.; Ooi, A.; Kawakami, K.; Minamoto, T. Inhibition of GSK-3β activity attenuates proliferation of human colon cancer cells in rodents. Cancer Sci. 2007, 98, 1388–1393. [Google Scholar] [CrossRef]
- Kenny, H.A.; Lal-Nag, M.; White, E.A.; Shen, M.; Chiang, C.-Y.; Mitra, A.K.; Zhang, Y.; Curtis, M.W.; Schryver, E.M.; Bettis, S.; et al. Quantitative high throughput screening using a primary human three-dimensional organotypic culture predicts in vivo efficacy. Nat. Commun. 2015, 6, 1–11. [Google Scholar] [CrossRef]
- Bhat, R.; Xue, Y.; Berg, S.; Hellberg, S.; Ormö, M.; Nilsson, Y.; Radesäter, A.-C.; Jerning, E.; Markgren, P.-O.; Borgegård, T.; et al. Structural Insights and Biological Effects of Glycogen Synthase Kinase 3-specific Inhibitor AR-A014418. J. Biol. Chem. 2003, 278, 45937–45945. [Google Scholar] [CrossRef] [PubMed]
- Diala, I.; Wagner, N.; Magdinier, F.; Shkreli, M.; Sirakov, M.; Bauwens, S.; Schluth-Bolard, C.; Simonet, T.; Renault, V.M.; Ye, J.; et al. Telomere protection and TRF2 expression are enhanced by the canonical Wnt signalling pathway. EMBO Rep. 2013, 14, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Wagner, K.-D.; Ying, Y.; Leong, W.; Jiang, J.; Hu, X.; Chen, Y.; Michiels, J.-F.; Lu, Y.; Gilson, E.; Wagner, N.; et al. The differential spatiotemporal expression pattern of shelterin genes throughout lifespan. Aging 2017, 9, 1219–1232. [Google Scholar] [CrossRef]
- Robin, J.D.; Burbano, M.J.; Peng, H.; Croce, O.; Thomas, J.L.; Laberthonniere, C.; Renault, V.; Lototska, L.; Pousse, M.; Tessier, F.; et al. Mitochondrial function in skeletal myofibers is controlled by a TRF2-SIRT3 axis over lifetime. Aging Cell 2020, 19, e13097. [Google Scholar] [CrossRef]
- Muñoz, P.; Blanco, R.; Flores, J.M.; Blasco, M.A. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat. Genet. 2005, 37, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Alder, J.K.; Barkauskas, C.E.; Limjunyawong, N.; Stanley, S.E.; Kembou, F.; Tuder, R.M.; Hogan, B.L.M.; Mitzner, W.; Armanios, M. Telomere dysfunction causes alveolar stem cell failure. Proc. Natl. Acad. Sci. USA 2015, 112, 5099–5104. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.G.; Walker, A.E.; Trott, D.W.; Machin, D.R.; Henson, G.D.; Reihl, K.D.; Cawthon, R.M.; Denchi, E.L.; Liu, Y.; Bloom, S.I.; et al. Induced Trf2 deletion leads to aging vascular phenotype in mice associated with arterial telomere uncapping, senescence signaling, and oxidative stress. J. Mol. Cell. Cardiol. 2019, 127, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Uryga, A.K.; Reinhold, J.; Figg, N.; Baker, L.; Finigan, A.J.; Gray, K.; Kumar, S.; Clarke, M.C.H.; Bennett, M.R. Vascular Smooth Muscle Cell Senescence Promotes Atherosclerosis and Features of Plaque Vulnerability. Circulation 2015, 132, 1909–1919. [Google Scholar] [CrossRef]

| Specificity | Company | Clone | Species | Isotype | Fluorochrome | Reference |
|---|---|---|---|---|---|---|
| anti CD107a FITC | BD Biosciences | 1D4B | Rat | IgG2a/k | FITC | 553793 |
| anti CD11b | BD Biosciences | M1/70 | Rat | IgG2b | APC-H7 | 550993 |
| Anti CD69 PE-Cy7 | BD Biosciences | H1.2F3 | Hamster | IgG1/K | PE-Cy7 | 552879 |
| Anti NKp46 CD335 | BD Biosciences | 29A1.4 | Rat | IgG2a | Alexa 647 | 560755 |
| Anti-CD3e | Biolegend | 145-C11 | Armenin Hamster | IgG | PerCP | 100302 |
| Anti-CD45 | BD Biosciences | 30-F11 | Rat | IgG2b, κ | AF700 | 560510 |
| Anti-F4/80 | Biolegend | BM8 | Rat | IgG2a, κ | BV510 | 123135 |
| Anti-Ly6C/Ly6G (Gr-1) | BD Biosciences | RB6-8C5 | Rat | IgG2b, κ | PE | 553128 |
| anti-CD31 | BD Biosciences | MEC 13.3 | Rat | IgG2a, κ | BV421 | 562939 |
| TRF2 | Imgenex | 4A794.15 | Mouse | IgG1, κ | N.A. | IMG-124A |
| Beta-Actin | Abcam | Rabbit | IgG | N.A. | Ab8227 | |
| Anti-mouse | Licor | Goat | IgG | IRDye 680 | 926-32220 | |
| Anti-Rabbit | Licor | Goat | IgG | IRDye 800CW | 926-32211 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Maï, M.; Janho dit Hreich, S.; Gaggioli, C.; Roisin, A.; Wagner, N.; Ye, J.; Jalinot, P.; Cherfils-Vicini, J.; Gilson, E. A Novel Screen for Expression Regulators of the Telomeric Protein TRF2 Identified Small Molecules That Impair TRF2 Dependent Immunosuppression and Tumor Growth. Cancers 2021, 13, 2998. https://doi.org/10.3390/cancers13122998
El Maï M, Janho dit Hreich S, Gaggioli C, Roisin A, Wagner N, Ye J, Jalinot P, Cherfils-Vicini J, Gilson E. A Novel Screen for Expression Regulators of the Telomeric Protein TRF2 Identified Small Molecules That Impair TRF2 Dependent Immunosuppression and Tumor Growth. Cancers. 2021; 13(12):2998. https://doi.org/10.3390/cancers13122998
Chicago/Turabian StyleEl Maï, Mounir, Serena Janho dit Hreich, Cedric Gaggioli, Armelle Roisin, Nicole Wagner, Jing Ye, Pierre Jalinot, Julien Cherfils-Vicini, and Eric Gilson. 2021. "A Novel Screen for Expression Regulators of the Telomeric Protein TRF2 Identified Small Molecules That Impair TRF2 Dependent Immunosuppression and Tumor Growth" Cancers 13, no. 12: 2998. https://doi.org/10.3390/cancers13122998
APA StyleEl Maï, M., Janho dit Hreich, S., Gaggioli, C., Roisin, A., Wagner, N., Ye, J., Jalinot, P., Cherfils-Vicini, J., & Gilson, E. (2021). A Novel Screen for Expression Regulators of the Telomeric Protein TRF2 Identified Small Molecules That Impair TRF2 Dependent Immunosuppression and Tumor Growth. Cancers, 13(12), 2998. https://doi.org/10.3390/cancers13122998







