Validation of Selected Head and Neck Cancer Prognostic Markers from the Pathology Atlas in an Oral Tongue Cancer Cohort
Abstract
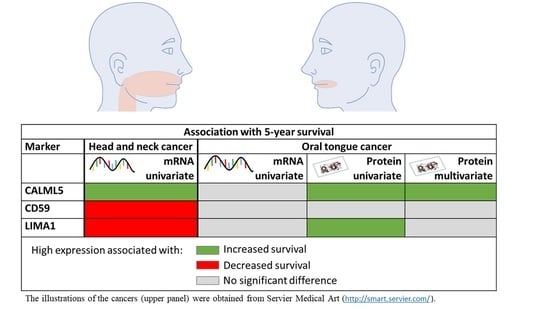
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Material
2.2. Tissue Microarray (TMA)
2.3. Selection of Markers
2.4. IHC Staining and Scoring
2.5. RNA Extraction and Quality Control
2.6. Reverse Transcription Quantitative PCR (RT-qPCR)
2.7. Statistical Analysis
3. Results
3.1. Immunohistochemical Staining and Scoring
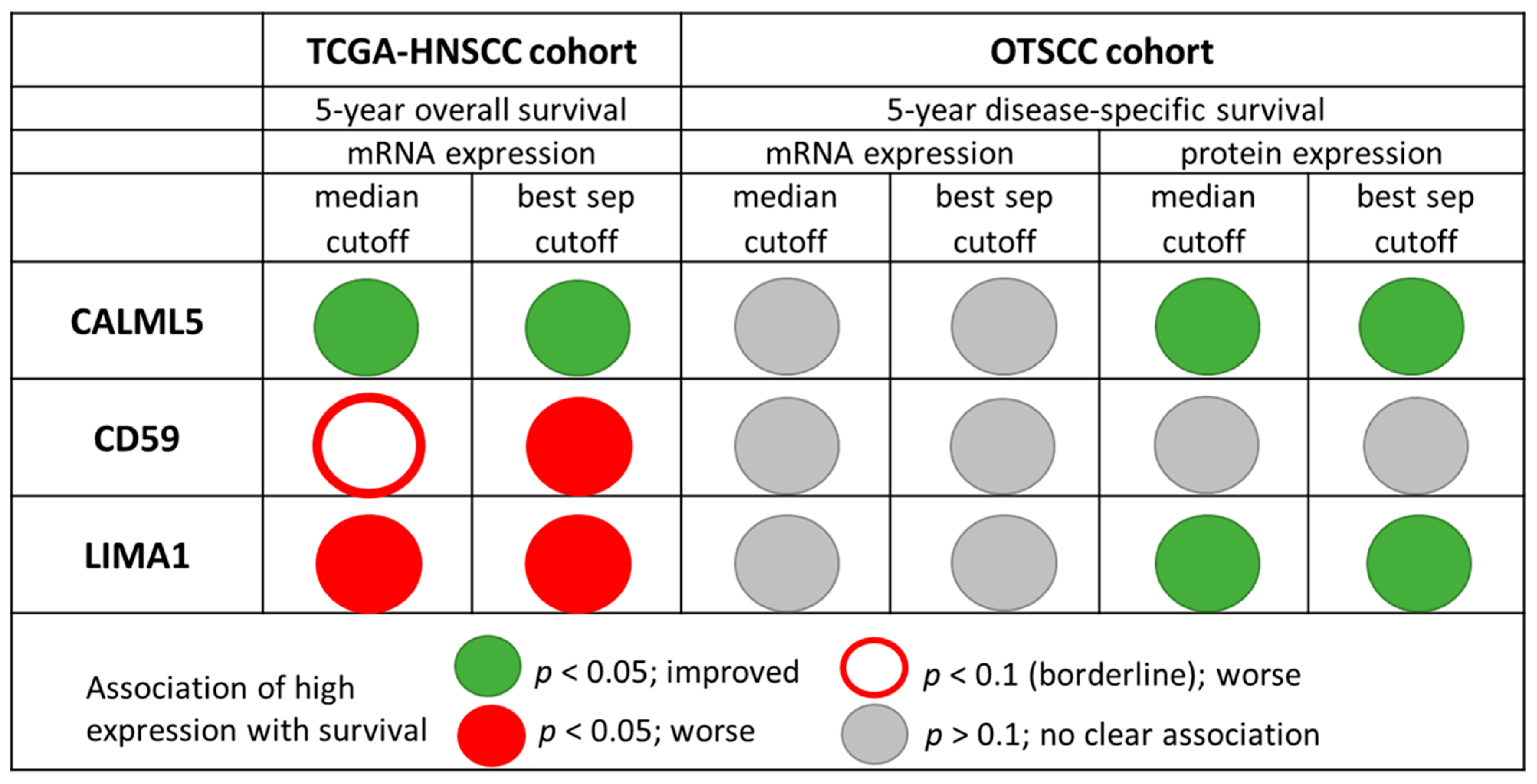
3.2. Univariate Analyses
3.3. Multivariate Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Waks, A.G.; Winer, E.P. Breast cancer treatment: A review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F. A pathology atlas of the human cancer transcriptome. Science 2017, 357. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, A.K.; Cahn, C.J.K.; Grandis, J.R.; Takata, T.; Slootweg, P.J. (Eds.) WHO Classification of Head and Neck Tumours, 4th ed.; International Agency for Research on Cancer (IARC) Press: Lyon, France, 2017. [Google Scholar]
- Petti, S. Lifestyle risk factors for oral cancer. Oral Oncol. 2009, 45, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Conway, D.; Purkayastha, M.; Chestnutt, I. The changing epidemiology of oral cancer: Definitions, trends, and risk factors. Br. Dent. J. 2018, 225, 867–873. [Google Scholar] [CrossRef]
- Wilms, T.; Khan, G.; Coates, P.J.; Sgaramella, N.; Fåhraeus, R.; Hassani, A.; Philip, P.S.; Norberg Spaak, L.; Califano, L.; Colella, G. No evidence for the presence of Epstein-Barr virus in squamous cell carcinoma of the mobile tongue. PLoS ONE 2017, 12, e0184201. [Google Scholar] [CrossRef]
- Network, C.G.A. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef]
- Gospodarowicz, M.K.; Brierley, J.D.; Wittekind, C. TNM Classification of Malignant Tumours; John Wiley & Sons: Hoboken, NJ, USA, 2017; pp. 18–21. [Google Scholar]
- Mroz, E.A.; Tward, A.M.; Hammon, R.J.; Ren, Y.; Rocco, J.W. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: Analysis of data from the Cancer Genome Atlas. PLoS Med. 2015, 12, e1001786. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.; Abba, M.C.; Molinolo, A.A.; Vitale-Cross, L.; Wang, Z.; Zaida, M.; Delic, N.C.; Samuels, Y.; Lyons, J.G.; Gutkind, J.S. The head and neck cancer cell oncogenome: A platform for the development of precision molecular therapies. Oncotarget 2014, 5, 8906. [Google Scholar] [CrossRef]
