Methylation Patterns of DKK1, DKK3 and GSK3β Are Accompanied with Different Expression Levels in Human Astrocytoma
Abstract
Simple Summary
Abstract
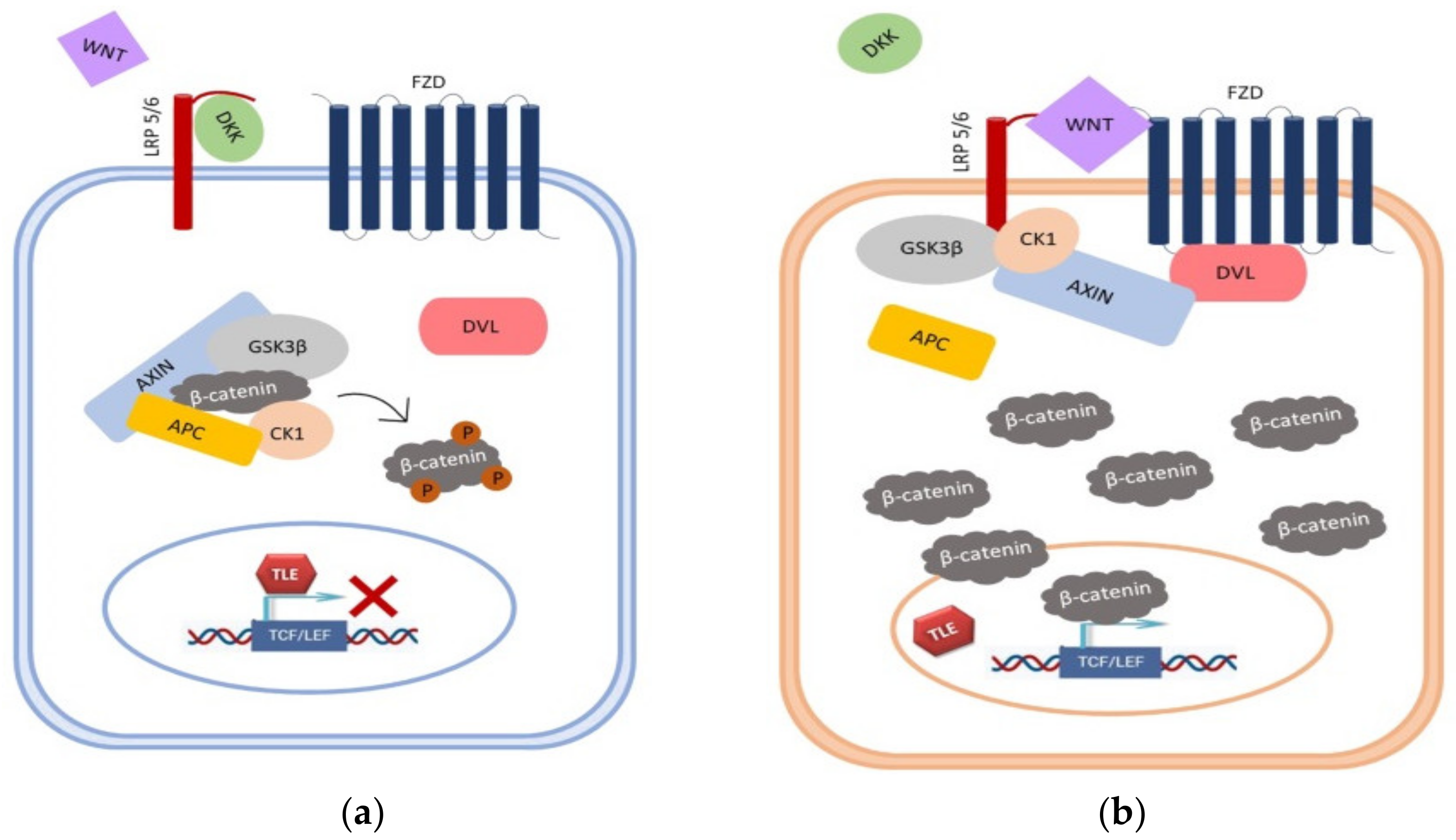
1. Introduction
2. Materials and Methods
2.1. Tissue Samples
2.2. DNA Extraction
2.3. Polymerase Chain Reaction (PCR), Restriction Fragment Length Polymorphism (RFLP), Loss of Heterozygosity (LOH)
2.3.1. Polymerase Chain Reaction
2.3.2. Restriction Fragment Length Polymorphism/Loss of Heterozygosity
2.3.3. Methylation-Specific PCR (MSP)
2.4. Immunohistochemistry (IHC)
2.5. Microscopic Analysis
2.6. Statistical Analysis
3. Results
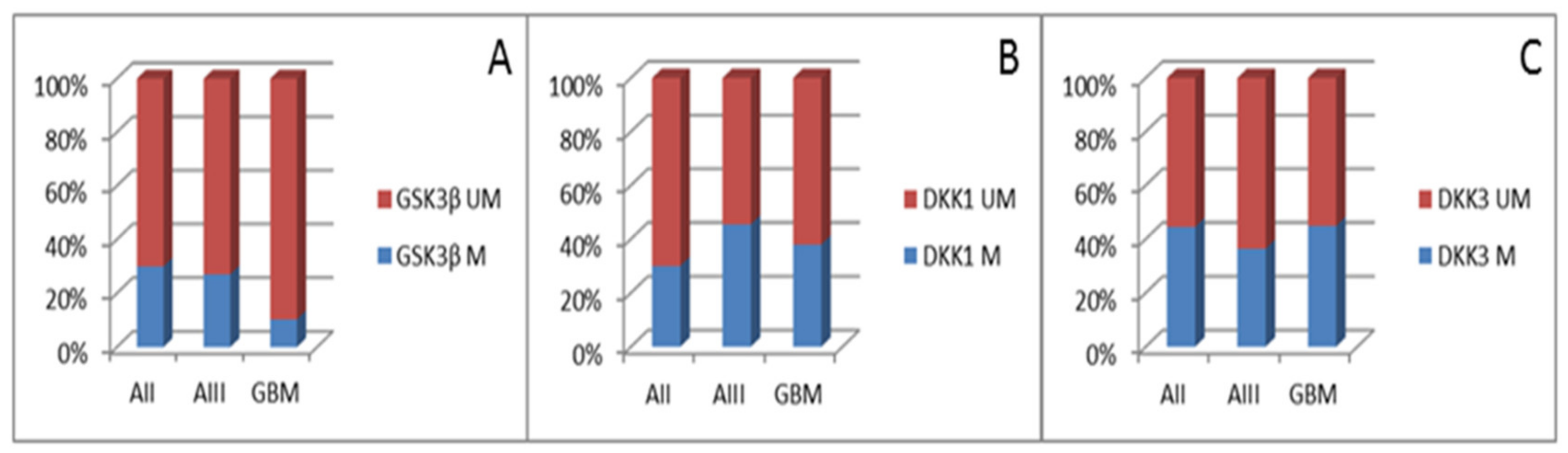
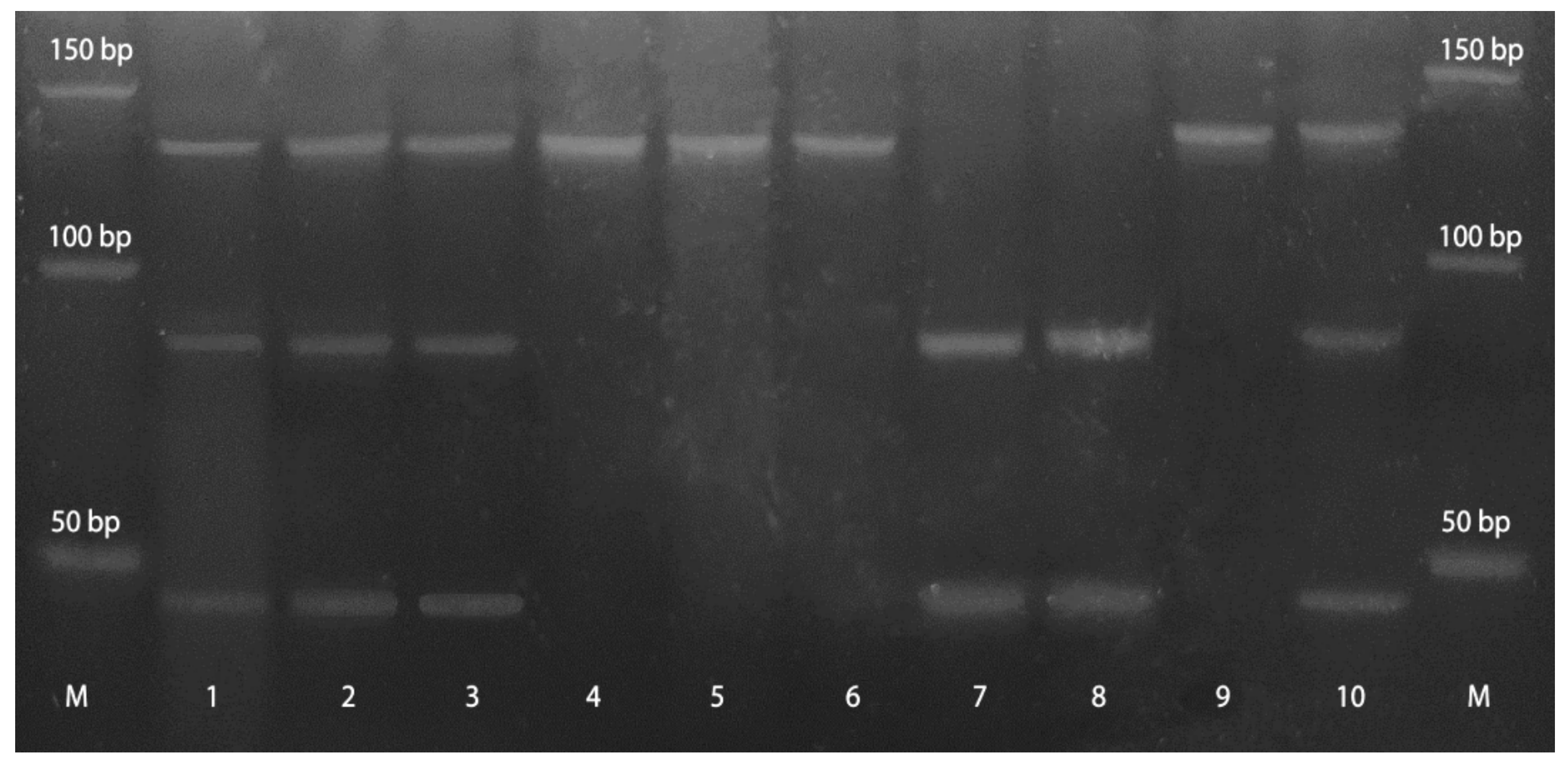
3.1. Methylation Status of Promoter Regions of GSK3β, DKK,1 and DKK3
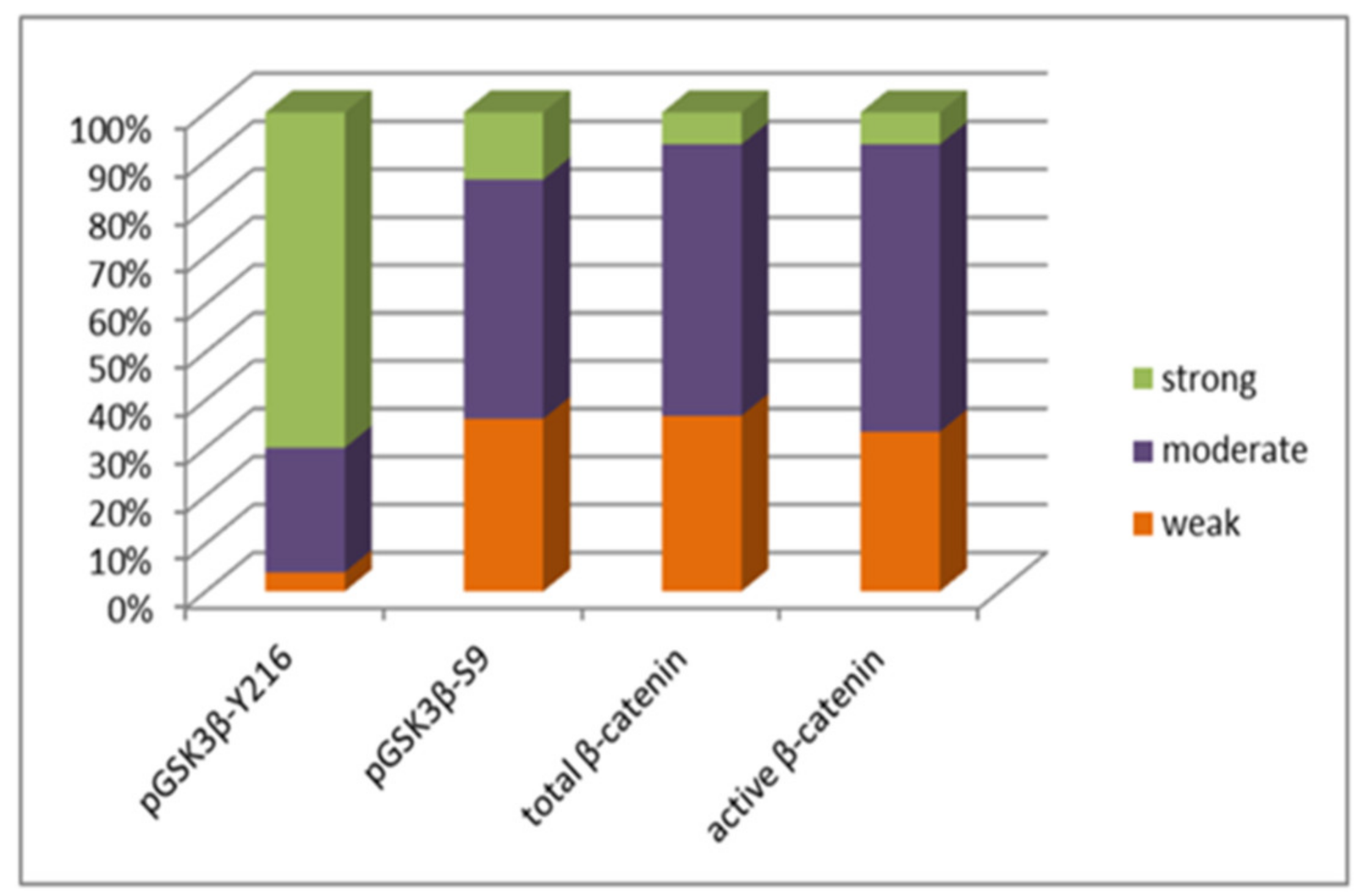
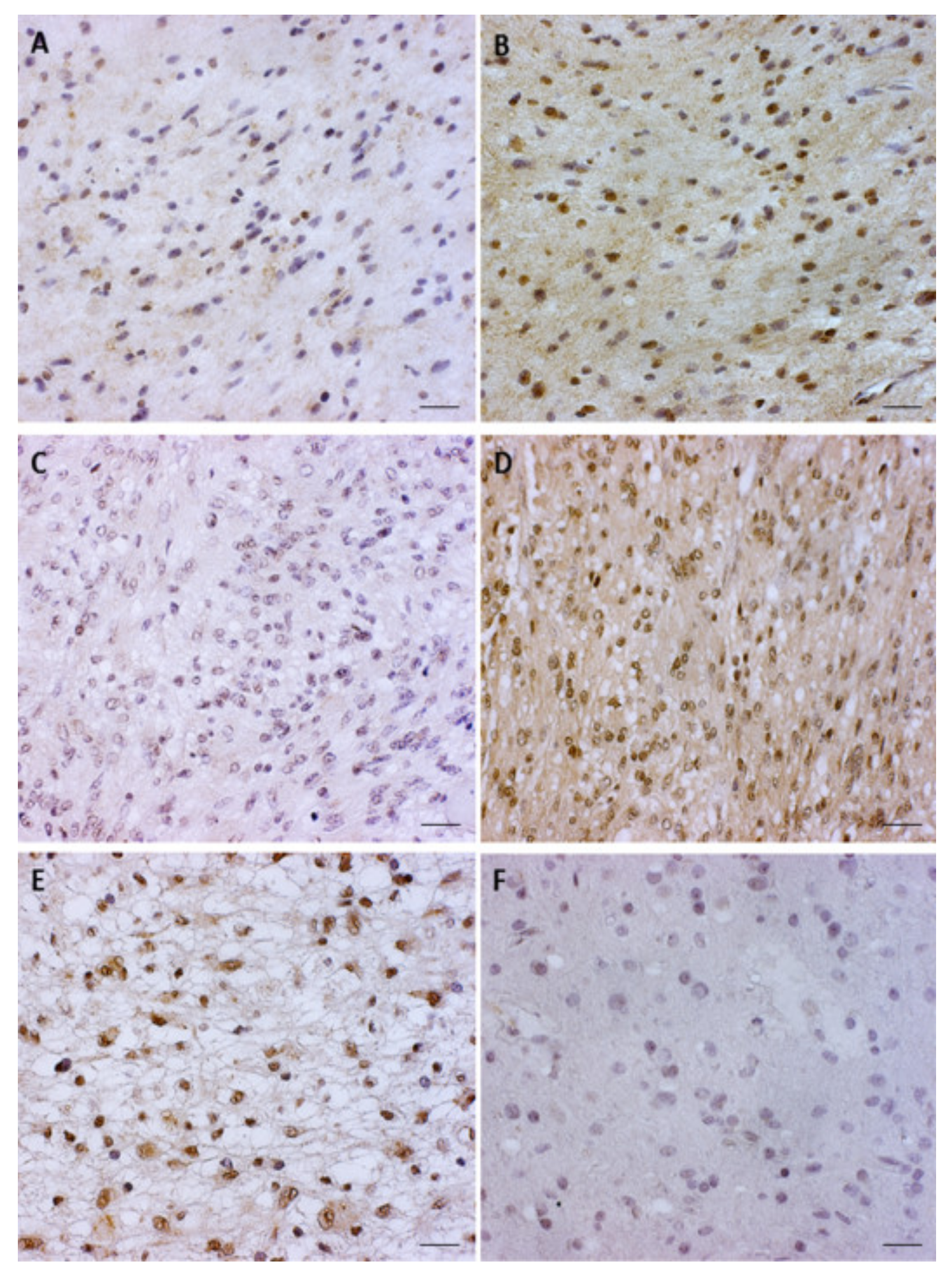
3.2. pGSK3β-S9 and pGSK3β-Y216 Expression Levels
3.3. Total β-Catenin and Unphosphorylated β-Catenin Expression Levels in Glioblastoma
3.4. APC Exon 11 Genetic Changes in Glioblastoma
3.5. The Correlations of Molecular Findings and Clinical Parameters
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Perry, A.; Wesseling, P. Histologic Classification of Gliomas. Handb. Clin. Neurol. 2016, 134, 71–95. [Google Scholar] [CrossRef] [PubMed]
- Wesseling, P.; Capper, D. WHO 2016 Classification of Gliomas. Neuropathol. Appl. Neurobiol. 2018, 44, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Kristensen, B.W.; Priesterbach-Ackley, L.P.; Petersen, J.K.; Wesseling, P. Molecular Pathology of Tumors of the Central Nervous System. Ann. Oncol. 2019, 30, 1265–1278. [Google Scholar] [CrossRef] [PubMed]
- Glibo, M.; Serman, A.; Karin-Kujundzic, V.; Bekavac Vlatkovic, I.; Miskovic, B.; Vranic, S.; Serman, L. The Role of Glycogen Synthase Kinase 3 (GSK3) in Cancer with Emphasis on Ovarian Cancer Development and Progression: A Comprehensive Review. Bosn. J. Basic Med. Sci. 2021, 21, 5–18. [Google Scholar] [CrossRef]
- Duda, P.; Akula, S.M.; Abrams, S.L.; Steelman, L.S.; Martelli, A.M.; Cocco, L.; Ratti, S.; Candido, S.; Libra, M.; Montalto, G.; et al. Targeting GSK3 and Associated Signaling Pathways Involved in Cancer. Cells 2020, 9, 1110. [Google Scholar] [CrossRef]
- Majewska, E.; Szeliga, M. AKT/GSK3β Signaling in Glioblastoma. Neurochem. Res. 2016, 42, 918–924. [Google Scholar] [CrossRef] [PubMed]
- Domoto, T.; Pyko, I.V.; Furuta, T.; Miyashita, K.; Uehara, M.; Shimasaki, T.; Nakada, M.; Minamoto, T. Glycogen Synthase Kinase-3β is a Pivotal Mediator of Cancer Invasion and Resistance to Therapy. Cancer Sci. 2016, 107, 1363–1372. [Google Scholar] [CrossRef] [PubMed]
