Targeting HIF-1α Regulatory Pathways as a Strategy to Hamper Tumor-Microenvironment Interactions in CLL
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients’ Samples
2.2. Cell Lines and Cell Culture
2.3. Western Blot (WB) Analysis
2.4. Akt and HIF-1α Activity
2.5. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
2.6. CXCL12 Quantification
2.7. Immunohistochemistry
2.8. Statistical Analysis
3. Results
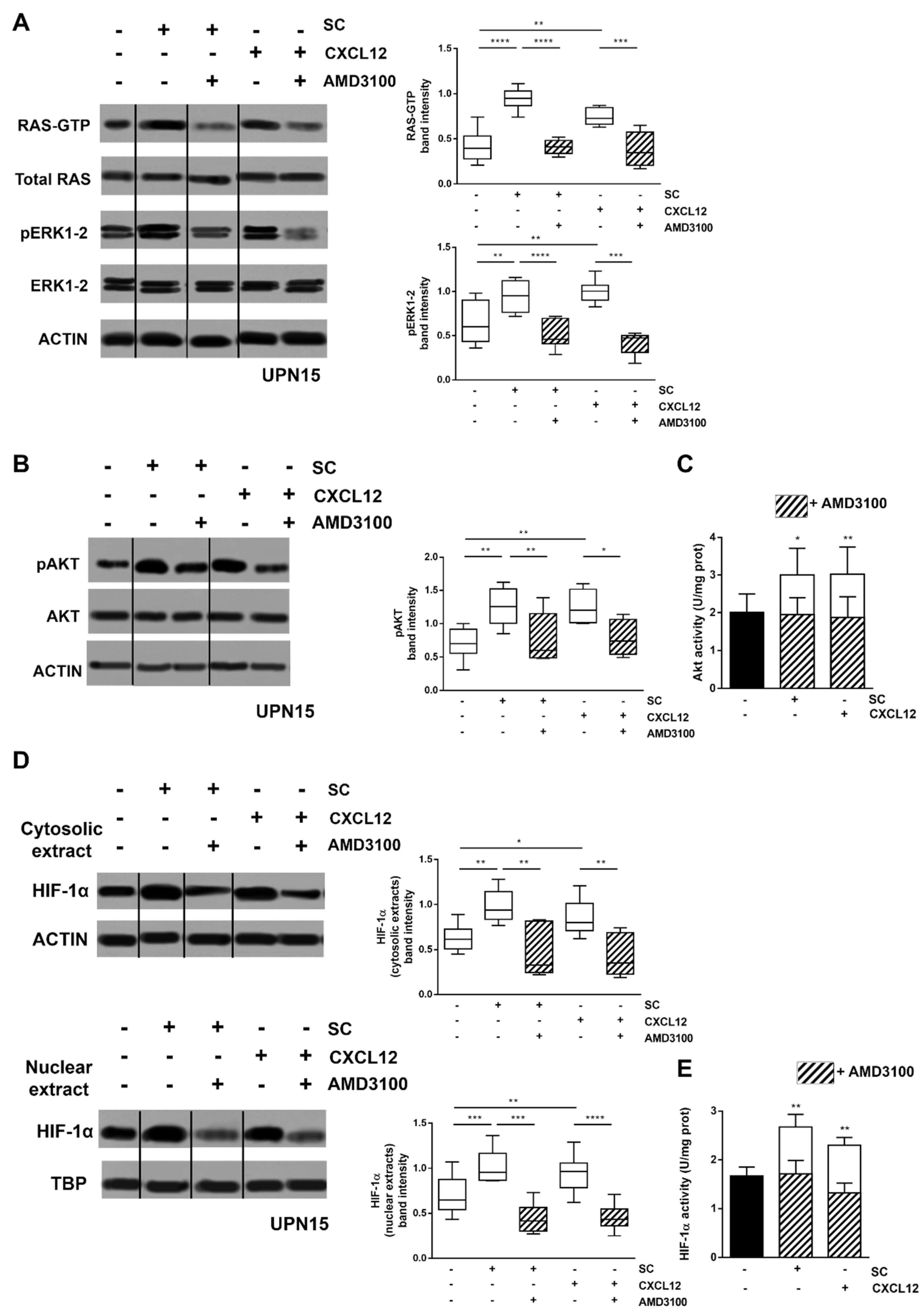
3.1. CXCL12/CXCR4 Axis Is a Main Regulator of SC-Induced HIF-1α Activation in CLL Cells
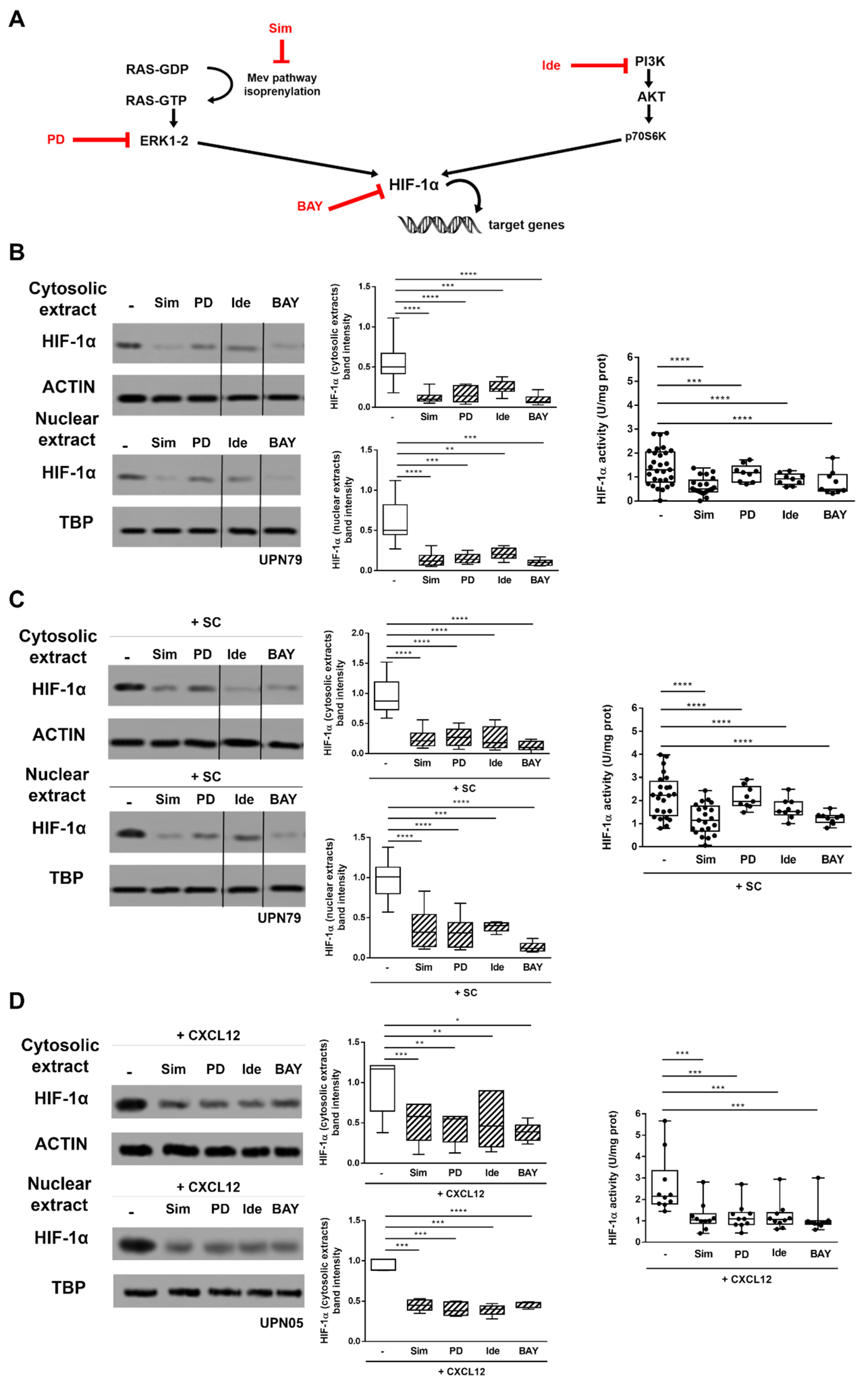
3.2. Inhibitors of CXCR4 Downstream Signalling Effectively Counteract SC- and CXCL12-Induced HIF-1α Upregulation in CLL Cells
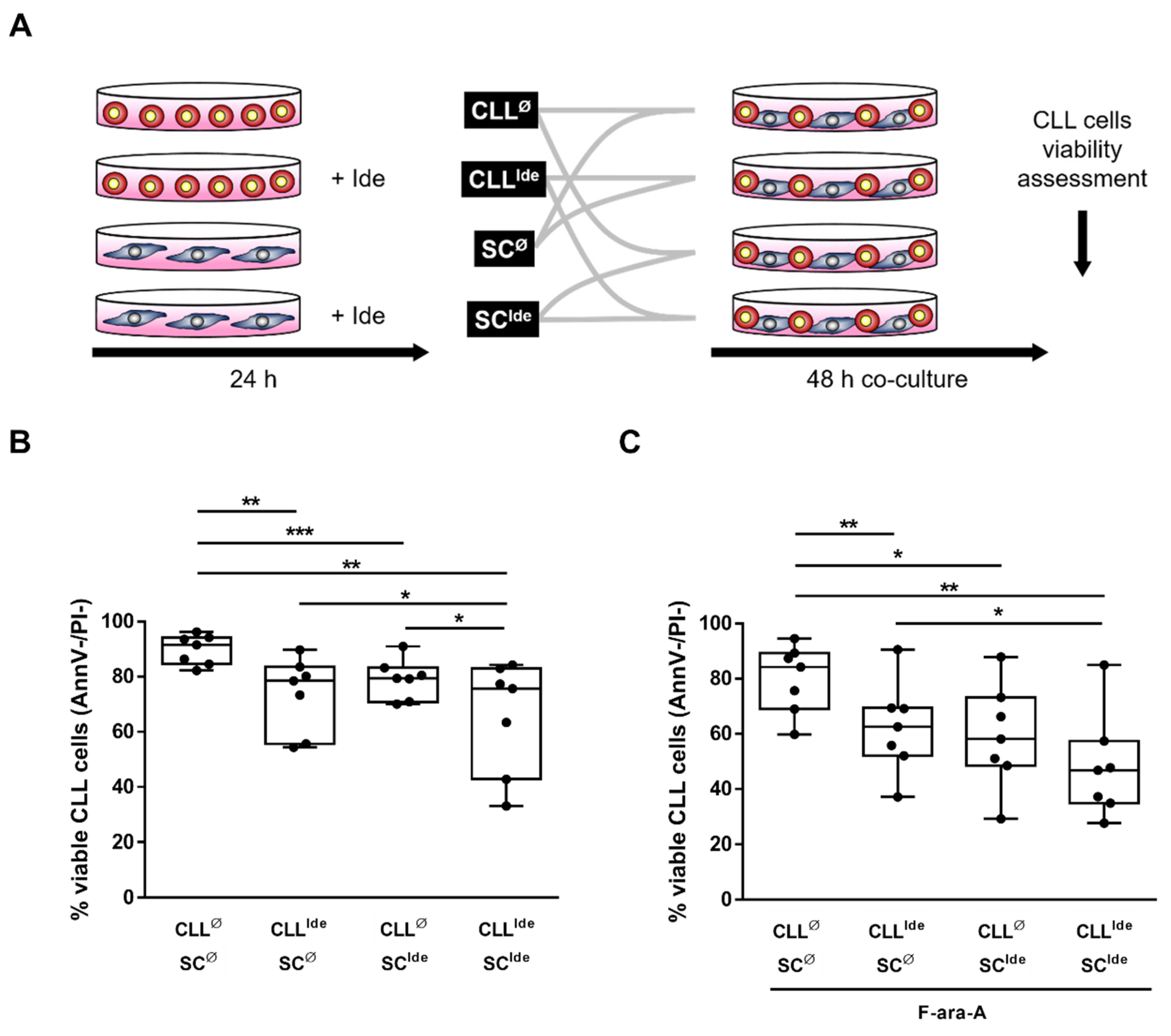
3.3. The Targeted Inhibition of HIF-1α Regulatory Pathways Hinders the SC-Mediated Protection from Spontaneous and Fludarabine-Induced Cell Death
3.4. The Inhibition of PI3K/AKT Pathway and Downstream HIF-1α Impairs CXCL12 Production in SC
3.5. Idelalisib Hampers Stroma-Derived Survival Signals by Targeting HIF-1α at the SC- and CLL Cell-Level
3.6. Treatment with Idelalisib Affects HIF-1α Expression and Activity in CLL Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, D.; Arora, R.; Kaur, P.; Singh, B.; Mannan, R.; Arora, S. Overexpression of hypoxia-inducible factor and metabolic pathways: Possible targets of cancer. Cell Biosci. 2017, 7, 62. [Google Scholar] [CrossRef]
- Wang, G.L.; Jiang, B.H.; Rue, E.A.; Semenza, G.L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 1995, 92, 5510–5514. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G. Signal transduction to hypoxia-inducible factor 1. Biochem. Pharmacol. 2002, 64, 993–998. [Google Scholar] [CrossRef]