- Nulton, T.J.; Kim, N.-K.; DiNardo, L.J.; Morgan, I.M.; Windle, B. Patients with integrated HPV16 in head and neck cancer show poor survival. Oral Oncol. 2018, 80, 52–55. [Google Scholar] [CrossRef]
- You, G.-R.; Cheng, A.-J.; Lee, L.-Y.; Huang, Y.-C.; Liu, H.; Chen, Y.-J.; Chang, J.T. Prognostic signature associated with radioresistance in head and neck cancer via transcriptomic and bioinformatic analyses. BMC Cancer 2019, 19, 1–11. [Google Scholar] [CrossRef]
- Bjerkli, I.-H.; Jetlund, O.; Karevold, G.; Karlsdóttir, Á.; Jaatun, E.; Uhlin-Hansen, L.; Rikardsen, O.G.; Hadler-Olsen, E.; Steigen, S.E. Characteristics and prognosis of primary treatment-naïve oral cavity squamous cell carcinoma in Norway, a descriptive retrospective study. PLoS ONE 2020, 15, e0227738. [Google Scholar] [CrossRef]
- Søland, T.M.; Bjerkli, I.H.; Georgsen, J.B.; Schreurs, O.; Jebsen, P.; Laurvik, H.; Sapkota, D. High-risk human papilloma virus was not detected in a Norwegian cohort of oral squamous cell carcinoma of the mobile tongue. Clin. Exp. Dent. Res. 2020, 7, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Wirsing, A.M.; Ervik, I.K.; Seppola, M.; Uhlin-Hansen, L.; Steigen, S.E.; Hadler-Olsen, E. Presence of high-endothelial venules correlates with a favorable immune microenvironment in oral squamous cell carcinoma. Mod. Pathol. 2018, 31, 910. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- McShane, L.M.; Altman, D.G.; Sauerbrei, W.; Taube, S.E.; Gion, M.; Clark, G.M. REporting recommendations for tumour MARKer prognostic studies (REMARK). Eur. J. Cancer 2005, 41, 1690–1696. [Google Scholar] [CrossRef] [PubMed]
- Altman, D.G.; McShane, L.M.; Sauerbrei, W.; Taube, S.E. Reporting recommendations for tumor marker prognostic studies (REMARK): Explanation and elaboration. BMC Med. 2012, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.K.; Boxer, L.D.; Ransohoff, J.D.; Siprashvili, Z.; Qu, K.; Lopez-Pajares, V.; Hollmig, S.T.; Khavari, P.A. CALML5 is a ZNF750-and TINCR-induced protein that binds stratifin to regulate epidermal differentiation. Genes Dev. 2015, 29, 2225–2230. [Google Scholar] [CrossRef]
- Debald, M.; Schildberg, F.A.; Linke, A.; Walgenbach, K.; Kuhn, W.; Hartmann, G.; Walgenbach-Bruenagel, G. Specific expression of k63-linked ubiquitination of calmodulin-like protein 5 in breast cancer of premenopausal patients. J. Cancer Res. Clin. Oncol. 2013, 139, 2125–2132. [Google Scholar] [CrossRef]