- Bugter, J.M.; Fenderico, N.; Maurice, M.M. Mutations and Mechanisms of WNT Pathway Tumour Suppressors in Cancer. Nat. Rev. Cancer 2021, 21, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Tamai, K.; Doble, B.; Li, S.; Huang, H.; Habas, R.; Okamura, H.; Woodgett, J.; He, X. A Dual-kinase Mechanism for Wnt Co-receptor Phosphorylation and Activation. Nature 2005, 438, 873–877. [Google Scholar] [CrossRef]
- Baetta, R.; Banfi, C. Dkk (Dickkopf) Proteins. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 1330–1342. [Google Scholar] [CrossRef]
- Zhang, K.; Watanabe, M.; Kashiwakura, Y.; Li, S.A.; Edamura, K.; Huang, P.; Yamaguchi, K.; Nasu, Y.; Kobayashi, Y.; Sakaguchi, M.; et al. Expression Pattern of REIC/Dkk-3 in Various Cell Types and the Implications of the Soluble Form in Prostatic Acinar Development. Int. J. Oncol. 2010, 37, 1495–1501. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.Y.; Yang, X.R.; Shen, Q.J.; Yang, L.X.; Xu, Y.; Qiu, S.J.; Sun, Y.F.; Zhang, X.; Wang, Z.; Zhu, K.; et al. Expression of Dickkopfrelated Protein 1 is Related to Lymphatic Metastasis and Indicates Poor Prognosis in Intrahepatic Cholangiocarcinoma Patients after Surgery. Cancer 2013, 119, 993–1003. [Google Scholar] [CrossRef]
- Begenik, H.; Kemik, A.S.; Emre, H.; Dulger, A.C.; Demirkiran, D.; Ebinc, S.; Kemik, O. The Association between Serum Dickkopf-1 Levels and Esophageal Squamous Cell Carcinoma. Hum. Exp. Toxicol. 2014, 33, 785–788. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhou, H.; Zhang, X.; Ma, X.; Liu, Z.; Liu, X. Elevated Levels of Dickkopf-1 are Associated with beta-catenin AccumuLation and Poor prognosis in Patients with Chondrosarcoma. PLoS ONE 2014, 9, e105414. [Google Scholar] [CrossRef]
- Rachner, T.D.; Thiele, S.; Göbel, A.; Browne, A.; Fuessel, S.; Erdmann, K.; Wirth, M.P.; Fröhner, M.; Todenhöfer, T.; Muders, M.H.; et al. High Serum Levels of Dickkopf-1 are Associated with a Poor Prognosis in Prostate Cancer Patients. BMC Cancer 2014, 14, 649. [Google Scholar] [CrossRef]
- Shi, Y.; Gong, H.L.; Zhou, L.; Tian, J.; Wang, Y. Dickkopf-1 is a Novel Prognostic Biomarker for Laryngeal Squamous Cell Carcinoma. Acta Otolaryngol. 2014, 134, 753–759. [Google Scholar] [CrossRef]
- Han, S.X.; Zhou, X.; Sui, X.; He, C.C.; Cai, M.J.; Ma, J.L.; Zhang, Y.Y.; Zhou, C.Y.; Ma, C.X.; Varela-Ramirez, A.; et al. Serum Dickkopf-1 is a Novel Serological Biomarker for the Diagnosis and Prognosis of Pancreatic Cancer. Oncotarget 2015, 6, 19907–19917. [Google Scholar] [CrossRef]
- Sun, D.K.; Wang, L.; Wang, J.M.; Zhang, P. Serum Dickkopf-1 Levels as a Clinical and Prognostic Factor in Patients with Bladder Cncer. Genet. Mol. Res. 2015, 14, 18181–18187. [Google Scholar] [CrossRef]
- Shi, X.D.; Yu, X.H.; Wu, W.R.; Xu, X.L.; Wang, J.Y.; Xu, L.B.; Zhang, R.; Liu, C. Dickkopf-1 Expression is Associated with Tumorigenity and Lymphatic Metastasis in Human Hilar Cholangiocarcinoma. Oncotarget 2016, 7, 70378–70387. [Google Scholar] [CrossRef]