- Dimova, E.Y.; Michiels, C.; Kietzmann, T. Kinases as upstream regulators of the HIF system: Their emerging potential as anti-cancer drug targets. Curr. Pharm. Des. 2009, 15, 3867–3877. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Shanafelt, T.D.; Cimmino, A.; Taccioli, C.; Volinia, S.; Liu, C.G.; Calin, G.A.; Croce, C.M.; Chan, D.A.; Giaccia, A.J.; et al. Aberrant regulation of pVHL levels by microRNA promotes the HIF/VEGF axis in CLL B cells. Blood 2009, 113, 5568–5574. [Google Scholar] [CrossRef]
- Rigoni, M.; Riganti, C.; Vitale, C.; Griggio, V.; Campia, I.; Robino, M.; Foglietta, M.; Castella, B.; Sciancalepore, P.; Buondonno, I.; et al. Simvastatin and downstream inhibitors circumvent constitutive and stromal cell-induced resistance to doxorubicin in IGHV unmutated CLL cells. Oncotarget 2015, 6, 29833–29846. [Google Scholar] [CrossRef]
- Valsecchi, R.; Coltella, N.; Belloni, D.; Ponente, M.; Hacken, E.T.; Scielzo, C.; Scarfo, L.; Bertilaccio, M.T.; Brambilla, P.; Lenti, E.; et al. HIF-1alpha regulates the interaction of chronic lymphocytic leukemia cells with the tumor microenvironment. Blood 2016, 127, 1987–1997. [Google Scholar] [CrossRef] [PubMed]
- Griggio, V.; Vitale, C.; Todaro, M.; Riganti, C.; Kopecka, J.; Salvetti, C.; Bomben, R.; Bo, M.D.; Magliulo, D.; Rossi, D.; et al. HIF-1alpha is over-expressed in leukemic cells from TP53-disrupted patients and is a promising therapeutic target in chronic lymphocytic leukemia. Haematologica 2020, 105, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A. The CLL cell microenvironment. Adv. Exp. Med. Biol. 2013, 792, 25–45. [Google Scholar] [CrossRef] [PubMed]
- Burger, M.; Hartmann, T.; Krome, M.; Rawluk, J.; Tamamura, H.; Fujii, N.; Kipps, T.J.; Burger, J.A. Small peptide inhibitors of the CXCR4 chemokine receptor (CD184) antagonize the activation, migration, and antiapoptotic responses of CXCL12 in chronic lymphocytic leukemia B cells. Blood 2005, 106, 1824–1830. [Google Scholar] [CrossRef]
- Burger, J.A.; Burger, M.; Kipps, T.J. Chronic lymphocytic leukemia B cells express functional CXCR4 chemokine receptors that mediate spontaneous migration beneath bone marrow stromal cells. Blood 1999, 94, 3658–3667. [Google Scholar] [CrossRef] [PubMed]