- Misawa, K.; Imai, A.; Matsui, H.; Kanai, A.; Misawa, Y.; Mochizuki, D.; Mima, M.; Yamada, S.; Kurokawa, T.; Nakagawa, T. Identification of novel methylation markers in HPV-associated oropharyngeal cancer: Genome-wide discovery, tissue verification and validation testing in ctDNA. Oncogene 2020, 39, 4741–4755. [Google Scholar] [CrossRef]
- Jiang, W.G.; Martin, T.A.; Lewis-Russell, J.M.; Douglas-Jones, A.; Ye, L.; Mansel, R.E. Eplin-alpha expression in human breast cancer, the impact on cellular migration and clinical outcome. Mol. Cancer 2008, 7, 1–10. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, X.; Osunkoya, A.O.; Iqbal, S.; Wang, Y.; Chen, Z.; Müller, S.; Josson, S.; Coleman, I.M.; Nelson, P.S. EPLIN downregulation promotes epithelial–mesenchymal transition in prostate cancer cells and correlates with clinical lymph node metastasis. Oncogene 2011, 30, 4941–4952. [Google Scholar] [CrossRef] [PubMed]
- Maul, R.S.; Chang, D.D. EPLIN, epithelial protein lost in neoplasm. Oncogene 1999, 18, 7838–7841. [Google Scholar] [CrossRef]
- Ohashi, T.; Idogawa, M.; Sasaki, Y.; Tokino, T. p53 mediates the suppression of cancer cell invasion by inducing LIMA1/EPLIN. Cancer Lett. 2017, 390, 58–66. [Google Scholar] [CrossRef]
- Kim, D.D.; Song, W.-C. Membrane complement regulatory proteins. Clin. Immunol. 2006, 118, 127–136. [Google Scholar] [CrossRef]
- Kesselring, R.; Thiel, A.; Pries, R.; Fichtner-Feigl, S.; Brunner, S.; Seidel, P.; Bruchhage, K.-L.; Wollenberg, B. The complement receptors CD46, CD55 and CD59 are regulated by the tumour microenvironment of head and neck cancer to facilitate escape of complement attack. Eur. J. Cancer 2014, 50, 2152–2161. [Google Scholar] [CrossRef] [PubMed]
- Leemans, C.R.; Snijders, P.J.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Barnes, L. World Health Organization Classification of Tumors: Pathology and Genetics of Head and Neck Tumors; WHO: Geneva, Switzerland, 2005. [Google Scholar]
- Sampath, K.; Ephrussi, A. CncRNAs: RNAs with both coding and non-coding roles in development. Development 2016, 143, 1234–1241. [Google Scholar] [CrossRef] [PubMed]