- Watany, M.; Badawi, R.; Elkhalawany, W.; Abd-Elsalam, S. Study of Dickkopf-1 (DKK-1) Gene Expression in Hepatocellular Carcinoma Patients. J. Clin. Diagn. Res. 2017, 11, Oc32–Oc34. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Wang, S.; Huang, L.; Zhang, S. Clinical Significance of Serum DKK-1 in Patients with Gynecological Cancer. Int. J. Gynecol. Cancer 2009, 19, 1177–1181. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, B.; Qi, L.; Li, Y.; Zhao, X.; Zhang, D.; Zhang, Y. Dickkopf-1 Expression is Down-regulated during the Colorectal Adenoma-carcinoma Sequence and Correlates with Reduced Microvessel Density and VEGF Expression. Histopathology 2015, 67, 158–166. [Google Scholar] [CrossRef]
- Zhao, Y.P.; Wang, W.; Wang, X.H.; Xu, Y.; Wang, Y.; Dong, Z.F.; Zhang, J.J. Downregulation of Serum DKK-1 Predicts Poor Prognosis in Patients with Papillary Thyroid Cancer. Genet. Mol. Res. 2015, 14, 18886–18894. [Google Scholar] [CrossRef]
- Mizobuchi, Y.; Matsuzaki, K.; Kuwayama, K.; Kitazato, K.; Mure, H.; Kageji, T.; Nagahiro, S. REIC/Dkk-3 Induces Cell Death in Human Malignant Glioma. Neuro Oncol. 2008, 10, 244–253. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, W.; Xu, Q.; Huang, Y. Elevated Expression of Dickkopf-1 Increases the Sensitivity of Human Glioma Cell Line SHG44 to BCNU. J. Exp. Clin. Cancer Res. 2010, 29, 131. [Google Scholar] [CrossRef]
- Götze, S.; Wolter, M.; Reifenberger, G.; Müller, O.; Sievers, S. Frequent Promoter Hypermethylation of Wnt Pathway Inhibitor Genes in Malignant Astrocytic Gliomas. Cancer Genet. 2010, 126, 2584–2593. [Google Scholar] [CrossRef]
- Oka, T.; Kurozumi, K.; Shimazu, Y.; Ichikawa, T.; Ishida, J.; Otani, Y.; Shimizu, T.; Tomita, Y.; Sakaguchi, M.; Watanabe, M.; et al. A Super Gene Expression System Enhances the Anti-glioma Effects of Adenovirus-mediated REIC/Dkk-3 Gene Therapy. Sci. Rep. 2016, 6, 33319. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Molecular Cloning—A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012. [Google Scholar]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A Simple Salting out Procedure for Extracting DNA from Human Nucleated Cells. Nucleic Acids Res. 1988, 16, 883–893. [Google Scholar] [CrossRef]
- Maehata, T.; Taniguchi, H.; Yamamoto, H.; Nosho, K.; Adachi, Y.; Miyamoto, N.; Miyamoto, C.; Akutsu, N.; Yamaoka, S.; Itoh, F. Transcriptional Silencing of Dickkopf Gene Family by CpG Island Hypermethylation in Human Gastrointestinal Cancer. World J. Gastroenterol. 2008, 14, 2702–2714. [Google Scholar] [CrossRef]
- Zhang, M.; Huang, M.; Cao, B.; Sheng, X.; Li, P. Methylation of the DKK3 Promoter is Associated with Poor Prognosis in Patients with Cervical Adenocarcinoma. Int. J. Clin. Exp. Pathol. 2018, 11, 788–794. [Google Scholar] [PubMed]