- Nishio, M.; Endo, T.; Tsukada, N.; Ohata, J.; Kitada, S.; Reed, J.C.; Zvaifler, N.J.; Kipps, T.J. Nurselike cells express BAFF and APRIL, which can promote survival of chronic lymphocytic leukemia cells via a paracrine pathway distinct from that of SDF-1alpha. Blood 2005, 106, 1012–1020. [Google Scholar] [CrossRef] [PubMed]
- Niedermeier, M.; Hennessy, B.T.; Knight, Z.A.; Henneberg, M.; Hu, J.; Kurtova, A.V.; Wierda, W.G.; Keating, M.J.; Shokat, K.M.; Burger, J.A. Isoform-selective phosphoinositide 3′-kinase inhibitors inhibit CXCR4 signaling and overcome stromal cell-mediated drug resistance in chronic lymphocytic leukemia: A novel therapeutic approach. Blood 2009, 113, 5549–5557. [Google Scholar] [CrossRef]
- Serra, S.; Vaisitti, T.; Audrito, V.; Bologna, C.; Buonincontri, R.; Chen, S.S.; Arruga, F.; Brusa, D.; Coscia, M.; Jaksic, O.; et al. Adenosine signaling mediates hypoxic responses in the chronic lymphocytic leukemia microenvironment. Blood Adv. 2016, 1, 47–61. [Google Scholar] [CrossRef]
- Sitkovsky, M.; Lukashev, D. Regulation of immune cells by local-tissue oxygen tension: HIF1 alpha and adenosine receptors. Nat. Rev. Immunol. 2005, 5, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Wang, L.; Esko, J.; Giordano, F.J.; Huang, Y.; Gerber, H.P.; Ferrara, N.; Johnson, R.S. Loss of HIF-1alpha in endothelial cells disrupts a hypoxia-driven VEGF autocrine loop necessary for tumorigenesis. Cancer Cell 2004, 6, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Kim, S.K.; Jung, B.J.; Choi, S.B.; Choi, E.Y.; Kim, C.S. Enhancing proliferation and optimizing the culture condition for human bone marrow stromal cells using hypoxia and fibroblast growth factor-2. Stem Cell Res. 2018, 28, 87–95. [Google Scholar] [CrossRef]