- Boxberg, M.; Leising, L.; Steiger, K.; Jesinghaus, M.; Alkhamas, A.; Mielke, M.; Pfarr, N.; Götz, C.; Wolff, K.D.; Weichert, W. Composition and Clinical Impact of the Immunologic Tumor Microenvironment in Oral Squamous Cell Carcinoma. J. Immunol. 2019, 202, 278–291. [Google Scholar] [CrossRef]
- Peltanova, B.; Raudenska, M.; Masarik, M. Effect of tumor microenvironment on pathogenesis of the head and neck squamous cell carcinoma: A systematic review. Mol. Cancer 2019, 18, 63. [Google Scholar] [CrossRef]
- Leemans, C.R.; Tiwari, R.; Nauta, J.J.; Waal, I.V.D.; Snow, G.B. Recurrence at the primary site in head and neck cancer and the significance of neck lymph node metastases as a prognostic factor. Cancer 1994, 73, 187–190. [Google Scholar] [CrossRef]
| The TCGA-HNSCC Cohort | n (%) |
|---|---|
| TCGA-HNSCC patients included in The Pathology Atlas analyses | 499 (100) |
| Gender | |
| Male | 366 (73) |
| Female | 133 (27) |
| Stage | |
| I | 25 (5) |
| II | 69 (14) |
| III | 78 (16) |
| IV | 259 (52) |
| Information missing | 68 (14) |
| Alive at data collection | |
| Yes | 281 (56) |
| No | 218 (44) |
| Location (original TCGA-HNSCC cohort) | 527 (100) |
| Other and unspecified parts of tongue | 132 (25) |
| Larynx | 117 (22) |
| Other and ill-defined sites in lip, oral cavity, and pharynx | 71 (13) |
| Floor of mouth | 56 (11) |
| Tonsil | 46 (9) |
| Other and unspecified parts of mouth | 43 (8) |
| Base of tongue | 24 (5) |
| Gum | 11 (2) |
| Oropharynx | 10 (2) |
| Hypopharynx | 9 (2) |
| Palate | 5 (1) |
| Lip | 3 (1) |
| Variable | n | 5-year DSS % | p-Value 1 | |
|---|---|---|---|---|
| Gender | Male | 75 | 66.7 | 0.789 |
| Female | 46 | 69.6 | ||
| Age at diagnosis, years | <65 | 60 | 66.7 | 0.776 |
| ≥65 | 61 | 68.9 | ||
| Smoking | Never | 30 | 73.3 | 0.521 |
| Current | 51 | 66.7 | ||
| Former | 29 | 58.6 | ||
| Missing | 11 | - | ||
| T status | T1 | 37 | 83.8 | 0.072 |
| T2 | 47 | 61.7 | ||
| T3 | 29 | 65.5 | ||
| Unknown | 8 | - | ||
| N status 2 | N0 | 84 | 81.0 | <0.001 |
| N+ | 36 | 36.1 | ||
| Missing | 1 | - | ||
| Stage | Low stage (stage I or II) | 62 | 82.3 | <0.001 |
| High stage (stage III or IV) | 55 | 50.9 | ||
| Nx/Unknown | 4 | - | ||
| Differentiation, whole tumor | Low-grade (well or moderate) | 106 | 73.6 | <0.001 |
| High-grade (poor) | 13 | 23.1 | ||
| Missing | 2 | - | ||
| Lymphocyte infiltration | Abundant | 77 | 74.0 | 0.029 |
| Little | 38 | 55.3 | ||
| Missing | 6 | - |