- Naghibalhossaini, F.; Zamani, M.; Mokarram, P.; Khalili, I.; Rasti, M.; Mostafavi-pour, Z. Epigenetic and Genetic Analysis of WNT Signaling Pathway in Sporadic Colorectal Cancer Patients from Iran. Mol. Biol. Rep. 2012, 39, 6171–6178. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M.; Katoh, M. Molecular Genetics and Targeted Therapy of WNT-related Human Diseases (Review). Int. J. Mol. Med. 2017, 40, 587–606. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, O.; Fraga, M.F.; Ballestar, E.; Paz, M.F.; Herranz, M.; Espada, J.; García, J.M.; Muñoz, A.; Esteller, M.; González-Sancho, J.M. Epigenetic Inactivation of the Wnt Antagonist DICKKOPF-1 (DKK-1) Gene in Human Colorectal Cancer. Oncogene 2006, 25, 4116–4121. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.C.; Zhang, X.L.; Yang, B.; Geng, J.; Peng, B.; Zheng, J.H. Decreased Expression of dkk1 and dkk3 in Human Clear Cell Renal Cell Carcinoma. Mol. Med. Rep. 2014, 9, 2367–2373. [Google Scholar] [CrossRef] [PubMed]
- Galamb, O.; Kalmar, A.; Peterfia, B.; Csabai, I.; Bodor, A.; Ribli, D.; Krenács, T.; Patai, Á.V.; Wichmann, B.; Barták, B.K.; et al. Aberrant DNA Methylation of WNT Pathway Genes in the Development and Progression of CIMP-negative Colorectal Cancer. Epigenetics 2016, 11, 588–602. [Google Scholar] [CrossRef]
- Wang, W.; He, Y.; Rui, J.; Xu, M.Q. miR-410 Acts as an Oncogene in Colorectal Cancer Cells by Targeting Dickkopf-related Protein 1 via the Wnt/β-catenin Signaling Pathway. Oncol. Lett. 2019, 17, 807–814. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Fan, J.; Yang, X.R.; Tan, Y.; Zhao, W.; Xu, Y.; Wang, N.; Niu, Y.; Wu, Z.; Zhou, J.; et al. Serum DKK1 as a Protein Biomarker for the Diagnosis of Hepatocellular Carcinoma: A Large-scale, Multicentre Study. Lancet Oncol. 2012, 13, 817–826. [Google Scholar] [CrossRef]
- Urakami, S.; Shiina, H.; Enokida, H.; Kawakami, T.; Kawamoto, K.; Hirata, H.; Tanaka, Y.; Kikuno, N.; Nakagawa, M.; Igawa, M. Combination Analysis of Hypermethylated Wnt-antagonist Family Genes as a Novel Epigenetic Biomarker Panel for Bladder Cancer Detection. Clin. Cancer Res. 2006, 12, 2109–2116. [Google Scholar] [CrossRef]
- Yue, W.; Sun, Q.; Dacic, S.; Landreneau, R.J.; Siegfried, J.M.; Yu, J.; Zhang, L. Downregulation of dkk3 Activates Beta-catenin/tcf-4 Signaling in Lung Cancer. Carcinogenesis 2008, 29, 84–92. [Google Scholar] [CrossRef]
- You, A.; Fokas, E.; Wang, L.F.; He, H.; Kleb, B.; Niederacher, D.; Engenhart-Cabillic, R.; An, H.X. Expression of the Wnt Antagonist dkk3 is Frequently Suppressed in Sporadic Epithelial Ovarian Cancer. J. Cancer Res. Clin. Oncol. 2011, 137, 621–627. [Google Scholar] [CrossRef]
- Park, J.M.; Kim, M.K.; Chi, K.C.; Kim, J.H.; Lee, S.H.; Lee, E.J. Aberrant Loss of dickkopf-3 in Gastric Cancer: Can it Predict Lymph Node Metastasis Preoperatively? World J. Surg. 2015, 39, 1018–1025. [Google Scholar] [CrossRef]