- Hallek, M.; Cheson, B.D.; Catovsky, D.; Caligaris-Cappio, F.; Dighiero, G.; Dohner, H.; Hillmen, P.; Keating, M.; Montserrat, E.; Chiorazzi, N.; et al. Guidelines for diagnosis, indications for treatment, response assessment and supportive management of chronic lymphocytic leukemia. Blood 2018, 131, 2745–2760. [Google Scholar] [CrossRef]
- Coscia, M.; Pantaleoni, F.; Riganti, C.; Vitale, C.; Rigoni, M.; Peola, S.; Castella, B.; Foglietta, M.; Griggio, V.; Drandi, D.; et al. IGHV unmutated CLL B cells are more prone to spontaneous apoptosis and subject to environmental prosurvival signals than mutated CLL B cells. Leukemia 2011, 25, 828–837. [Google Scholar] [CrossRef]
- Freed-Pastor, W.A.; Mizuno, H.; Zhao, X.; Langerød, A.; Moon, S.H.; Rodriguez-Barrueco, R.; Barsotti, A.; Chicas, A.; Li, W.; Polotskaia, A.; et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 2012, 148, 244–258. [Google Scholar] [CrossRef]
- Kurtova, A.V.; Balakrishnan, K.; Chen, R.; Ding, W.; Schnabl, S.; Quiroga, M.P.; Sivina, M.; Wierda, W.G.; Estrov, Z.; Keating, M.J.; et al. Diverse marrow stromal cells protect CLL cells from spontaneous and drug-induced apoptosis: Development of a reliable and reproducible system to assess stromal cell adhesion-mediated drug resistance. Blood 2009, 114, 4441–4450. [Google Scholar] [CrossRef]
- Riganti, C.; Orecchia, S.; Pescarmona, G.; Betta, P.G.; Ghigo, D.; Bosia, A. Statins revert doxorubicin resistance via nitric oxide in malignant mesothelioma. Int. J. Cancer 2006, 119, 17–27. [Google Scholar] [CrossRef]
- Ali, A.Y.; Guan, Q.; Wu, X.; Hou, S.; Banerji, V.; Johnston, J.B.; Wall, D.; Szwajcer, D.; Gibson, S.B.; Marshall, A.J. Expression and function of phosphoinositide 3-kinase delta in mesenchymal stromal cells from normal and leukaemic bone marrow. Br. J. Haematol. 2019, 185, 883–887. [Google Scholar] [CrossRef]