| Antibody | Antigen Retrieval | Blocking | Wash Buffer | Dilution | Incubation Time and Condition | Secondary Antibody | Positive Control |
|---|---|---|---|---|---|---|---|
| Anti-CD59, HPA026494, Sigma–Aldrich | Citrate buffer pH 6.0 | 1.5% goat serum (Dako X9070) | PBS | 1:100 | 30 min room temperature | Anti-rabbit HRP conjugated (Dako K-4011) | Human tonsils |
| Anti-CALML5, HPA040725, Sigma–Aldrich | Citrate buffer pH 6.0 | 1.5% goat serum (Dako X9070) | PBS | 1:2000 | Overnight 4 °C | Anti-rabbit HRP conjugated (Dako K-4011) | Hum. salivary glands |
| Anti-LIMA1, HPA052645, Sigma–Aldrich | Citrate buffer pH 6.0II | 1.5% goat serum (Dako X9070) | PBS | 1:200 | Overnight 4 °C | Anti-rabbit HRP conjugated (Dako K-4011) | Human intestine |
| Gene | Accession No. | Full Name | Primer Sequence (5′ to 3′) | Ampl/effic/corr | Size (bp) |
|---|---|---|---|---|---|
| Reference RNA | |||||
| eF1a | NM001402.5 | Elongation factor 1 alpha | F: TATCCACCTTTGGGTCGCTTT | 99.8/1.000 | 63 |
| R: TGATGACACCCACCGCAACT | |||||
| RPL27 | NM000988.3 | Ribosomal protein L27 | F: GCTGGACGCTACTCCGGAC | 96.8/0.998 | 64 |
| R: CGATCTGAGGTGCCATCATCA | |||||
| RPS13 | NM001017.2 | Ribosomal protein S13 | F: AGAGAGCCGGATTCACCGTTT | 95.1/0.999 | 62 |
| R: CAATTGGGAGGGAGGACTCG | |||||
| Target RNA | |||||
| CALML5 | NM017422.4 | Calmodulin-like 5 | F: CGGTGAGCTGACTCCTGAGG | 97.3/0.999 | 84 |
| R: GGCATTGATGGTGCCGTTT | |||||
| CD59 | NM000611.5; NM001127223.1; NM001127225.1; NM001127226.1; NM001127227.1; NM203329.2; NM203330.2; NM203331.2 | CD59 | F: GGGTGTCAGTCAGGGACAACA | 98.3/0.999 | 92 |
| R: TTCATGCCCTGCTATCTGGA | |||||
| LIMA1 | NM001113546.1; NM001113547.1; NM001243775.1; NM016357.4 | LIM domain and actin-binding 1 | F: GCCAAGGCCTCCTCTCAGC | 101.9/0.999 | 68 |
| R: CCAGGCGATCCTCAGCTTCT | |||||
| Oral Tongue Squamous Cell Carcinoma (OTSCC) Cohort | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mRNA Expression | Protein Expression | ||||||||||||
| Median Cut-off | Best Separation Cut-off | Median Cut-off | Best Separation Cut-off | ||||||||||
| n | DSS % | p-Value | n | DSS % | p-Value | n | DSS % | p-Value | n | DSS % | p-Value | ||
| CD59 | Low | 36 | 69.4 | 0.256 | = median | 44 | 61.4 | 0.493 | 18 | 55.6 | 0.398 1 | ||
| High | 28 | 57.1 | 74 | 70.3 | 100 | 69.0 | |||||||
| CALML5 | Low | 23 | 52.2 | 0.192 | = median | 66 | 57.6 | 0.016 | =median | ||||
| High | 24 | 75.0 | 52 | 78.8 | |||||||||