- Lorsy, E.; Topuz, A.S.; Geisler, C.; Stahl, S.; Garczyk, S.; von Stillfried, S.; Hoss, M.; Gluz, O.; Hartmann, A.; Knüchel, R. Loss of dickkopf 3 Promotes the Tumorigenesis of Basal Breast Cancer. PLoS ONE 2016, 11, e0160077. [Google Scholar] [CrossRef]
- Pei, Y.; Kano, J.; Iijima, T.; Morishita, Y.; Inadome, Y.; Noguchi, M. Overexpression of Dickkopf 3 in Hepatoblastomas and Hepatocellular Carcinomas. Virchows Arch. 2009, 454, 639–646. [Google Scholar] [CrossRef]
- Fujii, M.; Katase, N.; Lefeuvre, M.; Gunduz, M.; Buery, R.R.; Tamamura, R.; Tsujigiwa, H.; Nagatsuka, H. Dickkopf (dkk)-3 and β-catenin Expressions Increased in the Transition from Normal Oral Mucosal to Oral Squamous Cell Carcinoma. J. Mol. Histol. 2011, 42, 499–504. [Google Scholar] [CrossRef][Green Version]
- Wang, Z.; Lin, L.; Thomas, D.G.; Nadal, E.; Chang, A.C.; Beer, D.G.; Lin, J. The Role of Dickkopf-3 Overexpression in Esophageal Adenocarcinoma. J. Thorac. Cardiovasc. Surg. 2015, 150, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Valdora, F.; Banelli, B.; Stigliani, S.; Pfister, S.M.; Moretti, S.; Kool, M.; Remke, M.; Bai, A.H.C.; Brigati, C.; Hielscher, T.; et al. Epigenetic Silencing of DKK3 in Medulloblastoma. Int. J. Mol. Sci. 2013, 14, 7492–7505. [Google Scholar] [CrossRef] [PubMed]
- Huo, J.; Zhang, Y.; Li, R.; Wang, Y.; Wu, J.; Zhang, D. Upregulated MicroRNA-25 Mediates the Migration of Melanoma Cells by Targeting DKK3 through the WNT/β-Catenin Pathway. Int. J. Mol. Sci. 2016, 17, 1124. [Google Scholar] [CrossRef] [PubMed]
- Rui, Y.; Hu, M.; Wang, P.; Zhang, C.; Xu, H.; Li, Y.; Zhang, Y.; Gu, J.; Wang, Q. LncRNA HOTTIP Mediated DKK1 Downregulation Confers Metastasis and Invasion in Colorectal Cancer Cells. Histol. Histopathol. 2019, 34, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Yang, Z.; Zhou, H.; Yue, J.; Mu, T.; Zhang, Q.; Bi, X. Upregulation of DKK3 by miR-483-3p Plays an Important Role in the Chemoprevention of Colorectal Cancer Mediated by Black Raspberry Anthocyanins. Mol. Carcinog. 2020, 59, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Unnikrishnan, A.; Freeman, W.M.; Jackson, J.; Wren, J.D.; Porter, H.; Richardson, A. The Role of DNA Methylation in Epigenetics of Aging. Pharmacol. Ther. 2019, 195, 172–185. [Google Scholar] [CrossRef] [PubMed]
- Shakoori, A.; Ougolkov, A.; Yu, Z.W.; Zhang, B.; Modarressi, M.H.; Billadeau, D.D.; Mai, M.; Takahashi, Y.; Minalmoto, T. Deregulated GSK3beta Activity in Colorectal Cancer: Its Association with Tumor Cell Survival and Proliferation. Biochem. Biophys. Res. Commun. 2005, 334, 1365–1373. [Google Scholar] [CrossRef]