- Koczula, K.M.; Ludwig, C.; Hayden, R.; Cronin, L.; Pratt, G.; Parry, H.; Tennant, D.; Drayson, M.; Bunce, C.M.; Khanim, F.L.; et al. Metabolic plasticity in CLL: Adaptation to the hypoxic niche. Leukemia 2016, 30, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Vogler, M.; Butterworth, M.; Majid, A.; Walewska, R.J.; Sun, X.M.; Dyer, M.J.; Cohen, G.M. Concurrent up-regulation of BCL-XL and BCL2A1 induces approximately 1000-fold resistance to ABT-737 in chronic lymphocytic leukemia. Blood 2009, 113, 4403–4413. [Google Scholar] [CrossRef] [PubMed]
- Jayappa, K.D.; Portell, C.A.; Gordon, V.L.; Capaldo, B.J.; Bekiranov, S.; Axelrod, M.J.; Brett, L.K.; Wulfkuhle, J.D.; Gallagher, R.I.; Petricoin, E.F.; et al. Microenvironmental agonists generate de novo phenotypic resistance to combined ibrutinib plus venetoclax in CLL and MCL. Blood Adv. 2017, 1, 933–946. [Google Scholar] [CrossRef]
- Stamatopoulos, B.; Meuleman, N.; De Bruyn, C.; Pieters, K.; Mineur, P.; Le Roy, C.; Saint-Georges, S.; Varin-Blank, N.; Cymbalista, F.; Bron, D.; et al. AMD3100 disrupts the cross-talk between chronic lymphocytic leukemia cells and a mesenchymal stromal or nurse-like cell-based microenvironment: Pre-clinical evidence for its association with chronic lymphocytic leukemia treatments. Haematologica 2012, 97, 608–615. [Google Scholar] [CrossRef] [PubMed]
- O’Hayre, M.; Salanga, C.L.; Kipps, T.J.; Messmer, D.; Dorrestein, P.C.; Handel, T.M. Elucidating the CXCL12/CXCR4 signaling network in chronic lymphocytic leukemia through phosphoproteomics analysis. PLoS ONE 2010, 5, e11716. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A.; Tsukada, N.; Burger, M.; Zvaifler, N.J.; Dell’Aquila, M.; Kipps, T.J. Blood-derived nurse-like cells protect chronic lymphocytic leukemia B cells from spontaneous apoptosis through stromal cell-derived factor-1. Blood 2000, 96, 2655–2663. [Google Scholar] [CrossRef] [PubMed]
- Ceradini, D.J.; Kulkarni, A.R.; Callaghan, M.J.; Tepper, O.M.; Bastidas, N.; Kleinman, M.E.; Capla, J.M.; Galiano, R.D.; Levine, J.P.; Gurtner, G.C. Progenitor cell trafficking is regulated by hypoxic gradients through HIF-1 induction of SDF-1. Nat. Med. 2004, 10, 858–864. [Google Scholar] [CrossRef]