| LIMA1 | Low | 39 | 69.2 | 0.214 | = median | 52 | 57.7 | 0.043 | =median | ||||
| High | 25 | 56.0 | 64 | 75.0 | |||||||||
| Adjusted for CALML5 | Adjusted for LIMA1 | |||||
|---|---|---|---|---|---|---|
| Variable n (CALML5/LIMA1) | Hazard Ratio | 95% CI | p-Value | Hazard Ratio | 95% CI | p-Value |
| N status N0, n = 75/74 vs. N+, n = 30/29 | 0.337 | 0.159–0.714 | 0.005 | 0.302 | 0.143–0.636 | 0.002 |
| Differentiation, whole tumor Low-grade (well or moderate), n = 93/92 vs. high-grade (poor), n = 12/11 | 0.389 | 0.172–0.879 | 0.023 | 0.227 | 0.114–0.673 | 0.005 |
| T status T1, n = 32/32 vs. T2/T3, n = 73/71 | 0.458 | 0.177–1.184 | 0.107 | 0.508 | 0.201–1.288 | 0.154 |
| Lymphocyte infiltration Abundant, n = 69/68 vs. little, n = 36/35 | 0.529 | 0.262–1.067 | 0.075 | 0.738 | 0.331–1.644 | 0.457 |
| CALML5 protein expression Low, n = 57 vs. high, n = 48 | 2.363 | 1.011–5.523 | 0.047 | |||
| LIMA1 protein expression Low, n = 46 vs. high, n = 57 | 1.834 | 0.857–3.926 | 0.118 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wirsing, A.M.; Bjerkli, I.-H.; Steigen, S.E.; Rikardsen, O.; Magnussen, S.N.; Hegge, B.; Seppola, M.; Uhlin-Hansen, L.; Hadler-Olsen, E. Validation of Selected Head and Neck Cancer Prognostic Markers from the Pathology Atlas in an Oral Tongue Cancer Cohort. Cancers 2021, 13, 2387. https://doi.org/10.3390/cancers13102387
Wirsing AM, Bjerkli I-H, Steigen SE, Rikardsen O, Magnussen SN, Hegge B, Seppola M, Uhlin-Hansen L, Hadler-Olsen E. Validation of Selected Head and Neck Cancer Prognostic Markers from the Pathology Atlas in an Oral Tongue Cancer Cohort. Cancers. 2021; 13(10):2387. https://doi.org/10.3390/cancers13102387
Chicago/Turabian StyleWirsing, Anna Maria, Inger-Heidi Bjerkli, Sonja Eriksson Steigen, Oddveig Rikardsen, Synnøve Norvoll Magnussen, Beate Hegge, Marit Seppola, Lars Uhlin-Hansen, and Elin Hadler-Olsen. 2021. "Validation of Selected Head and Neck Cancer Prognostic Markers from the Pathology Atlas in an Oral Tongue Cancer Cohort" Cancers 13, no. 10: 2387. https://doi.org/10.3390/cancers13102387
APA StyleWirsing, A. M., Bjerkli, I.-H., Steigen, S. E., Rikardsen, O., Magnussen, S. N., Hegge, B., Seppola, M., Uhlin-Hansen, L., & Hadler-Olsen, E. (2021). Validation of Selected Head and Neck Cancer Prognostic Markers from the Pathology Atlas in an Oral Tongue Cancer Cohort. Cancers, 13(10), 2387. https://doi.org/10.3390/cancers13102387