- Shakoori, A.; Mai, W.; Miyashita, K.; Yasumoto, K.; Takahashi, Y.; Ooi, A. Inhibition of GSK-3 Beta Activity Attenuates Proliferation of Human Colon Cancer Cells in Rodents. Cancer Sci. 2007, 98, 1388–1393. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Wang, J.; Gao, Y.; Gao, T.W.; Chen, G.; Bower, K.A.; Odetallah, M.; Ding, M.; Ke, Z.; Luo, J. The Role of Glycogen Synthase Kinase 3beta in the Transformation of Epidermal Cells. Cancer Res. 2007, 67, 7756–7764. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Nagini, S.; Rana, A. Expression and Inactivation of Glycogen Synthase Kinase 3 Alpha/Beta and their Association with the Expression of Cyclin D1 and p53 in Oral Squamous Cell Carcinoma Progression. Mol. Cancer 2015, 14, 20. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Saito, H.; Masuda, S.; Yang, X.; Takano, Y. Phosphorylated GSK3beta-ser9 and EGFR are Good Prognostic Factors for Lung Carcinomas. Anticancer Res. 2007, 27, 3561–3569. [Google Scholar] [PubMed]
- Acikgoz, E.; Güler, G.; Camlar, M.; Oktem, G.; Aktug, H. Glycogen Synthase Kinase-3 Inhibition in Glioblastoma Multiforme Cells Induces Apoptosis, Cell Cycle Arrest and Changing Biomolecular Structure. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 209, 150–164. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Steelman, L.S.; Bertrand, F.E.; Davis, N.M.; Sokolosky, M.; Abrams, S.L.; Montalto, G.; D’Assoro, A.B.; Libra, M.; Nicoletti, F.; et al. GSK-3 as Potential Target for Therapeutic Intervention in Cancer. Oncotarget 2014, 5, 2881–2911. [Google Scholar] [CrossRef] [PubMed]
- Walz, A.; Ugolkov, A.; Chandra, S.; Kozikowski, A.; Carneiro, B.A.; O’Halloran, T.V.; Giles, F.J.; Billadeau, D.D.; Mazar, A.P. Molecular Pathways: Revisiting Glycogen Synthase Kinase-3β as a Target for the Treatment of Cancer. Clin. Cancer Res. 2017, 23, 1891–1897. [Google Scholar] [CrossRef]
- Sahin, I.; Eturi, A.; De Souza, A.; Pamarthy, S.; Tavora, F.; Giles, F.J.; Carneiro, B.A. Glycogen Synthase Kinase-3β Inhibitors as Novel Cancer Treatments and Modulators of Antitumor Immune Responses. Cancer Biol. Ther. 2019, 20, 1047–1056. [Google Scholar] [CrossRef]
- Kotliarova, S.; Pastorino, S.; Kovell, L.C.; Kotliarov, Y.; Song, H.; Zhang, W.; Bailey, R.; Maric, D.; Zenklusen, J.C.; Lee, J.; et al. Glycogen Synthase Kinase-3 Inhibition Induces Glioma Cell Death through c-MYC, Nuclear Factor-kappaB, and Glucose Regulation. Cancer Res. 2008, 68, 6643–6651. [Google Scholar] [CrossRef] [PubMed]
- Beurel, E.; Grieco, S.F.; Jope, R.S. Glycogen Synthase Kinase-3 (GSK3): Regulation, Actions, and Diseases. Pharmacol. Ther. 2015, 148, 114–131. [Google Scholar] [CrossRef]
- Utsuki, S.; Sato, Y.; Oka, H.; Tsuchiya, B.; Suzuki, S.; Fujii, K. Relationship between the Expression of E-, N-cadherins and Beta-catenin and Tumor Grade in Astrocytomas. J. Neurooncol. 2002, 57, 187–192. [Google Scholar] [CrossRef]
- Kahlert, U.D.; Suwala, A.K.; Koch, K.; Natsumeda, M.; Orr, B.A.; Hayashi, M.; Maciaczyk, J.; Eberhart, C.G. Pharmacologic Wnt Inhibition Reduces Proliferation, Survival, and Clonogenicity of Glioblastoma Cells. J. Neuropathol. Exp. Neurol. 2015, 74, 889–900. [Google Scholar] [CrossRef]