- Camnitz, W.; Burdick, M.D.; Strieter, R.M.; Mehrad, B.; Keeley, E.C. Dose-dependent Effect of Statin Therapy on Circulating CXCL12 Levels in Patients with Hyperlipidemia. Clin. Transl. Med. 2012, 1, 23. [Google Scholar] [CrossRef]
- Manso, B.A.; Zhang, H.; Mikkelson, M.G.; Gwin, K.A.; Secreto, C.R.; Ding, W.; Parikh, S.A.; Kay, N.E.; Medina, K.L. Bone marrow hematopoietic dysfunction in untreated chronic lymphocytic leukemia patients. Leukemia 2019, 33, 638–652. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vitale, C.; Griggio, V.; Riganti, C.; Todaro, M.; Kopecka, J.; Jones, R.; Salvetti, C.; Boccellato, E.; Perutelli, F.; Voena, C.; et al. Targeting HIF-1α Regulatory Pathways as a Strategy to Hamper Tumor-Microenvironment Interactions in CLL. Cancers 2021, 13, 2883. https://doi.org/10.3390/cancers13122883
Vitale C, Griggio V, Riganti C, Todaro M, Kopecka J, Jones R, Salvetti C, Boccellato E, Perutelli F, Voena C, et al. Targeting HIF-1α Regulatory Pathways as a Strategy to Hamper Tumor-Microenvironment Interactions in CLL. Cancers. 2021; 13(12):2883. https://doi.org/10.3390/cancers13122883
Chicago/Turabian StyleVitale, Candida, Valentina Griggio, Chiara Riganti, Maria Todaro, Joanna Kopecka, Rebecca Jones, Chiara Salvetti, Elia Boccellato, Francesca Perutelli, Claudia Voena, and et al. 2021. "Targeting HIF-1α Regulatory Pathways as a Strategy to Hamper Tumor-Microenvironment Interactions in CLL" Cancers 13, no. 12: 2883. https://doi.org/10.3390/cancers13122883
APA StyleVitale, C., Griggio, V., Riganti, C., Todaro, M., Kopecka, J., Jones, R., Salvetti, C., Boccellato, E., Perutelli, F., Voena, C., Godio, L., Boccadoro, M., & Coscia, M. (2021). Targeting HIF-1α Regulatory Pathways as a Strategy to Hamper Tumor-Microenvironment Interactions in CLL. Cancers, 13(12), 2883. https://doi.org/10.3390/cancers13122883