- Tompa, M.; Kalovits, F.; Nagy, A.; Kalman, B. Contribution of the Wnt Pathway to Defining Biology of Glioblastoma. Neuromolecular Med. 2018, 20, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, L.; Zhao, S.; Ji, X.; Luo, Y.; Ling, F. β-Catenin Overexpression in Malignant Glioma and Its Role in Proliferation and Apoptosis in Glioblastoma Cells. Med. Oncol. 2011, 28, 608–614. [Google Scholar] [CrossRef] [PubMed]
- Sareddy, G.R.; Panigrahi, M.; Challa, S.; Mahadevan, A.; Babu, P.P. Activation of Wnt/β-catenin/Tcf Signaling Pathway in Human Astrocytomas. Neurochem. Int. 2009, 55, 307–317. [Google Scholar] [CrossRef]
- Nikuševa-Martić, T.; Pećina-Šlaus, N.; Kušec, V.; Kokotović, T.; Mušinović, H.; Tomas, D.; Zeljko, M. Changes of AXIN-1 and Beta-catenin in Neuroepithelial Brain Tumors. Pathol. Oncol. Res. 2010, 16, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Kafka, A.; Bačić, M.; Tomas, D.; Žarković, K.; Bukovac, A.; Njirić, N.; Mrak, G.; Krsnik, Ž.; Pećina-Šlaus, N. Different Behaviour of DVL1, DVL2, DVL3 in Astrocytoma Malignancy Grades and their Association to TCF1 and LEF1 Upregulation. J. Cell. Mol. Med. 2019, 23, 641–655. [Google Scholar] [CrossRef]
- Woodgett, J.R. Judging a Protein by more than Its Name: GSK-3. Sci. STKE 2001, 2001, re12. [Google Scholar] [CrossRef]
- Ng, S.S.; Mahmoudi, T.; Danenberg, E.; Bejaoui, I.; de Lau, W.; Korswagen, H.C.; Schutte, M.; Clevers, H. Phosphatidylinositol 3-kinase Signaling does not Activate the Wnt Cascade. J. Biol. Chem. 2009, 284, 35308–35313. [Google Scholar] [CrossRef]
- Yun, S.H.; Park, J.I. PGC-1α Regulates Cell Proliferation and Invasion via AKT/GSK-3β/β-catenin Pathway in Human Colorectal Cancer SW620 and SW480 Cells. Anticancer Res. 2020, 40, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Veeck, J.; Dahl, E. Targeting the Wnt Pathway in Cancer: The Emerging Role of Dickkopf-3. Biochim. Biophys. Acta 2012, 1825, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Nikuševa-Martić, T.; Beroš, V.; Pećina-Šlaus, N.; Pećina, H.I.; Bulić-Jakuš, F. Genetic Changes of CDH1, APC, and CTNNB1 Found in Human Brain Tumors. Pathol. Res. Pract. 2007, 203, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Pećina-Šlaus, N.; Martić, T.N.; Zeljko, M.; Bulat, S. Brain Metastases Exhibit Gross Deletions of the APC Gene. Brain Tumor Pathol. 2011, 28, 223–228. [Google Scholar] [CrossRef]
- Pećina-Šlaus, N.; Kljaić, M.; Nikuševa-Martić, T. Loss of Heterozygosity of APC and CDH1 Genes in Laryngeal Squamous Cell Carcinoma. Pathol. Res. Pract. 2005, 201, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.G.; Ridsdale, J.; Tonks, A.; Darley, R.L. Factors Affecting the Nuclear Localization of β-Catenin in Normal and Malignant Tissue. J. Cell. Biochem. 2014, 115, 1351–1361. [Google Scholar] [CrossRef]
- Dar, M.S.; Singh, P.; Singh, G.; Jamwal, G.; Hussain, S.S.; Rana, A.; Akhter, Y.; Mong, S.P.; Dar, M.J. Terminal Regions of β-catenin are Critical for Regulating Its Adhesion and Transcription Functions. Biochim. Biophys. Acta 2016, 1863, 2345–2357. [Google Scholar] [CrossRef]

| Primer | Sequence | Product Size | |
|---|---|---|---|
| DKK1 | MR-F | 5′-CGTTCGTTGGTAGTTTTTATTTCGA-3′ | 175 bp |
| MR-R | 5′-GCGACTACCTTTATACCGCGAA-3′ | ||
| UMR-F | 5′-TGTTTGTTGGTAGTTTTTATTTTGA-3′ | 173 bp | |
| UMR-R | 5′-ACCACAACTACCTTTATACCACAAA-3′ | ||
| DKK3 | MR-F | 5′-CGGTTTTTTTTCGTTTTCGGG-3′ | 154 bp |
| MR-R | 5′-CAAACCGCTACATCTCCGCT-3′ | ||
| UMR-F | 5′-TTTTGGTTTTTTTTTGTTTTTGGG-3′ | 155 bp | |
| UMR-R | 5′-CCAA ACCACTACATCTCCACT-3′ | ||
| GSK3β | MR-F | 5′ CGTCGTTATCGTTATCGTTC 3′ | 135 bp |
| MR-R | 5′ AATAACTCGAAAATACGACG 3′ | ||
| UMR-F | 5′ GAGGAGTTGTTGTTATTGTTATTGTTT 3′ | 136 bp | |
| UMR-R | 5′ AAAAAAATAACTCAAAAATACAACA 3′ | ||
| Gene | Initial Denaturation | Cycle Conditions | Final Elongation | |||
|---|---|---|---|---|---|---|
| DKK1 | MR | 95 °C 5 min | 95 °C 30 s | 61 °C 30 s | 72 °C 30 s | 72 °C 7 min |
| UMR | 95 °C 5 min | 95 °C 30 s | 61 °C 30 s | 72 °C 30 s | 72 °C 7 min | |
| DKK3 | MR | 95 °C 5 min | 95 °C 30 s | 61 °C 30 s | 72 °C 30 s | 72 °C 7 min |
| UMR | 95 °C 5 min | 95 °C 30 s | 61 °C 30 s | 72 °C 30 s | 72 °C 7 min | |
| GSK3β | MR | 95 °C 5 min | 95 °C 30 s | 55 °C 30 s | 72 °C 30 s | 72 °C 7 min |
| UMR | 95 °C 5 min | 95 °C 30 s | 56,5 °C 30 s | 72 °C 30 s | 72 °C 7 min | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kafka, A.; Bukovac, A.; Brglez, E.; Jarmek, A.-M.; Poljak, K.; Brlek, P.; Žarković, K.; Njirić, N.; Pećina-Šlaus, N. Methylation Patterns of DKK1, DKK3 and GSK3β Are Accompanied with Different Expression Levels in Human Astrocytoma. Cancers 2021, 13, 2530. https://doi.org/10.3390/cancers13112530
Kafka A, Bukovac A, Brglez E, Jarmek A-M, Poljak K, Brlek P, Žarković K, Njirić N, Pećina-Šlaus N. Methylation Patterns of DKK1, DKK3 and GSK3β Are Accompanied with Different Expression Levels in Human Astrocytoma. Cancers. 2021; 13(11):2530. https://doi.org/10.3390/cancers13112530
Chicago/Turabian StyleKafka, Anja, Anja Bukovac, Emilija Brglez, Ana-Marija Jarmek, Karolina Poljak, Petar Brlek, Kamelija Žarković, Niko Njirić, and Nives Pećina-Šlaus. 2021. "Methylation Patterns of DKK1, DKK3 and GSK3β Are Accompanied with Different Expression Levels in Human Astrocytoma" Cancers 13, no. 11: 2530. https://doi.org/10.3390/cancers13112530
APA StyleKafka, A., Bukovac, A., Brglez, E., Jarmek, A.-M., Poljak, K., Brlek, P., Žarković, K., Njirić, N., & Pećina-Šlaus, N. (2021). Methylation Patterns of DKK1, DKK3 and GSK3β Are Accompanied with Different Expression Levels in Human Astrocytoma. Cancers, 13(11), 2530. https://doi.org/10.3390/cancers13112530








